Abstract
The uptake and distribution of Pi in plants requires multiple Pi transport systems that must function in concert to maintain homeostasis throughout growth and development. The Pi transporter PHT2;1 of Arabidopsis shares similarity with members of the Pi transporter family, which includes Na+/Pi symporters of fungal and animal origin and H+/Pi symporters of bacterial origin. Sequence comparisons between proteins of this family revealed that plant members possess extended N termini, which share features with chloroplast transit peptides. Localization of a PHT2;1–green fluorescent protein fusion protein indicates that it is present in the chloroplast envelope. A Pi transport function for PHT2;1 was confirmed in yeast using a truncated version of the protein lacking its transit peptide, which allowed targeting to the plasma membrane. To assess the in vivo role of PHT2;1 in phosphorus metabolism, we identified a null mutant, pht2;1-1. Analysis of the mutant reveals that PHT2;1 activity affects Pi allocation within the plant and modulates Pi-starvation responses, including the expression of Pi-starvation response genes and the translocation of Pi within leaves.
INTRODUCTION
Phosphorus is essential for all living organisms as a structural component of nucleic acids and phospholipids, a constituent of energy transfer reactions, and a regulator in signal transduction cascades. In plants, phosphorus also is of fundamental importance to photosynthesis. Within the chloroplast, phosphate is an essential substrate for photophosphorylation and also plays a central role in the partitioning of triose phosphates, the end products of photosynthesis, between the starch and Suc biosynthetic pathways.
Plants acquire phosphorus as Pi from the soil solution. Despite its widespread occurrence in the environment, Pi often is limiting for plant growth mainly because it exists in the soil in soluble organic and inorganic forms that cannot be used directly by the plant (Marschner, 1995). The use of Pi-rich fertilizer improves crop yields otherwise limited by Pi availability, but this practice is cost prohibitive in many parts of the world, and the leaching of excess Pi can be dangerous to aquatic ecosystems because of the resulting eutrophication. Therefore, an understanding of Pi use in plants is of significance to both agriculture and the environment.
The transport of Pi across membranes is a pivotal step in the regulation of Pi use. Plants require multiple Pi transport systems to mediate acquisition from diverse environments and to enable its subsequent transport to all of the cells and subcellular compartments of the plant (Bieleski, 1973; Marschner, 1995). Initially, Pi is transported into root epidermal cells and subsequently loaded into the xylem for distribution to the aerial portions of the plant. Under conditions of Pi deficiency, Pi also is retranslocated from shoot tissues to the roots via the phloem (Mimura, 1999; Raghothama, 1999).
Little is known of the transport mechanisms that operate in the vascular tissue or in the shoots. Within each cell, Pi is transported between intracellular compartments, including chloroplasts/plastids and mitochondria, where it is assimilated via photooxidative and oxidative phosphorylation. Pi also is transferred into and out of the vacuole, a process that plays an essential role in the homeostatic control of cytoplasmic Pi concentration (Mimura, 1999). During the past few years, some of the transporters that contribute to this complex array of transport processes have been identified. However, many still remain unknown.
The triose phosphate/phosphate translocator (TPT) was the first phosphate transporter to be cloned from plants (Flügge et al., 1989). This transporter resides on the inner membrane of the chloroplast envelope, where it mediates the counterexchange of triose phosphates or 3-phosphoglycerate and Pi between the stroma of the chloroplast and the cytosol of the cell (Flügge et al., 1989). TPT is relatively abundant and constitutes 10 to 12% of the protein of the inner envelope membrane. It was purified and cloned from spinach and subsequently from a range of plant species, including potato, pea, and maize (Flügge, 1999).
In addition to the TPT proteins, translocators that mediate phosphoenolpyruvate/phosphate exchange and Glc-6-phos-phate/phosphate exchange also are present in plastid inner envelope membranes (Fischer et al., 1997; Kammerer et al., 1998). The Glc-6-phosphate/phosphates are expressed exclusively in heterotrophic tissues such as potato tubers, whereas the phosphoenolpyruvate/phosphates are present in both photosynthetic and heterotrophic tissues. All of the plastid phosphate translocators are predicted to possess six membrane-spanning domains and to function as dimers (Flügge, 1999).
Phosphate is mobilized across the inner mitochondrial membrane by the mitochondrial phosphate transporter. Mitochondrial phosphate transporters belong to the mitochondrial carrier family, which is characterized by a six-transmembrane-domain structure that is composed of three repeated segments of two transmembrane α-helices separated by a hydrophilic loop (Laloi, 1999). Mitochondrial phosphate transporter genes have been cloned from several plant species, and like their mammalian counterparts, they are assumed to operate via Pi/H+ symport or Pi/OH− antiport and to catalyze Pi/Pi exchange between the matrix and the cytosol (Stappen and Kramer, 1994; Wohlrab and Briggs, 1994; Takabatake et al., 1999).
In addition to these organellar Pi transporters, two other phylogenetically distinct classes of Pi transporters have been identified in plants. The first class contains transporters sharing similarity with the high-affinity H+/Pi symporter PHO84 of Saccharomyces cerevisiae (Bun-ya et al., 1991). Several genes encoding members of this class have been isolated from plants (Muchhal et al., 1996; Kai et al., 1997; Leggewie et al., 1997; Mitsukawa et al., 1997; Smith et al., 1997; Daram et al., 1998; C. Liu et al., 1998; H. Liu et al., 1998; Okumura et al., 1998). These genes are expressed predominantly in the root, although some also show expression in shoot tissues. All of them are derepressed under Pi-limiting conditions, and at least two encode transporters that have cellular and subcellular locations, consistent with a role in the acquisition of Pi from the soil (Muchhal and Raghothama, 1999; Chiou et al., 2000).
Uptake experiments using heterologous expression systems indicate that the encoded transporters mediate high-affinity Pi transport and that H+/Pi symport is the most likely transport mechanism (Muchhal et al., 1996; Kai et al., 1997; Mitsukawa et al., 1997; Daram et al., 1998; H. Liu et al., 1998; Okumura et al., 1998). These plant Pi transporters, together with PHO84 and other related fungal H+/Pi symporters (Harrison and van Buuren, 1995; Versaw, 1995), constitute the Pi:H+ symporter (PHS) family within the major facilitator superfamily (Pao et al., 1998).
Transporters of the second class share similarity with the Na+/Pi symporter PHO-4 of Neurospora crassa (Mann et al., 1989) and the closely related transporter PHO89 of S. cerevisiae. PHO-4 and PHO89 are distinct from PHS transporters in both primary sequence and the ability to transport Pi via a Na+/Pi symport mechanism (Versaw and Metzenberg, 1995; Martinez and Persson, 1998) and are classified as members of the Pi transporter (PiT) family (Saier et al., 1999; Saier, 2000). Other members of the PiT family include mammalian type III Na+/Pi symporters (Kavanaugh et al., 1994; Olah et al., 1994), PitA and PitB of Escherichia coli (Harris et al., 2001), and several uncharacterized sequences representing all kingdoms of life (Saier et al., 1999; Saier, 2000).
Daram et al. (1999) identified a transporter gene, PHT2;1, from Arabidopsis that shares similarity with members of the PiT family, although PHT2;1 does not appear to function via a Na+/Pi symport mechanism. Unlike the plant PHS transporter genes, PHT2;1 is expressed primarily in green tissue with transcripts located in all cells of the leaf blade and central vascular tissue, where it is proposed to function in the transport of phosphate into leaves (Daram et al., 1999).
As part of a search for Na+/Pi transporters in plants, we also cloned PHT2;1. In contrast to Daram et al. (1999), our analysis of the protein sequence suggests that it contains an N-terminal transit peptide, and localization of a PHT2;1– green fluorescent protein (GFP) fusion protein indicates that it is present in the chloroplast envelope. The phenotype of a PHT2;1 null mutant, pht2;1-1, is consistent with reduced Pi transport into the chloroplast but also reveals links between PHT2;1 and Pi distribution within the plant and to components of the Pi-starvation response.
RESULTS
Expression and Localization Analyses in Arabidopsis
Several plant Pi transporter genes have been identified based on the similarity of their deduced amino acid sequences to those of fungal H+/Pi symporters. Therefore, we used a similar strategy to identify plant orthologs of the phylogenetically distinct class of fungal Na+/Pi symporters. A BLASTP database search (Altschul et al., 1990) for sequences similar to that of the N. crassa PHO-4 Na+/Pi symporter (Versaw and Metzenberg, 1995) yielded two closely related plant sequences, one from Arabidopsis and another from Mesembryanthemum crystallinum, as well as sequences of cross-kingdom origin, including those of the previously confirmed mammalian and fungal Na+/Pi symporters (Kavanaugh et al., 1994; Olah et al., 1994; Martinez and Persson, 1998).
The Arabidopsis nucleotide sequence corresponds to an open reading frame (ORF), ORF02, within an 81-kb contig from chromosome III (Quigley et al., 1996). This 1764-bp ORF codes for a 587–amino acid protein of 61.4 kD. Hydropathy profiles of the predicted protein sequence suggest the presence of 12 membrane-spanning domains (data not shown), which is consistent with the topology predicted for the related Na+/Pi symporters (Mann et al., 1989; Johann et al., 1992; Miller et al., 1994; Martinez and Persson, 1998). During the course of our investigation, Daram et al. (1999) described the cloning of this same ORF, which they named PHT2;1, and its expression in Arabidopsis and functional characterization in yeast.
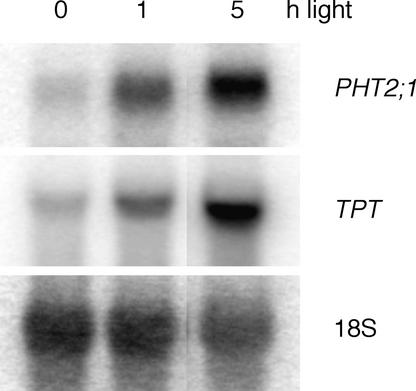
Our initial findings were similar to those reported. PHT2;1 is a single-copy gene that shows shoot-specific expression independent of external Pi concentrations (Figure 1). However, further analysis revealed that PHT2;1 expression varies during the photoperiod, with transcript levels 1.8-fold greater in shoot tissues harvested at the midpoint of the light phase than in tissues harvested at the midpoint of the dark phase (Figure 1). To determine whether PHT2;1 transcript levels are influenced by light, we monitored transcript levels in dark-treated plants and after exposure to light. After 6 days in darkness, PHT2;1 transcripts were barely detected, but after reexposure to light, induction occurred within 1 h (Figure 2). A similar pattern was detected for TPT, which was shown previously to be expressed in a light-dependent manner (Schulz et al., 1993).
Figure 1.
PHT2;1 Expression in Wild-Type Plants.
RNA gel blot analysis was performed on 5 μg of RNA isolated from root and leaf tissues of plants either deprived of Pi (−Pi) or grown under control conditions (+Pi) on washed sand. Plants were harvested between 1 and 2 h after the onset of the light phase of the photoperiod (lanes 1 to 4) or at the midpoints of the light and dark phases (lanes 5 and 6, respectively; 10 h of light/14 h of dark). Blots were hybridized with a PHT2;1 cDNA probe, stripped, and hybridized with an 18S rRNA probe to verify equivalent sample loading.
Figure 2.
Expression of PHT2;1 Is Induced in Response to Light.
RNA gel blot analysis was performed on 5 μg of total RNA isolated from plants held in complete darkness for 6 days with and without subsequent reexposure to light. Blots were hybridized with PHT2;1 and TPT cDNA probes, stripped, and hybridized with an 18S rRNA probe to verify equivalent sample loading.
PHT2;1 possesses a long N-terminal extension that is not present in the fungal Na+/Pi symporters, which prompted us to examine the PHT2;1 amino acid sequence for the presence of potential targeting sequences. Several protein-sorting prediction programs (Predotar version 0.5, TargetP version 1.0, PSORT, and ChloroP 1.1) were used, and the searches yielded complex results with positive scores for mitochondrial, chloroplast, and plasma membrane targeting. To distinguish these possibilities and confirm the subcellular localization of PHT2;1, its coding region was fused to that of the GFP (Chiu et al., 1996) and introduced into 4-week-old Arabidopsis leaves by particle bombardment.
After 24 h of incubation, green fluorescence was observed in the chloroplasts of transformed cells (Figure 3). Confocal microscopy revealed that the GFP signal is strongest at the chloroplast periphery, whereas the chlorophyll autofluorescence is uniform. This pattern, coupled with the fact that PHT2;1 is an integral membrane protein, is consistent with localization in the chloroplast envelope. Because the envelope inner membrane is the permeability barrier between the chloroplast stroma and the cytosol and is the location of other transport proteins, PHT2;1 most likely is located within the inner membrane.
Figure 3.
PHT2;1-GFP Fusion Protein Colocalizes with Chloroplasts.
A chimeric PHT2;1-GFP construct was introduced into Arabidopsis leaves by particle bombardment, and the localization of fluorescent signals was examined 24 h after transformation. All green signals detected colocalized with red signals. The green and red fluorescent signals indicate GFP and chlorophyll, respectively. Bar in (C) = 50 μm for (A) to (C).
Analysis of the TPT proteins, which are located in the chloroplast envelope inner membrane, resulted in subcellular targeting predictions similar to those obtained for PHT2;1. However, a program designed specifically to identify chloroplast proteins (ChloroP 1.1) predicted correctly that the TPTs are located in the chloroplast envelope and predicts transit peptide cleavage sites that match those derived experimentally (Fischer et al., 1994). The same analysis applied to PHT2;1 predicted cleavage resulting in a transit peptide of 71 amino acids.
Localization and Functional Analyses in Yeast
Daram et al. (1999) reported that PHT2;1 functions as a low-affinity H+/Pi symporter when expressed in yeast. An underlying assumption for Pi transport assays in yeast is that the transporter is located in the plasma membrane. For PHT2;1, this assumption is now questionable because of the potential for chloroplast proteins to be targeted to the mitochondria when expressed in fungal cells (Hurt et al., 1986; Pfaller et al., 1989; Brink et al., 1994). Therefore, we examined the location of PHT2;1 in yeast.
A PHT2;1-GFP fusion clone was constructed and introduced into the yeast mutant PAM2, which is defective in the high-affinity uptake of Pi (Martinez and Persson, 1998). Green fluorescence was detected within transformed cells, and the signal colocalized with that of the mitochondrion-selective dye MitoTracker Red, confirming that PHT2;1 is targeted to yeast mitochondria (Figures 4A to 4D). The putative chloroplast transit peptide appears to be responsible for the mitochondrial targeting, because deletion of this region from the fusion construct, PHT2;1(Δ1-71), resulted in localization of the green signal to the plasma membrane and to perinuclear structures that probably are a component of the endoplasmic reticulum (Figures 4E to 4H).
Figure 4.
Subcellular Localization of PHT2;1-GFP and PHT2;1(Δ1-71)-GFP Fusion Proteins in Yeast.
PAM2 yeast cells carrying PHT2;1-GFP ([A] to [D]) or PHT2;1(Δ1-71)-GFP ([E] to [H]) constructs were incubated with and without the mitochondrion-selective dye MitoTracker Red. The localization of green and red fluorescent signals, indicating GFP and mitochondria, respectively, was examined by confocal microscopy. Bar in (H) = 5 μm for (A) to (H).
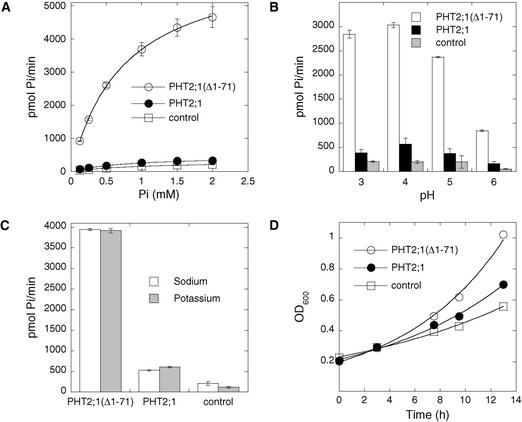
The Pi transport activities of PHT2;1 and PHT2;1(Δ1-71) then were compared in the yeast PAM2 strain. Both PHT2;1 and PHT2;1(Δ1-71) mediate saturable Pi transport in yeast with the same pH optimum of 4.0 and with similar apparent Km values of 870 ± 10 μM and 810 ± 40 μM, respectively (Figures 5A and 5B). This difference in apparent Km is not significant. Cells carrying the empty expression vector also exhibited saturable Pi uptake via the endogenous low-affinity transport system, with a Km value of 1010 ± 20 μM, which is significantly greater (P = 0.001) than that attributed to either PHT2;1 or PHT2;1(Δ1-71). In addition, the transport activity of cells expressing PHT2;1(Δ1-71) was approximately eightfold greater than that of cells expressing PHT2;1 under all assay conditions. This increased capacity for Pi transport also is reflected in cell growth.
Figure 5.
Transport Properties of PHT2;1 and PHT2;1(Δ1-71).
(A) Pi uptake rate of yeast strain PAM2 carrying PHT2;1, PHT2;1(Δ1-71), or a vector control plotted as a function of external Pi concentration at pH 4.0. Values for one of three independent experiments are shown (means ± se for three replicates).
(B) Effect of pH on the rate of Pi uptake. Uptake medium contained 25 mM citrate buffer to maintain the indicated pH and 1 mM Pi. Values from one of three independent experiments are shown (means ± se for three replicates).
(C) Effect of sodium on the rate of Pi uptake. Uptake medium contained 1 mM Pi and 25 mM citric acid and was titrated to pH 4.0 with either KOH or NaOH. Uptake values for each experiment are pmol Pi/min for 0.5 mL of yeast cell suspension with an absorbance of 1 unit at 600 nm. Values from one of three independent experiments are shown (means ± se for three replicates).
(D) Growth of transformed PAM2 cells in synthetic dextrose medium containing 25 mM citrate buffer, pH 4.0, and 0.22 mM Pi. OD600 was monitored as a measurement of growth.
When cultured in low-Pi medium (220 μM), the growth rate of cells expressing PHT2;1(Δ1-71) was greater than that of cells expressing PHT2;1, with doubling times of ∼5.5 and 7.5 h, respectively (Figure 5D). The transport activity of PHT2;1(Δ1-71) was highly specific for Pi, because a 40-fold excess of anions such as sulfate, nitrate, and chloride did not compete for Pi uptake (Table 1).
Table 1.
Specificity of PHT2;1
| Reagent | 33Pi Uptake Activity (%) |
|---|---|
| Water control | 100 |
| K2SO4 (10 mM) | 99 |
| KNO3 (10 mM) | 97 |
| KCl (10 mM) | 96 |
| KH2PO4 (0.5 mM) | 62 |
| KH2PO4 (2.5 mM) | 34 |
Anions tested as potential competitors in Pi uptake assays were added as potassium salts to yeast PAM2 cells expressing PHT2;1(Δ1-71), 30 s before the addition of labeled Pi (0.25 mM final concentration). Values were derived from three independent measurements per treatment with se < 3%.
Although we identified PHT2;1 based on its relatedness to a Na+/Pi symporter, we found that the Pi transport activity mediated by PHT2;1 and PHT2;1(Δ1-71) was unaffected regardless of whether Na+ was limited to that present solely as a contaminant in other media components or was present at the high concentration optimal for the related fungal Na+/Pi symporters (Figure 5C) (Versaw and Metzenberg, 1995; Martinez and Persson, 1998). Daram et al. (1999) also reported that PHT2;1 activity in yeast was independent of Na+. However, the maximum concentration of Na+ included in their transport assays was 250-fold less than the optimal concentration for the orthologous transporters, and Na+ dependence or enhancement of Pi transport activity may not have been detected under such conditions.
Isolation of a PHT2;1 Null Mutant
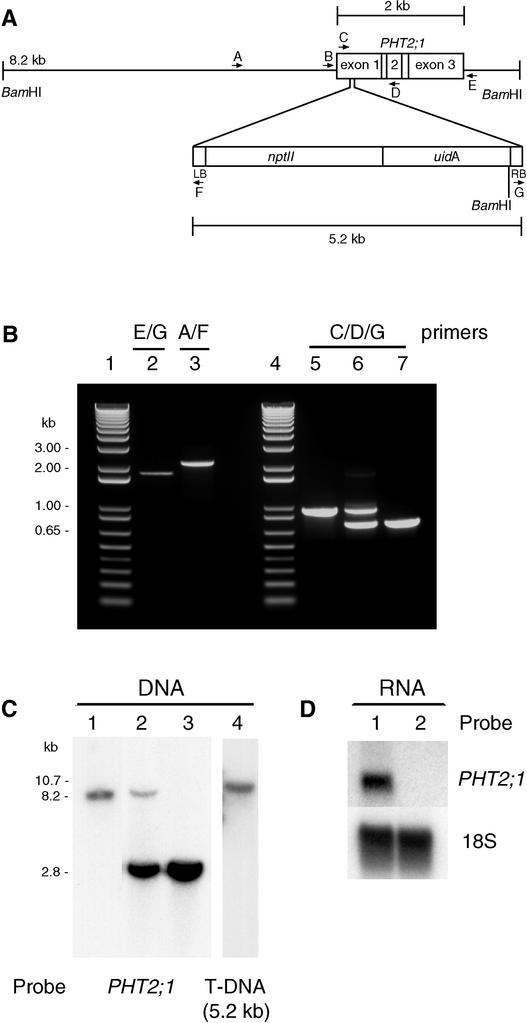
To obtain insight into the function of PHT2;1 and its role in phosphorus metabolism, we required a mutant that lacked a functional PHT2;1 allele. Therefore, we used a reverse-genetics approach to identify an insertion mutant. We screened a library of 12,000 independent T-DNA lines by PCR for a T-DNA insertion within the PHT2;1 coding region and found one pool that yielded distinct PHT2;1::T-DNA junction sequences. Reactions containing a primer combination specific for the T-DNA right border and the 3′ end of PHT2;1 yielded a single 1.8-kb junction fragment (Figure 6B, lane 2), and a primer combination specific for the T-DNA left border and the upstream region of the PHT2;1 locus yielded a single 2.2-kb junction fragment (Figure 6B, lane 3).
Figure 6.
Molecular Characterization of the pht2;1-1 locus.
(A) Scheme of the PHT2;1::T-DNA insertion characterized in pht2;1-1. The structure of the pht2;1-1 locus was deduced from PCR, DNA gel blot, and genomic sequence analyses.
(B) Agarose gel separation of PCR samples. DNA from the wild type (lane 5) and mutants containing the PHT2;1::T-DNA insertion (lanes 2, 3, 6, and 7) were amplified using the primer combinations indicated. Lanes 6 and 7 show the amplification products from plants hemizygous and homozygous for the PHT2;1::T-DNA insertion, respectively. Lanes 1 and 4 show 1-kb ladder standards.
(C) DNA gel blot analysis of BamHI-digested genomic DNA from the wild type (lane 1) and mutants hemizygous (lane 2) and homozygous (lanes 3 and 4) for the T-DNA insertion. Blots were hybridized with probes as indicated. Sizes of the detected bands are indicated in kb.
(D) RNA gel blot analysis of total RNA isolated from wild-type (lane 1) and pht2;1-1 (lane 2) leaf tissues hybridized with probes as indicated.
Sequencing of these junction fragments confirmed the presence of a T-DNA inserted within the PHT2;1 coding region, with left border and right border insertion sites 209 and 250 bp downstream, respectively, relative to the A of the ATG start codon (Figure 6A). Continued screening reduced the complexity of the population to a single seed pool representing 10 individual T-DNA lines. We screened individual plants from this seed pool and identified several positive for the PHT2;1 insertion. Plants hemizygous or homozygous for the PHT2;1::T-DNA locus were distinguished by PCR (Figure 6B, lanes 5 to 7) and verified by DNA gel blot analysis (Figure 6C). To determine whether the homozygous insertion line contains any additional T-DNA insertions elsewhere in the genome, we probed the same blot with the 5.2-kb T-DNA sequence.
Only one band was observed in the homozygous line (Figure 6C, lane 4), and its size is consistent with insertion of a single T-DNA. In addition, the F2 progeny of a cross between the wild type and the homozygous mutant line showed 3:1 segregation for resistance to kanamycin, consistent with the expected pattern for a single T-DNA insertion (236 kanamycin resistant:78 kanamycin susceptible; χ2 = 0.004, P = 0.94). RNA gel blot analysis of leaf RNA confirmed that the homozygous mutant lacks PHT2;1 transcripts (Figure 6D) and therefore is null. We named this line and the corresponding mutant allele pht2;1-1.
Growth and Pi Content of pht2;1-1 Plants
When plants were grown on Metro Mix 350, a complex, high-Pi growth medium, pht2;1-1 rosettes were smaller than wild-type rosettes (82% fresh weight [n = 18]) and had reduced Pi content (74% nmol Pi/mg fresh weight [n = 18]), indicating a defect that limits the accumulation of Pi in leaves and thus retards growth. To confirm that the phenotype of reduced Pi content correlates with the pht2;1-1 mutation, we examined random F2 progeny from a cross of wild-type and homozygous pht2;1-1 plants. As shown in Table 2, the homozygous mutant plants had an average leaf Pi content equal to 79% that of the wild type, whereas the leaf Pi content of hemizygous plants did not differ significantly from that of the wild type. This finding indicates that the pht2;1-1 mutation is recessive and that the reduced Pi content and mass correlate with the presence of the pht2;1-1 mutation.
Table 2.
Analysis of Wild Type × pht2;1-1 F2 Progeny
| Genotype | No. of Plants | Leaf Pi Content (nmol/mg fresh weight) |
|---|---|---|
| PHT2;1/PHT2;1 | 20 | 11.05 ± 0.09 |
| PHT2;1/pht2;1-1 | 27 | 10.89 ± 0.29 |
| pht2;1-1/pht2;1-1 | 17 | 8.76 ± 0.16 |
Plants were grown for 10 days on agar-solidified medium containing 1 mM Pi followed by 25 days on Metro Mix 350. Genotypes were determined by PCR as shown in Figure 2. Leaf Pi content was determined for 10 plants of each genotype. Values represent means ± se for two to three leaves from each plant.
To confirm that the pht2;1-1mutation rather than another closely linked mutation is responsible for the observed phenotype, we tested the ability of the wild-type PHT2;1 allele to complement the mutation. pht2;1-1 plants were transformed with either the binary vector pSKI089 as a control or PHT2;1/pSKI089, which carries a 4.2-kb segment of the genomic PHT2;1 locus spanning from 1.7 kb upstream to 0.3 kb downstream of the ORF.
The leaf Pi content of pht2;1-1 plants transformed with pSKI089 was 78% that of the wild type, whereas the leaf Pi content of pht2;1-1 plants transformed with PHT2;1/pSKI089 did not differ significantly from that of wild-type plants (Table 3). A subset of these plants was examined by reverse transcriptase–mediated PCR to confirm the presence/ absence of PHT2;1 transcripts, and as expected, pht2;1-1 plants transformed with pSKI089 lacked PHT2;1 transcripts. These results indicate that PHT2;1 is sufficient for complementation of the phenotype associated with the pht2;1-1 mutation. All noted differences are significant at a confidence level of >95% (Student's t test).
Table 3.
Complementation of the pht2;1-1 Leaf Pi Accumulation Defect
| Plant | Plasmid | No. of Plants | Leaf Pi Content (nmol/mg fresh weight) |
|---|---|---|---|
| Wild type | pSKI089 | 11 | 13.2 ± 0.5 |
| pht2;1-1 | pSKI089 | 8 | 10.2 ± 0.2 |
| pht2;1-1 | PHT2;1/pSKI089 | 12 | 13.0 ± 0.5 |
Wild-type and pht2;1-1 plants were transformed with either pSKI089 as a control or pSKI089 carrying PHT2;1. Leaf Pi content was determined for two to three leaves from each plant. Values shown are means ± se.
Effect of Pi Supply on Plant Growth and Pi Accumulation
To assess the effect of Pi supply on pht2;1-1 plants and to allow analysis of roots, we grew plants under defined nutrient conditions on agar medium and fertilized sand. As shown in Figure 7, plants grown on agar medium and sand had similar responses to Pi supply. Under high-Pi conditions, pht2;1-1 rosette fresh weight and Pi content were 80 and 70% those of the wild type, respectively, a phenotype similar to that observed in plants grown on Metro Mix 350. Under low-Pi conditions, we found no significant differences in the fresh weight of either roots or rosettes.
Figure 7.
Effect of Pi Supply on Plant Growth and Pi Accumulation.
Roots and rosette leaves from 8 to 10 plants grown under defined conditions were harvested and weighed ([A] and [B]). The Pi content was determined from three replicate samples ([C] and [D]). Values shown are means ± se for two independent experiments. WT, wild type.
The total phosphate content of whole plants also showed no significant difference (Table 4). However, pht2;1-1 root Pi content was 52 to 70% that of the wild type, and leaf Pi content was 129 to 156% that of the wild type, indicating that under low-Pi growth conditions, the pht2;1-1 mutation affects Pi allocation at the whole-plant level. The total phosphate content of wild-type and pht2;1-1 tissues showed differences equivalent to those observed for Pi, indicating that cellular Pi but not organic phosphate content is altered in the mutant (Table 4). In addition, when plants were grown under the same nutritional regimes but with exposure to continuous light, the Pi content of all tissues was diminished, yet the relative differences between the wild type and pht2;1-1 were maintained (data not shown). All noted differences between the wild type and pht2;1-1 are significant at a confidence level of >95% (Student's t test).
Table 4.
Effect of Pi Supply on the Accumulation of Pi and Total P in Plant Tissues
| Plant | Tissue | Pi in Medium (mM) |
Fresh Weight (mg/plant) |
Pi Content (nmol/mg fresh weight) |
Total P Content (nmol/mg fresh weight) |
|---|---|---|---|---|---|
| Wild type | Root | 0.2 | 22.3 ± 1.2 | 12.5 ± 1.0 | 24.8 ± 0.3 |
| pht2;1-1 | Root | 0.2 | 17.0 ± 2.0 | 9.9 ± 0.8 | 22.7 ± 0.8 |
| Wild type | Root | 4.0 | 19.2 ± 1.4 | 14.9 ± 0.8 | 26.8 ± 1.0 |
| pht2;1-1 | Root | 4.0 | 22.0 ± 0.8 | 17.7 ± 0.7 | 29.6 ± 1.4 |
| Wild type | Leaves | 0.2 | 80.4 ± 8.2 | 15.7 ± 0.5 | 27.7 ± 2.3 |
| pht2;1-1 | Leaves | 0.2 | 66.1 ± 4.3 | 21.5 ± 1.2 | 33.9 ± 1.5 |
| Wild type | Leaves | 4.0 | 216.4 ± 17.2 | 16.7 ± 0.6 | 24.9 ± 1.0 |
| pht2;1-1 | Leaves | 4.0 | 173.4 ± 7.5 | 12.2 ± 0.8 | 17.9 ± 1.2 |
Pi and total phosphorus content of roots and rosette leaves were determined for 8 to 10 plants grown for 4 weeks on agar-solidified medium. Values represent means ± se.
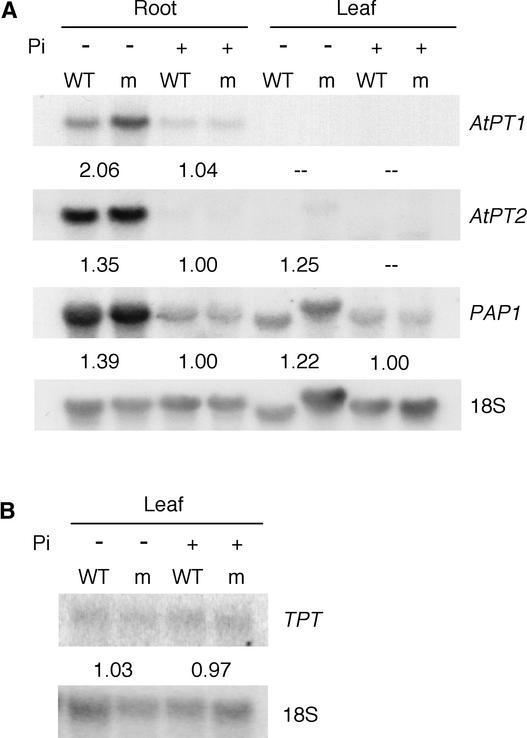
Increased Expression of Pi-Regulated Genes in the pht2;1-1 Background
We examined the effect of the pht2;1-1 mutation on the expression of several genes known to be upregulated in response to Pi deprivation. These included AtPT1 and AtPT2, which encode high-affinity Pi transporters that are expressed predominantly in the root (Muchhal et al., 1996), and PAP1, which encodes a purple acid phosphatase (Patel et al., 1997). Plants were grown on washed sand, fertilized with 1 mM Pi for 3 weeks, and then fertilized weekly with 0 or 1 mM Pi for an additional 3 weeks before harvest. RNA was isolated from pooled rosettes and roots, respectively, of eight plants grown under each condition.
RNA gel blot analysis showed that expression of each of the Pi-regulated genes was greater under Pi-deprived conditions in both wild-type and pht2;1-1 plants, indicating that the mechanisms responsible for this Pi deprivation response are functional in the mutant (Figure 8A). However, when the relative levels of expression of these genes in pht2;1-1 tissues were compared with those of the wild type, it was clear that expression was greater in the mutant under Pi-deprived conditions but not under Pi-sufficient conditions.
Figure 8.
Expression of Pi-Responsive Genes in Wild-Type and pht2;1-1 Mutant Plants Grown under Low-Pi and High-Pi Conditions.
Sequential RNA gel blot analyses were performed with 5 μg of total RNA from root and leaf tissues and hybridized with AtPT1, AtPT2, and PAP1 probes (A) and a TPT probe (B). Signal intensities were quantified using a PhosphorImager and normalized to that of the corresponding 18S rRNA. The numbers beneath each gel reflect the ratio (mutant to wild type) of the normalized signal intensities for the corresponding probe. Dashed lines indicate ratios that could not be determined because of signal intensities below the limit of detection. Separate blots were used in (A) and (B), but the samples were derived from the same tissues. The results shown represent one of two independent experiments. −Pi, low Pi; +Pi, high Pi; m, mutant; WT, wild type.
Although the increased level of expression for any individual gene was slight, collectively, the trend for increased expression under Pi-deprived conditions was significant (P = 0.02). Similar results were observed from an additional independent experiment as well as from plants grown for 4 weeks on agar medium containing 0.2 and 4 mM Pi (data not shown). By contrast, RNA gel blot analysis using the same RNA samples described above but hybridized with a TPT cDNA probe showed no significant difference in expression between the genetic backgrounds (Figure 8B).
Retranslocation of Pi in Leaves during Pi Deprivation
Under conditions of Pi deficiency, a redistribution of phosphate within the plant occurs that includes the translocation of Pi from older leaves to younger leaves (Mimura, 1995; Schachtman et al., 1998). To determine whether the redistribution process is functional in pht2;1-1 plants, we monitored the Pi content of young and old leaves during Pi deprivation. Young and old leaves were defined as the eight newest and eight oldest leaves in a rosette, respectively. Plants were grown on sand under high-Pi conditions (1 mM) for 5 weeks and then rinsed thoroughly with fertilizer lacking Pi to initiate Pi deprivation.
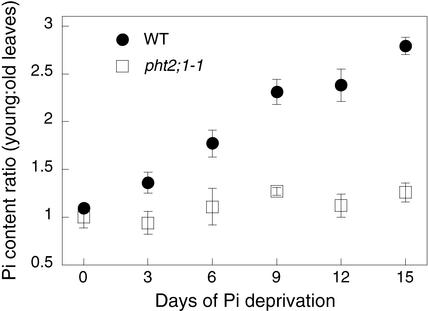
As shown in Figure 9, wild-type plants displayed the expected redistribution of Pi, that is, the ratio of young-to-old leaf Pi content increased with time of Pi deprivation. The increasing ratio represents both a reduction in the Pi content of old leaves and a modest increase in young leaves. By contrast, pht2;1-1 plants did not undergo a similar redistribution over this time course; the Pi content of both old and young leaves remained nearly constant.
Figure 9.
Pi Retranslocation in Wild-Type and pht2;1-1 Leaves during Pi Deprivation.
For each time point, young and old leaves, defined as the eight newest and eight oldest leaves in a rosette, respectively, were harvested from eight plants. The Pi content (nmol Pi/mg fresh weight) was determined from three to five replicate samples. The ratio of young-to-old leaf Pi content is plotted as a function of time after Pi deprivation. Values shown are means ± se. WT, wild type.
DISCUSSION
Although some members of the major facilitator superfamily are known to play a role in transport into organelles, until now, transporters of the PiT and PHS families have not been found in organellar membranes. The initial clue to the location of PHT2;1 came from its long N-terminal extension, which is a feature unique to the plant members of the PiT family. In other transport families, extended N-terminal regions have been associated with regulation (Harper et al., 1998; Pittman and Hirschi, 2001), and Daram et al. (1999) proposed this function for PHT2;1 as well. However, computer predictions indicated that the N-terminal region probably was a transit peptide, and localization of a PHT2;1-GFP fusion protein confirmed that PHT2;1 resides in the chloroplast. The ChloroP 1.1 program predicted correctly a chloroplast localization for PHT2;1 and suggested a cleavage site between amino acids 71 and 72, which results in a transit peptide similar in length to those of the TPTs (Fischer et al., 1994).
Some chloroplast transit peptides target their attached preproteins to mitochondria when expressed in fungal cells (Hurt et al., 1986; Pfaller et al., 1989; Brink et al., 1994), and this is true of PHT2;1 as well. The full-length protein is targeted to yeast mitochondria, whereas removal of its putative transit peptide results in localization to the plasma membrane and endoplasmic reticulum. Therefore, the report by Daram et al. (1999) that full-length PHT2;1 functions as a low-affinity H+/Pi symporter when expressed in yeast requires careful interpretation, because one requirement for such transport assays is that the transporter be located in the plasma membrane.
At least two possible explanations exist to account for the activity attributed to its presence. First, the uptake activity may reflect only a small, undetected portion of the expressed protein that has reached the plasma membrane via a nonspecific mechanism. Alternatively, insertion of PHT2;1 into mitochondrial membranes, where it may be functional, could have pleiotropic effects on the control of Pi homeostasis. Thus, Pi transport across the plasma membrane may be affected indirectly, which presumably would involve the endogenous low-affinity Pi transporters (Tamai et al., 1985; Wykoff and O'Shea, 2001).
To investigate PHT2;1 Pi transport activity, we compared transport mediated by PHT2;1 with that mediated by PHT2;1(Δ1-71), the truncated transporter lacking the transit peptide. If the former hypothesis is correct, then yeast bearing PHT2;1(Δ1-71) should have similar transport properties to yeast bearing the full-length transporter, but with greater activity as a result of the increased amount of protein targeted to the plasma membrane. Our findings are consistent with this hypothesis. Pi transport mediated by the full-length and truncated transporters displayed similar pH optima and apparent Km values, but the activity of cells bearing PHT2;1(Δ1-71) was approximately eightfold greater than that of cells carrying the full-length transporter under all conditions. In addition, the apparent Km values for the full-length and truncated transporter were significantly less than those for cells carrying the empty vector.
Based on the in planta localization of the full-length transporter, features of the transit peptide, and functional analyses in yeast, we conclude that PHT2;1 is a low-affinity Pi transporter located in the chloroplast inner envelope membrane. The pH dependence of Pi transport and the inhibitory effect of protonophores (Daram et al., 1999) suggest that PHT2;1 is a H+/Pi symporter. Therefore, the pH difference that is maintained across the inner envelope membrane could be used to energize Pi import into the stroma.
The low affinity for Pi displayed by PHT2;1 is consistent with the relatively high Pi concentration expected within the cytoplasm. Although measurement of the Pi content of subcellular compartments is difficult and may not accurately represent the metabolically active concentration, estimates of 10 to 15 mM for the cytoplasm (Mimura, 1999) and 20 to 35 mM for the chloroplast (Dietz and Heber, 1984) suggest that the chloroplast may require mechanisms capable of concentrating Pi. Pi import into the chloroplast generally is cited as occurring mainly via TPT; however, TPT mediates the stoichiometric counterexchange of triose phosphate or 3-phosphoglycerate for Pi and thus would have limited obvious potential for concentrating Pi.
Most transgenic plants with diminished levels of TPT lack a visible phenotype (Riesmeier et al., 1993; Barnes et al., 1994; Heineke et al., 1994; Häusler et al., 1998), and it has been proposed that this is attributable to a compensatory increase in starch turnover and Glc export to the cytosol (Heineke et al., 1994; Häusler et al., 1998). Although carbon metabolism is altered is these plants, the reduction in TPT levels does not result in reduced photosynthetic rates under ambient conditions (Riesmeier et al., 1993; Barnes et al., 1994; Heineke et al., 1994; Häusler et al., 1998), which is consistent with the presence of an alternative mechanism for Pi import.
Our investigation of the null mutant pht2;1-1 reveals that PHT2;1 function affects the accumulation of Pi in leaves and the allocation of Pi throughout the plant. Under high-Pi growth conditions, the reduction in leaf Pi content and the overall reduction in growth are consistent with Pi limitation of photosynthesis (Walker and Sivak, 1986), presumably as a result of diminished Pi import into chloroplasts. Given the viability of the mutant, it is clear that other mechanisms for transporting Pi into the chloroplast are sufficient to allow growth, albeit at a reduced level.
It is possible that TPT is solely responsible for the remaining import activity. However, other uncharacterized transporters also may share this function. Considering this possibility, it is interesting that at least five sequences exist within the Arabidopsis genome that encode proteins with similarity to mammalian type I Na+/Pi symporters, three of which are predicted to be targeted to the chloroplast.
It might be expected that under low-Pi growth conditions, the pht2;1-1 phenotypes would be similar to or even more extreme than those observed under high-Pi conditions. However, although the Pi content of pht2;1-1 roots was diminished during growth under low-Pi conditions, the root and rosette mass were comparable to those of the wild type, whereas the Pi content of the leaves was significantly greater than that of the wild type. These data indicate that under low-Pi conditions, the allocation of Pi within pht2;1-1 is altered, and apparently, the increased levels of Pi in leaves is sufficient to enable growth comparable to that of the wild type.
It is known that Pi deficiency is accompanied by increases in the translocation and retranslocation of Pi between shoots and roots (Bieleski, 1973; Drew and Saker, 1984; Jeschke et al., 1997), and it is possible that these processes do not function correctly in pht2;1-1. Alterations in Pi translocation are just one part of a complex array of responses that are triggered in plants during Pi deprivation. Other responses include the increased expression of genes that encode products involved in the acquisition of Pi, such as phosphatases and Pi transporters, and modifications in photosynthesis and metabolism. The molecular mechanisms that underlie the regulation of these Pi-starvation responses in plants are largely unknown, and although a number of Pi-starvation response mutants have been isolated (Poirier et al., 1991; Delhaize and Randall, 1995; Trull and Deikman, 1998; Chen et al., 2000), the first regulatory protein was identified only recently (Wykoff et al., 1999; Rubio et al., 2001).
By contrast, in microorganisms, Pi-starvation response systems and the mechanisms of their regulation have been well characterized (Wanner, 1993; Lenburg and O'Shea, 1996; Metzenberg, 1998). To examine other Pi-deprivation responses in pht2;1-1, we analyzed the expression of genes encoding two Pi transporters and a phosphatase during growth under both low-Pi and high-Pi conditions. For each of these genes, the level of expression in both the wild type and pht2;1-1 was greater under low-Pi conditions, indicating that the mechanisms responsible for increased expression under low-Pi conditions are functional in pht2;1-1. However, when the level of expression in mutant tissues was compared with that of the wild type, expression was greater in the mutant background under low-Pi conditions but not under high-Pi conditions. This derepression of Pi-regulated genes to a level greater than that observed in the wild type indicates that the regulation of this Pi-starvation response also is altered in the mutant.
The phosphate status of the shoot appears to control Pi uptake and other Pi-starvation responses in the roots, and although the mechanism is unknown, it is thought to involve retranslocation of Pi from the shoots to the root (Drew and Saker, 1984; Schjørring and Jensén, 1984; Marschner and Cakmak, 1986; C. Liu et al., 1998). An Arabidopsis mutant, pho2, that overaccumulates Pi in the shoots and shows a partial defect in the translocation of Pi between the shoots and roots (Delhaize and Randall, 1995; Dong et al., 1998) defines one component of this regulatory system, but to date, pho2 has not been cloned.
Similarly, one possible explanation for the phenotypes observed in pht2;1-1 is that under low-Pi growth conditions, retranslocation of Pi between the shoot and the root is impaired, resulting in higher Pi concentrations in the leaves and enhanced expression of Pi-starvation response genes in the roots. It is not known whether pht2;1-1 is impaired in its ability to retranslocate Pi to the roots; however, it is clear that pht2;1-1 fails to retranslocate Pi efficiently from old leaves to young leaves, a phenotype that is consistent with this hypothesis.
The mechanisms by which plants coordinate Pi acquisition and allocation to meet the varying demands of both photosynthetic and nonphotosynthetic tissues remain largely unknown. The pht2;1-1 mutant reveals that PHT2;1 is an important component of this process and demonstrates that this novel chloroplast transporter influences the allocation of Pi throughout the plant and affects the expression of Pi-starvation responses. The mechanisms by which PHT2;1 affects these processes will be the subject of future investigations.
It is possible that the involvement is indirect and that the distribution of Pi within the plant is controlled in response to the relative levels of Pi in the various subcellular compartments, including the chloroplast. Alternatively, PHT2;1 might be involved directly, as in yeast and E. coli, in which protein complexes that include Pi transporters are involved in the sensing and signaling of phosphate status and affect downstream responses through interaction with regulatory proteins (Webb and Cox, 1994; Yompakdee et al., 1996).
METHODS
Plasmids
PHT2;1 cDNA was amplified from Arabidopsis thaliana ecotype Columbia RNA by reverse transcriptase–mediated (RT)–PCR using an oligonucleotide-(dT)18 primer in the reverse transcriptase reaction, followed by PCR with gene-specific primers based on the ATORF02 sequence. PCR primers were designed to introduce unique BamHI and XhoI sites at the 5′ and 3′ ends of the gene, respectively (primer B, 5′-CGTGGATCCATGACTCTTCCTTATCG-3′; primer E, 5′-GTCCTCGAGCATGTTCTTCGTATAAC-3′). The amplified 1.8-kb cDNA was cloned into the yeast expression vector pWV3, creating PHT2;1/pWV3. The vector pWV3 is a derivative of pGAD GH (Clontech, Palo Alto, CA), in which the region encoding the GAL4 activation domain has been deleted. Both strands of the PHT2;1 insert were sequenced to ensure the integrity of the amplified sequence.
For localization experiments in Arabidopsis, a PCR-generated SalI fragment containing the complete open reading frame (ORF) of PHT2;1 was cloned in frame to the 5′ end of green fluorescent protein (GFP) in the plasmid CaMV35S-sGFP(S65T)-nos (Chiu et al., 1996) to generate PHT2;1-GFP. The plasmid PHT2;1(Δ1-71)-GFP was created similarly but lacks the first 213 bp of the PHT2;1 ORF, which corresponds to the encoded 71–amino acid putative transit peptide, and an ATG start codon has been inserted. For localization experiments in yeast, PHT2;1-GFP and PHT2;1(Δ1-71)-GFP were amplified and cloned as BamHI-XhoI fragments into pWV3 to give PHT2;1-GFP/pWV3 and PHT2;1(Δ1-71)-GFP/pWV3, respectively.
For complementation of pht2;1-1, a 4.2-kb segment of the PHT2;1 locus, spanning from 1.7 kb upstream to 0.3 kb downstream of the ORF, was amplified by PCR from Arabidopsis ecotype Columbia genomic DNA. The 4.2-kb segment was digested with SpeI and cloned into the binary vector pSKI089 to create PHT2;1/pSKI089. The vector pSKI089 is a derivative of pSKI006 (Weigel et al., 2000) in which a unique SpeI restriction site has been introduced and the HindIII site has been replaced with ClaI.
Localization of GFP Fusion Proteins
Programs used to predict protein subcellular localization include Predotar version 0.5 (http://www.inra.fr/Internet/Produits/Predotar/), TargetP version 1.0 (http://www.cbs.dtu.dk/services/TargetP/) (Emanuelsson et al., 2000), PSORT (http://psort.nibb.ac.jp/form.html), and ChloroP 1.1 (http://www.cbs.dtu.dk/services/ChloroP) (Emanuelsson et al., 1999). For localization experiments in Arabidopsis, the chimeric construct PHT2;1-GFP was introduced into 4-week-old leaves by particle bombardment using the Biolistic PDS-1000/He particle-delivery system (Bio-Rad). Bombardment parameters were as follows: pressure rupture disks rated at 900 p.s.i.; a vacuum at 28 inches of Hg; and gold particles 1.0 μm in diameter. After bombardment, leaves were incubated at room temperature for 24 h before imaging with confocal microscopy. For routine examination of transformation efficiency, we screened leaves for GFP-expressing cells using an Olympus SMZ-12 stereofluorescence microscope (Tokyo, Japan) equipped with a 100-W mercury epifluorescent light source and a standard fluorescein isothiocyanate filter set.
For localization experiments in yeast, the chimeric constructs PHT2;1-GFP/pWV3 and PHT2;1(Δ1-71)-GFP/pWV3 were introduced into the yeast mutant PAM2 (Martinez and Persson, 1998). Transformed cells were grown to the midlogarithmic phase in synthetic dextrose (SD)-Leu liquid medium, then incubated with and without the mitochondrion-selective dye MitoTracker Red CM-H2XRos (Molecular Probes, Eugene, OR) as described (Burgess et al., 1994), and then examined immediately by confocal microscopy.
Arabidopsis and yeast cells were imaged with a Bio-Rad 1024 ES confocal laser scanning microscope equipped with a ×63 water-immersion objective (numerical aperture of 1.2). Optical sections were scanned three times and the signals were averaged according to the Kalman equation (LaserSharp MRC-1024 software, Bio-Rad, Hercules, CA). Fluorescence from GFP was imaged using the 488-nm line of the krypton-argon laser, and emission was detected at 522 nm, whereas chlorophyll autofluorescence was excited at 488 nm, and emission was detected at 680 nm. Fluorescence from MitoTracker Red was excited by light at 568 nm, and emission was detected at 590 nm. Concurrent transmitted light images were recorded with differential interference contrast optics and the transmitted light detector.
Yeast Pi Transport and Growth Assays
Constructs were tested for their ability to complement the Pi uptake defect of yeast mutant PAM2 (Martinez and Persson, 1998). Pi transport assays were conducted essentially as described by Bun-ya et al. (1991), except that 25 mM Na-citrate buffer was included in the growth/uptake medium (SD-Leu) to maintain pH at the desired value. Kinetic data were analyzed by nonlinear regression to determine Michaelis-Menten parameters. For experiments testing the effect of Na+ on Pi transport, uptake assays were performed at pH 4.0 in uptake medium in which the NaCl normally present was replaced with KCl and K-citrate was substituted for Na-citrate. The final concentration of K+ ions was ∼30 mM, as determined by titration of the medium to the desired pH with KOH.
The standard growth/uptake medium contained an equivalent concentration of Na+ ions at the same pH value, as determined by titration with NaOH. Competition assays were conducted in 25 mM Na-citrate buffer, pH 4.0, containing 2% Glc. For growth comparisons, transformed cells were grown to the midlogarithmic phase in liquid low-Pi medium (SD-Leu medium containing 220 μM KH2PO4 and 25 mM Na-citrate, pH 4.0), harvested by centrifugation, and suspended in fresh medium. Cultures were incubated at 30°C with agitation (150 rpm), and OD600 was monitored over time as a measurement of growth.
Plant Growth Conditions
All plants were grown in growth rooms or chambers at 21 to 23°C with a light intensity of 100 μmol·m−2·s−1. For general propagation, plants were grown on Metro Mix 350 (Scotts, Marysville, OH) with a 16-h-light/8-h-dark cycle. For dark-treated plants, surface-sterilized seeds were germinated in flasks containing half-strength Murashige and Skoog (1962) medium, pH 5.7, and 1% Suc for 4 days with constant agitation (150 rpm) and constant light, covered with foil wrap, and grown for another 6 days. Under the growth regimes described below, plants had rosette leaves with little or no other green tissues present at the time of harvest.
For growth on agar-solidified medium, surface-sterilized seeds were grown for 4 weeks on half-strength Murashige and Skoog (1962) medium, pH 5.7, solidified with 0.7% (w/v) agar and containing either 0.2 or 4.0 mM Pi. Plants were grown in GA-7 boxes (four plants per box; Magenta Corp., Chicago, IL) with a 16-h-light/8-h-dark cycle. For all other growth regimes, surface-sterilized seeds were germinated for 10 days on agar-solidified medium containing 1 mM Pi, washed with distilled water, and transferred to the desired growth medium with a 10-h-light/14-h-dark cycle. For growth on Metro Mix 350, plants were transferred to 36-cell packs (one plant per cell; TFI Plastics, Thanesville, Ontario, Canada) for an additional 3 to 4 weeks before tissues were harvested. Plants were fertilized weekly with half-strength Hoagland solution (Arnon and Hoagland, 1940), pH 6.0, containing 1.0 mM Pi.
For growth on sand, plants were transferred to 36-cell packs (one plant per cell) containing acid-washed sand for an additional 3 weeks and fertilized weekly with half-strength Hoagland solution containing either 0.01 or 1.0 mM Pi. For Pi-starvation experiments, plants were grown on washed sand and fertilized weekly for 3 weeks with half-strength Hoagland solution containing 1 mM Pi, then fertilized weekly with half-strength Hoagland solution containing 0 or 1 mM Pi for an additional 3 weeks before tissues were harvested. A single lot of agar, which contributed 7.6 μM Pi to the medium when used at 0.7% (w/v), was used throughout this study. Pi was added to medium and fertilizer as KH2PO4. When the Pi concentration was varied, K2SO4 was added to maintain a constant K+ concentration.
PHT2;1 Mutant Screen
Pooled genomic DNA samples representing a library of 12,000 individual T-DNA lines (ABRC, Ohio State University, Columbus) were screened by PCR for T-DNA insertion mutations within the coding region of PHT2;1. T-DNA border primers read in opposite directions toward the T-DNA surrounding sequences, and PHT2;1::T-DNA junction sequences were amplified from reactions containing primer combinations specific for regions flanking both T-DNA borders. A 1.8-kb junction sequence was amplified in reactions containing the T-DNA right border primer (primer G, 5′-TCGGGCCTAACTTTT-GGTG-3′) and a PHT2;1 3′-specific primer (primer E; see above). A 2.2-kb junction sequence was amplified in reactions containing the T-DNA left border primer (primer F, 5′-GAACATCGGTCTCAATGC-A-3′) and a PHT2;1 5′-specific primer (primer A, 5′-GTCACTAGT-CGGAAGAAGCTATCATATGTATTG-3′).
Both strands of the amplified junction fragments were sequenced to confirm the presence of PHT2;1 sequence and to determine the T-DNA insertion site. Seeds from the positive pool of T-DNA lines were grown, and individual plants were screened by PCR as described above. Plants hemizygous or homozygous for the PHT2;1::T-DNA locus were distinguished by PCR using a combination of two PHT2;1-specific primers (primer C, 5′-CTTCCTTATCGTTTCTCTTCCG-3′; primer D, 5′-ATATACAAGCCCAAACCCAACC-3′) and the right border primer (primer G). An individual line homozygous for the T-DNA insertion was identified and verified by DNA gel blot analysis. We named this line and the corresponding mutant allele pht2;1-1.
Complementation of pht2;1-1
pht2;1-1 plants were transformed using the floral dip method (Clough and Bent, 1998) with either pSKI089 as a control or PHT2;1/pSKI089, which contains a 4.2-kb segment of the genomic PHT2;1 locus spanning from 1.7 kb upstream to 0.3 kb downstream of the ORF. Phosphinothricin-resistant transformants were screened by PCR to confirm the presence of the bar gene using primers described previously (Trieu et al., 2000) and to confirm the presence/absence of a wild-type PHT2;1 allele, as described above. Leaf Pi content (nmol Pi/mg fresh weight) was determined for each plant (11 wild-type plants, 8 pSKI089-transformed plants, and 12 PHT2;1/pSKI089-transformed plants).
A subset of the transformed plants were analyzed further by RT-PCR to confirm the presence of PHT2;1 transcripts. RT-PCR was conducted on 1 μg of total RNA isolated from leaves of individual plants. A 582-bp fragment of the PHT2;1 gene was amplified using 5′-CTCCTCTCTTCCTTAGCTGCAGCTG-3′ and 5′-GCGAATGAC-ATGAAGCAAGCGGAGAG-3′. As a control, a 492-bp EIF-4A2 gene fragment was amplified using 5′-GCAAGAGAATCTTCTTAGGGGTATCTATGC-3′ and 5′-GGTGGGAGAAGCTGGAATATGTCATAG-3′. The primers chosen for the amplification of PHT2;1 and EIF-4A2 gene fragments were from separate exons, and all primers were included in each PCR procedure.
Pi Content Determination
Tissues were rinsed in distilled water, blotted dry, frozen, and ground to a fine powder in liquid nitrogen. The ground tissues were suspended in 1% glacial acetic acid and mixed thoroughly. After a brief centrifugation to pellet cellular debris, aliquots of the solution were assayed for Pi using a phosphomolybdate colorimetric assay as described previously (Ames, 1966). Total phosphate in ashed and hydrolyzed samples was determined by colorimetric assay (Ames, 1966) or by inductively coupled plasma mass spectrometry, which was performed at the Soil and Plant Analysis Laboratory of the University of Wisconsin-Madison.
Plant Nucleic Acid Isolation and Blotting
DNA and RNA were isolated from plant tissues by standard methods (Dellaporta et al., 1983; Chomczynski and Sacchi, 1987). Unless indicated otherwise, tissues were harvested for RNA isolation between 1 and 2 h after the onset of the daylight portion of the photoperiod. Blotting and hybridization were performed as described previously (Church and Gilbert, 1984). A PhosphorImager (Molecular Dynamics, Sunnyvale, CA) was used to quantify signal intensities on RNA gel blots and to confirm complete probe removal for sequential hybridizations.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Numbers
The GenBank accession numbers for the sequences described in this article are X97484 (ATORF02), U84890 (Mesembryanthemum crystallinum cDNA), M31364 (PHO-4), AF515591 (PHT2;1), AJ302645 (PHT2;1), and AF386923 (EIF-4A2).
Acknowledgments
We thank G. May and W. Schneider for critical review of the manuscript and the members of the Plant Biology Division for helpful discussions. This work was supported by The Samuel Roberts Noble Foundation.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.002220.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Ames, B.N. (1966). Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 8, 115–118. [Google Scholar]
- Arnon, D.I., and Hoagland, D.R. (1940). Crop production in artificial culture solutions and in soils with special reference to factors influencing yields and absorption of inorganic nutrients. Soil Sci. 50, 463–483. [Google Scholar]
- Barnes, S.A., Knight, J.S., and Gray, J.C. (1994). Alteration of the amount of the chloroplast phosphate translocator in transgenic tobacco affects the distribution of assimilate between starch and sugar. Plant Physiol. 106, 1123–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieleski, R.L. (1973). Phosphate pools, phosphate transport and phosphate availability. Annu. Rev. Plant Physiol. 24, 225–252. [Google Scholar]
- Brink, S., Flugge, U.I., Chaumont, F., Boutry, M., Emmermann, M., Schmitz, U., Becker, K., and Pfanner, N. (1994). Preproteins of chloroplast envelope inner membrane contain targeting information for receptor-dependent import into fungal mitochondria. J. Biol. Chem. 269, 16478–16485. [PubMed] [Google Scholar]
- Bun-ya, M., Nishimura, M., Harashima, S., and Oshima, Y. (1991). The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol. Cell. Biol. 11, 3229–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, S.M., Delannoy, M., and Jensen, R.E. (1994). MMM1 encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J. Cell Biol. 126, 1375–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D.L., Delatorre, C.A., Bakker, A., and Abel, S. (2000). Conditional identification of phosphate-starvation-response mutants in Arabidopsis thaliana. Planta 211, 13–22. [DOI] [PubMed] [Google Scholar]
- Chiou, T.J., Liu, H., and Harrison, M.J. (2000). The spatial expression patterns of a phosphate transporter (MtPT1) from Medicago truncatula indicate a role in phosphate transport at the root/soil interface. Plant J. 25, 1–15. [DOI] [PubMed] [Google Scholar]
- Chiu, W., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6, 325–330. [DOI] [PubMed] [Google Scholar]
- Chomczynski, P., and Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159. [DOI] [PubMed] [Google Scholar]
- Church, G.M., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Daram, P., Brunner, S., Persson, B.L., Amrhein, N., and Bucher, M. (1998). Functional analysis and cell-specific expression of a phosphate transporter from tomato. Planta 206, 225–233. [DOI] [PubMed] [Google Scholar]
- Daram, P., Brunner, S., Rausch, C., Steiner, C., Amrhein, N., and Bucher, M. (1999). Pht2;1 encodes a low-affinity phosphate transporter from Arabidopsis. Plant Cell 11, 2153–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize, E., and Randall, P.J. (1995). Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol. 107, 207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta, S.L., Wood, J., and Hicks, J.B. (1983). A plant DNA minipreparation. Plant Mol. Biol. Rep. 1, 19–22. [Google Scholar]
- Dietz, K., and Heber, U. (1984). Rate-limiting factors in leaf photosynthesis. I. Carbon fluxes in the Calvin cycle. Biochim. Biophys. Acta 767, 432–443. [Google Scholar]
- Dong, B., Rengel, Z., and Delhaize, E. (1998). Uptake and translocation of phosphate by pho2 mutant and wild-type seedlings of Arabidopsis thaliana. Planta 205, 251–256. [DOI] [PubMed] [Google Scholar]
- Drew, M.C., and Saker, L.R. (1984). Uptake and long-distance transport of phosphate, potassium and chloride in relation to internal ion concentrations in barley: Evidence of non-allosteric regulation. Planta 160, 500–507. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., and von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, K., Arbinger, B., Kammerer, B., Busch, C., Brink, S., Wallmeier, H., Sauer, N., Eckerskorn, C., and Flügge, U.I. (1994). Cloning and in vivo expression of functional triose phosphate/phosphate translocators from C3- and C4-plants: Evidence for the putative participation of specific amino acid residues in the recognition of phosphoenolpyruvate. Plant J. 5, 215–226. [DOI] [PubMed] [Google Scholar]
- Fischer, K., Kammerer, B., Gutensohn, M., Arbinger, B., Weber, A., Häusler, R.E., and Flügge, U.I. (1997). A new class of plastidic phosphate translocators: A putative link between primary and secondary metabolism by the phosphoenolpyruvate/phosphate antiporter. Plant Cell 9, 453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flügge, U.I. (1999). Phosphate translocators in plastids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 27–45. [DOI] [PubMed] [Google Scholar]
- Flügge, U.I., Fischer, K., Gross, A., Sebald, W., Lottspeich, F., and Eckerskorn, C. (1989). The triose phosphate-3-phosphoglycerate-phosphate translocator from spinach chloroplasts: Nucleotide sequence of a full-length cDNA clone and import of the in vitro synthesized precursor protein into chloroplasts. EMBO J. 8, 39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, J.F., Hong, B., Hwang, I., Guo, H.Q., Stoddard, R., Huang, J.F., Palmgren, M.G., and Sze, H. (1998). A novel calmodulin-regulated Ca2+-ATPase (ACA2) from Arabidopsis with an N-terminal autoinhibitory domain. J. Biol. Chem. 273, 1099–1106. [DOI] [PubMed] [Google Scholar]
- Harris, R.M., Webb, D.C., Howitt, S.M., and Cox, G.B. (2001). Characterization of PitA and PitB from Escherichia coli. J. Bacteriol. 183, 5008–5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, M.J., and van Buuren, M.L. (1995). A phosphate transporter from the mycorrhizal fungus Glomus versiforme. Nature 378, 626–629. [DOI] [PubMed] [Google Scholar]
- Häusler, R.E., Schlieben, N.H., Schultz, B., and Flügge, U. (1998). Compensation of the decreased triose phosphate/phosphate translocator activity by accelerated starch turnover and glucose transport in transgenic tobacco. Planta 204, 366–376. [DOI] [PubMed] [Google Scholar]
- Heineke, D., Kruse, A., Flügge, U., Frommer, W.B., Riesmeier, J.W., Willmitzer, L., and Heldt, H.W. (1994). Effect of antisense repression of the chloroplast triose-phosphate translocator on photosynthetic metabolism in transgenic potato plants. Planta 193, 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt, E.C., Sotanifar, N., Goldschmidt-Clermont, M., Rochaix, J., and Schatz, G. (1986). The cleavable pre-sequence of an imported chloroplast protein directs attached polypeptides into yeast mitochondria. EMBO J. 5, 1343–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeschke, W.D., Kirkby, E.A., Peuke, A.D., Pate, J.S., and Hartung, W. (1997). Effects of P deficiency on assimilation and transport of nitrate and phosphate in intact plants of castor bean (Ricinus communis L.). J. Exp. Bot. 48, 75–91. [Google Scholar]
- Johann, S.V., Gibbons, J.J., and O'Hara, B. (1992). GLVR1, a receptor for gibbon ape leukemia virus, is homologous to a phosphate permease of Neurospora crassa and is expressed at high levels in the brain and thymus. J. Virol. 66, 1635–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai, M., Masuda, Y., Kikuchi, Y., Osaki, M., and Tadano, T. (1997). Isolation and characterization of a cDNA from Catharanthus roseus which is highly homologous with phosphate transporter. Soil Sci. Plant Nutr. 43, 227–235. [Google Scholar]
- Kammerer, B., Fischer, K., Hilpert, B., Schubert, S., Gutensohn, M., Weber, A., and Flügge, U.I. (1998). Molecular characterization of a carbon transporter in plastids from heterotrophic tissues: The glucose 6-phosphate/phosphate antiporter. Plant Cell 10, 105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanaugh, M.P., Miller, D.G., Zhang, W., Law, W., Kozak, S.L., Kabat, D., and Miller, A.D. (1994). Cell-surface receptors for gibbon ape leukemia virus and amphotropic murine retrovirus are inducible sodium-dependent phosphate symporters. Proc. Natl. Acad. Sci. USA 91, 7071–7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloi, M. (1999). Plant mitochondrial carriers: An overview. Cell. Mol. Life Sci. 56, 918–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggewie, G., Willmitzer, L., and Riesmeier, J.W. (1997). Two cDNAs from potato are able to complement a phosphate uptake-deficient yeast mutant: Identification of phosphate transporters from higher plants. Plant Cell 9, 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenburg, M.E., and O'Shea, E.K. (1996). Signaling phosphate starvation. Trends Biochem. Sci. 21, 383–387. [PubMed] [Google Scholar]
- Liu, C., Muchhal, U.S., Uthappa, M., Kononowicz, A.K., and Raghothama, K.G. (1998). Tomato phosphate transporter genes are differentially regulated in plant tissues by phosphorus. Plant Physiol. 116, 91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Trieu, A.T., Blaylock, L.A., and Harrison, M.J. (1998). Cloning and characterization of two phosphate transporters from Medicago truncatula roots: Regulation in response to phosphate and to colonization by arbuscular mycorrhizal (AM) fungi. Mol. Plant-Microbe Interact. 11, 14–22. [DOI] [PubMed] [Google Scholar]
- Mann, B.J., Bowman, B.J., Grotelueschen, J., and Metzenberg, R.L. (1989). Nucleotide sequence of pho-4+, encoding a phosphate-repressible phosphate permease of Neurospora crassa. Gene 83, 281–289. [DOI] [PubMed] [Google Scholar]
- Marschner, H. (1995). Mineral Nutrition of Higher Plants. (London: Academic Press).
- Marschner, H., and Cakmak, I. (1986). Mechanism of phosphorus-induced zinc deficiency in cotton. II. Evidence for impaired shoot control of phosphorus uptake and translocation under zinc deficiency. Physiol. Plant. 68, 491–496. [Google Scholar]
- Martinez, P., and Persson, B.L. (1998). Identification, cloning and characterization of a derepressible Na+-coupled phosphate transporter in Saccharomyces cerevisiae. Mol. Gen. Genet. 258, 628–638. [DOI] [PubMed] [Google Scholar]
- Metzenberg, R.L. (1998). How Neurospora crassa gets its phosphorus. In Phosphorus in Plant Biology: Regulatory Roles in Molecular, Cellular, Organismic, and Ecosystem Processes, J.P. Lynch and J. Deikman, eds (Rockville, MD: American Society of Plant Physiologists), pp. 181–191.
- Miller, D.G., Edwards, R.H., and Miller, A.D. (1994). Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc. Natl. Acad. Sci. USA 91, 78–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura, T. (1995). Homeostasis and transport of inorganic phosphate in plants. Plant Cell Physiol. 36, 1–7. [Google Scholar]
- Mimura, T. (1999). Regulation of phosphate transport and homeostasis in plant cells. Int. Rev. Cytol. 191, 149–200. [Google Scholar]
- Mitsukawa, N., Okumura, S., Shirano, Y., Sato, S., Kato, T., Harashima, S., and Shibata, D. (1997). Overexpression of an Arabidopsis thaliana high-affinity phosphate transporter gene in tobacco cultured cells enhances cell growth under phosphate-limited conditions. Proc. Natl. Acad. Sci. USA 94, 7098–7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchhal, U.S., Pardo, J.M., and Raghathama, K.G. (1996). Phosphate transporters from the higher plant Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 93, 10519–10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchhal, U.S., and Raghothama, K.G. (1999). Transcriptional regulation of plant phosphate transporters. Proc. Natl. Acad. Sci. USA 96, 5868–5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. [Google Scholar]
- Okumura, S., Mitsukawa, N., Shirano, Y., and Shibata, D. (1998). Phosphate transporter gene family of Arabidopsis thaliana. DNA Res. 5, 261–269. [DOI] [PubMed] [Google Scholar]
- Olah, Z., Lehel, C., Anderson, W.B., Eiden, M.V., and Wilson, C.A. (1994). The cellular receptor for gibbon ape leukemia virus is a novel high affinity sodium-dependent phosphate transporter. J. Biol. Chem. 269, 25426–25431. [PubMed] [Google Scholar]
- Pao, S.S., Paulsen, I.T., and Saier, M.H., Jr. (1998). Major facilitator superfamily. Microbiol. Mol. Biol. 62, 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, K., Lockless, S., Thomas, B., and McKnight, T. (1997). A secreted purple acid phosphatase from Arabidopsis. Plant Physiol. 111, 81. [Google Scholar]
- Pfaller, R., Pfanner, N., and Neupert, W. (1989). Mitochondrial protein import: Bypass of proteinaceous surface receptors can occur with low specificity and efficiency. J. Biol. Chem. 264, 34–39. [PubMed] [Google Scholar]
- Pittman, J.K., and Hirschi, K.D. (2001). Regulation of CAX1, an Arabidopsis Ca2+/H+ antiporter: Identification of an N-terminal autoinhibitory domain. Plant Physiol. 127, 1020–1029. [PMC free article] [PubMed] [Google Scholar]
- Poirier, Y., Thoma, S., Somerville, C., and Schiefelbein, J. (1991). A mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol. 97, 1087–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley, F., Dao, P., Cottet, A., and Mache, R. (1996). Sequence analysis of an 81 kb contig from Arabidopsis thaliana chromosome III. Nucleic Acids Res. 24, 4313–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama, K. (1999). Phosphate acquisition. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 665–693. [DOI] [PubMed] [Google Scholar]
- Riesmeier, J.W., Flügge, U.I., Schulz, B., Heineke, D., Heldt, H.W., Willmitzer, L., and Frommer, W.B. (1993). Antisense repression of the chloroplast triose phosphate translocator affects carbon partitioning in transgenic potato plants. Proc. Natl. Acad. Sci. USA 90, 6160–6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio, V., Linhares, F., Solano, R., Martin, A.C., Iglesias, J., Leyva, A., and Paz-Ares, J. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15, 2122–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier, M.H., Jr. (2000). A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. 64, 354–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier, M.H., Jr., et al. (1999). Phylogenetic characterization of novel transport protein families revealed by genome analyses. Biochim. Biophys. Acta 1422, 1–56. [DOI] [PubMed] [Google Scholar]
- Schachtman, D.P., Reid, R.J., and Ayling, S.M. (1998). Phosphorus uptake by plants: From soil to cell. Plant Physiol. 116, 447–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjørring, J.K., and Jensén, P. (1984). Phosphorus nutrition of barley, buckwheat and rape seedlings. I. Influence of seed-borne P and external P levels on growth, P content and 32P/31P-fractionation in shoots and roots. Physiol. Plant. 61, 577–583. [Google Scholar]
- Schulz, B., Frommer, W.B., Flugge, U.I., Hummel, S., Fischer, K., and Willmitzer, L. (1993). Expression of the triose phosphate translocator gene from potato is light dependent and restricted to green tissues. Mol. Gen. Genet. 238, 357–361. [DOI] [PubMed] [Google Scholar]
- Smith, F.W., Ealing, P.M., Dong, B., and Delhaize, E. (1997). The cloning of two Arabidopsis genes belonging to a phosphate transporter family. Plant J. 11, 83–92. [DOI] [PubMed] [Google Scholar]
- Stappen, R., and Kramer, R. (1994). Kinetic mechanism of phosphate/phosphate and phosphate/OH− antiports catalyzed by reconstituted phosphate carrier from beef heart mitochondria. J. Biol. Chem. 269, 11240–11246. [PubMed] [Google Scholar]
- Takabatake, R., Hata, S., Taniguchi, M., Kouchi, H., Sugiyama, T., and Izui, K. (1999). Isolation and characterization of cDNAs encoding mitochondrial phosphate transporters in soybean, maize, rice, and Arabidopsis. Plant Mol. Biol. 40, 479–486. [DOI] [PubMed] [Google Scholar]
- Tamai, Y., Toh-e, A., and Oshima, Y. (1985). Regulation of inorganic phosphate transport systems in Saccharomyces cerevisiae. J. Bacteriol. 164, 964–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu, A.T., Burleigh, S.H., Kardailsky, I.V., Maldonado-Mendoza, I.E., Versaw, W.K., Blaylock, L.A., Shin, H., Chiou, T.-J., Katagi, H., Dewbre, G.R., Weigel, D., and Harrison, M.J. (2000). Transformation of Medicago truncatula via infiltration of seedlings or flowering plants with Agrobacterium. Plant J. 22, 531–542. [DOI] [PubMed] [Google Scholar]
- Trull, M.C., and Deikman, J. (1998). An Arabidopsis mutant missing one acid phosphatase isoform. Planta 206, 544–550. [DOI] [PubMed] [Google Scholar]
- Versaw, W.K. (1995). A phosphate-repressible, high-affinity phosphate permease is encoded by the pho-5+ gene of Neurospora crassa. Gene 153, 135–139. [DOI] [PubMed] [Google Scholar]
- Versaw, W.K., and Metzenberg, R.L. (1995). Repressible cation-phosphate symporters in Neurospora crassa. Proc. Natl. Acad. Sci. USA 92, 3884–3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, D.A., and Sivak, M.N. (1986). Photosynthesis and phosphate: A cellular affair? Trends Biochem. Sci. 11, 176–179. [Google Scholar]
- Wanner, B.L. (1993). Gene regulation by phosphate in enteric bacteria. J. Cell. Biochem. 51, 47–54. [DOI] [PubMed] [Google Scholar]
- Webb, D.C., and Cox, G.B. (1994). Proposed mechanism for phosphate translocation by the phosphate-specific transport (Pst) system and role of the Pst system in phosphate regulation. In Phosphates in Microorganisms: Cellular and Molecular Biology, A. Torriani-Gorini, E. Yagil, and S. Silver, eds (Washington, DC: American Society for Microbiology), pp. 37–42.
- Weigel, D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122, 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlrab, H., and Briggs, C. (1994). Yeast mitochondrial phosphate transport protein expressed in Escherichia coli: Site-directed mutations at threonine-43 and at a similar location in the second tandem repeat (isoleucine-141). Biochemistry 33, 9371–9375. [DOI] [PubMed] [Google Scholar]
- Wykoff, D.D., Grossman, A.R., Weeks, D.P., Usuda, H., and Shimogawara, K. (1999). Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proc. Natl. Acad. Sci. USA 96, 15336–15342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff, D.D., and O'Shea, E.K. (2001). Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics 159, 1491–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yompakdee, C., Ogawa, N., Harashima, S., and Oshima, Y. (1996). A putative membrane protein, Pho88p, involved in inorganic phosphate transport in Saccharomyces cerevisiae. Mol. Gen. Genet. 251, 580–590. [DOI] [PubMed] [Google Scholar]