Abstract
Mutants able to germinate and perform early growth in medium containing a high NaCl concentration were identified during the course of two independent screenings and named salt resistant (sre) and salobreño (sañ). The sre and sañ mutants also were able to germinate in high-osmoticum medium, indicating that they are osmotolerant in a germination assay. Complementation analyses revealed that sre1-1, sre1-2, sañ3-1, and sañ3-2 were alleles of the abscisic acid (ABA) biosynthesis ABA2 gene. A map-based cloning strategy allowed the identification of the ABA2 gene and molecular characterization of four new aba2 alleles. The ABA2 gene product belongs to the family of short-chain dehydrogenases/reductases, which are known to be NAD- or NADP-dependent oxidoreductases. Recombinant ABA2 protein produced in Escherichia coli exhibits a Km value for xanthoxin of 19 μM and catalyzes in a NAD-dependent manner the conversion of xanthoxin to abscisic aldehyde, as determined by HPLC–mass spectrometry. The ABA2 mRNA is expressed constitutively in all plant organs examined and is not upregulated in response to osmotic stress. The results of this work are discussed in the context of previous genetic and biochemical evidence regarding ABA biosynthesis, confirming the xanthoxin→abscisic aldehyde→ABA transition as the last steps of the major ABA biosynthetic pathway.
INTRODUCTION
ABA is a sesquiterpenoid plant hormone involved in the control of a wide range of essential physiological processes, mainly seed development and germination as well as plant responses to different stresses (Zeevaart and Creelman, 1988; Koornneef and Karssen, 1994; Rock and Quatrano, 1995; Leung and Giraudat, 1998). The level of abscisic acid (ABA) increases in plants during seed development and under many environmental stresses, particularly drought and salinity. Thus, understanding of the ABA biosynthetic pathway and its regulation is of crucial importance in elucidating the factors that mediate both plant development and stress responses.
During embryogenesis and seed formation, ABA is involved in the control of many events, such as embryo morphogenesis, storage protein synthesis, desiccation tolerance, and the onset and maintenance of dormancy (Koornneef and Karssen, 1994; Rock and Quatrano, 1995). During seed development, ABA levels increase at the end of embryogenesis, are maximal during mid development when storage reserves are accumulated, and then decline during desiccation. Therefore, the peak of ABA precedes the desiccation step, and it is assumed that it plays a crucial role in allowing the embryo to withstand dehydration. Thus, water deficit is a normal component of seed development, and the natural process of seed development involves physiological and biochemical changes that may be similar to responses observed in adult plants exposed to drought stress.
During vegetative growth, ABA has been shown to be essential in plant responses to environmental stresses involving water deficit, such as the regulation of stomatal aperture and gene expression (Skriver and Mundy, 1990; Chandler and Robertson, 1994; Leung and Giraudat, 1998). Plant water deficit occurs when the rate of transpiration exceeds the rate of water uptake. Therefore, ABA regulation of the transpiration flow through stomatal pores is a crucial response of the plant to water deficit. ABA is synthesized in response to drought stress and triggers a calcium-activated signaling cascade in guard cells that results in stomatal closing (Schroeder et al., 2001).
ABA-deficient mutants are known in maize, Arabidopsis, and a few other species, and they are affected in different steps of ABA biosynthesis. For instance, the conversion of zeaxanthin to all-trans-violaxanthin by a two-step epoxidation is impaired in the Arabidopsis aba1 and Nicotiana plumbaginifolia aba2 mutants (Marin et al., 1996; Audran et al., 2001). Both loci encode a zeaxanthin epoxidase (ZEP), which was the first enzyme to be identified as an ABA biosynthetic enzyme (Marin et al., 1996). The next step in ABA biosynthesis involves the conversion of all-trans-violaxanthin to 9-cis-violaxanthin or 9′-cis-neoxanthin (Schwartz et al., 1997b). Mutants affected in this step have not been identified (Tan et al., 1997; Seo and Koshiba, 2002). The oxidative cleavage of 9-cis-violaxanthin and/or 9′-cis-neoxanthin to produce xanthoxin is catalyzed by a 9-cis-epoxycarotenoid dioxygenase (NCED), which appears to be the key regulatory step of ABA biosynthesis (Schwartz et al., 1997b; Tan et al., 1997; Qin and Zeevaart, 1999). The maize viviparous14 and the tomato notabilis mutants both are impaired in the reaction catalyzed by NCED (Tan et al., 1997; Burbidge et al., 1999). The steps downstream of the cleavage reaction are proposed to be the conversion of xanthoxin to ABA-aldehyde, which is defective in the Arabidopsis aba2 mutant (Leon-Kloosterziel et al., 1996; Schwartz et al., 1997a), and the oxidation of ABA-aldehyde to ABA, which involves the recently characterized ABA3 and abscisic aldehyde oxidase (AAO3) genes (Seo et al., 2000a, 2000b; Bittner et al., 2001; Xiong et al., 2001). Alternatively, a different model involving the oxidation of xanthoxin to xanthoxic acid, and further oxidation and rearrangement to ABA, also has been proposed (Milborrow, 2001).
During the course of a screening for mutants able to germinate under high salt concentration, we (M.G.-G., R.S., and P.L.R.) isolated several candidates that upon genetic analysis fell into four complementation groups (data not shown); these were named sre1 to sre4 for salt resistant in a germination assay. In particular, the sre1-1 and sre1-2 mutants were found to be alleles of the ABA2 gene. Previously, Quesada et al. (2000) reported the isolation of salobreño (sañ) mutants, which are able to germinate under 250 mM NaCl. In this work, sañ3-1 and sañ3-2 were found to be alleles of the ABA2 gene. Map-based cloning of sre1-1 revealed that the corresponding gene encodes a putative short-chain alcohol dehydrogenase.
A preliminary report describing the identification of the Arabidopsis At1g52340 gene as ABA2 was published previously (Rook et al., 2001). Our data confirm and extend this result, because several aba2 alleles were characterized molecularly in this work, whereas only the aba2-1 allele was characterized unequivocally by Rook et al. (2001). The other putative aba2 allele (isi4) complemented the aba2-1 phenotype, and Rook et al. (2001) discussed the possibility that isi4 represents a different closely linked locus. Alternatively, the possibility of interallelic complementation between the aba2-1 and isi4 gene products also was discussed. The multimeric nature of short-chain dehydrogenase/reductase (SDR) enzymes allows for the possibility of interallelic complementation; however, this hypothesis was not addressed experimentally (Rook et al., 2001). This option might explain the apparent contradiction observed, that is, that the Glc-insensitive gin1-1 mutant (a putative null aba2 allele [J. Sheen, personal communication]) was allelic to both isi4 and aba2-1, whereas the latter two mutants complemented each other (Rook et al., 2001). No biochemical characterization of the ABA2 gene product was reported by Rook et al. (2001).
In this work, we report the cloning of the ABA2 cDNA and show that recombinant ABA2 protein was able to efficiently oxidize xanthoxin in a NAD-dependent manner; the reaction product was determined by HPLC–mass spectrometry to be abscisic aldehyde. This result demonstrates that the conversion of xanthoxin to abscisic aldehyde is catalyzed by a single enzyme. Together with previous genetic and biochemical evidence (Sindhu and Walton, 1987, 1988; Rock and Zeevaart, 1990; Schwartz et al., 1997a; revised in Cutler and Krochko, 1999), these results confirm that the conversion of xanthoxin to abscisic aldehyde catalyzed by ABA2 and then to ABA by AAO3 (Seo et al., 2000a, 2000b) represent the last steps of the major ABA biosynthetic pathway.
RESULTS
Identification of New aba2 Alleles
During the course of a screening for mutants able to germinate and perform early seedling growth under inhibitory concentrations of NaCl, we identified a mutant (in a Columbia [Col] genetic background), sre1, that additionally showed reduced dormancy and a dramatic wilty phenotype (data not shown). It has been reported that alleles of both ABA-insensitive and ABA-deficient loci can be recovered in germination-based screenings for mutants resistant to high salt or high sugar concentrations (Leon-Kloosterziel et al., 1996; Huijser et al., 2000; Laby et al., 2000; Quesada et al., 2000). Therefore, to determine whether sre1 plants are insensitive to ABA or deficient in ABA, we tested the ABA and paclobutrazol sensitivity of the mutant in a seed germination assay (Table 1).
Table 1.
Percentage of Seed Germination in the Presence of ABA and Paclobutrazol
| Seed | 3 μM ABA | Paclobutrazol |
|---|---|---|
| Col | 0 | 0a |
| Ler | 0 | 0a |
| sre1-1 | 0 | 90 ± 5a |
| sre1-2 | 0 | 87 ± 6a |
| aba1 | 0 | 88 ± 4a |
| aba2-1 | 0 | 89 ± 6a |
| aba3-2 | 0 | 62 ± 4a |
| abi1 | 92 ± 4 | 65 ± 5a |
| sañ3-1 | 0 | 36 ± 7b |
| sañ3-2 | 0 | 68 ± 3b |
Approximately 200 seeds of each line were sown on MS plates supplemented with 3 μM ABA or paclobutrazol. sd values were calculated from three independent experiments. Ler, Landsberg erecta.
Medium supplemented with 10 μM paclobutrazol.
Medium supplemented with 100 μM paclobutrazol.
sre1 was not able to germinate in the presence of ABA; therefore, it is not an abi mutant. Instead, sre1 showed >85% germination in the presence of the gibberellin biosynthesis inhibitor paclobutrazol (Koornneef et al., 1982). These data, as well as the reduced dormancy and dramatic wilty phenotype, suggest that sre1 is an ABA-deficient mutant. Indeed, complementation tests revealed that sre1 was allelic to aba2-1 (Table 2). In addition, the sañ mutants sañ3-1 (in a Landsberg erecta background) and sañ3-2 (in a Col background) (Quesada et al., 2000) also were found to be resistant to paclobutrazol (Table 1) and alleles of ABA2 (Table 2).
Table 2.
Complementation Tests of Arabidopsis sre1, sañ3, and aba Mutants
| Cross | Generation | Total Seeds Sown |
Germinated |
|---|---|---|---|
| sre1-1/sre1-1 × sre1-2/sre1-2 | F1 | 111 | 106 |
| sre1-1/sre1-1 × aba1-1/aba1-1 | F1 | 70 | 0 |
| sre1-1/sre1-1 × aba2-1/aba2-1 | F1 | 147 | 145 |
| sre1-1/sre1-1 × aba3-2/aba3-2 | F1 | 92 | 0 |
| sañ3-1/sañ3-1 × sañ3-2/sañ3-2 | F1 | 561 | 228 |
| sañ3-2/sañ3-2 × aba2-1/aba2-1 | F1 | 54 | 44 |
Complementation tests were performed by analyzing intercrosses among sre1, sañ3, and aba homozygous mutants. F1 seeds were scored for germination in 150 mM NaCl at 5 days after sowing (crosses involving sre1), 250 mM NaCl (cross sañ3-1/sañ3-1 × sañ3-2/sañ3-2), or 100 μM paclobutrazol (cross sañ3-2/sañ3-2 × aba2-1/aba2-1) at 7 days after sowing.
Five aba2 alleles have been described previously (Leon-Kloosterziel et al., 1996; Nambara et al., 1998; Laby et al., 2000; Rook et al., 2001), and several new aba2 alleles have been isolated by J. Sheen (personal communication). We have resolved the designation of our aba2 alleles by initiating a new allelic series. Two alleles of sre1 (sre1-1 and sre1-2) were isolated in this work (Table 2), and we have renamed them aba2-11 and aba2-12, respectively. Accordingly, the two alleles of sañ3, sañ3-1 and sañ3-2 (Quesada et al., 2000), were renamed aba2-13 and aba2-14, respectively.
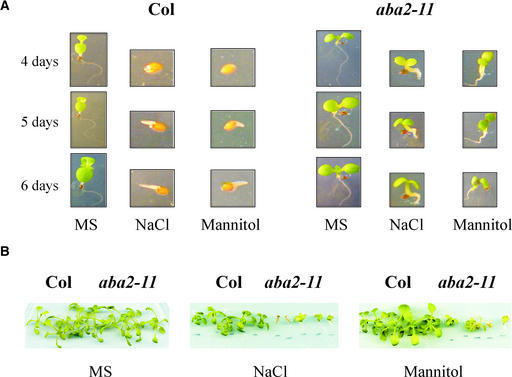
The identification of aba2 alleles through screenings based on the ability to germinate and perform early growth under high-salt concentration highlights the role of ABA in promoting the inhibition of both seed germination and early seedling development under low-water-potential conditions. Four days after sowing in medium supplemented with 200 mM NaCl or 400 mM mannitol, wild-type seeds did not even show radicle emergence, whereas aba2-11 seeds had already green and almost fully developed cotyledons (Figure 1A). Under such conditions, wild-type seeds were severely delayed in germination with respect to unstressed conditions (cf. germination at 4 days in Murashige and Skoog [1962] medium, 200 mM NaCl, and 400 mM mannitol). Additionally, after radicle emergence, further growth was arrested.
Figure 1.
Phenotype of aba2-11 Plants with Respect to Seed Germination and Seedling Development in the Presence of Either NaCl or Mannitol.
(A) Seed germination in the presence of 200 mM NaCl and 400 mM mannitol. Wild-type (Col; left) and aba2-11 (right) seeds were germinated on Murashige and Skoog (1962) medium (MS), medium supplemented with 200 mM NaCl, or 400 mM mannitol. The photographs were taken at 4, 5, and 6 days after sowing.
(B) Osmotic stress sensitivity of aba2-11 plants. Six-day-old aba2-11 or wild-type seedlings were transferred from MS plates to medium lacking Suc and supplemented with either 100 mM NaCl or 200 mM mannitol. Sealing of the plates was done with Micropore tape to allow gas exchange with the environment. The photograph was taken at 2 weeks after the seedlings were transferred to MS plates (left) or plates were supplemented with NaCl (center) or mannitol (right).
By contrast, aba2-11 seeds were relatively insensitive to such osmotic stress, germinated well, and were able to green and expand cotyledons. This property also was observed in ABA-insensitive (abi1 and abi2) and other ABA-defective (aba1-1, aba2-1, aba3-2, and aba3-4) mutants, which also were able to germinate and develop green cotyledons under inhibitory concentrations of NaCl or mannitol (Werner and Finkelstein, 1995; Leon-Kloosterziel et al., 1996; Xiong et al., 2001; this work). Although aba2-11 seeds were more tolerant of osmotic stress at germination than wild-type seedlings, aba2-11 plants were more sensitive to salt or water stress at later stages of development. For instance, aba2-11 seedlings showed higher sensitivity to concentrations of NaCl or mannitol that cause osmotic stress than wild-type seedlings (Figure 1B). Indeed, aba2-11 seedlings were killed by prolonged exposure to 100 mM NaCl or 200 mM mannitol, whereas wild-type plants survived (Figure 1B).
ABA Synthesis Is Reduced Severely in aba2-11 Plants
aba2-11 is likely a null ABA2 allele (data presented below); therefore, we were interested in investigating the effect of the aba2-11 mutation on ABA levels. To this end, leaves of mock-treated, salt-treated, or drought-stressed aba2-11 mutant plants were analyzed for ABA content, and the results were compared with ABA levels in wild-type and aba2-1 plants. ABA levels of aba2-1 have been measured (Leon-Kloosterziel et al., 1996; Rook et al., 2001). Their data, together with those of Schwartz et al. (1997a), indicate that aba2-1 plants still retain some capacity to convert xanthoxin to ABA, suggesting that aba2-1 might be leaky to some extent (Leon-Kloosterziel et al., 1996). Indeed, in vitro biochemical analysis of the aba2-1 protein revealed residual activity (4 to 6%) compared with wild-type ABA2 (data presented below).
Table 3 shows that unstressed rosette leaves of aba2-11 as well as aba2-1 plants contained ∼20 to 25% of the wild-type level of ABA. ABA levels increased approximately eightfold in wild-type leaves upon water or salt stress. By contrast, ABA levels in the aba2-11 mutant remained approximately constant and increased twofold in aba2-1 plants (Leon-Kloosterziel et al., 1996; this work). Thus, ABA content in aba2-11 and aba2-1 rosette leaves exposed to water or salt stress was between 3 and 6% compared with that of wild-type plants.
Table 3.
ABA Content
| Leaf | Control | Salt Stress | Water Stress |
|---|---|---|---|
| Col | 103 ± 15 | 828 ± 70 | 811 ± 76 |
| aba2-1 | 22 ± 3 | 45 ± 4 | 46 ± 4 |
| aba2-11 | 25 ± 3 | 35 ± 5 | 27 ± 5 |
ABA levels (ng ABA/g dry weight) were measured in rosette leaves of wild-type, aba2-1, and aba2-11 plants.
Map-Based Cloning of the ABA2 Gene
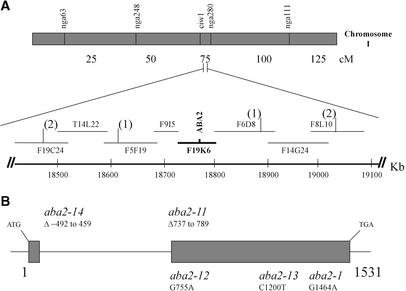
The aba2 locus was mapped to the lower arm of chromosome I by Leon-Kloosterziel et al. (1996), linked to the morphological marker ch1. To establish the molecular basis of the phenotype of aba2 mutants, positional cloning of the ABA2 gene was performed. Homozygous aba2-11 mutant plants (in a Col genetic background) were crossed to wild-type plants (Landsberg erecta ecotype). The resulting F1 plants were self-pollinated, and F2 seeds were obtained. From the segregating F2 generation, homozygous aba2-11 individuals were selected on the basis of their ability to germinate under 200 mM NaCl, and subsequently, DNA was prepared from them.
Mapping of the aba2-11 locus was performed by testing linkage to simple sequence length polymorphism (SSLP) markers (Bell and Ecker, 1994; Lukowitz et al., 2000). Initially, both ciw1 and nga280 were identified as cosegregating flanking markers in the centromeric and telomeric vicinity of the target locus, respectively (Figure 2A). New SSLP markers were developed by surveying released genomic DNA sequences for simple repeats and small insertions/deletions (http://www.Arabidopsis.org/Cereon). A BAC contig covering the ABA2 locus was established according to the recombination breakpoints defined by SSLP markers derived from the F19C24, F5F19, F6D8, and F8L10 sequences (Figure 2A). Detailed examination of the genes on BAC clones in this region identified several candidates that could function in ABA biosynthesis (http://mips.gsf.de/cgi-bin/proj/thal/bac_cosmid?).
Figure 2.
Map-Based Cloning and Genomic Structure of the ABA2 Gene.
(A) Mapping of ABA2 to the lower arm of chromosome I and localization on BAC clone F19K6. The SSLP markers ciw1 and nga280 were identified initially as cosegregating flanking markers in the centromeric and telomeric vicinity of the target locus, respectively. Vertical bars represent polymorphic SSLP markers used to localize recombination break points (number of break points per marker is indicated in parentheses) and to establish a BAC contig covering the ABA2 locus. The position of the BAC contig with respect to the Arabidopsis Genome Initiative map (http://www.arabidopsis.org/servlets/mapper) is indicated. cM, centimorgan.
(B) Structure of the ABA2 gene and mutations in different aba2 alleles. The numbering begins at the ATG translation start codon. Closed boxes represent the open reading frame.
Among them, the At1g52340 gene, which is located on BAC F19K6 and encodes a member of the short-chain alcohol dehydrogenase family, appeared to be a good candidate, according to the biochemical defect of aba2 plants (Schwartz et al., 1997a). Genomic DNA corresponding to the At1g52340 gene was amplified by PCR from wild-type and aba2-11 plants. Gel electrophoresis analysis revealed that the PCR product amplified from aba2-11 plants showed a size reduction compared with the wild-type product.
Sequencing of the At1g52340 gene in the aba2-11 mutant revealed a 53-bp deletion comprising nucleotides 737 to 789, which also leads to a frameshift in the open reading frame (Figure 2B; see also supplemental data). Analysis of the F2 chromosomes of homozygous aba2-11 individuals revealed cosegregation of the small deletion observed in the At1g52340 gene with the ABA-deficient phenotype. These data, together with the identification of mutations in the other aba2 alleles analyzed (Figure 2B, Table 4), unequivocally confirm the notion that At1g52340 represents the ABA2 gene.
Table 4.
Molecular Characterization of aba2 Mutations
| Allele | Mutagen | Nucleotide Change | Protein Change |
|---|---|---|---|
| aba2-1 | EMS | G1464A | Ser-264 to Asn |
| aba2-11 (sre1-1) | T-DNA | 53 bp, from 737 to 789 | Deletion/frameshift |
| aba2-12 (sre1-2) | T-DNA | G755A | Gly-28 to Arg |
| aba2-13 (sañ3-1) | EMS | C1200T | Ser-176 to Phe |
| aba2-14 (sañ3-2) | Fast neutrons | 951 bp, from −492 to 459 | Deletion/frameshift |
Nucleotide numbering refers to the ATG start codon. EMS, ethyl methanesulfonate.
Both aba2-11 and aba2-14 are likely to be null alleles of the ABA2 gene (Table 4; see also supplemental data). The 53-bp deletion of the aba2-11 allele leads to a truncated peptide of only 22 amino acid residues encoded by the ABA2 open reading frame plus 10 erroneous residues created by the frameshift. aba2-14 contains a deletion of 951 bp comprising nucleotides −492 to 459 of the ABA2 gene (numbering refers to the ATG start codon), which abolishes the transcription of the ABA2 gene (data not shown). The other new aba2 alleles, aba2-12 and aba2-13, as well as aba2-1 (Rook et al., 2001), contain mutations that are predicted to cause amino acid changes in the ABA2 gene product (Table 4).
ABA2 Encodes a Short-Chain Alcohol Dehydrogenase
To obtain the cDNA sequence of ABA2, reverse transcriptase–mediated PCR was performed using mRNA extracted from wild-type Col plants and primers derived from the ABA2 gene sequence. The resulting product was cloned and sequenced. Comparison of the cDNA with the genomic sequence confirmed that the ABA2 gene consists of two exons and one intron (Figure 2B; see also supplemental data). The ABA2 cDNA contains an 858-bp open reading frame that putatively encodes a protein of 285 amino acids with a predicted molecular mass of 30.2 kD. The PSORT program (http://psort.nibb.ac.jp) predicts a putative chloroplast transit peptide at the N-terminal sequence of the putative gene product; however, the CHLOROP program (http://www.cbs.dtu.dk/services) does not predict such a transit peptide. Additionally, cleavage of the N-terminal targeting signal predicted by PSORT would affect the coenzyme binding site of ABA2 (see below), which makes the PSORT prediction unlikely.
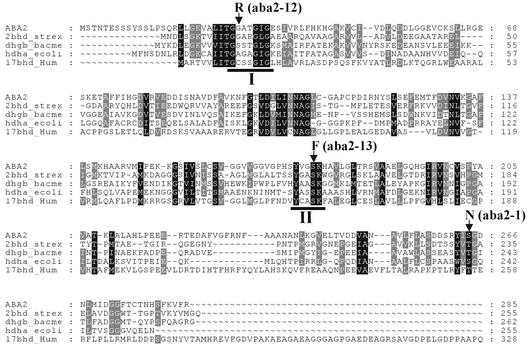
Database searches showed that ABA2 is a member of the SDR family (Figure 3). This family of enzymes contains prokaryotic dehydrogenases, insect alcohol dehydrogenase, and mammalian enzymes such as 15-hydroxyprostaglandin dehydrogenase and 17β-hydroxysteroid dehydrogenase (Persson et al., 1991; Jornvall et al., 1995). The ABA2 primary structure shows two sequence motifs that are common to members of this family. These are the N-terminal Gly27-X-X-X-Gly31-X-Gly33 coenzyme binding motif and the Tyr173-X-X-X-Lys177 motif, which are structurally located in the active site of these enzymes (Jornvall et al., 1995) (Figure 3). Typically, the enzymes belonging to the SDR family exhibit residue identities of ∼15 to 30%. However, tertiary structures of some members of the family reveal a similar one-domain structure with a seven/eight β-strand pattern in an α/β arrangement (Jornvall et al., 1995). This pattern is typical of the coenzyme binding fold of dehydrogenases in general (Rossmann and Argos, 1978); however, the one-domain structure constitutes a clear difference from the two-domain structure of the well-studied yeast and mam-malian alcohol, lactate, malate, and glyceraldehyde 3-phosphate dehydrogenases (also known as medium-chain, classic, or in some cases zinc-dependent dehydrogenases).
Figure 3.
Alignment of ABA2 with Other Members of the SDR Family.
The sequences of Arabidopsis ABA2 (At1g52340), Streptomyces exfoliatus 3α,20β-hydroxysteroid dehydrogenase (2bhd strex), Bacillus megaterium Glc 1-dehydrogenase (dhgb bacme), E. coli 7α-hydroxysteroid dehydrogenase (hdha ecoli), and human 17β-hydroxysteroid dehydrogenase (17bhd Hum) were aligned using the PileUp program and Genedoc software. Positions with identical amino acid residues or conservative substitutions are highlighted in black when they match in all proteins or gray in the case of four matches. The GXXXGXG residues of the coenzyme binding site (I) and the TyrXXXLys sequence motif (II) are underlined. The amino acid changes resulting from aba2-1, aba2-12, and aba2-13 mutations are indicated.
The aba2-1, aba2-12, and aba2-13 mutations are predicted to affect crucial domains for the functionality of the ABA2 enzyme (Figure 4). For instance, the aba2-12 mutation presumably affects the binding of the coenzyme, because this mutation results in an amino acid change from Gly-28 to Arg (Table 4). The aba2-13 mutation results in the substitution of Ser-176 by Phe and likely affects the catalytic center of the enzyme, which involves Tyr-173 and Lys-177 (Figure 4).
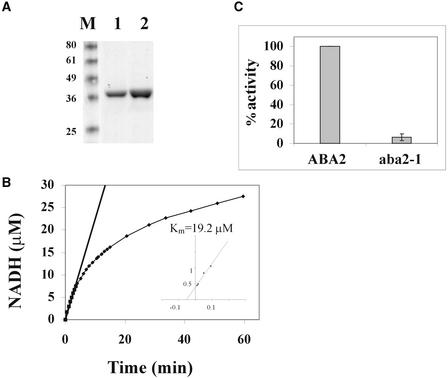
Figure 4.
Purification and Assay of ABA2 Activity.
(A) SDS-PAGE analysis of purified His-tagged ABA2 and aba2-1. Lanes 1 and 2 contain 3 and 6 μg of recombinant ABA2 or aba2-1 protein, respectively. Molecular mass standards (M) are indicated in kD.
(B) Enzyme kinetics of the recombinant ABA2 protein. ABA2 (10 ng/μL) was incubated in a buffer containing 100 mM K2HPO4, pH 7.2, 100 μM xanthoxin as a substrate, and 100 μM NAD as coenzyme. The reduction rate of NAD to NADH was monitored by measuring A340. Regression analysis of the data (R2 = 0.99) was performed using Microsoft Excel software to calculate the initial velocity (Vo) of the reaction (224 μM NADH·min−1·mg−1) to this substrate (S) concentration. The inset shows a double reciprocal plot of 1/Vo versus 1/S at different concentrations of xanthoxin (10, 20, 60, and 100 μM).
(C) aba2-1 activity compared with ABA2 activity. The assay conditions were as described in (B), and the reaction was incubated for 20 min. The error bar indicates sd values and was calculated from three independent experiments.
Additionally, the Ser-176 residue lies in a long helix (αF), which constitutes a subunit interacting area in the structures determined for other members of the short-chain alcohol dehydrogenase family (Ghosh et al., 1991; Jornvall et al., 1995). Therefore, the aba2-13 mutation also might affect the subunit association and quaternary structure of the enzyme. Finally, the aba2-1 mutation results in an amino acid substitution from Ser-264 to Asn in the C-terminal part of the enzyme. This amino acid change might affect the presumed substrate binding site, which lies in the C-terminal part of the short-chain alcohol dehydrogenases (Ghosh et al., 1991; Jornvall et al., 1995).
Biochemical Characterization of ABA2
ABA2 was expressed in Escherichia coli as a His-tagged protein, and the heterologously expressed protein was purified by nickel–nitrilotriacetic acid agarose affinity chromatography (Figure 4A). To determine the effect of the aba2-1 mutation on the activity of the protein product, the S264A aba2-1 protein also was purified (Figure 4A). ABA2 was able to catalyze the NAD-dependent oxidation of xanthoxin with a Km value of 19 μM (Figure 4B). NADP could not replace NAD as coenzyme, and other alcohols, such as ethanol, isopropanol, butanol, cyclohexanol, and 2,6-dimethylcyclohexanol, were not oxidized by ABA2 (data not shown). The 3,5,5′-trimethylcyclohexanol alcohol was oxidized by ABA2, although with a Km value of 10 mM (data not shown). The aba2-1 protein exhibited reduced ability to catalyze the NAD-dependent oxidation of xanthoxin (Figure 4C).
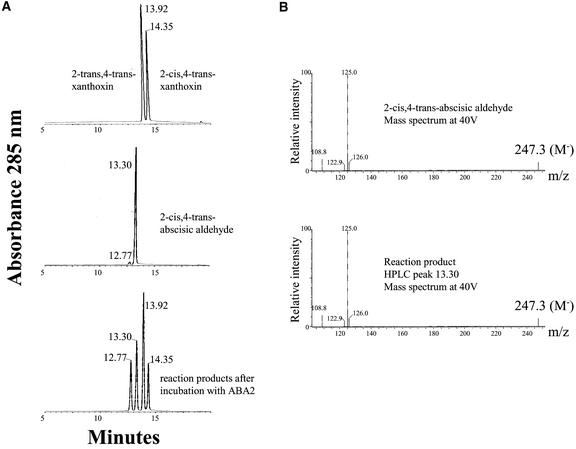
The xanthoxin substrate used was a mixture of the 2-cis, 4-trans and 2-trans,4-trans isomers, with slightly more of the latter than of the former (Figure 5A). Exposure of a methanolic solution of xanthoxin to sunlight leads to the rapid formation of a 1:1 equilibrium mixture of both isomers (Burden and Taylor, 1970). The abscisic aldehyde standard, mostly 2-cis,4-trans, also contains a very minor peak corresponding to the 2-trans,4-trans isomer. After incubation of the xanthoxin substrate with ABA2, the reaction products were analyzed by HPLC–mass spectrometry. The products were identified as 2-cis,4-trans–abscisic aldehyde (HPLC peak with a retention time of 13.30 min) and 2-trans,4-trans–abscisic aldehyde (HPLC peak with a retention time of 12.77 min) by comparison with the HPLC profile of the abscisic aldehyde standard (Figure 5A) and mass spectrometry (Figure 5B).
Figure 5.
Identification by HPLC–Mass Spectrometry Analysis of the Reaction Products after Incubation of Xanthoxin with ABA2.
(A) Chromatograms of the xanthoxin substrate (top), abscisic aldehyde standard (middle), and reaction products (bottom) after incubation with ABA2 as described in Methods.
(B) Mass spectra of the 2-cis,4-trans–abscisic aldehyde standard (top) and the reaction product eluted at the retention time of 13.30 min (bottom). The molecular anion of the compound (M−) is indicated. A similar spectrum was obtained for the reaction product eluted at the retention time of 12.77 min, which corresponds to the 2-trans,4-trans isomer produced from 2-trans,4-trans–xanthoxin.
ABA2 was able to convert each isomer of xanthoxin to its corresponding abscisic aldehyde isomer, although 2-cis,4-trans–xanthoxin was used more efficiently because the ratio of trans,trans–xanthoxin to cis,trans–xanthoxin was twofold higher after incubation with ABA2 than in the original substrate. Cleavage of 9-cis-epoxy–carotenoids results in the production of 2-cis,4-trans–xanthoxin (Schwartz et al., 1997b); therefore, we assume the biological substrate of ABA2 to be 2-cis,4-trans–xanthoxin.
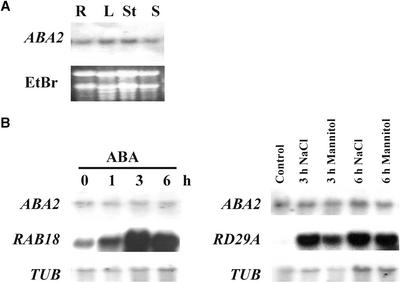
Expression of ABA2 Is Not Upregulated by Osmotic Stress
To analyze the expression pattern of ABA2, total RNA from different parts of wild-type plants was extracted, and RNA gel blot analysis was performed (Figure 6A). The results indicate that ABA2 is expressed constitutively (at low levels) in all plant organs examined. The transcript level of ABA2 did not increase significantly in response to ABA, NaCl, or mannitol treatment (Figure 6B). As a control in the experiment, the induction of both RAB18 by ABA and RD29A by salt/mannitol treatment was analyzed. The expression pattern of ABA2 was in agreement with previous results showing that the enzyme encoded by ABA2 is expressed constitutively (Sindhu and Walton, 1987) and is not induced by water stress conditions (Schwartz et al., 1997a).
Figure 6.
ABA2 Gene Expression.
(A) ABA2 mRNA expression in various plant organs. The blot contained ∼10 μg of total RNA prepared from roots (R), leaves (L), stems (St), and siliques (S). The loading of the gel was visualized by ethidium bromide (EtBr) staining.
(B) ABA2 transcript level did not change significantly in response to 50 μM ABA (left), 150 mM NaCl (right), or 300 mM mannitol (right). As a control, the transcript levels of RAB18 and RD29A genes also were analyzed. The loading of the gel was visualized by hybridization with a tubulin probe (TUB).
DISCUSSION
The role of ABA in the initiation and maintenance of seed dormancy (i.e., the inhibition of seed germination under favorable environmental conditions) is well known (Koornneef and Karssen, 1994; Bewley, 1997). ABA also serves to arrest early seedling growth under adverse environmental conditions (Lopez-Molina et al., 2001). Germination commences with the uptake of water by the dry seed, and it is completed when a part of the embryo, usually the radicle, extends to penetrate the surrounding structures (Bewley, 1997). In this work, we show that radicle emergence of nondormant seeds is delayed by ABA under osmotic stress (Figure 1A; cf. germination at 4 days in wild-type and aba2-11 seeds).
Therefore, the apparent paradox that plants with decreased levels or reduced response to ABA appear to be salt tolerant during germination and early seedling establishment can be explained as a lack of the physiological brake to germination under unfavorable water conditions. The failure of wild-type seeds to germinate and grow under these conditions might be attributable to the inaccessibility of energy resources in the embryo. Indeed, the ABA-induced protein kinase PKABA1 mediates the accessibility of the germinating embryo to the starchy endosperm in barley aleurone layers (Gomez-Cadenas et al., 1999).
ABA-deficient mutants have been identified on the basis of their wilty phenotype or seed germination characteristics (Koornneef, 1986; Leon-Kloosterziel et al., 1996). For instance, Arabidopsis ABA-deficient mutants have been identified by virtue of their ability to germinate in the presence of the gibberellin biosynthesis inhibitor paclobutrazol (Leon-Kloosterziel et al., 1996) or as revertants of nongerminating gibberellin-deficient mutants (Koornneef et al., 1982). Biochemical characterization of the mutants has revealed the biosynthetic defect in most of the ABA-deficient loci known (Seo and Koshiba, 2002). Biochemical analysis of the Arabidopsis aba2-1 mutant was reported by Schwartz et al. (1997a). This work revealed that aba2-1 cell-free extracts showed a substantially reduced ability to convert xanthoxin to ABA. Conversion of xanthoxin to ABA involves ring modifications as well as the oxidation of the 4′-hydroxyl to a ketone group and of the 1-aldehyde to a carboxylic acid group. Particularly, the aba2-1 mutant was blocked in the conversion of xanthoxin to abscisic aldehyde (Schwartz et al., 1997a). However, because three chemical reactions are required to convert xanthoxin to abscisic aldehyde, it was unclear whether other enzymes, in addition to ABA2, would be involved in this reaction. Our analysis of the reaction catalyzed by ABA2 establishes that a single enzyme is able to efficiently convert xanthoxin to abscisic aldehyde.
These data, together with the biochemical analysis of the AAO3 enzyme (Seo et al., 2000a, 2000b), as well as previous genetic and biochemical characterization of Arabidopsis aba2-1 (Schwartz et al., 1997a) and mutants affected in the oxidation of abscisic aldehyde to ABA (Arabidopsis aba3 and aao3, dr in potato, nar2a in barley, aba1 in tobacco, and flc and sit in tomato; revised in Cutler and Krochko, 1999), support the xanthoxin→abscisic aldehyde→ABA route as the final steps of the major ABA biosynthetic pathway (Sindhu and Walton, 1987, 1988; Rock and Zeevaart, 1990; Sindhu et al., 1990).
Conversion of xanthoxin to ABA by cell-free extracts from bean leaves was reported to be localized in the cytosol (Sindhu and Walton, 1987). On the other hand, the experiments of Milborrow and Lee (1998) with intact spinach leaf chloroplasts show that the entire pathway of ABA biosynthesis can occur within the chloroplasts. Determination of the in vivo subcellular localization of ABA2 and AAO3 is required to establish definitively whether the last steps of ABA biosynthesis are chloroplastic or cytosolic.
Previous work has shown that the enzyme encoded by ABA2 is expressed constitutively and that the conversion of xanthoxin to ABA is not increased significantly under water stress conditions (Sindhu and Walton, 1987; Schwartz et al., 1997a). Our analysis of the ABA2 transcript under unstressed or osmotic stress conditions confirms this previous finding at the mRNA level (Figure 6). Other genes involved in ABA biosynthesis, such as ZEP, NCED, ABA3, and AAO3, are induced by water stress (Tan et al., 1997; Audran et al., 1998; Seo et al., 2000a; Xiong et al., 2001). However, only in the case of NCED has the induction been confirmed at the protein level (Qin and Zeevaart, 1999). In addition, although the expression of AAO3 mRNA in leaves increases under water stress conditions, the level of protein or its activity does not change significantly (Seo et al., 2000a).
Although ABA2 does not catalyze a rate-limiting step in ABA biosynthesis, it is crucial for the basal level of ABA (Leon-Kloosterziel et al., 1996) (Table 3). Considering the severe reduction in ABA levels measured in aba2-1 (Leon-Kloosterziel et al., 1996) and aba2-11 (Table 3), ABA2 appears to be the main player in the biosynthesis of abscisic aldehyde. However, a residual ABA level is measured even in the aba2-11 null mutant, suggesting that either other minor ABA biosynthetic pathways exist (Seo and Koshiba, 2002) or other SDR genes might substitute for the absence of ABA2 with low efficiency. Analysis of the Arabidopsis genome revealed several SDR genes (At3g51680, At2g47130, At3g29250, At3g26770, At4g03140, and additional in-tandem gene duplications) exhibiting sequence similarity to ABA2. Work is under way to determine whether these SDRs play a role in ABA biosynthesis.
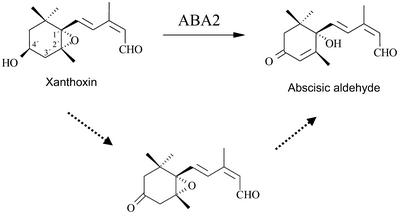
SDR enzymes usually catalyze alcohol-ketone/aldehyde substrate transitions; however, there also exist SDR forms that exhibit dehydratase, epimerase, or isomerase activity (Jornvall et al., 1999). Conversion of xanthoxin to ABA-aldehyde by ABA2 involves three chemical modifications: oxidation of the 4′-hydroxyl to a ketone, desaturation of a 2′,3′ bond, and opening of the epoxide ring (Figure 7). As suggested previously (Schwartz et al., 1997a), the oxidation of the ABA precursor 4′-hydroxyl group to ketone by ABA2 alcohol dehydrogenase activity might result in a subsequent spontaneous epoxide rearrangement. Alternatively, a second ABA2-catalyzed enzymatic step might be required to transform the 1′,2′-epoxy group into an allylic alcohol (1′-hydroxy-2′-ene) (Figure 7). Allylic alcohols can be generated from epoxide rearrangement through basic catalysis (Södergren et al., 2000). Several examples of this reaction are known in organic chemistry, but in all cases a chemical catalyst is required (Kee et al., 2000; Södergren et al., 2000, and references therein). In that case, key residues of the ABA2 active site might provide, in addition to the alcohol dehydrogenase activity, a second isomerase activity required to rearrange the epoxide ring.
Figure 7.
Reaction Catalyzed by ABA2.
The conversion of xanthoxin to ABA-aldehyde involves the oxidation of the 4′-hydroxyl to a ketone group as well as the opening of the 1′,2′-epoxide group to give rise to a desaturated 2′,3′ bond and a 1′-hydroxyl group. A putative two-step reaction mechanism is discussed in the text and indicated by dashed arrows.
Many examples of peptides carrying two or more enzymatic activities are known. For instance, the Arabidopsis ent-kaurene oxidase catalyzes three steps of gibberellin biosynthesis (Helliwell et al., 1999). Determination of the crystal structure of ABA2 will elucidate the reaction mechanism of the xanthoxin conversion to abscisic aldehyde and the enzymatic properties of this plant SDR.
METHODS
Plant Material
Arabidopsis thaliana plants (ecotype Columbia [Col]) were grown routinely under greenhouse conditions in pots containing a 1:3 perlite:soil mixture. For in vitro culture, seeds were surface-sterilized by treatment with 70% ethanol for 20 min, followed by commercial bleach (2.5%) containing 0.05% Triton X-100 for 10 min, and finally, four washes with sterile distilled water. Stratification of the seeds was conducted during 3 days at 4°C.
Afterwards, seeds were sown on Murashige and Skoog (1962) (MS) plates containing solid medium composed of MS basal salts and 1% Suc, solidified with 1% agar, and pH adjusted to 5.7 with KOH before autoclaving. Different concentrations of NaCl and mannitol were made by adding appropriate amounts of reagents to the basal medium. Plates were sealed and incubated in a controlled-environment growth chamber at 22°C under a 16-h-light/8-h-dark photoperiod at 80 to 100 μmol·m−2·s−1.
Screening Conditions
T-DNA lines were constructed in the laboratory (Detlef Weigel, Salk Institute, La Jolla, CA) using the pSKI15 vector. Approximately 17,000 lines (stock numbers N21995 and N21991) were provided by the ABRC (Columbus, OH). The distributed T-DNA pools consisted of T4 seeds. Approximately 2 × 105 seeds were screened at high seed density (50 Petri plates of 14 cm diameter containing ∼4000 seeds per plate) on MS medium (plus 1% Suc) containing 200 mM NaCl. Seeds were considered to be salt resistant only after they produced fully green expanded cotyledons. Selected salt-resistant candidates (T4) were grown in soil to obtain the T5 progeny for further studies. The T5 progeny of the candidates were retested at low seed density (up to 200 seeds per 9-cm-diameter Petri plate) under 150 to 200 mM NaCl. Even though the screening was performed using a T-DNA collection, the sre1-1 and sre1-2 mutations were not linked to a T-DNA insertion.
Genetic Analysis and Map-Based Cloning of ABA2
Backcrosses of sre mutants to the wild type and intercrosses among sre mutants as well as those of sre with aba mutants were performed by transferring pollen to the stigmas of emasculated flowers. F1 and F2 seeds were scored for germination in 150 to 200 mM NaCl. To map the ABA2 locus, homozygous aba2-11 plants (in a Col background) were crossed to wild-type plants of the Landsberg erecta background. From the segregating F2 generation, 410 homozygous aba2-11 individuals were selected, and DNA was extracted individually.
Mapping of the ABA2 locus was performed by testing linkage with simple sequence length polymorphism (SSLP) markers (Bell and Ecker, 1994; Lukowitz et al., 2000). ABA2 was found to be linked to the SSLP markers cw1 and nga280 on the lower arm of chromosome I. New SSLP markers were developed by surveying released genomic DNA sequences for simple repeats and small insertions/deletions. Thus, SSLP markers were developed based on the sequences of the BAC clones F11M15, F5F19, F6D8, F12M16, F15I1, T15A14, F16N3, and F7F22.
Germination Assays
Seeds were plated on solid medium composed of MS basal salts, 1% Suc, and different concentrations of NaCl or mannitol. After 5 days of incubation as described above, the percentage of seeds that had germinated and developed fully green expanded cotyledons was determined. To measure abscisic acid (ABA) sensitivity, seeds were plated on solid medium composed of MS basal salts, 1% Suc, and 3 μM ABA. After 8 days of incubation at 22°C under a 16-h-light/8-h-dark photoperiod at 80 to 100 μmol·m−2·s−1, the percentage of seeds that had germinated and developed fully green expanded cotyledons was determined. To measure paclobutrazol sensitivity, seeds were plated on medium containing the indicated concentrations of paclobutrazol (Table 1) and germination was determined after 7 days.
ABA Extraction and Determination
Lyophilized samples (rosette leaves) of mock-treated, salt-treated (200 mM NaCl for 5 days), or drought-stressed (unwatered for 1 week) plants were ground to a fine powder with mortar and pestle. Duplicate samples (50 mg dry weight each) were extracted with 5 mL of 80% acetone containing 100 mg/L 2,6-ditert-butyl-methyl phenol and 500 mg/L citric acid for 16 h at 4°C in the dark. The extracts were homogenized further in a Polytron at maximum speed for 1 min and centrifuged at 3000g for 5 min.
A 1-mL aliquot of the extract was evaporated in a Speedvac concentrator. The sample was resuspended in 100 μL of Tris saline buffer (50 mM Tris, 1 mM MgCl2, and 150 mM NaCl, pH 7.8) and analyzed directly or diluted with Tris saline buffer to fit the ABA content of the extracts within the linear range of the ABA standard curve of the assay. Quantitative analysis of ABA was performed by the indirect ELISA method, essentially as described by Walker-Simmons et al. (2000). The monoclonal antibodies against ABA were purchased from Idex Laboratories (San Bruno, CA).
Cloning of ABA2 cDNA
The ABA2 cDNA was obtained by reverse transcriptase–mediated PCR amplification. RNA from Col wild-type plants was used as a template for the synthesis of the first-strand cDNA using Moloney murine leukemia virus reverse transcriptase and the gene-specific primer 3ABA2 (5′-AGACATGATAAATTGGCGGAC-3′). One tenth of the reaction product was used as a template for PCR amplification of the full-length ABA2 cDNA using the primers 5ABA2 (5′-GTCTGTGTC-AATAGTGAGGCA-3′) and 3ABA2. The PCR product was cloned subsequently into the EcoRV site of pBluescript SK+ (Stratagene), generating the pSKABA2 construct.
Molecular Characterization of aba2 Alleles
Synthetic oligonucleotide primers were designed according to the At1g52340 gene sequence, and overlapping fragments encompassing the entire gene were amplified by PCR from aba2 mutants. The amplified products were sequenced on both strands. To avoid errors caused by PCR, three independent PCR samples were mixed and sequenced in batches.
Expression and Purification of His-Tagged ABA2 and aba2-1
The coding region of the ABA2 cDNA was amplified by PCR from the pSKABA2 construct using primers BRI (5′-CGCGGATCCGAATTC-ATGTCAACGAACACTGAATCT-3′) and B (5′-CGCGGATCCTCA-TCTGAAGACTTTAAAGGA-3′). The PCR product was subcloned into the BamHI site of pET28a vector (Novagen, Madison, WI), generating the pET28a.ABA2 construct. A DNA fragment containing the aba2-1 mutation was amplified by PCR from genomic DNA using primers 795 (5′-AGGAGACTATGGTGCGATTGG-3′) and 3ABA2. The PCR product was cloned subsequently into the EcoRV site of pBluescript SK (Stratagene). Then, a SphI-XbaI DNA fragment containing the aba2-1 mutation was isolated to replace the corresponding fragment in the wild-type ABA2 cDNA, generating the pET28a.aba2-1 construct.
Once the pET28a.ABA2 and pET28a.aba2-1 constructs were verified by sequencing, they were used to transform Escherichia coli strain BL21(DE3). Expression of His-tagged ABA2 or aba2-1 was induced with 1 mM isopropylthio-β-galactoside in E. coli BL21(DE3) cells transformed with pET28a.ABA2 or pET28a.aba2-1, respectively. The recombinant protein was purified by nickel–nitrilotriacetic acid agarose affinity chromatography according to the manufacturer's instructions (Qiagen, Valencia, CA).
Assay of ABA2 Activity
Enzyme assays contained 100 mM K2HPO4, pH 7.2, 100 μM NAD, different concentrations of xanthoxin, and the indicated amounts of ABA2 protein (Figure 4) . Assays were incubated for the indicated periods of time (Figure 4) at 25°C in a total volume of 1 mL. The rate of reduction of NAD to NADH was followed by scoring the increase in A340. To determine the reaction products, the reaction mixture was extracted twice with 1 mL of ethyl acetate, and the ethyl acetate fractions were combined, concentrated, and analyzed by HPLC–mass spectrometry.
HPLC–Mass Spectrometry Analyses
HPLC–mass spectrometry analyses were performed using a Waters 1525 chromatograph coupled directly to a ZMD Micromass mass spectrometer (Waters, Milford, MA) in the negative electrospray ionization mode with the following source parameters: capillary voltage of 2500 V; cone voltage of 40 V; source block temperature of 100°C; desolvation temperature of 300°C. Samples were injected on a reverse-phase Symmetry C18 column (5-μm particle size, 4.6 × 150 mm; Waters) equilibrated with 1% acetic acid. The column was eluted with a linear gradient between solvent A (1% acetic acid) and solvent B (methanol) at a flow rate of 1 mL/min applied for 20 min. The elution from the column was monitored with a Waters 486 absorbance detector at a wavelength of 285 nm. Liquid chromatography–mass spectrometry spectra were recorded in the mass-to-charge range between 100 and 300.
RNA Analysis
Approximately 20 to 25 7-day-old seedlings were transferred from MS plates to 125-mL flasks containing 25 mL of MS solution and 1% Suc. The flasks were shaken at 130 rpm under cool fluorescent light. After 10 days, seedlings were mock treated or treated with ABA, NaCl, or mannitol as indicated (Figure 6). Plant material was collected and frozen in liquid nitrogen. Total RNA was extracted as described (Verwoerd et al., 1989), separated on formaldehyde-agarose gels, and blotted to a nylon membrane. Blots were hybridized with random-priming 32P-labeled probes.
The RAB18, RD29A, and TUB probes were prepared by PCR amplification from genomic DNA of wild-type Col plants. Primers were RAB18 (5′-ATGGCGTCTTACCAGAACCGT-3′ and 5′-CCAGATCCGGAGCGGTGAAGC-3′), RD29A (5′-TAATATGGAAGTGACTGATGAGTCT-3′ and 5′-TTAAAGCTCCTTCTGCACCGGAAC-3′), and TUB (5′-CCTGATAACTTCGTCTTTGG-3′ and 5′-GTGAACTCCATCTCGTCCAT-3′).
Upon request, all novel material described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Numbers
The accession numbers for the sequences shown in Figure 3 are as follows: Arabidopsis ABA2 (At1g52340), Q9C826; Streptomyces exfoliatus 3α,20β-hydroxysteroid dehydrogenase (2bhd strex), S10707; Bacillus megaterium Glc 1-dehydrogenase (dhgb bacme), S01227; E. coli 7α-hydroxysteroid dehydrogenase (hdha ecoli), P25529; and human 17β-hydroxysteroid dehydrogenase (17bhd Hum), P14061.
Supplementary Material
Acknowledgments
We thank J. Sheen (Boston, MA) and T. Koshiba (Tokyo, Japan) for communicating unpublished results. We are indebted to J. Zeevaart (East Lansing, MI) for providing xanthoxin and abscisic aldehyde. We thank the group of Lorenzo Zacarias (Instituto de Agroquímica y Tecnología de Alimentos, Valencia, Spain) for invaluable help in determining ABA content as well as Marcelo E. Domine and Miguel Angel Miranda (Instituto de Tecnología Química, Valencia, Spain) for chemical support. We also thank Julio Salinas (Instituto Nacional de Investigación Agraria, Madrid, Spain) for helpful suggestions, the ABRC for providing seed stocks, and J.M. Serrano and S. Gerbert for technical assistance. M.G.-G., J.M.B., and P.P. were supported by Ministerio de Educación y Cultura, Ministerio de Ciencia y Tecnología, and Generalitat Valenciana fellowships, respectively. P.L.R. was supported by a Ramón y Cajal research contract. This work was supported by Grants GV-CAPA-00-13-C02-02 and GV-01-511 from the Generalitat Valenciana and by Grant BIO 2000-1082 from the Ministerio de Ciencia y Tecnología.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.002477.
Footnotes
Online version contains Web-only data.
References
- Audran, C., Borel, C., Frey, A., Sotta, B., Meyer, C., Simonneau, T., and Marion-Poll, A. (1998). Expression studies of the zeaxanthin epoxidase gene in Nicotiana plumbaginifolia. Plant Physiol. 118, 1021–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audran, C., Liotenberg, S., Gonneau, M., North, H., Frey, A., Tap-Waksman, K., Vartanian, N., and Marion-Poll, A. (2001). Localisation and expression of zeaxanthin epoxidase mRNA in Arabidopsis in response to drought stress and during seed development. Aust. J. Plant Physiol. 28, 1161–1173. [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Bewley, J.D. (1997). Seed germination and dormancy. Plant Cell 9, 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner, F., Oreb, M., and Mendel, R.R. (2001). ABA3 is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine dehydrogenase in Arabidopsis thaliana. J. Biol. Chem. 276, 40381–40384. [DOI] [PubMed] [Google Scholar]
- Burbidge, A., Grieve, T.M., Jackson, A., Thompson, A., McCarty, D.R., and Taylor, I.B. (1999). Characterization of the ABA-deficient tomato mutant notabilis and its relationship with maize vp14. Plant J. 17, 427–431. [DOI] [PubMed] [Google Scholar]
- Burden, R.S., and Taylor, H.F. (1970). The structure and chemical transformations of xanthoxin. Tetrahedron Lett. 47, 4071–4074. [DOI] [PubMed] [Google Scholar]
- Chandler, P.M., and Robertson, M. (1994). Gene expression regulated by abscisic acid and its relation to stress tolerance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45, 113–141. [Google Scholar]
- Cutler, A.J., and Krochko, J.E. (1999). Formation and breakdown of ABA. Trends Plant Sci. 4, 472–478. [DOI] [PubMed] [Google Scholar]
- Ghosh, D., Weeks, C.M., Grochulski, P., Duax, W.L., Erman, M., Rimsay, R.L., and Orr, J.C. (1991). Three-dimensional structure of holo 3α,20β-hydroxysteroid dehydrogenase: A member of a short-chain dehydrogenase family. Proc. Natl. Acad. Sci. USA 88, 10064–10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cadenas, A., Verhey, S.D., Holappa, L.D., Shen, Q., Ho, T.H., and Walker-Simmons, M.K. (1999). An abscisic acid-induced protein kinase, PKABA1, mediates abscisic acid-suppressed gene expression in barley aleurone layers. Proc. Natl. Acad. Sci. USA 96, 1767–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell, C.A., Poole, A., Peacock, W.J., and Dennis, E.S. (1999). Arabidopsis ent-kaurene oxidase catalyses three steps of gibberellin biosynthesis. Plant Physiol. 119, 507–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser, C., Kortstee, A., Pego, J., Weisbeek, P., Wisman, E., and Smeekens, S. (2000). The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: Involvement of abscisic acid in sugar responses. Plant J. 23, 577–585. [DOI] [PubMed] [Google Scholar]
- Jornvall, H., Hoog, J.O., and Persson, B. (1999). SDR and MDR: Completed genome sequences show these protein families to be large, of old origin, and of complex nature. FEBS Lett. 445, 261–264. [DOI] [PubMed] [Google Scholar]
- Jornvall, H., Persson, B., Krook, M., Atrian, S., Gonzalez-Duarte, R., Jeffery, J., and Ghosh, D. (1995). Short-chain dehydrogenases/reductases (SDR). Biochemistry 34, 6003–6013. [DOI] [PubMed] [Google Scholar]
- Kee, A., O'Brien, P., Pilgram, C., and Watson, S.T. (2000). Diastereoselective epoxide rearrangements using lithium amide bases: First stereocontrolled synthesis of 4-deoxyconduritols. Chem. Commun. 16, 1521–1522. [Google Scholar]
- Koornneef, M. (1986). Genetic aspects of abscisic acid. In A Genetic Approach to Plant Biochemistry, A.D. Blonstein and P.J. King, eds (Vienna: Springer Verlag), pp. 35–54.
- Koornneef, M., Jorna, M.L., Brinkhorst-Van der Swan, D.L.C., and Karssen, C.M. (1982). The isolation of abscisic acid (ABA)-deficient mutants by selection of induced revertants in non-germinating gibberellin-sensitive lines of Arabidopsis thaliana. Theor. Appl. Genet. 61, 385–393. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., and Karssen, C.M. (1994). Seed dormancy and germination. In Arabidopsis, C. Somerville and E. Meyerowitz, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 313–334.
- Laby, R.J., Kincaid, M.S., Kim, D., and Gibson, S.I. (2000). The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J. 23, 587–596. [DOI] [PubMed] [Google Scholar]
- Leon-Kloosterziel, K.M., Gil, M.A., Ruijs, G.J., Jacobsen, S.E., Olszewski, N.E., Schwartz, S.H., Zeevaart, J.A., and Koornneef, M. (1996). Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J. 10, 655–661. [DOI] [PubMed] [Google Scholar]
- Leung, J., and Giraudat, J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 199–222. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina, L., Mongrand, S., and Chua, N.H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 98, 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz, W., Gillmor, C.S., and Scheible, W.R. (2000). Positional cloning in Arabidopsis: Why it feels good to have a genome initiative working for you. Plant Physiol. 123, 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin, E., Nussaume, L., Quesada, A., Gonneau, M., Sotta, B., Hugueney, P., Frey, A., and Marion-Poll, A. (1996). Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J. 15, 2331–2342. [PMC free article] [PubMed] [Google Scholar]
- Milborrow, B.V. (2001). The pathway of biosynthesis of abscisic acid in vascular plants: A review of the present state of knowledge of ABA biosynthesis. J. Exp. Bot. 52, 1145–1164. [PubMed] [Google Scholar]
- Milborrow, B.V., and Lee, H.S. (1998). Endogenous biosynthetic precursors of (+)abscisic acid. VI. Carotenoids and ABA are formed by the non-mevalonate triose-pyruvate pathway in chloroplasts. Aust. J. Plant Physiol. 25, 507–512. [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant 15, 473–497. [Google Scholar]
- Nambara, E., Kawaide, H., Kamiya, Y., and Naito, S. (1998). Characterization of an Arabidopsis thaliana mutant that has a defect in ABA accumulation: ABA-dependent and ABA-independent accumulation of free amino acids during dehydration. Plant Cell Physiol. 39, 853–858. [DOI] [PubMed] [Google Scholar]
- Persson, B., Krook, M., and Jornvall, H. (1991). Characteristics of short-chain alcohol dehydrogenases and related enzymes. Eur. J. Biochem. 200, 537–543. [DOI] [PubMed] [Google Scholar]
- Qin, X., and Zeevaart, J.A. (1999). The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc. Natl. Acad. Sci. USA 96, 15354–15361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada, V., Ponce, M.R., and Micol, J.L. (2000). Genetic analysis of salt-tolerant mutants in Arabidopsis thaliana. Genetics 154, 421–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock, C.D., and Quatrano, R.S. (1995). The role of hormones during seed development. In Plant Hormones, P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 671–697.
- Rock, C.D., and Zeevaart, J.A. (1990). Abscisic (ABA)-aldehyde is a precursor to, and 1′,4′-trans-ABA-diol, a catabolite of ABA in apple. Plant Physiol. 93, 915–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook, F., Corke, F., Card, R., Munz, G., Smith, C., and Bevan, M.W. (2001). Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J. 26, 421–433. [DOI] [PubMed] [Google Scholar]
- Rossmann, M.G., and Argos, P. (1978). The taxonomy of binding sites in proteins. Mol. Cell. Biochem. 21, 161–182. [DOI] [PubMed] [Google Scholar]
- Schroeder, J.I., Allen, G.J., Hugouvieux, V., Kwak, J.M., and Waner, D. (2001). Guard cell signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 627–658. [DOI] [PubMed] [Google Scholar]
- Schwartz, S.H., Leon-Kloosterziel, K.M., Koornneef, M., and Zeevaart, J.A. (1997. a). Biochemical characterization of the aba2 and aba3 mutants in Arabidopsis thaliana. Plant Physiol. 114, 161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S.H., Tan, B.C., Gage, D.A., Zeevaart, J.A., and McCarty, D.R. (1997. b). Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276, 1872–1874. [DOI] [PubMed] [Google Scholar]
- Seo, M., Koiwai, H., Akaba, S., Komano, T., Oritani, T., Kamiya, Y., and Koshiba, T. (2000. a). Abscisic aldehyde oxidase in leaves of Arabidopsis thaliana. Plant J. 23, 481–488. [DOI] [PubMed] [Google Scholar]
- Seo, M., and Koshiba, T. (2002). Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 7, 41–48. [DOI] [PubMed] [Google Scholar]
- Seo, M., Peeters, A.J., Koiwai, H., Oritani, T., Marion-Poll, A., Zeevaart, J.A., Koornneef, M., Kamiya, Y., and Koshiba, T. (2000. b). The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc. Natl. Acad. Sci. USA 97, 12908–12913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindhu, R.K., Griffin, D.H., and Walton, D.C. (1990). Abscisic aldehyde is an intermediate in the enzymatic conversion of xanthoxin to abscisic acid in Phaseolus vulgaris L. leaves. Plant Physiol. 93, 689–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindhu, R.K., and Walton, D.C. (1987). Conversion of xanthoxin to abscisic acid by cell-free preparations from bean leaves. Plant Physiol. 85, 916–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindhu, R.K., and Walton, D.C. (1988). Xanthoxin metabolism in cell-free preparations from wild type and wilty mutants of tomato. Plant Physiol. 88, 178–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skriver, K., and Mundy, J. (1990). Gene expression in response to abscisic acid and osmotic stress. Plant Cell 2, 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Södergren, M.J., Bertilsson, S.K., and Andersson, P.G. (2000). Allylic alcohols via catalytic asymmetric epoxide rearrangement. J. Am. Chem. Soc. 122, 6610–6618. [Google Scholar]
- Tan, B.C., Schwartz, S.H., Zeevaart, J.A., and McCarty, D.R. (1997). Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. USA 94, 12235–12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd, T.C., Dekker, B.M., and Hoekema, A. (1989). A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 17, 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker-Simmons, M.K., Rose, P.A., Hogge, L.R., and Abrams, S.R. (2000). Abscisic acid: ABA immunoassay and gas chromatography/mass spectrometry verification. Methods Mol. Biol. 141, 33–47. [DOI] [PubMed] [Google Scholar]
- Werner, W.E., and Finkelstein, R.R. (1995). Arabidopsis mutants with reduced response to NaCl and osmotic stress. Physiol. Plant. 93, 659–666. [Google Scholar]
- Xiong, L., Ishitani, M., Lee, H., and Zhu, J.K. (2001). The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress. Plant Cell 13, 2063–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart, J.A., and Creelman, R.A. (1988). Metabolism and physiology of abscisic acid. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39, 439–473. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.