Abstract
The mode of neural stem cell division in the forebrain proliferative zones profoundly influences neocortical growth by regulating the number and diversity of neurons and glia. Long-term time-lapse multiphoton microscopy of embryonic mouse cortex reveals new details of the complex three-dimensional rotation and oscillation of the mitotic spindle before stem cell division. Importantly, the duration and amplitude of spindle movement predicts and specifies the eventual mode of mitotic division. These technological advances have provided dramatic data and insights into the kinetics of neural stem cell division by elucidating the involvement of spindle rotation in selection of the cleavage plane and the mode of neural stem cell division that together determine the size of the mammalian neocortex.
The number and diversity of cells in the cerebral cortex is due, in great measure, to the mode of progenitor cell division in the proliferative ventricular zone (VZ) before birth (1–4). At each mitotic division, the proliferative fate of each daughter cell, either reentering or exiting the cell cycle, dramatically influences neocortical growth (5). Symmetrical founder cell divisions, which predominate early, yield more progenitors and lead to an exponential expansion of the VZ population. In contrast, asymmetrical divisions, which prevail during later stages of neurogenesis, lead to cell commitment and diversity of the cortical architecture (1). Critical parameters of cortical growth are, therefore, the duration of the early symmetrical division phase and the time and extent of the transition to the asymmetrical mode of division (1–3). Despite the importance of this developmental strategy for brain growth, it is unknown how conversion between these major types of mitotic divisions is achieved in the VZ of the mammalian forebrain.
The current model for how different daughter cell fates result from asymmetric stem cell divisions assumes that cytoplasmic fate-determining molecules are unequally distributed during mother cell mitosis (6–9). This asymmetrical segregation can initiate diverse cellular programs during development of the daughter cells, including restriction of potential and differentiation (10–13). A prerequisite for producing asymmetric allocation of intracellular contents, observed in rodent telencephalon, is rotation and alignment of the mitotic spindle before division (14). Supporting such a causative role in cell fate determination, spindle movement before anaphase and the choice of the final cleavage plane has been correlated with daughter cell behavior in both invertebrates (6, 15–19) and vertebrates (refs. 10, 13, 20, and 21; see Fig. 1). In mammalian studies, however, determination of the molecular mechanisms linking spindle orientation, parcellation of intracellular factors, and fate determination has suffered from the lack of an intact model system. We sought to develop the capability for real-time analysis of these issues in living tissue that preserves the three-dimensional relationship between stem and progenitor cells in the neocortical VZ.
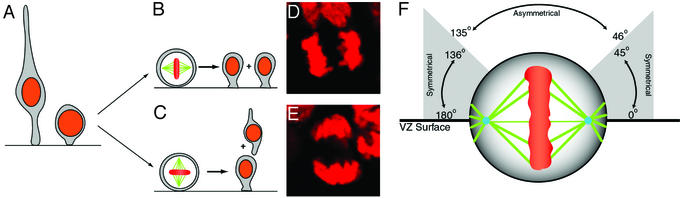
Figure 1.
Cleavage plane changes as neurogenesis proceeds. As VZ cells near mitosis, they round up at the VZ surface (A). During early neurogenesis, most divisions occur with vertical cleavage planes and presumably yield two symmetrical progenitor daughter cells (B and D). Later in neurogenesis, horizontal cleavage planes are thought to generate basal daughters that are postmitotic neurons (C and E). The VZ surface is at the bottom of the images in D and E. A scheme proposed by Chenn and McConnell (10) correlates the orientation of mitotic cleavage with daughter cell fate (F). Live imaging experiments (see Movie 3) confirm both the relationship between final cleavage plane and daughter cell behavior as well as the existence of boundaries at the 45° and 135° orientations.
Here, we have used a combination of time-lapse multiphoton microscopy and vital nucleic acid staining in organotypic slices to address how mitotic spindle movement may regulate neocortical neurogenesis. By using these methods, we were able to image deep in organotypic tissue to directly assay the intricate movements of cortical progenitors as they divided in their native milieu. We performed this procedure for multiple days spanning the cortical neurogenic period and were able to monitor cell divisions live for up to 48 continuous hours. We also correlated the parameters of this rotation with eventual cleavage plane orientation and found that cleavage plane does not necessarily predict cell fate, especially at later stages of neurogenesis when symmetrical divisions yield postmitotic neurons. The kinetic results indicate extensive oscillation and three-dimensional rotation of the mitotic spindle during metaphase that is developmentally regulated during cortical neurogenesis. Moreover, the pattern of spindle rotation preceding division consistently predicts the angle of the eventual cleavage plane. Daughter cell behavior therefore is intimately related to an integrative process of spindle alignment during metaphase. We speculate that short-range extrinsic and/or intrinsic signaling mechanisms provide for alternative spindle orientations and that the mitotic plate rotation is the opportunity for the spindle to be trapped in a position appropriate for that particular developmental time point.
Materials and Methods
Organotypic Slices, Explants, and Whole Brain Preparations.
Coronal slices at the level of future sensorimotor cortex in mouse (ICR strain) embryos were obtained as described (22) by using methods approved by the Yale Animal Care and Use Committee. We collected and analyzed cortical tissue from embryonic day (E)12 to E16, bracketing the course of murine neocortical neurogenesis. We chose to perform the studies on coronal organotypic slices since VZ cells in cortical explants have been demonstrated to behave differently than in whole slices (23), because abnormal substrate contact (e.g., with the coverslip on inverted en face cultures) with the ventricular surface may have altered proliferative dynamics, and because z-axis resolution was poor compared with x- and y-axis resolution. Briefly, freshly isolated whole brains in MEM were cut into 300-μm slices with a tissue chopper and then transferred into serum-free medium (SFM; neurobasal medium supplemented with N2, B27, and Glutamax) containing 2.5 nM SYTO 82 (Molecular Probes) for 1 h. Slices were destained for 0.5 h in fresh SFM and then embedded in growth factor-reduced Matrigel on a 15-mm coverslip that was then hardened in a 37°C incubator for 30 min. Coverslips with embedded tissue then were mounted in an RC-25F imaging chamber and bolted into a series 20 platform heater that was stably heated to 37°C by using a dual channel heater controller (Warner Instruments, Hamden, CT). SFM was preheated with an SF-28 in-line heater (Warner Instruments) and superfused through the chamber at 0.1 ml/min with a P720 miniperistaltic pump (Instek, Melrose, MA). Because the cut edges of the slice contain damaged cells and because the outside (cut) edges of the VZ partially collapse during the culturing period, we used multiphoton imaging to excite only deep layers of the slice that were undamaged and contained a sharp ventricular border.
Multiphoton Imaging.
Stable tissue preparations were imaged by using a Zeiss LSM 510 NLO system with an 8-W-pumped Coherent Mira 900F laser (Coherent Laser Group, Santa Clara, CA) direct-coupled to an Axiovert 100M microscope (Zeiss). All time-lapse studies were conducted with either an oil-immersed 25 × 0.8 numerical aperture (n.a.) Plan-NeoFluar lens or an oil-immersed 1.3 n.a. 100X Fluar lens. Tissue viability was ensured by minimal laser throughput (typically <5% of 1.2 W mode-locked power) and fast bidirectional scanning. SYTO 82-stained slices were excited at 780 nm and emission was collected with rhodamine isothiocyanate band pass optics with internal detectors. We routinely collected 15–30 μm-thick image stacks 80–120 μm deep within the tissue of the coronal slices. Time series experiments were conducted by automatically collecting identical stacks of averaged frames (2–4 line averages) at the same tissue position every 5 min for up to 48 h. The imaging preparation was stable in all planes throughout the experiments due to tight thermostatic control of the micro- and macroenvironments.
Image Analysis.
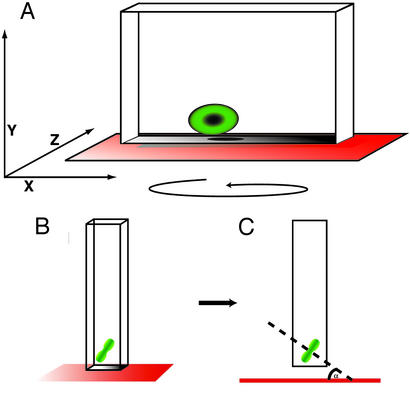
Time series experiments were analyzed by using Zeiss lsm software. Briefly, each z-stack of images, representing one time point, was reconstructed into three dimensions and then projected into a 180° rotated series. The reconstructed stack then was manually rotated until the edge of each metaphase plate was made visible, and a perpendicular line then was drawn through the middle of the mitotic plate (i.e., along the spindle axis). The angle of the mitotic plate was calculated in reference to a straight line drawn parallel to the ventricular surface (see Fig. 3). This procedure was repeated for every time point and for every cell in metaphase during the series. Movies 1–5, which are published as supporting information on the PNAS web site, www.pnas.org, were made in Adobe PREMIERE 5.0 from series of flattened images from each time point.
Figure 3.
Method for projecting image stacks to measure spindle angle. Stacks of 1-μm-thick optical images were projected into 3D images (A) by using Zeiss lsm software. These reconstructions were then rotated around the y axis until the edge of the metaphase plate (green) was resolved (B). This rotation brings the spindle axis parallel to the computer screen. Finally, a line through the spindle axis (dashed black line) was projected to another (red) line drawn parallel to the ventricular surface and the angle between the two lines was measured (C).
Results
The developing neocortical wall was maintained in long-term slice cultures in minimal defined media (22) on the stage of a multiphoton microscope. The reduced photo-damage and increased penetration of living tissue inherent to multiphoton microscopy (24–28) enabled us to collect four-dimensional (X, Y, and Z planes + time) image series for up to 2 days on the microscope. This system maintains the architectonic nature of the neocortical wall and is conducive to the study of proliferative kinetics during neocortical growth. The time-lapse procedure elucidated the remarkable and dynamic nature of the embryonic VZ, with all VZ progenitors exhibiting interkinetic movements (3, 29–35) and cell division simultaneously in each field (see Movies 1 and 2).
We first conducted time-lapse studies of dividing progenitors to determine whether the slice cultures maintained the important causal relationship between the plane of mother cell cleavage and the fate of the resulting daughter cells (9) during murine neocortical neurogenesis (Fig. 1 B–F). We indeed found, during early and midcorticogenesis, that cleavage planes vertical to the ventricle yielded daughter cells that remained closely opposed and confined to the VZ. In contrast, horizontal cleavages resulted in basal daughter cells that migrated quickly away from the ventricular surface (see Movie 3). Moreover, the relationship between the actual angle of the final cleavage and daughter cell behavior confirmed that cleavage planes above 45° from the ventricular surface yielded presumptive postmitotic neurons (10, 36–38) (see Fig. 1F). Therefore, in our slice preparation, vertical cleavages resulted in “symmetrical” daughter cell behavior and horizontal division planes yielded divergent, or “asymmetrical,” behavior. Nevertheless, time-lapse microscopy also revealed many startling features of the developing neocortical wall, the most remarkable of which was the significant rotation and oscillation of mitotic figures throughout metaphase (Fig. 2; see Movies 4 and 5). We thus developed a method for measurement and analysis of this four-dimensional process.
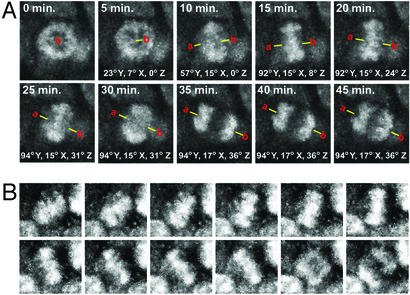
Figure 2.
Mitotic spindles rotate during neocortical neurogenesis. SYTO 82-labeled mitotic figures were studied over time by using 2-photon microscopy. Metaphase plates in E14 (A) and E15 (B) VZ rotate during metaphase. The frames are single optical sections taken from a coronal plane 100 μm deep in a cortical slice. The VZ surface is at the bottom of each frame. The cumulative degrees of spindle rotation are noted for each plane of rotation in each frame in A. This mitotic spindle (denoted with a line between points a and b) rotated through a total of 147° in three dimensions during the 45-min-long metaphase.
We measured and recorded spindle orientation in all four dimensions, but to better present the data, we removed one of the dimensions from the analysis by rotating three-dimensional reconstructed stacks of images around the y axis to bring the edge of the metaphase plate into alignment (Fig. 3). This procedure canceled out y-axis rotation and allowed us to measure the angle of the spindle with respect to the surface of the lateral ventricle over time. Because of this method of analysis, it is important to note that VZ spindles did rotate around the y axis before division and that the presented results are therefore an underestimate of the actual amount of rotation.
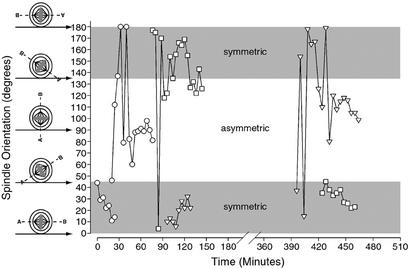
Metaphase plates exhibited varied types of movement ranging from short bidirectional oscillatory movements to prolonged unidirectional rotation before division. Plots of spindle rotation (Fig. 4), analyzed for every dividing cell in all of the four-dimensional experiments (62 slices, n = 428 total cells), uncovered two significantly different types of spindle behavior preceding VZ progenitor division. Dividing cells either progressed through metaphase quickly with little or no rotation, or wide oscillations in orientation occurred during early metaphase that increased in frequency and became tuned (i.e., clustered) around the future cleavage angle during later metaphase (Fig. 4). In all cases, the spindle rotation ceased at the onset of anaphase in dividing cells.
Figure 4.
History plots of mitotic spindle orientation. The mitotic spindle rotation in six representative divisions illustrates the different behavior of individual mitotic figures over time. Spindles either exhibited wide oscillations before anaphase or proceeded through mitosis quickly with little or no rotation. Drawings along the y axis illustrate the appearance of mitotic spindles and metaphase plates at different orientations. Regions within the plot are shaded with respect to symmetrical (gray) or asymmetrical (white) orientations. There was no correlation between the starting and ending orientations.
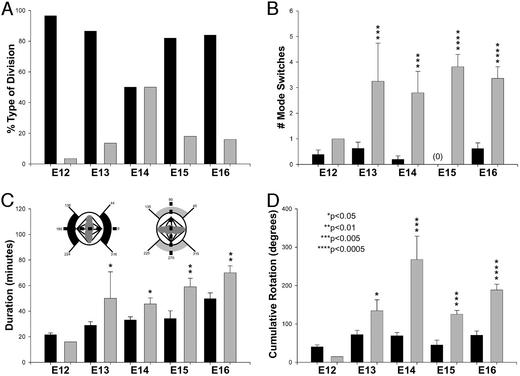
To determine the relationship between the spindle rotation and the eventual cleavage plane at anaphase, we conducted a post hoc analysis of spindle behavior after grouping cells with respect to their final cleavage plane orientation (Fig. 1F). The analysis revealed major differences between cells dividing vertically (symmetrically) and horizontally (asymmetrically) in all parameters measured (Fig. 5). Progenitors that eventually divided horizontally exhibited increased cumulative angular rotation during metaphase (Fig. 5D). Moreover, the number of oscillations over the 45° and 135° boundaries illustrated in Fig. 1 (termed “mode switches”) was larger in cells with eventual horizontal cleavage planes (Fig. 5B). Presumably due to the increased spindle rotation in these cells, the duration of metaphase preceding asymmetric cleavages was on average 15 min, or 26% longer (Fig. 5C). Thus, when compared with symmetric cleavages, the increased spindle rotation and number of fate switches in asymmetric cleavages suggests that more prolonged and/or complex integration is required before division.
Figure 5.
Symmetric and asymmetric cleavages are predicted by mitotic spindle rotation. The mode of cell division was determined by final cleavage orientation as depicted in Fig. 1F and Inset in C. (A) During early periods of neurogenesis on E12 and E13, most divisions were symmetrical (i.e., with vertical cleavage planes, black bars). There was a gradual increase in the number of asymmetrical cleavage orientations (gray bars) until E14, when the ratio of symmetrical to asymmetrical divisions was 50:50. After E14, cleavage planes reverted back to symmetrical orientations. The number of mode switches (i.e., rotations over the 45° and 135° boundary lines) (B), duration of metaphase (C), and cumulative angles rotated through during metaphase (D) were all increased in cells with eventual asymmetrical cleavage planes.
Neurogenesis in the mammalian neocortical VZ begins with few neurons born initially; there is a gradual increase in the rate of neuronal production so that the bulk of neurons are generated during later stages of neurogenesis (4). Interestingly, cleavage plane orientations in dividing VZ cells also change over the course of neurogenesis (10, 37, 38) in step with the graded neuronal commitment and therefore may be directly related to the changing VZ output. To determine the developmental change in VZ cleavage planes, we performed time-lapse imaging of the ventricular surface over the course of murine cortical neurogenesis from E12 to E16 (Fig. 5A). As expected, we found higher proportions of vertical (symmetrical) divisions early and an increase in horizontal (asymmetrical) divisions until midneurogenesis on E14, when there were equal numbers of vertical and horizontal cleavages. Surprisingly, however, cleavage profiles at later stages of neurogenesis reverted back to vertical orientations, suggesting that VZ progenitor divisions on E15 and E16 are predominately symmetrical. Moreover, the vertical cleavages later in neurogenesis were very similar to early vertical divisions in several key parameters such as the amount of rotation and the relative duration of metaphase.
Discussion
The present study provides comprehensive real-time analysis of the dynamic rotation of the metaphase plate with the graded nature of final cleavage orientations in the mammalian neocortical VZ. The only previous study of VZ cell spindle dynamics was conducted on en face cultures and many of the spindles rotating around the z axis (i.e., the axis parallel to the ventricular surface, resulting in horizontal cleavage planes) were therefore likely not recorded (14). The present study, which takes advantage of deep sequential optical sectioning of coronal preparations, allowed us to follow the behavior of metaphase plates rotating in all three dimensions. This long-term time-lapse imaging and four-dimensional reconstruction elucidated several instructive connections between spindle rotation and eventual cleavage plane orientation. Overall, the results suggest that VZ neurogenesis is related to rearrangement of the mitotic plate before division and that spindle movement during metaphase is a prerequisite for modulating the mode of division. Thus, spindle rotation in the dividing cell is likely to be an integrative mechanism sensing the changing intracellular molecular milieu and thereby providing the potential to respond to changes in the external environment.
Spindle Rotation Differs During Symmetric and Asymmetric Cleavages.
We found that cells dividing with vertical cleavage planes rotate less during metaphase and reach anaphase more quickly than do cells dividing horizontally (Figs. 4 and 5). This finding indicates that vertical cleavages may be a default orientation and that horizontal divisions require longer and more complicated reorganization of the spindle during which cytoplasmic signals are presumably being integrated. During early and late neurogenesis, most VZ divisions occur vertically and the mitotic spindle must therefore be trapped along the horizontal axis of the mother cell. In contrast, the transition to a large proportion of horizontal cleavage orientations by E14 indicates that, in those cells, the molecules that trap the spindle astral microtubules are repositioned to the apico-basal axis of the mother cell.
Studies in species ranging from fungus to fruit flies have implicated cortical domains of the actin cytoskeleton as necessary for the positioning of the mitotic spindle, and pharmacological and laser photolysis experiments have shown that spindle movement is accomplished by reorganization of contacts between astral microtubules and the actin cytoskeleton (39, 40). Further, molecules such as dynactin and dynein (15, 17, 41), cdc12p (42), the PAR proteins (43, 44) and the Inscutable/Pins proteins (reviewed in ref. 18) are enriched in these cortical specializations, are necessary for cell polarity, and are involved in spindle orientation. However, the wide oscillatory behavior of the spindle evident in the VZ progenitors (Fig. 4) indicates that these microtubule tethers may themselves not be solidly captured by the actin cytoskeleton at the onset of metaphase, as suggested previously by Chang (42), or perhaps that there are multiple cortical tethers that provide for spindle rotation and constant seeking of the final cleavage orientation.
The most parsimonious explanation of our results on cleavage orientation is that there is a default (most favorable) orientation, vertical to the surface of the ventricle, and the dividing cell “learns” at the onset of metaphase whether that orientation is to be adopted. If so, acquisition of the default orientation is rapid and does not require substantial spindle movement. In contrast, if the default position is not to be assumed, the spindle is set in motion to find the next most favored orientation.
Cleavage Plane Orientation Does Not Always Predict Daughter Cell Fate.
Our data on the temporal distribution of cleavage plane orientations and the resulting behavior of early VZ daughter cells is direct confirmation of the correlation first published in 1995 by Chenn and McConnell (10). In that study, it was suggested that daughter cells of vertical cleavages remain closely opposed and are likely to continue proliferating whereas basal daughter cells of horizontal divisions migrate away from their apical siblings and from their site of origin. In the present study, daughter cells from preparations isolated during early neurogenesis behaved as predicted, with basal daughters migrating quickly away from their ventricular-remaining apical siblings after horizontal cleavages (see Movie 3). Thus, cleavage plane orientation and resulting daughter cell behavior do change in a manner consistent with the predicted gradual increase in the number of neurogenic divisions during early neurogenesis (1, 4). However, mitotic divisions later in the neurogenic interval suggest a previously unrecognized method for proliferative fate specification.
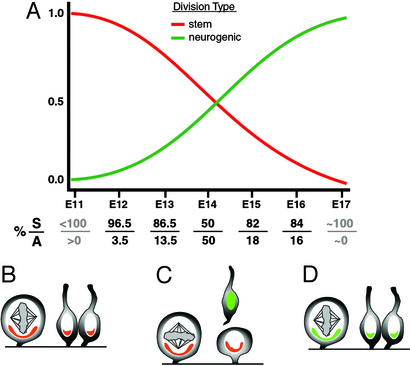
Our analysis revealed that neural stem cells in the VZ revert back to vertical cleavage orientations at later stages of cortical neurogenesis. Therefore, just as with their early generated cousins, these later divisions can be classified as symmetrical based both on their cleavage orientation and the similar behavior of the resulting daughter cells. However, in contrast to the stem cell renewal that occurs from early VZ divisions, it has been demonstrated in vivo that the majority of VZ divisions later in neurogenesis yield postmitotic neurons (4). Thus, it follows that these late vertical alignments generate two neuronal daughter cells (Fig. 6). Although the morphoregulatory molecules involved in neuronal production through asymmetric divisions are becoming elucidated, the present study suggests a mechanism by which two postmitotic neurons can be generated from a symmetrical division, seen here predominantly during later stages of cortical neurogenesis. Because we found that the spindle rotation and final alignments occur as quickly in late symmetrical cleavages as in the early symmetrical divisions (Fig. 5), the symmetrical neurogenic division is likely to be a default state at later stages of cortical neurogenesis. Moreover, most late divisions occur in this mode and result in termination of the stem cell lineage. Taken together then, vertical cleavage orientations from early and late VZ progenitors are likely to generate different types of daughters; cleavage orientation and initial behavior therefore may be instructive as to whether the daughter cells are similar or dissimilar with respect to proliferative fate (i.e., whether or not cells reenter or exit the cell cycle). However, cleavage orientation is not a valid predictor of cell phenotype as disparate daughters can be generated by the same spindle behavior and orientation depending on the developmental age.
Figure 6.
Model of neurogenesis and cleavage plane orientation. (A) The ratio of stem (red lines) and neurogenic (green lines) divisions changes over the course of neocortical neurogenesis. Most divisions at early stages of neurogenesis are regenerative with both daughter cells reentering the cell cycle. In contrast, most divisions at the end of neurogenesis yield postmitotic cells (adapted from ref 4). The percentage of symmetrical (S) and asymmetrical (A) cleavages is noted below the x axis for the ages measured. Cleavage plane orientation data illustrate that vertical cleavages predominate during early neurogenesis (B), when most daughter cells remain proliferative after mitosis (red cells). At the middle of neurogenesis on E14 (C), there is an equal proportion of asymmetrical divisions with horizontal cleavage planes that yield postmitotic neurons (green cells). At the end of neurogenesis (D), however, cleavages revert back to vertical orientations. Given the type of cells generated during late neurogenesis depicted in A, these later symmetrical orientations likely yield two postmitotic neuronal daughter cells.
A current theory for the generation of fate asymmetry is based on the discrete localization of cytoplasmic fate-determining molecules within the mother cell and their unequal distribution to daughter cells during division. The molecules so far identified, such as Notch, Numb, Miranda, Staufen, and Prospero (10, 20, 45, 46), have been shown to exhibit either basal or apical “capping” before division in static studies. Importantly, the fate molecule parcellation model only works in conjunction with variable orientations of the final cleavage plane. Assuming that the localization of fate molecule is fixed, only if the plane of cleavage can be modified can divergent cell fates be generated. If, on the other hand, fate molecules are not tethered in the same intracellular location throughout neurogenesis, or if there are multiple signaling pathways as the late vertical neurogenic divisions suggest, then spindle rotation is an even more critical requirement for modulating mode of division.
The nature and location of the signals that specify the developmentally regulated spindle rotation and alignment, and thereby the mode of division, are not known. One possibility is that a factor(s) in the external environment, e.g., a diffusible molecule, controls the cleavage orientation and mode of division over time. Similarly, signaling mechanisms intrinsic to the dividing VZ cells may specify the rate of neuronal and glial generation. In all of our experiments, we were able to detect closely associated, and in some cases directly apposed, vertical and horizontal divisions (Movie 3). There were no “hot spots” where one type of cleavage orientation was favored to the exclusion of another. Thus, our data are incompatible with long-range or broad external controls and suggest that either short range (i.e., cell-contact-mediated) or intrinsic pathways specify the change in mode of division during neurogenesis. Furthermore, several studies have shown that contiguous VZ cells are likely clonally related (10, 20, 45–48) and are gap junction-coupled (49). Examples such as in Movie 3, where neighboring cells divide in opposite orientations, therefore suggest the possibility that clonally related cells, even those synchronized with respect to cell cycle phase, possess the ability to divide in different modes.
The present results take advantage of advances in imaging and optical stabilization of living preparations to demonstrate that spindle rotation is controlled differently before vertical and horizontal divisions and that the proportions of vertical and horizontal cleavages change over the course of neurogenesis. Moreover, temporal analysis of final cleavage plane orientations suggests that vertical divisions present during early times of neurogenesis yield different types of daughters than do vertical divisions found during late neurogenesis. A recent examination of retrovirally labeled VZ cells (5) has suggested that all modes of division likely coexist within the VZ throughout neurogenesis. In particular, based on clade size distribution, it is probable that both types of symmetrical divisions (i.e., yielding two proliferative or two postmitotic daughters) coexist at each age of development. By using cleavage plane orientation, mitotic spindle kinetics, and initial migratory behavior, our study clearly illustrates a shift to asymmetrical divisions in the middle of neurogenesis. However, without independent proliferative-stage-specific markers, we are unable to classify the individual symmetrical divisions as yielding proliferative or postmitotic daughters. Interestingly, the data presented here in Fig. 5A is very similar to the results predicted from Model 1 in Cai et al. (5). Together, these results suggest that multiple modes of division coexist in the neocortical VZ, and that mitotic spindle rotation plays a “central” role in specifying the proliferative capacity of cells at each division event. The precise correlation between mitotic cell behavior and the resulting multitude of possible cell fates awaits robust probes for cell fate determinants during division as well as for cell phenotype markers expressed soon after cell division.
Supplementary Material
Acknowledgments
This work was supported by U.S. Public Health Service grants (to P.R.) and by NS10729 and Cure Autism Now (to T.F.H.).
Abbreviations
- VZ
ventricular zone
- En
embryonic day n
References
- 1.Rakic P. Science. 1988;241:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- 2.Caviness V S, Jr, Takahashi T, Nowakowski R S. Trends Neurosci. 1995;18:379–383. doi: 10.1016/0166-2236(95)93933-o. [DOI] [PubMed] [Google Scholar]
- 3.Rakic P. Trends Neurosci. 1995;18:383–388. doi: 10.1016/0166-2236(95)93934-p. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi T, Nowakowski R S, Caviness V S., Jr J Neurosci. 1996;16:6183–6196. doi: 10.1523/JNEUROSCI.16-19-06183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai L, Hayes N L, Takahashi T, Caviness V S, Jr, Nowakowski R S. J Neurosci Res. 2002;69:731–744. doi: 10.1002/jnr.10398. [DOI] [PubMed] [Google Scholar]
- 6.Horvitz H R, Herskowitz I. Cell. 1992;68:237–255. doi: 10.1016/0092-8674(92)90468-r. [DOI] [PubMed] [Google Scholar]
- 7.Chang F, Drubin D G. Curr Biol. 1996;6:651–654. doi: 10.1016/s0960-9822(09)00440-0. [DOI] [PubMed] [Google Scholar]
- 8.Shapiro L, Losick R. Science. 1997;276:712–718. doi: 10.1126/science.276.5313.712. [DOI] [PubMed] [Google Scholar]
- 9.Jan Y N, Jan L Y. Nature. 1998;392:775–778. doi: 10.1038/33854. [DOI] [PubMed] [Google Scholar]
- 10.Chenn A, McConnell S K. Cell. 1995;82:631–641. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- 11.Guo M, Jan L Y, Jan Y N. Neuron. 1996;17:27–41. doi: 10.1016/s0896-6273(00)80278-0. [DOI] [PubMed] [Google Scholar]
- 12.Lu B, Rothenberg M, Jan L Y, Jan Y N. Cell. 1998;95:225–235. doi: 10.1016/s0092-8674(00)81753-5. [DOI] [PubMed] [Google Scholar]
- 13.Shen Q, Zhong W, Jan Y N, Temple S. Development (Cambridge, UK) 2002;129:4843–4853. doi: 10.1242/dev.129.20.4843. [DOI] [PubMed] [Google Scholar]
- 14.Adams R J. J Neurosci. 1996;16:7610–7618. doi: 10.1523/JNEUROSCI.16-23-07610.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skop A R, White J G. Curr Biol. 1998;8:1110–1116. doi: 10.1016/s0960-9822(98)70465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Theesfeld C L, Irazoqui J E, Bloom K, Lew D J. J Cell Biol. 1999;146:1019–1032. doi: 10.1083/jcb.146.5.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Connell C B, Wang Y L. Mol Biol Cell. 2000;11:1765–1774. doi: 10.1091/mbc.11.5.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaltschmidt J A, Davidson C M, Brown N H, Brand A H. Nat Cell Biol. 2000;2:7–12. doi: 10.1038/71323. [DOI] [PubMed] [Google Scholar]
- 19.Doe C Q, Bowerman B. Curr Opin Cell Biol. 2001;13:68–75. doi: 10.1016/s0955-0674(00)00176-9. [DOI] [PubMed] [Google Scholar]
- 20.Zhong W, Feder J N, Jiang M M, Jan L Y, Jan Y N. Neuron. 1996;17:43–53. doi: 10.1016/s0896-6273(00)80279-2. [DOI] [PubMed] [Google Scholar]
- 21.Cayouette M, Whitmore A V, Jeffery G, Raff M. J Neurosci. 2001;21:5643–5651. doi: 10.1523/JNEUROSCI.21-15-05643.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haydar T F, Bambrick L L, Krueger B K, Rakic P. Brain Res Brain Res Protoc. 1999;4:425–437. doi: 10.1016/s1385-299x(99)00033-1. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi T, Bhide P G, Goto T, Miyama S, Caviness V S., Jr Exp Neurol. 1999;156:407–417. doi: 10.1006/exnr.1999.7023. [DOI] [PubMed] [Google Scholar]
- 24.Svoboda K, Denk W, Kleinfeld D, Tank D W. Nature. 1997;385:161–165. doi: 10.1038/385161a0. [DOI] [PubMed] [Google Scholar]
- 25.Potter S M. Curr Biol. 1996;6:1595–1598. doi: 10.1016/s0960-9822(02)70782-3. [DOI] [PubMed] [Google Scholar]
- 26.Centonze V E, White J G. Biophys J. 1998;75:2015–2024. doi: 10.1016/S0006-3495(98)77643-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Squirrell J M, Wokosin D L, White J G, Bavister B D. Nat Biotechnol. 1999;17:763–767. doi: 10.1038/11698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cannell M B, Soeller C. Microscopy Today. 2000. June 20–26. [Google Scholar]
- 29.Sauer M E, Walker B E. Proc Soc Exp Biol Med. 1959;101:557–560. doi: 10.3181/00379727-101-25014. [DOI] [PubMed] [Google Scholar]
- 30.Angevine J B, Sidman R L. Nature. 1961;192:766–768. doi: 10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- 31.Rakic P. Science. 1974;183:425–427. doi: 10.1126/science.183.4123.425. [DOI] [PubMed] [Google Scholar]
- 32.Caviness V S, Jr, Sidman R L. J Comp Neurol. 1973;148:141–151. doi: 10.1002/cne.901480202. [DOI] [PubMed] [Google Scholar]
- 33.Shoukimas G M, Hinds J W. J Comp Neurol. 1978;179:795–830. doi: 10.1002/cne.901790407. [DOI] [PubMed] [Google Scholar]
- 34.Nowakowski R S, Rakic P. J Comp Neurol. 1981;196:129–154. doi: 10.1002/cne.901960110. [DOI] [PubMed] [Google Scholar]
- 35.Caviness V S., Jr Brain Res. 1982;256:293–302. doi: 10.1016/0165-3806(82)90141-9. [DOI] [PubMed] [Google Scholar]
- 36.Smart I H. J Anat. 1972;113:109–129. [PMC free article] [PubMed] [Google Scholar]
- 37.Zamenhof S. Brain Res. 1985;352:306–309. doi: 10.1016/0165-3806(85)90120-8. [DOI] [PubMed] [Google Scholar]
- 38.Zamenhof S. Brain Res. 1987;428:143–146. doi: 10.1016/0165-3806(87)90094-0. [DOI] [PubMed] [Google Scholar]
- 39.Hyman A A, White J G. J Cell Biol. 1987;105:2123–2135. doi: 10.1083/jcb.105.5.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hyman A A. J Cell Biol. 1989;109:1185–1193. doi: 10.1083/jcb.109.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Busson S, Dujardin D, Moreau A, Dompierre J, De Mey J R. Curr Biol. 1998;8:541–544. doi: 10.1016/s0960-9822(98)70208-8. [DOI] [PubMed] [Google Scholar]
- 42.Chang F. Curr Biol. 1999;9:849–852. doi: 10.1016/s0960-9822(99)80372-8. [DOI] [PubMed] [Google Scholar]
- 43.Petronczki M, Knoblich J A. Nat Cell Biol. 2001;3:43–49. doi: 10.1038/35050550. [DOI] [PubMed] [Google Scholar]
- 44.Wodarz A. Curr Biol. 2001;11:R975–R978. doi: 10.1016/s0960-9822(01)00578-4. [DOI] [PubMed] [Google Scholar]
- 45.Matsuzaki F, Ohshiro T, Ikeshima-Kataoka H, Izumi H. Development (Cambridge, UK) 1998;125:4089–4098. doi: 10.1242/dev.125.20.4089. [DOI] [PubMed] [Google Scholar]
- 46.Wakamatsu Y, Maynard T M, Jones S U, Weston J A. Neuron. 1999;23:71–81. doi: 10.1016/s0896-6273(00)80754-0. [DOI] [PubMed] [Google Scholar]
- 47.Cai L, Hayes N L, Nowakowski R S. J Neurosci. 1997;17:2088–2100. doi: 10.1523/JNEUROSCI.17-06-02088.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cai L, Hayes N L, Nowakowski R S. J Neurosci. 1997;17:2079–2087. doi: 10.1523/JNEUROSCI.17-06-02079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bittman K, Owens D F, Kriegstein A R, LoTurco J J. J Neurosci. 1997;17:7037–7044. doi: 10.1523/JNEUROSCI.17-18-07037.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.