Abstract
The structure of the voltage sensor and the detailed physical basis of voltage-dependent activation in ion channels have not been determined. We now have identified conserved molecular rearrangements underlying two major voltage-dependent conformational changes during activation of divergent K+ channels, ether-à-go-go (eag) and Shaker. Two conserved arginines of the S4 voltage sensor move sequentially into an extracellular gating pocket, where they interact with an acidic residue in S2. In eag, these transitions are modulated by a divalent ion that binds in the gating pocket. Conservation of key molecular details in the activation mechanism confirms that voltage sensors in divergent K+ channels share a common structure. Molecular modeling reveals that structural constraints derived from eag and Shaker specify the unique packing arrangement of transmembrane segments S2, S3, and S4 within the voltage sensor.
From functional and structural analyses, permeation through K+ channels is understood at the atomic level (1). Much less is known about the mechanism of voltage-dependent activation. In voltage-dependent K+, Na+, and Ca2+ channels, the fourth transmembrane segment, S4, plays a vital role in transducing changes in membrane potential into pore opening (2, 3). In K+ channels, biophysical analysis indicates that each of the four subunits undergoes a minimum of two voltage-dependent conformational changes that result in a state that is permissive for pore opening. Then, a less voltage-dependent transition that is likely to involve all of the subunits opens the channel (4, 5). The structure of the voltage sensor and the detailed molecular rearrangements that underlie the voltage-dependent transitions of activation have not been determined. We now have used the unique functional properties of a mutant of the ether-à-go-go K+ channel (eag) to identify conserved molecular rearrangements that are characteristic of the two major voltage-dependent conformational changes in K+ channels. Furthermore, using structural constraints from eag and Shaker, we have determined the specific packing arrangement of transmembrane segments S2, S3, and S4 in the voltage sensor.
Methods
Wild-type and mutant eag channels were expressed in Xenopus oocytes and analyzed by using a two-electrode voltage clamp (6). The bath solution contained 118 mM NaCl, 1.8 mM CaCl2, and 10 mM Hepes, pH 7.2, and the indicated concentrations of Ni2+ were added as NiCl2. From a holding potential of −80 mV, depolarizing test pulses were applied in the absence or presence of Ni2+ as indicated. The time to half-peak current amplitude was measured and normalized as fold change in t1/2.
Molecular dynamics calculations were performed essentially as described (7). A variety of distance constraints based on experimental data were modeled as artificial distance-energy restraints in the form of a quadratic function with harmonic force constants of 5–20 kcal⋅[mol Å2]−1 (7). Models were based on the following reasonable assumptions: (i) the central pore in voltage-gated K+ channels is similar in structure to KcsA (1), which is consistent with the high degree of sequence similarity between Shaker and KcsA, with toxin-binding data, and with the generation of functional chimeric channels containing the Shaker voltage sensor and the KcsA pore (8, 9); (ii) S1–S4 form transmembrane α-helices that are oriented roughly perpendicular to the plane of the membrane, which is consistent with alanine and tryptophan scanning mutagenesis of these segments in voltage-gated K+ channels (10–12); (iii) the voltage sensor and pore domains interact closely in the structure, which is supported by single-particle electron microscope images of Shaker channels at 25-Å resolution (13), by perturbation analysis of the pore domain (14), by distance measurements using lanthanide-based resonance energy transfer (15) and tethered quaternary ammonium pore blockers (16), and by electrostatic calculations (17).
Results
Ni2+ Modulation of eag Activation Kinetics.
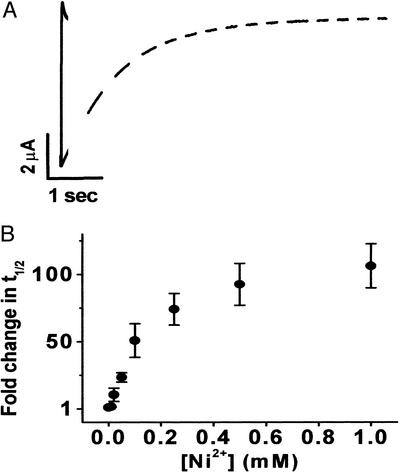
A variety of extracellular cations, including H+, Mg2+, Mn2+, and Ni2+, slow activation gating in voltage-dependent eag K+ channels (18, 19). Ni2+ has a particularly dramatic effect. At a near-saturating concentration (1 mM), Ni2+ increased the time to half-maximal current amplitude by more than 100-fold (Fig. 1). Application of long, depolarizing pulses revealed that Ni2+ does not significantly reduce the amplitude of eag currents (Fig. 1A). Therefore, Ni2+ exclusively modulates the kinetics of activation and channel opening with no effect on K+ permeation. The half-maximal concentration for Ni2+ action was ≈0.1 mM (Fig. 1B). This is significantly lower than the half-maximal concentration for Mg2+ (3.5 mM) (6). The higher apparent affinity for Ni2+ may reflect the fact that Ni2+ has a more flexible coordination geometry than Mg2+ (20).
Figure 1.
Ni2+ dramatically slows activation of wild-type eag. (A) Currents shown were elicited by depolarizing to +60 mV from a holding potential of −80 mV in the absence (solid trace) or presence (dashed trace) of 500 μM Ni2+. (B) The time to half-maximal current amplitude (t1/2) was measured at +60 mV in various Ni2+ concentrations as indicated, normalized to the control condition, and plotted as fold change in t1/2 vs. Ni2+ concentration (n = 3). In this and subsequent figures, graph symbols represent the mean ± SEM.
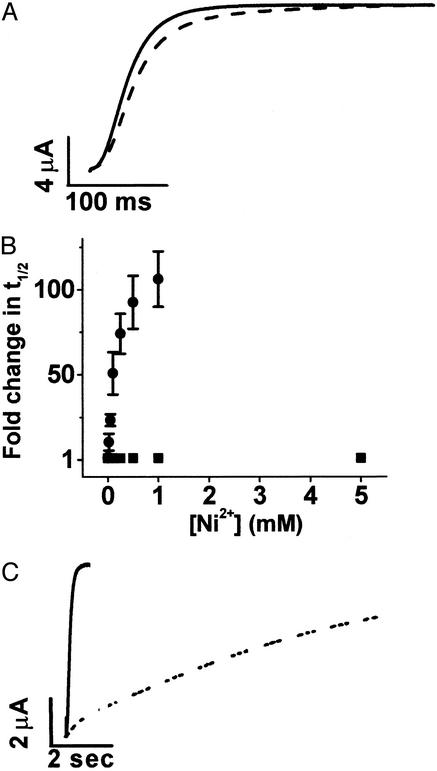
Previously, we showed that Mg2+ modulates activation kinetics by binding in an extracellular-facing pocket between transmembrane segments S2 and S3, where it interacts with two acidic residues, D278 in S2 and D327 in S3 (6). Acidic residues are located in analogous positions throughout the eag subfamily of K+ channels but are replaced by uncharged residues in all other voltage-dependent K+ channels (6). This explains why modulation by Mg2+ is restricted to eag and its homologues. To determine whether Ni2+ modulates eag gating by binding to the same site as Mg2+, we investigated the effect of Ni2+ on activation kinetics in two mutant channels, D278V and D327A (Fig. 2). Ionic current kinetics in the D278V channel were insensitive to Ni2+, consistent with the idea that D278 coordinates bound Ni2+ (Fig. 2 A and B). Surprisingly, D327A channels retained Ni2+ sensitivity (Fig. 2C). Taken together, these results indicate that Ni2+ binds in the same pocket as Mg2+ but suggest that Ni2+ is not situated identically within it. Because D327 is not required for Ni2+ binding, Ni2+ may be coordinated by one or more alternative ligands located in the pocket.
Figure 2.
D278 is required for Ni2+ sensitivity, but D327 is not. (A) D278V currents were evoked by a pulse to +60 mV in the absence (solid trace) or presence (dashed trace) of 5 mM Ni2+. (B) The time to half-maximal current amplitude (t1/2) at +60 mV was measured in various concentrations of Ni2+ for D278V (■) or wild-type eag (●), expressed as fold change and plotted as a function of Ni2+ concentration (n = 3–4). Wild-type data are the same as in Fig. 1B. (C) D327A currents were evoked by a pulse to +60 mV in the absence (solid trace) or presence (dashed trace) of 500 μM Ni2+.
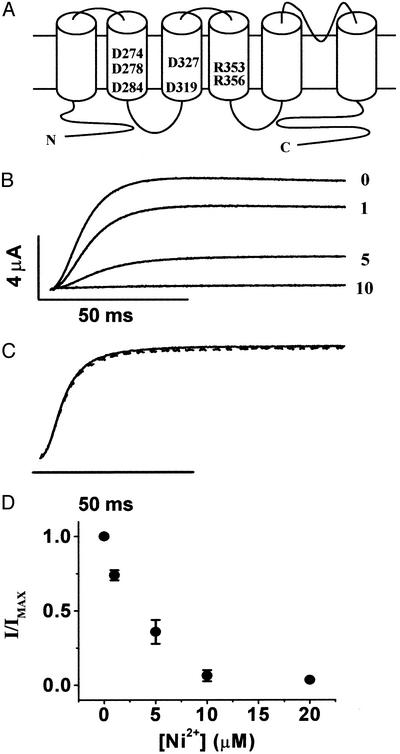
The eag S2 segment contains two acidic residues, D274 and D284, in addition to D278 (Fig. 3A). These are the eag counterparts of highly conserved acidic residues present in analogous locations in the S2 segments of voltage-dependent K+, Na+, and Ca2+ channels, as well as cyclic nucleotide-gated channels (21). If, as expected from perturbation analysis (10, 11), S2 adopts an α-helical structure, then D274 and D278 are located on the same face of the helix, one rung apart. We have shown previously that D274 does not contribute to the Mg2+-binding site but is located nearby (6). Mg2+ modulates activation gating in the D274A mutant channel with the same kinetic effects and apparent affinity as the wild type (6). However, mutating D274 to glutamate and, thereby, lengthening the side chain by one methylene group increases the apparent affinity of eag for Mg2+ (6). These data suggest that E274 is long enough to interact with the bound ion. We concluded from these results that D274 is exposed in the extracellular-facing pocket that contains the ion-binding site but is unable to coordinate the ion unless the side chain is lengthened.
Figure 3.
Ni2+ prevents activation gating in D274A channels. (A) Topology of eag showing six transmembrane segments and a reentrant P loop. The approximate locations of acidic residues in S2 and S3 and two of the basic residues in S4 are indicated. (B) Representative traces of D274A from the same oocyte in the presence of 0, 1, 5, or 10 μM Ni2+ as indicated. Currents were elicited by a pulse to +60 mV from a holding potential of −80 mV. (C) Scaled currents from 0 (solid trace) and 10 μM Ni2+ (dashed trace) demonstrate that activation kinetics are unaltered. (D) Peak current amplitudes at +60 mV were measured in various concentrations of Ni2+, normalized to the control condition, and plotted vs. Ni2+ concentration (n = 4).
Ni2+ Prevents Opening of D274A Channels.
We now report that application of Ni2+ to D274A channels revealed a novel phenotype. D274A channels were highly sensitive to Ni2+, but, rather than changing the time course of channel opening, Ni2+ decreased the current amplitude with no detectable change in activation kinetics (Fig. 3 B and C). We were unable to detect a slow component of activation during very long (10-s) depolarizing pulses (data not shown). These results indicate that Ni2+ prevents activation gating in D274A channels. According to this idea, activation kinetics are unaffected because residual current detected in the presence of Ni2+ results from unbound channels. Ni2+ inhibited activation gating with high affinity (Fig. 3D). The half-maximal concentration for the decline in current amplitude in D274A channels was ≈3 μM, which is significantly lower than the half-maximal concentration for modulating activation kinetics in wild-type eag. These values are not directly comparable, however, because the effects of Ni2+ on wild-type and D274A channels were assessed in different conformations, the open state and a closed state, respectively.
Effect of Ni2+ on D274A Channels Reversed by Neutralizing R353 in S4.
Previous work in Shaker channels suggests a structural basis for the novel effect of Ni2+ on D274A channels (22, 23). D274 is the homologue of E283 in the S2 segment of Shaker (Fig. 4A). Using a second-site suppressor strategy, we have presented evidence that E283 is involved in short-range electrostatic structural interactions with R368 and R371, two positively charged residues in the S4 segment that participate in the voltage-dependent conformational changes of S4 during activation gating of Shaker channels (22). Similar to D274A in eag, a neutralization mutation of E283, E283Q, does not eliminate functional expression of Shaker (15). In contrast, the charge-reversal mutation, E283R, disrupts folding of the Shaker protein. The folding defect of E283R is suppressed specifically by R368E or R371E (22). Functional analysis suggests that during voltage-dependent activation of Shaker channels, first R368 and, subsequently, R371 enter an extracellular-facing gating pocket, where they interact with E283 (22, 23). We now propose that analogous conformational changes occur during activation of eag channels and that in D274A mutant channels, the presence of Ni2+ prevents the movement of one or more S4 residues into the extracellular pocket that contains both the D274A mutation and the ion-binding site.
Figure 4.
R353Q restores activation gating of D274A mutant channels in Ni2+. (A) Sequences of the S2 and S4 transmembrane segments from Drosophila eag (d-eag), Shaker (d-ShB), and other representative voltage-gated K+ channels have been aligned. Noted on the S2 alignment are the locations of conserved acidic residues (bold) and a completely conserved phenylalanine (bold italic). Noted on the S4 alignment are regularly spaced basic residues (bold) and the highly conserved out-of-register histidine/arginine corresponding to position 363 in eag (bold italic). Numbers above the eag sequence refer to residues in eag, whereas numbers below the Shaker sequence refer to residues in Shaker. Numbers at the bottom of the alignment define a generic nomenclature for conserved charged residues in S2 and S4. D/E1 in S2 corresponds to the homologous residues D274 in eag and E283 in Shaker (d-ShB). R3 and R4 in S4 correspond to the homologous residues R353/R368 (eag/Shaker) and R356/R371, respectively. Other sequences shown are from the eag-like K+ channel (elk), the human (h) eag-related gene (erg), and Kv2.1, Kv3.1, and Kv5.1 from rat (r). (B) D274A+R353Q currents were evoked at +60 mV in the absence (solid trace) or presence (dashed trace) of 500 μM Ni2+. (C) The time to half-maximal current amplitude (t1/2) in D274A+R353Q channels was measured at +60 mV in various Ni2+ concentrations as indicated, normalized to the control condition, and plotted as fold change in t1/2 vs. Ni2+ concentration (n = 3). (D Left) Representative traces of D274A+R356Q from the same oocyte in the presence of 0, 5, 10, or 20 μM Ni2+, as indicated. Currents were elicited by a pulse to +60 mV from a holding potential of −80 mV. (Right) Scaled currents from 0 (solid trace) and 10 μM Ni2+ (dashed trace) demonstrate that activation kinetics are unaltered. (E) Peak current amplitudes of D274A+R356Q channels at +60 mV were measured in various concentrations of Ni2+, normalized to the control condition, and plotted vs. Ni2+ concentration; n = 4. (F) Representative traces of D274A+R353Q+R356Q from the same oocyte in the presence of 0, 20, or 50 μM Ni2+, as indicated. Currents were elicited by a pulse to +60 mV from a holding potential of −80 mV.
An alignment of eag and Kv channels indicates that R353 and R356 in the eag S4 segment are homologous to R368 and R371 in the Shaker channel (Fig. 4A). We propose that the combined presence of Ni2+ and a mutation that neutralizes their electrostatic partner, D274, prevents the movement of the positively charged R353 or R356 residues into the extracellularly facing pocket, thereby preventing channel opening. To test this hypothesis, we made the neutralization mutations R353Q and R356Q, paired them individually with D274A, and investigated the effect of extracellular Ni2+ on the double-mutant channels.
Combining R353Q with D274A restored the ability of the eag channel to open in the presence of Ni2+ (Fig. 4B). This result strongly suggests that Ni2+ specifically prevents movement of positively charged R353 into the gating pocket in the D274A channel. As in wild-type eag, Ni2+ significantly slowed activation of D274A+R353Q channels. At saturating concentrations, Ni2+ increased the time to half-maximal current amplitude by 95-fold. The half-maximal concentration for this effect was ≈0.05 mM, which is within 2-fold that of wild-type eag (Fig. 4C).
In contrast, Ni2+ prevented opening of D274A+R356Q channels (Fig. 4D). As in D274A, application of Ni2+ resulted in a decline in current amplitude with no significant change in activation kinetics. The concentration of Ni2+ that gave a half-maximal effect was ≈7 μM (Fig. 4E). That Ni2+ prevents opening of D274A+R356Q channels suggests that movement of R353 into the pocket is a prerequisite for the movement of R356; that is, there is sequential movement of R353 and R356 into the extracellular pocket.
If R353 and R356 move sequentially into the extracellular gating pocket, then addition of the R353Q mutation to D274A+R356Q should restore channel opening in the presence of Ni2+. Indeed, Ni2+ did not prevent opening of D274A+R353Q+R356Q channels. Rather, Ni2+ slowed activation kinetics significantly (Fig. 4F).
Discussion
Modulation of eag Activation by Divalent Ions.
Analysis of eag ionic and gating currents indicates that divalent ions decelerate the kinetics of at least two voltage-dependent conformational changes during activation, including a rate-limiting transition between closed states that occurs at hyperpolarized potentials and a subsequent transition that is closely linked to opening of the pore (19). Our results indicate that Ni2+ prevents opening of D274A channels by preventing the movement of R353 into the extracellular gating pocket, presumably by electrostatic repulsion. Movement of R353 therefore is likely to correspond to the rate-determining transition between closed states that occurs early in the gating pathway (19). We observe that Ni2+ dramatically slows activation in D274A+R353Q channels, as it does in wild type (Fig. 4 B and C), consistent with the idea that divalent ions modulate at least two steps along the gating pathway (19). A reasonable proposal is that Ni2+ also modulates the kinetics of the subsequent transition, in which R356 moves into the pocket. This transition is likely to be closely linked to opening (19). In Shaker channels, movement of the analogous residue, R371, into the pocket results in a conformation that is permissive for pore opening (23).
Model for Structural Rearrangements Underlying Two Stages of Voltage-Dependent Activation.
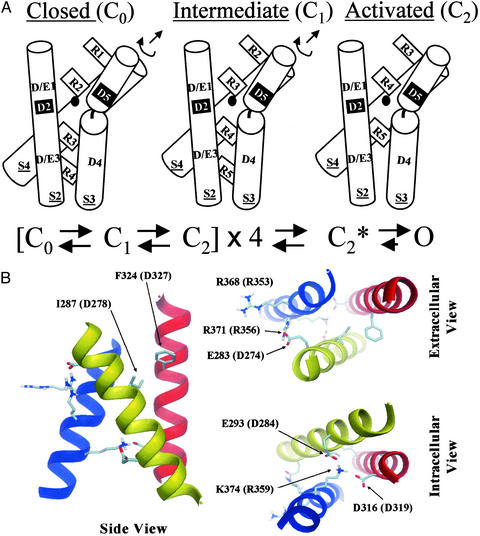
In Fig. 5A, we propose a model for two key molecular rearrangements that underlie the mechanism of voltage-dependent activation in K+ channels. We use a generic numbering system for conserved residues in S2 and S4, in which D274 in eag and E283 in Shaker correspond to D/E1 in S2, R353/R368 (Shaker/eag residues) to R3 in S4, and R356/R371 to R4 in S4 (see Fig. 4A). In the resting conformation, R3 and R4 are located near the intracellular surface of the protein (24, 25). During the first major voltage-dependent transition of activation, R3 in S4 moves into proximity with D/E1 in S2. The resulting electrostatic structural interaction is a key feature of the intermediate closed conformation. In our model, movement of R3 corresponds to the transition between closed states that is rate-limiting during activation of eag but not Shaker channels (19). In eag, binding of Mg2+ or Ni2+ in the extracellular pocket further slows this transition. We propose that in D274A, Ni2+ specifically blocks the transition in which R3 moves into the pocket. During the second major voltage-dependent transition, R4 moves into the pocket, where it interacts with D/E1 in S2, generating a conformation that is permissive for pore opening. We propose that divalent cations also modulate this step in eag. Although the two major voltage-dependent conformational changes of activation no doubt involve the movement of a number of additional amino acids in the protein, we propose that the specific molecular rearrangements involving D/E1 in S2 and R3 and R4 in S4 are characteristic of these transitions. After each of the subunits undergoes the second transition, opening would occur in a highly biased, forward reaction (4, 5).
Figure 5.
Structural constraints determine the unique packing arrangement of S2, S3, and S4 in the voltage sensor in K+ channels. (A Upper) A model for key molecular rearrangements underlying the two major voltage-dependent conformational changes of activation in voltage-dependent K+ channels. Our data indicate that two specific S4 residues, corresponding to R3 and R4 (generic nomenclature; see text), move sequentially into an extracellular gating pocket, where they interact with D/E1 in S2. During the first voltage-dependent transition, R3 moves into proximity with D/E1, resulting in an intermediate, closed conformation (C1). During the second voltage-dependent transition, R4 moves into proximity with D/E1, generating an activated conformation that permits pore opening (C2). Clockwise rotation and translation of S4 could contribute to these conformational changes, and movements of other parts of the protein, including S2 and S3, are not precluded. In the model, D2 and D5 (shown in black boxes) correspond to the metal ion-binding site (D278 in S2 and D327 in S3) in eag, and the black circle represents the bound metal ion. (Lower) A simplified scheme for K+ channel gating includes two voltage-dependent conformational changes per subunit. After all four subunits have undergone these transitions, the channel is in a conformation (C2*) permissive for pore opening. (B) The figure shows a structural model for the packing of transmembrane segments S2 (yellow helix, representing residues 280–299 in Shaker), S3 (red helix, residues 312–331), and S4 (blue helix, residues 362–379) in the activated conformation of the voltage sensor of K+ channels. The model was obtained by a conformational search procedure constrained by experimental data as described previously, with the exception that the ion-binding site in eag was included as a structural constraint (7). Side chains are shown to indicate neighboring residues. However, side chain conformations are speculative. Residues are labeled according to the Shaker sequence, with the corresponding eag residues in parentheses (see alignment, Fig. 4A). S1 is not shown in the model because of the lack of experimental constraints for this segment. In addition to a side view (Left), views from the extracellular (Upper Right) and intracellular (Lower Right) sides of the membrane are depicted.
Interestingly, a large family of gating modifier toxins appears to bind to the extracellular pocket that contains the ion-binding site in eag (26, 27). This includes hanatoxin, a specific inhibitor of the Kv2.1 K+ channel, plus related toxins that affect voltage-dependent Ca2+ and Na+ channels (28). These toxins do not block the pore; rather, they bind in the vicinity of the voltage sensor to modify gating. Hanatoxin inhibits Kv2.1 by binding to S3 in the region extracellular to the conserved proline residue (26). The structure of hanatoxin has been determined and includes six surface basic residues (29). We speculate that one or more of these residues sticks into the extracellular gating pocket and prevents entry of the charged S4 residues, thereby blocking activation gating in Kv2.1 similarly to Ni2+ in the D274A eag mutant channel.
Structural Organization of the Voltage Sensor in K+ Channels.
Evidence for conservation of key molecular details in the mechanism of voltage-dependent activation in eag and Shaker, two divergent members of the K+ channel superfamily, confirms that their voltage sensors share a common structure. Although a detailed atomic structure of a voltage-gated K+ channel is not yet available, data from second-site suppressor analysis of Shaker and identification of the ion-binding site in eag put specific constraints on the structural organization of the voltage sensor (6, 22, 23, 30). We previously suggested that these constraints uniquely specify the packing of S2, S3, and S4 in the voltage sensor (6). This conclusion now has been supported by structural modeling based on a molecular dynamics high-temperature simulated annealing procedure (Fig. 5B). Recently, the ensemble of plausible structural models for the Shaker channel was explored by using a conformational search constrained by a homology model of the pore based on the KcsA structure and experimental data from intragenic suppression, perturbation analysis, resonance energy transfer, and the use of tethered pore blockers (7). The eag constraint was not included, however. Two possible packing arrangements of S2, S3, and S4 were obtained that are consistent with second-site suppressor results in Shaker, suggesting that E283 in S2 interacts with R368 and R371 in S4, and K374 in S4 interacts with E293 in S2 and D316 in S3 (22, 23, 30). Identification of the ion-binding site in eag provides an additional structural constraint between S2 and S3 (6), and strong experimental justification now has been provided for use of Shaker and eag constraints in the same model. When voltage-sensor constraints from both Shaker and eag are incorporated, the same computational approach reveals a single possible packing arrangement for transmembrane segments S2, S3, and S4 in the voltage sensor (Fig. 5B). Viewed from the extracellular side of the membrane, S2–S4 are arranged sequentially in a counterclockwise direction. The position of S1 is uncertain because existing experimental data do not define its location relative to the other transmembrane segments. However, S1 is not essential for our conclusions. The significant finding is identification of the unique packing arrangement of S2, S3, and S4, the functional core of the voltage sensor (2, 22, 23, 30).
Thus, the packing of S2, S3, and S4 in Fig. 5A was not chosen arbitrarily. It conforms to the single arrangement of these segments that is compatible with known constraints. It is important to note that the Shaker and eag data do not enable us to determine the crossing angles of these transmembrane segments. This feature of the model in Fig. 5A is uncertain. An additional speculative feature is the kink shown in the S3 helix near a highly conserved proline residue. A break in the secondary structure near this residue is consistent with perturbation analysis of S3 in Kv2.1 and Shaker (11, 12). In eag, the proline residue is P325, located just 2 amino acids before D327. This proline may be important for generating the extracellular pocket that contains D274 and the binding site for Ni2+ and Mg2+ ions.
Acknowledgments
We are grateful to Drs. Sally Krasne, Joanna Jen, and Dorine Starace and members of the Papazian laboratory for comments on the manuscript. This work was supported by National Institutes of Health Grants GM43459 (to D.M.P.) and GM62342 (to B.R.).
Abbreviation
- eag
ether-à-go-go
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Zhou Y, Morais-Cabral J H, Kaufman A, MacKinnon R. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 2.Seoh S A, Sigg D, Papazian D M, Bezanilla F. Neuron. 1996;16:1159–1167. doi: 10.1016/s0896-6273(00)80142-7. [DOI] [PubMed] [Google Scholar]
- 3.Aggarwal S K, MacKinnon R. Neuron. 1996;16:1169–1177. doi: 10.1016/s0896-6273(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 4.Bezanilla F, Perozo E, Stefani E. Biophys J. 1994;66:1011–1021. doi: 10.1016/S0006-3495(94)80882-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zagotta W N, Hoshi T, Aldrich R W. J Gen Physiol. 1994;103:321–362. doi: 10.1085/jgp.103.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silverman W R, Tang C Y, Mock A F, Huh K B, Papazian D M. J Gen Physiol. 2000;116:663–678. doi: 10.1085/jgp.116.5.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roux B. Novartis Found Symp. 2002;245:84–101. [PubMed] [Google Scholar]
- 8.MacKinnon R, Cohen S L, Kuo A, Lee A, Chait B T. Science. 1998;280:106–109. doi: 10.1126/science.280.5360.106. [DOI] [PubMed] [Google Scholar]
- 9.Lu Z, Klem A M, Ramu Y. Nature. 2001;413:809–813. doi: 10.1038/35101535. [DOI] [PubMed] [Google Scholar]
- 10.Monks S A, Needleman D J, Miller C. J Gen Physiol. 1999;113:415–423. doi: 10.1085/jgp.113.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li-Smerin Y, Hackos D H, Swartz K J. J Gen Physiol. 2000;115:33–49. doi: 10.1085/jgp.115.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong K H, Miller C. J Gen Physiol. 2000;115:51–58. doi: 10.1085/jgp.115.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sokolova O, Kolmakova-Partensky L, Grigorieff N. Structure (London) 2001;9:215–220. doi: 10.1016/s0969-2126(01)00578-0. [DOI] [PubMed] [Google Scholar]
- 14.Li-Smerin Y, Hackos D H, Swartz K J. Neuron. 2000;25:411–423. doi: 10.1016/s0896-6273(00)80904-6. [DOI] [PubMed] [Google Scholar]
- 15.Cha A, Snyder G E, Selvin P R, Bezanilla F. Nature. 1999;402:809–813. doi: 10.1038/45552. [DOI] [PubMed] [Google Scholar]
- 16.Blaustein R O, Cole P A, Williams C, Miller C. Nat Struct Biol. 2000;7:309–311. doi: 10.1038/74076. [DOI] [PubMed] [Google Scholar]
- 17.Elinder F, Mannikko R, Larsson H P. J Gen Physiol. 2001;118:1–10. doi: 10.1085/jgp.118.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terlau H, Ludwig J, Steffan R, Pongs O, Stuhmer W, Heinemann S H. Pflügers Arch. 1996;432:301–312. doi: 10.1007/s004240050137. [DOI] [PubMed] [Google Scholar]
- 19.Tang C Y, Bezanilla F, Papazian D M. J Gen Physiol. 2000;115:319–338. doi: 10.1085/jgp.115.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.da Silva J J R F, Williams R J P. The Biological Chemistry of the Elements: The Inorganic Chemistry of Life. New York: Oxford Univ. Press; 2001. p. 561. [Google Scholar]
- 21.Montal M. FASEB J. 1990;4:2623–2635. doi: 10.1096/fasebj.4.9.1693348. [DOI] [PubMed] [Google Scholar]
- 22.Tiwari-Woodruff S K, Schulteis C T, Mock A F, Papazian D M. Biophys J. 1997;72:1489–1500. doi: 10.1016/S0006-3495(97)78797-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tiwari-Woodruff S K, Lin M A, Schulteis C T, Papazian D M. J Gen Physiol. 2000;115:123–138. doi: 10.1085/jgp.115.2.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Starace D M, Stefani E, Bezanilla F. Neuron. 1997;19:1319–1327. doi: 10.1016/s0896-6273(00)80422-5. [DOI] [PubMed] [Google Scholar]
- 25.Starace D M, Bezanilla F. J Gen Physiol. 2001;117:469–490. doi: 10.1085/jgp.117.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li-Smerin Y, Swartz K J. J Gen Physiol. 2000;115:673–684. doi: 10.1085/jgp.115.6.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cestele S, Catterall W A. Biochimie. 2000;82:883–892. doi: 10.1016/s0300-9084(00)01174-3. [DOI] [PubMed] [Google Scholar]
- 28.Li-Smerin Y, Swartz K J. Proc Natl Acad Sci USA. 1998;95:8585–8589. doi: 10.1073/pnas.95.15.8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi H, Kim J I, Min H J, Sato K, Swartz K J, Shimida I. J Mol Biol. 2000;297:771–780. doi: 10.1006/jmbi.2000.3609. [DOI] [PubMed] [Google Scholar]
- 30.Papazian D M, Shao X M, Seoh S A, Mock A F, Huang Y, Wainstock D H. Neuron. 1995;14:1293–1301. doi: 10.1016/0896-6273(95)90276-7. [DOI] [PubMed] [Google Scholar]