Abstract
Acanthamoeba myosin I heavy chain kinase (MIHCK) phosphorylates the heavy chains of amoeba myosins I, increasing their actin-activated ATPase activities. The activity of MIHCK is increased by binding to acidic phospholipids or membranes and by autophosphorylation at multiple sites. Phosphorylation at a single site is necessary and sufficient for full activation of the expressed catalytic domain. The rate of autophosphorylation of native MIHCK is controlled by a region N-terminal to the catalytic domain. By its substrate specificity and the sequence of its C-terminal catalytic domain, MIHCK was identified as a p21-activated kinase (PAK). We have now cloned the full-length genomic DNA and cDNA of MIHCK and have shown it to contain the conserved p21-binding site common to many members of the PAK family. Like some mammalian PAKs, MIHCK is activated by Rac and Cdc42, and this activation is GTP-dependent and accompanied by autophosphorylation. In contrast to mammalian PAKs, activation of MIHCK by Rac and Cdc42 requires the presence of acidic lipids. Also unlike mammalian PAK, MIHCK is not activated by sphingosine or other non-negatively charged lipids. The acidic lipid-binding site is near the N terminus followed by the p21-binding region. The N-terminal regulatory domain of MIHCK contains alternating strongly positive and strongly negative regions. and the extremely Pro-rich middle region of MIHCK has a strongly acidic N-terminal segment and a strongly basic C-terminal segment. We propose that autophosphorylation activates MIHCK by neutralizing the basic segment of the Pro-rich region, thus unfolding the regulatory domain and abolishing its inhibition of the catalytic domain.
Myosin I heavy chain kinase (MIHCK), originally purified from Acanthamoeba castellanii (1, 2), phosphorylates a serine or threonine residue approximately at the middle of the catalytic domain of the single heavy chain of the amoeba myosins I (3) and enhances their actin-activated MgATPase activity (1, 4). The regulation of the activity of native, purified MIHCK has been studied extensively (for review, see ref. 5). Activity is greatly enhanced by binding to acidic phospholipids (6–8) or native plasma membranes (9) and even further by acidic phospholipid- or membrane-enhanced, intermolecular autophosphorylation at multiple sites (6, 9); calcium/calmodulin inhibits activation by lipids (10). Studies performed on proteolytic fragments of the purified protein (10–12) and on the expressed catalytic domain (13–16) established the presence of two regulatory regions: a single serine in the catalytic domain [which spans the C-terminal 35 kDa (12)] whose phosphorylation is necessary and sufficient for activity (15) and a short region at the N terminus of the 97-kDa native enzyme whose binding to acidic phospholipids or removal by proteolysis enhances kinase activity and autophosphorylation (10, 11).
The previously determined sequence of its catalytic domain (13) and its substrate specificity in vitro (14, 17) identified MIHCK as a member of the p21-activated kinase (PAK) family, of which the best characterized members are mammalian PAKs and yeast STE20. Mammalian PAKs were originally discovered as targets for two small GTP-binding proteins (p21s)—Rac and Cdc42—which, in their GTP-bound state, bind to and activate PAKs (18). As with MIHCK, activation of PAK and STE20 is accompanied by kinase autophosphorylation at multiple sites including one within the catalytic domain (18–23).
Although all members of the PAK family have highly homologous catalytic domains, not all PAKs, despite their names, bind p21s (for reviews on PAK family see refs. 24–26). The ability to bind p21s is determined by the presence of a conserved p21-binding domain (PBD) located outside of the catalytic domain. The p21-binding domain consists of a short Cdc42/Rac interactive binding (CRIB) sequence, which contains 8 highly conserved residues (27) followed by a less conserved, ≈40-aa region (28–30).
In this paper, we report the complete amino acid sequence of MIHCK as deduced from the full-length genomic DNA and cDNA. MIHCK has a p21-binding site, with six of eight conserved CRIB residues, near its N terminus. As with some mammalian PAKs, the activity of MIHCK is stimulated by a GTP-dependent interaction with Rac and Cdc42 and not by Rho, but unlike the mammalian PAKs, this activation requires the presence of acidic lipids. From the sequence and previous experiments with proteolytic fragments of native enzyme, the putative acidic lipid-binding site is tentatively identified, and the implications of the highly polarized distribution of positive and negative amino acids for the mechanism of regulation of kinase activity are discussed.
MATERIALS AND METHODS
Isolation of MIHCK Genomic DNA.
Genomic DNA was purified by using the Qiagen (Chatsworth, CA) Genomic DNA kit and digested with either SalI, XhoI, XbaI, or BamHI restriction enzymes (Stratagene). The digests were electrophoresed on agarose, transferred to nitrocellulose, and probed with probe A (31). Probe A (358 bp, residues 1470–1827 in the cDNA sequence) was generated by PCR using MIHCK catalytic domain cDNA as the template. Because the XbaI digest had the largest band (4.5 kb) by Southern blotting, a preparative XbaI digest was prepared. The region of the gel containing DNA fragments between 4.3 and 4.7 kb was electroeluted, and the DNA was ligated to the ZAP Express vector (Stratagene), digested with XbaI, and packaged by using Gigapack II XL packaging extract (Stratagene) according to the manufacturer’s protocol. The sublibrary was plated by using XL1-Blue MRF′ competent cells, titered, and screened by using probe A. The excised pBK-CMV phagemid vector containing possible clones of the desired insert were analyzed by using restriction digests. One clone was subjected to automated sequencing (Seqwright, Houston).
Isolation of MIHCK cDNA.
Total Acanthamoeba RNA was isolated from washed cells by using the RNA isolation kit (Stratagene), and mRNA was isolated by using the Poly(A) Quik mRNA isolation kit (Stratagene). A cDNA library was constructed by using the ZAP Express cDNA synthesis kit (Stratagene). First-strand cDNA was synthesized by using a mixture of primers A, B, and C [Poly(A)]. The library was packaged by using the Gigapack III packaging extract (Stratagene), plated by using XL1-Blue MRF′ competent cells, titered, and amplified according to the supplied protocols. Two probes were designed to screen the cDNA library. Probe N (226 bp, residues 120–345 in the cDNA sequence) was generated by PCR using the genomic DNA clone as a template. Probe C (306 bp, residues 1946–2251 in the cDNA sequence) was generated by PCR using the MIHCK catalytic domain cDNA as the template. The cDNA library was initially screened by using probe N. The positive plaques were replated and screened a second time with probe N and probe C to ensure a full-length clone. The phagemids were excised; potential positive clones were analyzed by restriction digests, and four (clones 26, 43, 45 and 49) were sequenced (Seqwright). Clone 26 was full-length but had three mutations relative to the genomic DNA: T was substituted for C at position 1095, which still coded for Arg; GCG was inserted following position 1144, which added an Ala residue; and G was substituted for A at position 1168, which changed Thr to Ala. Clone 43 was missing 23 bp at the 5′ end but continued through to the Poly(A) at the 3′ end. Clone 45 was full length, but residues 611–613 were deleted, resulting in the loss of a Gln residue. Clone 49 was missing 23 bp at the 5′ end but continued through the Poly(A). In clone 49, the regions that were mutated in clones 26 and 45 were the same as in the genomic DNA sequence.
Proteins and Lipids.
MIHCK was purified from Acanthamoeba castellanii as described (10, 32). Human Rac1, Cdc42, and Rho were expressed in E. coli with His tag or glutathione S-transferase (GST) at the N terminus and purified on Ni-NTA or glutathione-agarose, respectively. The lipid sources were as follows: phosphatidylserine, phosphatidylinositol, phosphatidylcholine, and ceramide were from Avanti Polar Lipids (catalogue nos. 840032, 840042, 840053, and 860052, respectively); phosphatidic acid, oleic acid, linoleic acid, and sphingosine were from Sigma (catalogue nos. P-9511, O-7501, L-8134, and S-6879, respectively). Lipid vesicles and fatty acid micelles were prepared as described (6) by drying a chloroform solution of lipid under argon and then sonicating in water in a bath sonicator until the mixture became translucent.
Enzymatic Assays.
MIHCK activity was measured by using as substrate synthetic peptide PC9 [GRGRSSVYS, the sequence around the phosphorylation site (italicized Ser) of Acanthamoeba myosin IC (3)] as described (7). Briefly, reactions were started by adding 0.24 μg (final concentration 30 nM) of MIHCK to 0.1 ml (final volume) of reaction mixture containing (final concentrations) 50 mM imidazole (pH 7.5), 3.5 mM MgCl2, 2.5 mM [γ-32P]ATP (30,000 cpm/nmol), 1 mM EGTA, 0.2 mg/ml BSA, and 200 μM PC9. When specified, the reaction mixture included 10 μl of GTP-binding proteins in exchange buffer (see below) and/or lipids. Reactions were stopped with acetic acid for measurements of PC9 phosphorylation and with SDS sample buffer for measurements of kinase autophosphorylation. To ensure that Rac, Cdc42, and Rho were in the guanosine 5′-[γ-thio]triphosphate- (GTP[γS])-bound or GDP-bound state, the proteins (0.1–0.4 mg/ml) were incubated for 10 min at 30°C in exchange buffer containing 12.5 mM Tris (pH 7.5), 5 mM EDTA, 0.5 mM DTT, and 2 mM GDP or GTP[γS]. MgCl2 was then added to a final concentration of 10 mM. Samples were kept on ice and used within 30 min. The GTP-binding proteins were added to the assay cocktail just before the kinase at final concentrations of 1–2 μg of His-tagged and 2–4 μg of GST-coupled proteins in 100 μl of reaction cocktail. The extent of kinase activation was the same within this range and did not change with further increase in the concentrations of GTP-binding proteins. In contrast to mammalian PAKs (18), Acanthamoeba MIHCK slowly but completely autophosphorylates in vitro in the absence of activating factors, ultimately (under appropriate conditions) reaching full enzymatic activity (6). Acidic lipids increase the rate of kinase autophosphorylation and enhance its ability to phosphorylate myosin I and peptide substrates (6, 7). Activation by lipids is greatest under conditions in which autophosphorylation in the absence of lipids is lowest and acidic lipids do not activate fully phosphorylated kinase (7). Therefore, short incubation times and low temperatures generally were used to study activation of MIHCK by lipids and p21s.
RESULTS
Cloning of Full-Length MIHCK DNA.
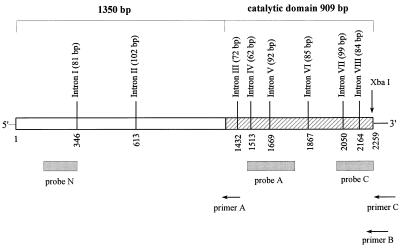
To obtain a highly specific probe for cDNA screening, MIHCK genomic DNA was first cloned from a MIHCK-enriched Acanthamoeba genomic library. The 4.5-kb genomic DNA clone contained 1,911 bp 5′-terminal to the first coding ATG and 75 bp 3′-terminal to the stop codon. Full-length MIHCK cDNA was obtained by screening a MIHCK-enriched Acanthamoeba cDNA library with a 225-bp probe corresponding to the 5′-end segment of the MIHCK gene. The cloning procedure and the positions and lengths of the introns in the genomic DNA are shown in Fig. 1.
Figure 1.
Schematic illustration of cloning of Acanthamoeba MIHCK genomic DNA and cDNA. Genomic DNA was cut with XbaI and the digestion mixture was probed with probe A. A genomic DNA library constructed from the positive band was screened with probe A, and one of the positive clones (4.5 kb) was fully sequenced. A cDNA library was constructed by using poly(A) primer (primer C) and two gene-specific primers (primer B and primer A), screened with probes N and C, and 2 of over 50 positive clones were fully sequenced, and 2 were partially sequenced. For further details see Materials and Methods. The numbering in the figure corresponds to the cDNA beginning at the first coding ATG. The positions and sizes of the introns in the genomic DNA are marked. The diagonal lines define the previously cloned catalytic domain. The complete genomic and cDNA sequences are available in GenBank under accession numbers AF104910 and U67056, respectively.
The Deduced Amino Acid Sequence of MIHCK.
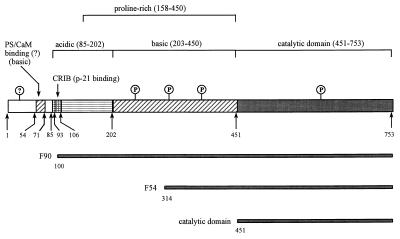
MIHCK contains 753 residues with a calculated molecular mass of 79.3 kDa (Fig. 2), which is substantially lower than the apparent mass of 97 kDa estimated from the mobility of native MIHCK on SDS/PAGE (6). This difference could be explained if some of the 5′-terminal sequence were missing from the cDNA, but this is highly unlikely as neither the cDNA nor the genomic DNA (which had 1.9 kb 5′ to the first ATG in the cDNA sequence) contained any obvious translation start sites other than the first ATG in the ORF of the cDNA. Also, the difference between the SDS/PAGE masses of the 97-kDa full-length native kinase and its 90-kDa C-terminal chymotryptic fragment (Fig. 3, F90) (11) is more than accounted for by the calculated mass of residues 1–100 (10.5 kDa) that are removed by chymotryptic cleavage. The more probable reason for the differences between the apparent and calculated masses is the high proline content (≈40%) between residues 158 and 449 of MIHCK (Fig. 2). This is consistent with the observation that all of the C-terminal proteolytic peptides derived from native kinase that contain some or all of the Pro-rich region migrate on SDS/PAGE (11, 12) with apparent masses higher than their calculated masses (12, 13). For example, the calculated masses of the previously described (11) C-terminal fragments that migrate in SDS/PAGE as proteins of 90 kDa and 54 kDa (see Fig. 3) are 69 kDa and 47 kDa, respectively, whereas the calculated and apparent masses (12, 13) of the C-terminal catalytic domain, which does not contain any of the Pro-rich region, are identical.
Figure 2.
Deduced protein sequence of MIHCK. The C-terminal catalytic domain and the p21-binding region near the N terminus are boxed; the CRIB motif is underlined; acidic residues are in red, basic in green, and prolines in blue. Residues 451–753 are identical to the sequence of the previously cloned catalytic domain (13) and sequences 102–117, 198–212, 255–262, 285–299, 314–328, 437–462, and 626–640 correspond to the N-terminal sequences of seven proteolytic fragments of native kinase (11, 12).
Figure 3.
Schematic representation of the MIHCK molecule. The numbers correspond to the residue numbers in full-length kinase. The regions of interest discussed in the text are indicated at the top of the figure. In addition to the essential autophosphorylation site (P) shown in the catalytic domain, there are multiple other autophosphorylation sites (P), especially in the central, basic, Pro-rich region (ref. 12; and see text) and possibly in the N-terminal region (?). The origins of the proteolytic fragments described previously (11, 12), and whose properties are discussed in the text, are shown at the bottom of the figure.
Detailed Analysis of the MIHCK Sequence.
Residues 93–149 of MIHCK (Figs. 2 and 4) correspond to the PBD, which is characteristic of many members of the PAK family and other p21-binding proteins (24, 25, 28, 33). The PBD consists of the CRIB motif, residues 93–106 in MIHCK [as defined by Burbello et al. (27)], and the next 43 residues (residues 107–149 in MIHCK). Recently, the CRIB region and the following 16 residues of αPAK (Fig. 4) were shown to be required for binding of p21s (29, 30). Although MIHCK has 14 of the 18 residues that typically are conserved in the entire PBD (30), including 6 of the 8 typically conserved CRIB residues, the differences in this region between MIHCK and other PAKs may underlie the significant differences in their interactions with p21s (see below). For example, the highly conserved His-83 in the CRIB region of αPAK that is thought to be important for its binding of, and regulation by, p21s (29, 34) is replaced by Arg-101 in MIHCK (Fig. 4).
Figure 4.
(A) Comparison of the PBDs of MIHCK and α-PAK. The CRIB region (27) is marked at the top of the figure. The conserved residues from comparisons of the sequences of many p21-binding proteins (28, 30, 33) are in bold at the top of the figure and, when present, also in bold in the MIHCK and PAK sequences. (B) Comparison of regions of high sequence identity in the Pro, Gly, Ala-rich regions of MIHCK and myosin IC.
The region between the p21-binding domain and the catalytic domain of MIHCK, residues 158–449, is extremely Pro-rich and includes multiple PXXP motifs that have been shown to bind to SH3 domains (35, 36). This is of special interest because the Acanthamoeba myosins I—the substrates of MIHCK—contain SH3 domains in their C-terminal tails (37). There are three stretches of 5–7 prolines between residues 158 and 200. In addition to these stretches, there are 20 PXXP motifs (some of them overlapping) between residues 200 and 450, of which 8 are PPRP. One, 284RGLPPRPGA292, matches exactly the consensus sequence of peptides that bind with high affinity to the SH3 domain of phosphatidylinositol-3-kinase—RXLPPRPXX, where X is any residue other than cysteine, residues critical for SH3 binding are in boldface, and residues that contribute less importantly are in italics (35, 36). This specific peptide is one of the class I group of SH3-binding motifs that occur in at least 10 proteins including human dynamin, phosphatidylinositol-3-kinase, and GTPase activating protein (GAP) for Cdc42 (35).
Another striking feature of the MIHCK sequence is the alternating clusters of positive and negative charges in the 450 residues N-terminal to the catalytic domain (Figs. 2 and 3). Residues 59–71, which may contain the binding site(s) for acidic phospholipids and calmodulin (refs. 24 and 25; see Discussion), contain 8 lysine residues and only 1 aspartic acid recidue. Not far from this highly basic region, a cluster of 5 acidic amino acids, residues 85–89, occurs just before the CRIB sequence, and the entire region between residues 85 and 202 (which includes the CRIB sequence, the p21-binding region, and the beginning of the Pro-rich region) is strongly acidic, with 24 acidic and 8 basic amino acids. Acidic amino acids are particularly concentrated in the Pro-rich 172–202 region (10 acidic residues, 2 basic residues, and 14 proline residues). The rest of the central Pro-rich region (residues 203–450) up to the C-terminal catalytic domain is extremely basic with 23 Arg, 1 Lys, and just 2 Asp residues. Only the very N terminus of MIHCK (residues 1–52) and the entire C-terminal catalytic domain (residues 451–753) have normal distributions of charges.
Interestingly, the tail regions of the three Acanthamoeba myosins I also contain a basic Pro-rich segment similar to that in the central region of MIHCK (37–39). For example, residues 1020–1168 of myosin IC (39) and residues 203–450 of MIHCK have similarly high contents of Arg, Pro, and Ala/Gly, and short stretches of sequences are very similar. For example, residues 312–348 of MIHCK are about 70% identical in sequence to residues 1095–1127 of myosin IC (Fig. 4B). It is tempting to speculate that these regions have similar functions in the two enzymes. The main difference between the basic Pro-rich regions of myosin I and MIHCK is the high Ser/Thr content in MIHCK. Regions with amino acid compositions even more similar than those of myosin I to that of the basic Pro, Arg, Ala/Gly, Ser/Thr-rich region of MIHCK occur in synapsin and dynamin (40, 41).
Only the catalytic domain and the p21-binding region of MIHCK show significant sequence similarities to other members of the PAK family. The very N terminus of MIHCK (i.e., the 92 residues before the CRIB motif) and the Pro-rich central region are very unlike other PAKs and also have no significant similarity to any other kinases. In particular, and most surprisingly, there is no sequence similarity in these regions even to Dictyostelium MIHCK, which not only has (presumably) homologous substrates in vivo (myosins I with phosphorylation sites homologous to those of the Acanthamoeba myosins I) but also is similarly regulated in vitro, i.e., activated by acidic phospholipid-enhanced, calcium/calmodulin-inhibited autophosphorylation of multiple sites (42). Finally, other than the sequence around the single regulatory phosphorylation site (Ser-627) in the catalytic domain (15), there is no sequence in MIHCK that resembles the sequence of the single site in the myosins I that is phosphorylated by MIHCK (3, 7, 15).
Activation of MIHCK by Small GTP-Binding Proteins.
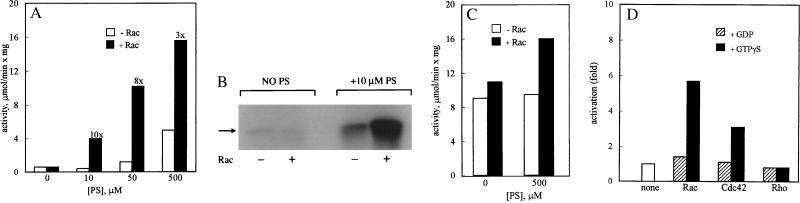
Although MIHCK contains a highly conserved p21-binding region, it was not activated by Rac alone, but Rac did activate MIHCK when phosphatidylserine also was present (Fig. 5A). Activation by Rac was accompanied by an increase in kinase autophosphorylation (Fig. 5B). The extent of Rac activation depended on the assay conditions (see Materials and Methods) and was more pronounced under conditions that minimized activity in the presence of phospholipid alone (low phosphatidylserine concentration and short incubation times). Under the assay conditions described in Fig. 5A, maximum activation varied between 7- and 14-fold with different kinase, phospholipid, and Rac preparations. Rac had no significant effect on the activity of fully phosphorylated kinase in the absence of phospholipid but increased the activity of fully phosphorylated kinase about 70% when the assay was done in the presence of phospholipid (Fig. 5C). Rac did not activate partially autophosphorylated MIHCK in the absence of phosphatidylserine (data not shown). The results were the same when the kinase was autophosphorylated in the absence or presence of phosphatidylserine (data not shown). MIHCK was also activated by Cdc42, and activation by Rac and Cdc42 was GTP-dependent (Fig. 5D). Activation was consistently greater with Rac than with Cdc42, and both Rac and Cdc42 were better activators when expressed with an N-terminal poly(His) tag than were the GST-fusion proteins (data not shown). Rho did not activate MIHCK (Fig. 5D).
Figure 5.
Activation of MIHCK by Rac and Cdc42. (A) Phosphatidylserine-dependent activation of unphosphorylated MIHCK by Rac. MIHCK was incubated for 1 min at 20°C with synthetic peptide PC9 in the absence or presence of phosphatidylserine, at the indicated concentrations, and in the absence (open bars) or presence (closed bars) of GST–Rac–GTP[γS]. The fold-activation by Rac is shown at the top of the bars. (B) Autophosphorylation of MIHCK under identical conditions as in A. (C) Effect of Rac on the activity of phosphorylated MIHCK. MIHCK was fully autophosphorylated by incubation with ATP in the absence of phosphatidylserine and then assayed for 1 min at 20°C in the absence or presence of 500 μM phosphatidylserine and in the absence or presence of GST–Rac–GTP[γS], as indicated. (D) GTP[γS] dependence of activation of MIHCK by Rac and Cdc42. Unphosphorylated MIHCK was assayed in the presence of 500 mM phosphatidylserine and in the absence or presence of Rac, Cdc42, and Rho. All GTP-binding proteins were His-tagged at the N terminus and either in GTP[γS] or in GDP-bound form, as indicated in the figure. Activity was measured at 20°C for 1 min with Cdc42 and Rho and for 2 min with Rac. All values were normalized to the activity of MIHCK in the absence of p21s.
Lipid Specificity for Activation of MIHCK.
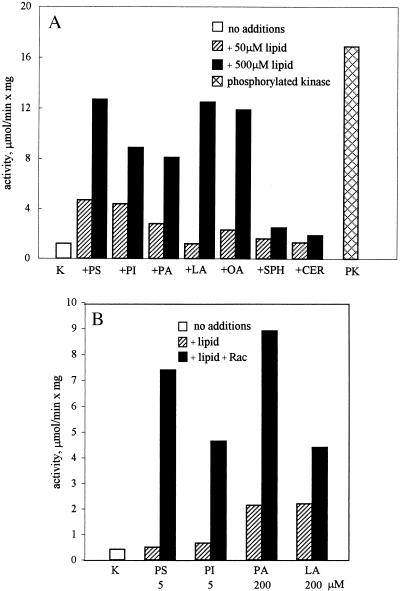
We previously showed that MIHCK is activated by phosphatidylserine and phosphatidylinositol but not by phosphatidylcholine or phosphatidylethanolamine and concluded that MIHCK is activated only by acidic phospholipids (7). We extended these studies because of the recent demonstration that mammalian PAK can be activated in the absence of p21s by sphingosine and phosphatidylinositol but not by phosphatidylserine (43). As shown in Fig. 6A, MIHCK was not significantly activated by basic sphingosine or neutral ceramide but was substantially activated by acidic lipids including, in addition to those previously tested, phosphatidic acid and linoleic and oleic acids. The lipid concentration required for half-maximum activation was consistently higher for fatty acids than for phospholipids (data not shown), but the exact values varied somewhat with different lipid preparations. All of the lipids that activated MIHCK also supported lipid-dependent activation by Rac (Fig. 6B), whereas phosphatidylcholine, sphingosine, and ceramide, which did not activate MIHCK, also did not support Rac activation (data not shown). As with phosphatidylserine, Rac activation in the presence of phosphatidylinositol and phosphatidic acid was greatest at low lipid concentrations (about 5 μM), whereas at least 50 μM linoleic acid was required to support activation by Rac, which was never greater than 2- to 3-fold (Fig. 6B).
Figure 6.
Effect of various lipids on the activity of unphosphorylated MIHCK with and without Rac. (A) The activity of unphosphorylated MIHCK was assayed at 30°C in the absence (open bars) or presence of 50 μM (crossed bars) or 500 μM (black bars) phosphatidylserine (PS), phosphatidylinositol (PI), phosphatidic acid (PA), linoleic acid (LA), oleic acid (OA), sphingosine (SPH), or ceramide (CER). The cross-hatched bar (PK) shows the activity of fully phosphorylated (maximally activated) MIHCK under the same conditions. (B) Activation of unphosphorylated MIHCK by Rac in the presence of various lipids. Activity was measured for 1 min at 20°C in the presence of phosphatidylserine (PS), phosphatidylinisitol (PI), phosphatidic acid (PA), or linoleic acid (LA) at the indicated concentrations in the absence (hatched bars) or presence (solid bars) of GST–Rac–GTP[γS]. The open bar (K) shows the activity of unphosphorylated MIHCK in the absence of lipid and Rac.
DISCUSSION
The data presented in this paper place MIHCK in the subfamily of PAKs activated by small GTP-binding proteins but with several important differences compared with p21-activated mammalian PAKs. (i) The basal activity of MIHCK (6, 7) is much higher than the basal activity of mammalian PAKs (18, 28, 44) and higher than the activity of fully active mammalian PAK expressed in E. coli (H.B. and U.K., unpublished data). This and some of the other differences between MIHCK and mammalian PAKs (see item ii) may be rationalized if MIHCK were already partially activated as purified from the amoeba. Any putative activation could not, however, be caused by proteolytic removal of the inhibitory N-terminal region because the N terminus of native MIHCK is blocked to Edman degradation (11) and is unlikely to be caused by partial phosphorylation because the essential regulatory site within the catalytic domain is not detectably phosphorylated (15). (ii) MIHCK (but not mammalian PAKs) can be maximally autophosphorylated (but more slowly) and thereby fully activated in the absence of lipids or p21s (6). (iii) Autophosphorylation (and activation) of mammalian PAKs is stimulated by basic as well as acidic lipids except for phosphatidylserine (43), whereas only acidic lipids, including phosphatidylserine, enhance autophosphorylation of MIHCK (ref. 7, and herein). The inability of phosphatidylserine to activate mammalian PAK may be the result of the high Mg2+ concentration (10 mM) in the PAK assay; similar concentrations of Mg2+ aggregate phosphatidylserine and inhibit its activation of MIHCK (data not shown). (iv) p21 activation of MIHCK requires the presence of acidic lipids (data herein), whereas lipids are not required for p21 activation of mammalian PAKs (18, 28, 44). The absence of a lipid requirement for p21 activation of mammalian PAKs may relate to the differences in the CRIB and p21-binding regions of MIHCK and PAK (Fig. 4). (v) Rac is a more effective activator than Cdc42 of MIHCK (data herein), whereas Cdc42 activates mammalian PAKs more than Rac (18, 28, 44).
From the sequence of the full-length kinase (data herein) and the properties of the expressed C-terminal catalytic domain (13, 15, 16) and proteolytic fragments of native kinase (10–12), some reasonable inferences can be made about the structural basis of the regulation of MIHCK activity. Full-length, native kinase is partially active and the rate of its autophosphorylation, which results in full activation, is greatly enhanced by calcium/calmodulin-inhibited binding to acidic lipids (6, 7, 10). In contrast, proteolytic fragment F90, which lacks only the first 100 aa of the native kinase (Fig. 3) and all shorter fragments that include the full C-terminal catalytic domain do not bind to acidic lipids or calmodulin. Thus, it seems highly likely that the stretch of basic amino acids (residues 59–71; Fig. 2) that precedes the CRIB sequence (Figs. 2 and 3) is the region to which acidic lipids and calmodulin (which also is acidic) bind. According to both the Chou–Fasman and Garnier secondary structure predictions, residues 54–71 of MIHCK form an α-helix with basic residues clustered together similar to the regions known to bind calmodulin in other calmodulin-binding proteins (for review, see ref. 45). The observation that calcium/calmodulin is a competitive inhibitor of activation by acidic lipids, rather than an activator, suggests that hydrophobic interactions involving the fatty acyl groups of the lipid micelles may be important for activation. The requirement for acidic lipids for p21-activation of MIHCK may be explained if the lipids competitively reverse ionic interactions between the basic, putative lipid-binding site and neighboring acidic residues that block p21-binding.
The catalytic activities of the MIHCK catalytic domain and proteolytic fragment F54 (Fig. 3) are enhanced by phospholipid-independent autophosphorylation, which is as rapid as autophosphorylation of native, full-length MIHCK in the presence of phospholipid (11, 15). This is probably also true for proteolytic fragment F90 (Fig. 3), but F90 has been studied only in digest mixtures containing other fragments (11). These data indicate that the N-terminal region inhibits MIHCK autophosphorylation (including autophosphorylation of the regulatory site within the catalytic domain) and, consequently, catalytic activity. Moreover, at least a portion, if not all, of the inhibitory region of MIHCK is almost certainly N-terminal to F90, i.e., within the first 99 residues (Figs. 3 and 4), which is different from mammalian PAK whose activity was shown recently (29) to be inhibited by a peptide corresponding to the PBD region.
The data presented here and in previous papers (discussed above) suggest that MIHCK is maintained in a state of low activity by ionic interactions between its oppositely charged segments [which include the basic, putative acidic lipid-binding site, the Glu/Asp stretch just before the CRIB motif, the acidic Pro-rich, basic segment following the CRIB motif, and the basic, Pro-rich segment after the CRIB motif (Figs. 2 and 3)] and that inhibition of activity is relieved by autophosphorylation of multiple sites. If the Pro-rich region were held in a folded position by ionic interactions between its acidic and basic regions (perhaps facilitated by interactions between the acidic and basic regions N-terminal to the CRIB region), the inhibitory N-terminal region might be brought sufficiently near the C-terminal catalytic domain to inhibit activity either by specific interactions or simply by blocking substrate access. Binding of acidic lipids to the strongly basic α-helix preceding the CRIB motif may partially disrupt these ionic interactions (and also uncover the p21-binding site allowing p21 to bind and further disrupt, perhaps sterically, the inhibitory, folded structure) facilitating autophosphorylation. Autophosphorylation of the strongly basic, Pro-rich segment, which becomes heavily phosphorylated in native kinase (12), would then completely disrupt the ionic interactions, and the kinase would unfold, removing the inhibitory N-terminal region from the catalytic domain. This hypothesis is consistent with the fact that MIHCK is activated simply by proteolytic removal of the N-terminal segment. Unfolding of MIHCK by autophosphorylation may also expose the central Pro-rich region and allow it to interact with other proteins, in particular, perhaps those with SH3 domains such as myosin I, which is its substrate, and cytoskeletal proteins.
The proposal that multiple phosphorylations activate MIHCK simply by neutralizing ionic interactions by changing the net charge of regions rich in basic amino acids may be applicable to other kinases. For example, despite the complete lack of sequence homology outside of their catalytic domains and p21-binding regions, Dictyostelium and Acanthamoeba MIHCK are similarly regulated. Dictyostelium MIHCK also is activated by acidic phospholipids and small GTP-binding proteins associated with autophosphorylation of multiple sites (46). The regulatory region of Dictyostelium MIHCK also has polarized charged regions, but the order of the charged segments is reversed with the basic region being N-terminal to the acidic region (46). Although mammalian PAKs do not have long, highly charged segments within their regulatory regions, they do have short stretches of strongly charged basic and acidic residues suggesting that their inhibitory conformations may be also maintained by strong ionic interactions (18, 28).
Acknowledgments
We thank Dr. S. Gutkind (National Institutes of Health) for the GST-coupled Rac and Cdc42 cDNA and Drs. J. A. Hammer (National Institutes of Health) and T. Bzdega (Georgetown University) for helpful discussions on cloning strategy.
ABBREVIATIONS
- MIHCK
myosin I heavy chain kinase
- PAK
p21-activated kinase
- CRIB
Cdc42/Rac interactive binding
- PBD
p21-binding domain
- GST
glutathione S-transferase
Footnotes
References
- 1.Pollard T D, Korn E D. J Biol Chem. 1973;248:4691–4697. [PubMed] [Google Scholar]
- 2.Maruta H, Korn E D. J Biol Chem. 1977;252:8329–8332. [PubMed] [Google Scholar]
- 3.Brzeska H, Lynch T J, Martin B M, Korn E D. J Biol Chem. 1989;264:19340–19348. [PubMed] [Google Scholar]
- 4.Lynch T J, Brzeska H, Miyata H, Korn E D. J Biol Chem. 1989;264:19333–19339. [PubMed] [Google Scholar]
- 5.Brzeska H, Korn E D. J Biol Chem. 1996;271:16983–16986. doi: 10.1074/jbc.271.29.16983. [DOI] [PubMed] [Google Scholar]
- 6.Brzeska H, Lynch T J, Korn E D. J Biol Chem. 1990;265:3591–3594. [PubMed] [Google Scholar]
- 7.Brzeska H, Lynch T J, Martin B M, Corigliano-Murphy A, Korn E D. J Biol Chem. 1990;265:16138–16144. [PubMed] [Google Scholar]
- 8.Wang Z, Brzeska H, Baines I C, Korn E D. J Biol Chem. 1995;270:27969–27976. doi: 10.1074/jbc.270.46.27969. [DOI] [PubMed] [Google Scholar]
- 9.Kulesza-Lipka D, Brzeska H, Baines I C, Korn E D. J Biol Chem. 1993;268:17995–18001. [PubMed] [Google Scholar]
- 10.Brzeska H, Kulesza-Lipka D, Korn E D. J Biol Chem. 1992;267:23870–23875. [PubMed] [Google Scholar]
- 11.Brzeska H, Martin B M, Kulesza-Lipka D, Baines I C, Korn E D. J Biol Chem. 1992;267:4949–4956. [PubMed] [Google Scholar]
- 12.Brzeska H, Martin B M, Korn E D. J Biol Chem. 1996;271:27049–27055. doi: 10.1074/jbc.271.43.27049. [DOI] [PubMed] [Google Scholar]
- 13.Brzeska H, Szczepanowska J, Korn E D. J Biol Chem. 1996;271:27056–27062. doi: 10.1074/jbc.271.43.27056. [DOI] [PubMed] [Google Scholar]
- 14.Brzeska H, Knaus U G, Wang Z, Bokoch G M, Korn E D. Proc Natl Acad Sci USA. 1997;94:1092–1095. doi: 10.1073/pnas.94.4.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szczepanowska J, Zhang X, Herring C J, Qin J, Korn E D, Brzeska H. Proc Natl Acad Sci USA. 1997;94:8503–8508. doi: 10.1073/pnas.94.16.8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szczepanowska J, Ramaachandran U, Herring C J, Gruschus J M, Qin J, Korn E D, Brzeska H. Proc Natl Acad Sci USA. 1998;95:4146–4151. doi: 10.1073/pnas.95.8.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu C, Lee S, Furmaniak-Kazmierczak E, Côté G P, Thomas D Y, Leberer E. J Biol Chem. 1996;271:31787–31790. doi: 10.1074/jbc.271.50.31787. [DOI] [PubMed] [Google Scholar]
- 18.Manser E, Leung T, Salihuddin H, Zhao Z, Lim L. Nature (London) 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 19.Benner G E, Dennis P B, Masaracchia R A. J Biol Chem. 1995;270:21121–21128. doi: 10.1074/jbc.270.36.21121. [DOI] [PubMed] [Google Scholar]
- 20.Manser E, Huang H, Loo T, Chen X, Dong J, Leung T, Lim L. Mol Cell Biol. 1997;17:1129–1143. doi: 10.1128/mcb.17.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu C, Whiteway M, Thomas D Y, Leberer E. J Biol Chem. 1995;270:15984–15992. doi: 10.1074/jbc.270.27.15984. [DOI] [PubMed] [Google Scholar]
- 22.Martin G A, Bollag G, McCormick F, Abo A. EMBO J. 1995;14:1970–1978. doi: 10.1002/j.1460-2075.1995.tb07189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu J S, Chen W J, Ni M H, Chan W H, Yang S D. Biochem J. 1998;334:121–131. doi: 10.1042/bj3340121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim L, Manser E, Leung T, Hall C. Eur J Biochem. 1996;242:171–185. doi: 10.1111/j.1432-1033.1996.0171r.x. [DOI] [PubMed] [Google Scholar]
- 25.Sells M A, Chernoff J. Trends Cell Biol. 1997;7:162–167. doi: 10.1016/S0962-8924(97)01003-9. [DOI] [PubMed] [Google Scholar]
- 26.Knaus U G, Bokoch G M. Int J Biochem Cell Biol. 1998;30:857–862. doi: 10.1016/s1357-2725(98)00059-4. [DOI] [PubMed] [Google Scholar]
- 27.Burbelo P D, Drechsel D, Hall A. J Biol Chem. 1995;270:29071–29074. doi: 10.1074/jbc.270.49.29071. [DOI] [PubMed] [Google Scholar]
- 28.Manser E, Chong C, Zhao Z, Leung T, Michael G, Hall C, Lim L. J Biol Chem. 1995;270:25070–25078. doi: 10.1074/jbc.270.42.25070. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Z, Manser E, Chen X, Chong C, Leung T, Lim L. Mol Cell Biol. 1998;18:2153–2163. doi: 10.1128/mcb.18.4.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson G, Owen D, Chalk P A, Lowe P N. Biochemistry. 1998;37:7885–7891. doi: 10.1021/bi980140+. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 32.Lynch T J, Brzeska H, Baines I C, Korn E D. Methods Enzymol. 1990;196:12–23. doi: 10.1016/0076-6879(91)96004-b. [DOI] [PubMed] [Google Scholar]
- 33.Symons M, Derry J M J, Karkak B, Jiang S, Lemachieu V, McCormick F, Francke U, Abo A. Cell. 1998;84:723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- 34.Sells M A, Knaus U G, Bagrodia S, Ambrose D, Bokoch G M, Chernoff J. Curr Biol. 1997;7:202–210. doi: 10.1016/s0960-9822(97)70091-5. [DOI] [PubMed] [Google Scholar]
- 35.Yu H, Chen J K, Feng S, Dalgarno D C, Brauer A W, Schreiber S L. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- 36.Cohen G B, Ren R, Baltimore D. Cell. 1995;80:237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- 37.Hammer J A I. Trends Cell Biol. 1991;1:50–56. doi: 10.1016/0962-8924(91)90089-r. [DOI] [PubMed] [Google Scholar]
- 38.Pollard T D, Doberstein S K, Zot H G. Annu Rev Physiol. 1991;53:653–681. doi: 10.1146/annurev.ph.53.030191.003253. [DOI] [PubMed] [Google Scholar]
- 39.Jung G, Schmidt C J, Hammer J A I. Gene. 1989;82:269–280. doi: 10.1016/0378-1119(89)90052-8. [DOI] [PubMed] [Google Scholar]
- 40.Sudhof T C, Czernik A J, Kao H T, Takei K, Johnston P A, Horiuchi A, Kanazir S D, Wagner M A, Perin M S, De Camilli P. Science. 1989;245:1474–1480. doi: 10.1126/science.2506642. [DOI] [PubMed] [Google Scholar]
- 41.Obar R A, Collins C A, Hammarback J A, Shpetner H S, Vallee R B. Nature (London) 1990;347:256–261. doi: 10.1038/347256a0. [DOI] [PubMed] [Google Scholar]
- 42.Lee S, Mahasneh A, DeLaRoche M, Côté G P. J Biol Chem. 1998;273:27911–27917. doi: 10.1074/jbc.273.43.27911. [DOI] [PubMed] [Google Scholar]
- 43.Bokoch G M, Reilly A M, Daniels R H, King C C, Olivera A, Spiegel S, Knaus U G. J Biol Chem. 1998;273:8137–8144. doi: 10.1074/jbc.273.14.8137. [DOI] [PubMed] [Google Scholar]
- 44.Jakobi R, Chen C, Tuazon P T, Traugh J A. J Biol Chem. 1996;271:6206–6211. doi: 10.1074/jbc.271.11.6206. [DOI] [PubMed] [Google Scholar]
- 45.James P, Vorherr T, Carafoli E. Trends Biochem Sci. 1995;20:38–42. doi: 10.1016/s0968-0004(00)88949-5. [DOI] [PubMed] [Google Scholar]
- 46.Lee S, Egelhoff T T, Mahasneh A, Côté G P. J Biol Chem. 1996;271:27044–27048. doi: 10.1074/jbc.271.43.27044. [DOI] [PubMed] [Google Scholar]