Abstract
Menin is a 70-kDa protein encoded by MEN1, the tumor suppressor gene disrupted in multiple endocrine neoplasia type 1. In a yeast two-hybrid system based on reconstitution of Ras signaling, menin was found to interact with the 32-kDa subunit (RPA2) of replication protein A (RPA), a heterotrimeric protein required for DNA replication, recombination, and repair. The menin-RPA2 interaction was confirmed in a conventional yeast two-hybrid system and by direct interaction between purified proteins. Menin-RPA2 binding was inhibited by a number of menin missense mutations found in individuals with multiple endocrine neoplasia type 1, and the interacting regions were mapped to the N-terminal portion of menin and amino acids 43 to 171 of RPA2. This region of RPA2 contains a weak single-stranded DNA-binding domain, but menin had no detectable effect on RPA-DNA binding in vitro. Menin bound preferentially in vitro to free RPA2 rather than the RPA heterotrimer or a subcomplex consisting of RPA2 bound to the 14-kDa subunit (RPA3). However, the 70-kDa subunit (RPA1) was coprecipitated from HeLa cell extracts along with RPA2 by menin-specific antibodies, suggesting that menin binds to the RPA heterotrimer or a novel RPA1-RPA2-containing complex in vivo. This finding was consistent with the extensive overlap in the nuclear localization patterns of endogenous menin, RPA2, and RPA1 observed by immunofluorescence.
Multiple endocrine neoplasia type 1 is a rare, autosomal dominant tumor syndrome, typically defined by the presence of tumors in at least two of the following three tissues: the parathyroid, enteropancreatic endocrine tissue, and the anterior pituitary (reviewed in reference 46). The human MEN1 gene was identified by positional cloning in 1997 (13) and shown to encode a 610-amino-acid product (menin) with predominantly nuclear localization (24). Loss of heterozygosity in the region containing the MEN1 locus has been observed in tumor tissue obtained from individuals with multiple endocrine neoplasia type 1 (39) and from mice with an engineered deletion in one of their Men1 alleles (16), predictive of a tumor suppressor function. This is supported by the detection of MEN1 frameshift or nonsense mutations in approximately 70% of human multiple endocrine neoplasia type 1 tumors (46).
Menin overexpression has also been shown to diminish the tumorigenic phenotype of Ras-transformed NIH 3T3 cells (36), consistent with its putative tumor suppressor function. Knockout of both Men1 alleles in mice has been shown to result in embryonic lethality (16), suggesting that menin is also important for early development. However, the absence of significant homology to other proteins has complicated efforts to elucidate the function(s) of menin and/or the mechanisms of its tumor suppressor activity.
A number of menin-interacting proteins have been identified in an effort to obtain clues about menin function, including the AP-1 transcription factor JunD (2), the putative tumor metastasis suppressor/nucleoside diphosphate kinase nm23 (54), the homeobox protein Pem (placenta and embryonic expression gene product [40]), and the transcription factor NF-κB (27). Coprecipitation of menin with antibodies specific for the transforming growth factor beta-activated transcription factor Smad3 has also been demonstrated, and reducing menin levels via an antisense strategy has been shown to inhibit transforming growth factor beta-mediated induction of Smad3 activity (31). Although menin seems to facilitate transforming growth factor beta signal transduction through Smad3, it has been shown to repress JunD (2) and NF-κB (27) activity. Since most yeast two-hybrid systems rely on reporter gene activation for the identification of interacting proteins, the finding that menin can repress such transcription raised the possibility that other menin interactors could be masked by menin-mediated repression of the acidic activation domains fused to the prey.
To address this issue, a search for additional interactors was performed in a yeast two-hybrid system based on changes in localization of a member of the Ras signaling pathway (5) rather than transcriptional activation. This system, called the Sos recruitment system, is based on the observation that human Sos, a guanyl nucleotide exchange factor for Ras, can rescue the temperature-sensitive phenotype of a mutant allele (cdc25-2) of the orthologous yeast gene. Membrane recruitment of Sos is necessary to achieve this effect. Fusion of a bait protein to a portion of Sos that retains guanyl nucleotide exchange activity but lacks a membrane localization signal thus allows it to be screened for interaction with myristoylated prey fusion proteins, which results in membrane recruitment of Sos and yeast growth at the nonpermissive temperature.
One of the menin-specific interactors identified by using this system was the 32-kDa subunit of RPA, a heterotrimeric protein consisting of two additional subunits of 70 and 14 kDa. In order of decreasing size, these three subunits have been designated RPA1, RPA2, and RPA3. The RPA heterotrimer has been shown to bind single-stranded DNA (ssDNA) with high affinity and appears to be the eukaryotic equivalent of the Escherichia coli ssDNA-binding protein SSB. Both SSB and RPA have been shown to play important roles in DNA replication, recombination, and repair; however, RPA has also been implicated in the regulation of apoptosis and gene expression (reviewed in reference 28). The importance of RPA in a number of processes related to cell growth, survival, and genome integrity makes it an interesting candidate for interaction with a tumor suppressor protein such as menin.
MATERIALS AND METHODS
Sos recruitment system.
The bait plasmid pSos menin was subcloned by ligating the appropriate NcoI-BstEII and BstEII-NotI fragments of pCMV Sport menin (13, 23), which expresses full-length human menin, into the NcoI and NotI sites of pSos (Stratagene). The human breast cell line HBL-100 cDNA library ne-2 (68) was a gift from Richard Baer. This library was amplified once by inoculating two 250-ml-volumes of Luria-Bertani (LB) broth containing 100 μg of ampicillin per ml with 3 μl (approximately 9 × 107 CFU) each of the library stock, dividing the resulting cultures into 200 aliquots of 2.5 ml each, and growing them overnight at 37°C with shaking. The cultures were then pooled and divided into four 125-ml aliquots, each of which was then diluted with 500 ml of fresh LB broth containing ampicillin and grown for another 4 h at 37°C. DNA was then extracted from these cells with a Qiagen Plasmid Mega Kit.
Selection of menin interactors was performed by transforming a cdc25H yeast strain (Stratagene; MATα ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3,112 cdc25-2 GAL+) expressing Sos-menin with ne-2 library DNA according to the protocol supplied by the manufacturer.
Half of the transformations were plated on 15-cm plates of complete minimal (CM)/glucose medium lacking leucine and uracil, and half were plated on 15-cm CM/galactose plates lacking leucine and uracil. After 48 h of growth at 25°C, transformants on the galactose-containing plates were shifted to 37°C, while transformants on the glucose-containing plates were replica plated onto 15-cm CM/glucose plates lacking leucine and uracil and CM/galactose plates lacking leucine and uracil prior to incubation at 37°C. Prey plasmids from colonies that exhibited galactose-dependent growth at 37°C were extracted with a yeast DNA isolation kit from Stratagene and transformed into supercompetent XL1-B E. coli cells for plasmid DNA amplification, purification, and sequencing. The identities of the cDNA inserts were determined by a BLAST (3) search of the GenBank databases.
Confirmation of menin-RPA2 interaction in a conventional yeast two-hybrid system.
pAS2-1 menin, which expresses full-length human menin fused to the Saccharomyces cerevisiae Gal4 DNA-binding domain, and pGAD10-JD1, which expresses amino acids 8 to 340 of human JunD fused to the S. cerevisiae Gal4 activation domain, have been described previously (2). pVA3-1, which expresses amino acids 72 to 390 of mouse p53 fused to the Gal4 DNA-binding domain, and pTD1-1, which expresses amino acids 87 to 708 of the simian virus 40 T antigen fused to the Gal4 activation domain, were obtained from Clontech.
pACT2 rpa2-59 and pACT2 rpa2-70, which contain the Gal4 activation domain coding sequence fused to two independent RPA2 cDNAs that expressed menin-interacting products in the Sos recruitment system (clones 59 and 70), were subcloned by ligating the EcoRI-XhoI fragments of the corresponding ne-2 library prey plasmids into the EcoRI and XhoI sites of pACT2 (Clontech). pACT2 rpa2, which expresses amino acids 1 to 271 of RPA2 fused directly to the Gal4 activation domain, was constructed by PCR amplification of the RPA2- coding region of clone 70 with primers rpa2-Nterm (5′-GAAGGATCCCCAAGATGTGGAACAGTG-3′) and rpa2-Cterm (5′-TAGCTCGAGTTATTCTGCATCTGTG-3′), followed by digestion of the resulting product with BamHI and XhoI and ligation into the BamHI and XhoI sites of pACT2.
pAS2-1 rpa1 was subcloned by ligating the NdeI-BamHI fragment of pET11a rpa1 into the corresponding sites of pAS2-1 (Clontech). pET11a rpa1 was subcloned by PCR amplification of the RPA1-coding region of pcDNA3.1(+) rpa1 with primers rpa1-Nterm2 (5′-CGACATATGGTCGGCCAGCTGAGC-3′) and rpa1-Cterm3 (5′-ATAGGATCCACTAGTTCACATCAATGCACT-3′), digestion of the resulting product with NdeI and BamHI, and ligation into the NdeI and BamHI sites of pET11a (Novagen). pcDNA3.1(+) rpa1 and pACT2 rpa1 were subcloned by PCR amplification of Marathon-Ready leukocyte cDNA (Clontech) with primers rpa1-Nterm (5′-TTGGGATCCGAGCCATGGTCGGCCAGCTGA-3′) and rpa1-Cterm2 (5′-CTGGAATTCTCACATCAATGCACTTCTC-3′), followed by digestion of the resulting product with BamHI and EcoRI and ligation into the corresponding sites of pcDNA3.1(+) (Invitrogen) and pACT2.
pACT2 rpa3 and pAS2-1 rpa3 were subcloned by inserting the BamHI-XhoI fragment of pCDNA3.1(+) rpa3 into the BamHI and XhoI or BamHI and SalI sites, respectively, of pACT2 or pAS2-1. pcDNA3.1(+) rpa3 was constructed by PCR amplification of the RPA3-coding region from American Type Culture Collection (ATCC) clone 814353 with primers rpa3-Nterm (5′-CTTGGATCCTAATCATGGTGGACATGATGGAC-3′) and rpa3-Cterm (5′-CCACTCGAGTCAATCATGTTGCACAATC-3′), followed by digestion of the resulting product with BamHI-XhoI and ligation into the BamHI and XhoI sites of pcDNA3.1(+). pACT2 rpa4 was subcloned by inserting the BamHI-XhoI fragment of pCDNA3.1(+) rpa4 into the BamHI and XhoI sites of pACT2.
pcDNA3.1(+) rpa4 was cloned by nested PCR amplification of Marathon-Ready small intestine cDNA (Clontech) with primers rpa4-5′UTR (5′-CTTTCGGACAGCTCGAAGCCCTTCT-3′) and rpa4-3′UTR (5′-TCACAGCTGGATGCAAGGGTCTTCG-3′), followed by reamplification with primers rpa4-Nterm (5′-GAGGGATCCTGAAGATGAGTAAGAGTGG-3′) and rpa4-Cterm (5′-TCCTCGAGTCAATCAGCAGACTTAAAATGC-3′), digestion of the resulting product with BamHI and XhoI, and ligation into the BamHI and XhoI sites of pcDNA3.1(+).
pAS2-1 xpa was subcloned by ligating the BamHI-XhoI fragment of pcDNA3.1(+) xpa into the BamHI and SalI sites of pAS2-1. pcDNA3.1(+) xpa, which contains the full-length xeroderma pigmentosum complementation group A (XPA) protein-coding sequence, was subcloned by PCR amplification of ATCC expressed sequence tag clone 546444 and pcDNA3.1(+) xpaΔ, which contains an internal deletion in the XPA-coding sequence between nucleotides 70 and 174, with primer pairs xpa-SacIIF (5′-AACCCGCGGAGCTGCCTGCCTCGGTGCGGGCGAGTATC-3′) and xpa-AclIR (5′-TTTAACGTTAGCCATGCCTCCAGTAGCCGCAG-3′) or xpa-AclIF (5′-ATGGCTAACGTTAAAGCAGCCCCAAAG-3′) and xpa-Cterm (5′-GAACTCGAGAATCACATTTTTTCATATGTC-3′), respectively. The resulting products were then digested with SacII and AclI or AclI and XhoI, as appropriate, and ligated into the SacII and XhoI sites of pCDNA3.1(+) xpaΔ.
pcDNA3.1(+) xpaΔ was cloned by nested PCR amplification of Marathon-Ready ovary cDNA (Clontech) with PCR primers xpa-5′UTR (5′-AGCTAGGTCCTCGGAGTGGGCCAGAG-3′) and xpa-3′UTR (5′-AAATATAAAATTCTATAAAACAGGTCACTG-3′), followed by reamplification with primers xpa-Nterm (5′-AGTGGATCCGAGATGGCGGCGGCCGACGGG-3′) and xpa-Cterm (5′-GAACTCGAGAATCACATTTTTTCATATGTC-3′). The resulting product was then digested with BamHI and XhoI and inserted into the BamHI and XhoI sites of pcDNA3.1(+).
All of the above plasmids were verified by sequencing through the PCR-derived regions.
Yeast strain Y190 was transformed with the indicated bait and prey plasmids by a lithium acetate-based method with a YEASTMAKER yeast transformation system (Clontech) and plated on CM/glucose medium lacking leucine and tryptophan. After incubation for 3 to 5 days at 30°C, β-galactosidase activity was measured by a colony filter lift assay, according to the instructions in Clontech's MATCHMAKER yeast two-hybrid kit.
Analysis of menin missense mutations for interaction with RPA2.
pAS2-1 menin plasmids with a L22R, H139D, A176P, or W436R mutation were constructed by PCR amplification of pCMV Sport menin mutants (2) with primers Y1f (5′-GCCGAATTCGCCGCCATGGGGCTGAAGGCCGC-3′) and Y4r (5′-GCCGGATCCCCGAAGTCCCCAGTAGTTCAGAG-3′), followed by digestion with EcoRI and BamHI and insertion into the EcoRI and BamHI sites of pAS2-1. Menin containing missense mutation P12L, H139Y, F144V, F159C, A160P, A164D, or W183S was introduced into pAS2-1 by ligating the NcoI-KpnI fragment of pCMV Sport menin containing the corresponding mutation and the KpnI-BamHI fragment of pAS2-1 menin into the NcoI and BamHI sites of pAS2-1. The L286P, A309P, T344R, and F447S mutations were introduced into pAS2-1 menin by ligating the NcoI-KpnI fragments of pCMV Sport menin mutants into the NcoI and XmaI sites of pAS2-1 menin. The resulting plasmids were verified by sequencing through the regions containing the mutations.
Mapping the interaction domains of RPA2 and menin.
pAS2-1 plasmids encoding amino acids 1 to 448, 1 to 502, and 178 to 610 of menin have been described previously (2). pAS2-1 plasmids encoding amino acids 1 to 286, 41 to 610, 280 to 610, or 402 to 610 of menin were constructed by PCR amplification of pCMV Sport menin with appropriate primers containing engineered EcoRI and BamHI sites (Y1f, Y2r [5′-GCCGGATCCTAGATCTGCCAGGTTCCCTAAGGC-3′], Y15f [5′-GCCGAATTCCTGGGCTTCGTGGAGCATTTTCTGGCT-3′], Y4r, Y3f [5′-GCCGAATTCTTAGGGAACCTGGCAGATCTAGAG-3′], and Y8f [5′-GCCGAATTCTCCGCCCTCCAGGACCCTGAGTGC-3′]), followed by insertion of the restricted PCR products into the EcoRI and BamHI sites of pAS2-1. pAS2-1 menin1-102 was subcloned by inserting the EcoRI-BamHI fragment of EGFP2 (24), which expresses amino acids 1 to 102 of menin fused to the C terminus of green fluorescent protein (GFP), into the EcoRI and BamHI sites of pAS2-1.
pACT2 plasmids encoding amino acids 1 to 42, 43 to 171, 172 to 271, 1 to 171, or 43 to 271 of RPA2 were constructed by amplification of RPA2 clone 59 or 70 with appropriate pairings of the following primers: rpa2-Nterm, rpa2-Cterm42 (5′-AATCTCGAGTTATCGGGCTCTTGATTTC-3′), rpa2-Nterm43 (5′-AGAGGATCCGAGCCATGGCCCAGCACATTGTGCC-3′), rpa2-Cterm171 (5′-CTGCTCGAGTTATTTGCTTAGTACCATGTG-3′), rpa2-Nterm172 (5′-ACAGGATCCTAAGCATGGCCAACAGCCAGCCC-3′), and rpa2-Cterm, followed by digestion with BamHI and XhoI and ligation into the BamHI and XhoI sites of pACT2. All of the PCR-derived inserts were sequenced to verify the absence of errors.
Gel filtration experiments.
The tricistronic plasmid pET11a rpa1 · 3 · 2, which coexpresses human RPA1, RPA2, and His-RPA3, was constructed by a multistep method similar to that described by Bochkareva et al. (10), culminating in insertion of the EcoRI-XbaI fragment of pET11a rpa2 into the EcoRI and SpeI sites of pET11a rpa1 · 3. pET11a rpa1 · 3 was constructed by insertion of the EcoRI-XbaI fragment of pET15b rpa3 into the EcoRI and SpeI sites of pET11a rpa1. pET15b rpa3 was subcloned by PCR amplification of the RPA3-coding sequence from pcDNA3.1(+) rpa3 with primers rpa3-Nterm2 (5′-CTACATATGGTGGACATGATGGAC-3′) and rpa3-Cterm2 (5′-AGAGGATCCACTAGTTCAATCATGTTGCAC-3′), followed by digestion with NdeI and BamHI and insertion into the NdeI and BamHI sites of pET15b (Novagen).
pET11a rpa2 was subcloned by PCR amplification of the RPA2-coding sequence from pACT2 rpa2 with the primers rpa2-Nterm2 (5′-CCCCATATGTGGAACAGTGGATTC-3′) and rpa2-Cterm2 (5′-AGAGGATCCTTATTCTGCATCTGT-3′), followed by ligation of NdeI-NcoI and NcoI-BamHI fragments into the NdeI and BamHI sites of pET11a. The bicistronic plasmid pET15b rpa3 · 2 was constructed by ligation of the HindIII-XbaI fragments of pET11a rpa2 into the HindIII and SpeI sites of pET15b rpa3. pGEX-4T-1 rpa2 was subcloned by insertion of the BamHI-XhoI fragment of pACT2 rpa2 into the corresponding sites of pGEX-4T-1 (Amersham Pharmacia Biotech). All of the PCR-derived inserts were sequenced to verify the absence of errors.
Recombinant human menin was purified as previously described (37). For purification of the RPA heterotrimer, E. coli BL21(DE3) transformed with pET11a rpa1 · 3 · 2 was induced with 0.5 mM isopropyl β-d-thiogalactopyranoside and grown at room temperature with shaking for 7 h. Cells were pelleted by centrifugation, frozen in a dry ice-methanol bath, and stored at −80°C until needed. The pellet was then thawed on ice and resuspended in 10 ml of cold lysis buffer (50 mM NaH2PO4 [pH 7], 0.3 M NaCl plus a protease inhibitor cocktail [Roche Biochemicals]) per liter of the original culture; the salt concentration was adjusted to 0.8 M, and the cells were lysed in a French press. The lysate was diluted fourfold with cold lysis buffer and clarified by centrifugation at 4°C for 30 min at 20,000 × g, and His-RPA3-containing complexes were purified from the supernatant by batch affinity chromatography on Co2+-based Talon metal affinity resin (Clontech).
Bound proteins were eluted with buffer containing 50 mM imidazole and diluted fivefold with TGED (10 mM Tris-Cl [pH 8], 5% glycerol, 0.1 mM EDTA, 0.1 mM dithiothreitol) containing 0.2 M NaCl, followed by a second affinity purification step on an ssDNA-agarose (Amersham Pharmacia Biotech) column. The column was washed in TGED containing increasing amounts of salt (0.2, 0.3, and 0.75 M), followed by elution of bound RPA in TGED containing 1.5 M NaCl. The eluate was dialyzed in TGED containing 0.2 M NaCl and concentrated with a Centricon 30 concentrator (Amicon) at 4°C, followed by storage at −80°C.
The RPA2-RPA3 complex was purified from E. coli BL21(DE3) transformed with pET15b rpa3 · 2 as described above, except that the steps involving adjustment of the initial NaCl concentration to 0.8 M and affinity chromatography on ssDNA agarose were omitted.
E. coli BL21(DE3) transformed with pET15b rpa3 was lysed and subjected to batch chromatography on metal affinity resin as described for RPA2-RPA3, but after fivefold dilution with TGED, the eluate was further separated by ion exchange chromatography on a Resource-Q column (Amersham Pharmacia Biotech), with a 0.04 to 0.8 M NaCl gradient in TGED. The peak RPA3-containing fractions, which eluted between 0.14 M and 0.21 M NaCl, were then concentrated with a Centricon 3 concentrator and stored at −80°C.
RPA2 expression in E. coli BL21(DE3) transformed with pGEX-4T-1 rpa2 and lysis were performed as described above for RPA except that the cell pellet was resuspended in cold phosphate-buffered saline (PBS) containing 1% Triton X-100 and a protease inhibitor cocktail (Roche Biochemicals). The lysate was clarified as described above, and glutathione S-transferase (GST)-RPA2 was purified from the supernatant by batch affinity chromatography on glutathione-Sepharose 4B (Amersham Pharmacia Biotech). Bound proteins were eluted three to four times with 50 mM Tris-Cl (pH 8) containing 10 mM reduced glutathione and dialyzed overnight at 4°C against TGED containing 0.2 M NaCl and 10 U of thrombin (Sigma) per mg of GST-RPA2 to remove the GST tag. The protein sample was then passed through a combination benzamidine-glutathione-Sepharose column to remove the thrombin, uncleaved GST-RPA2, and the cleaved GST tag. The flowthrough was then concentrated with a Centricon 10 concentrator.
Gel filtration experiments were performed by mixing desired molar ratios of the protein samples to be analyzed, incubating them at 4°C for 30 min, and separating the mixtures on an analytical gel filtration Superdex 200 PC 3.2/30 column at 4°C in TGED containing 0.2 M NaCl with a SMART system (Amersham Pharmacia Biotech). Fractions of 0.2 ml were collected and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 4 to 20% gradient minigels, followed by chemiluminescent Western blotting.
Electrophoretic mobility shift assay.
The probe was prepared by incubation of 50 ng of ssDNA oligonucleotide (dC)30 with approximately 40 μCi of [γ-32P]ATP (specific activity, 3,000 Ci/mmol) and 10 U of T4 polynucleotide kinase in a total volume of 20 μl for 30 min at 37°C. The reaction was terminated with 5 μl of 1% SDS-100 mM EDTA, 75 μl of double-distilled H2O was added, and unincorporated label was removed by gel filtration on a Micro-Spin G-25 column (Amersham Pharmacia Biotech) according to the manufacturer's directions.
Binding reactions were performed in 25 mM HEPES, pH 7.4, 50 to 200 mM NaCl, 1 mM dithiothreitol, 4% glycerol, and 0.1% Igepal in the presence or absence of 50 μg of bovine serum albumin per ml with 30 fmol of labeled (dC)30 and purified proteins, as indicated in the legend to Fig. 5. RPA, wild-type and mutant menin, and GST were purified as described for the gel filtration and/or coprecipitation experiments.
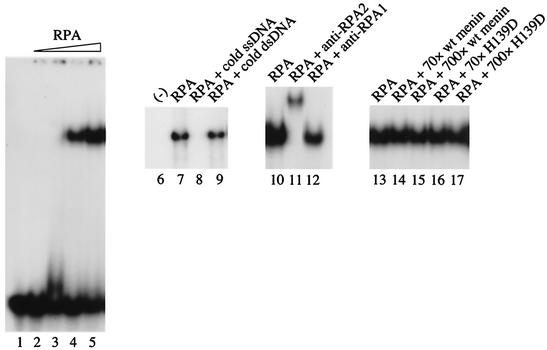
FIG. 5.
Ability of RPA to bind ssDNA in vitro is not specifically influenced by menin. Binding reactions were performed by mixing purified wild-type or mutant menin (H139D) with RPA as indicated, followed by the addition of 32P-labeled (dC)30 and incubation for 20 min at room temperature. With the exception of the sample in lane 1, GST was also added to all of the binding reaction mixtures as a negative control/nonspecific stabilizing protein. Loading dye was then added, and the samples were analyzed by electrophoresis at 150 V on an 8% polyacrylamide gel in TBE (45 mM Tris-Cl, 45 mM boric acid, 1 mM EDTA) at room temperature, followed by vacuum drying at 70°C and autoradiography with an intensifying screen at −80°C. The components of the binding reactions were as follows: 25 fmol of (dC)30 (all lanes), ≈0.3 (lane 3), 3 (lane 4), or 30 (lanes 5 and 7 to 17) fmol of RPA, 2.5 pmol of unlabeled ssDNA (lane 8), 2 pmol of unlabeled dsDNA (lane 9), 0.4 μg of anti-RPA2 (αSSB34A; lane 11), 0.4 μg of anti-RPA1 (αSSB70C; lane 12), and/or 2.1 (70-fold excess) or 21 pmol (700-fold excess) of menin or H139D (lanes 14 to 17), as indicated.
Cell culture and transfections.
HeLa cells were obtained from the ATCC and cultivated in Dulbecco's modified Eagle's medium (Invitrogen) containing high glucose, glutamine, 10% fetal bovine serum (Biofluids), and a 1× concentration of antibiotic-antimycotic mix (Invitrogen) at 37°C in a humidified chamber containing 5% CO2.
The mouse embryo fibroblast cell lines Men10 and Men17, which are wild-type and menin-null, respectively, were derived (details to be published elsewhere) from embryos produced by mating mice heterozygous for a deletion in the Men1 allele (Men1ΔN3-8/+) (16).
The GFP-menin-expressing cell line M10A was prepared by stable transfection of two 10-cm dishes of HeLa cells with 20 μg each of linearized EGFP1 (24), followed by isolation of G418-resistant colonies by trypsinization in cloning cylinders, clonal expansion, and maintenance in medium containing 1 mg of active G418 (Invitrogen) per ml. Expression of the GFP-menin fusion protein was confirmed by Western blotting.
Transient transfections were performed with 1 μg each of pCDNA3.1(+) menin [subcloned by inserting the EcoRI fragment of pCMV Sport menin into the EcoRI site of pcDNA3.1(+)] or pCMV Sport menin and pcDNA3.1(+) rpa2 [subcloned by inserting the BamHI-XhoI fragment of pACT2 rpa2 into the corresponding sites of pcDNA3.1(+)]. SuperFect reagent (Qiagen) was used for both stable and transient transfections according to the manufacturer's directions.
Antibodies.
The menin-specific antibodies SQV and AEA have been described previously (24). Antibodies α-menin1 and α-menin3 were generated by injecting rabbits with human menin purified from E. coli as previously described (37); VRQW1 was raised in rabbits against the RPA2-based peptide NH2-VRQWVDTDDTSSENTVVPPETYV-COOH, and all three antibodies were affinity purified as described by Goldsmith et al. (21). Monoclonal antibodies that recognize RPA2 (αSSB34A) and RPA1 (αSSB70C) were purchased from the Lab Vision Corporation. Monoclonal anti-RPA2 antibody RPA34-20 and anti-RPA3 antibodies were obtained from Oncogene Research Products and GeneTex, respectively.
Coprecipitation experiments.
For preparation of total cell extracts, cells were rinsed twice in cold PBS and lysed in cold NETN (50 mM Tris-Cl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.5% Igepal) or radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-Cl [pH 7.5], 150 mM NaCl, 1% Igepal, 1% deoxycholate, 0.1% SDS) containing a protease inhibitor cocktail (Roche Biochemicals). After incubation for 10 min on ice, cellular debris was removed by microcentrifugation for 15 min at 4°C, and the supernatants were incubated with 5 μg of the indicated antibodies at 4°C for 4 to 8 h or overnight with rocking. Fifty microliters of a 1:1 suspension of protein A- or G-Sepharose that had been washed in the same buffer used for lysis were then added, and the immunoprecipitates were incubated for an additional 1.5 to 2.5 h at 4°C with rocking. After washing four times with cold lysis buffer, immunoprecipitated proteins were eluted from the Sepharose beads by the addition of protein sample buffer and heating for 10 min at 75°C and analyzed by SDS-PAGE, followed by chemiluminescent Western blotting.
Nuclear extracts were prepared with the NE-PER kit (Pierce), and their protein concentrations were determined with a detergent-compatible protein assay (Bio-Rad). Equal aliquots were then diluted two- to threefold with cold Tris-buffered saline containing 10% glycerol and 0.1% Igepal and immunoprecipitated as described above.
Pulldown experiments with purified proteins were performed according to the procedure of Dutta et al. (19) with 40 μl of glutathione-Sepharose or anti-FLAG agarose beads containing similar amounts of bound GST or FLAG fusion proteins and 0.5 to 1 μg of purified RPA, RPA2-RPA3, menin, or free GST or GST-RPA2 proteins, as indicated. RPA, RPA2-RPA3, and menin (both wild-type and mutant forms) were purified as described for the gel filtration experiments. Free GST-RPA2 was prepared as described for untagged RPA2 except that the steps involving thrombin cleavage and removal were omitted. GST was purified from E. coli BL21(DE3) transformed with pGEX-3X (Amersham Pharmacia Biotech) as described for GST-RPA2. Glutathione-Sepharose-bound proteins were prepared as described for free GST and GST-RPA2 except that purification was halted prior to the elution step. FLAG-BAP and FLAG-menin proteins bound to anti-Flag-agarose were prepared as described by Knapp et al. (37).
Immunofluorescence.
Cells were seeded on two-well glass chamber slides and incubated for 24 h at 37°C in a humidified chamber containing 5% CO2. After two rinses with PBS, the cells were fixed in 3% paraformaldehyde-2% sucrose or in PBS containing 4% paraformaldehyde for 15 to 20 min at room temperature, rinsed three times with PBS, permeabilized for 5 min on ice in CSK buffer (20 mM HEPES [pH 7.3 to 7.5], 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose, 0.5% Triton X-100), rinsed three times with PBS, and stored overnight at 4°C. The cells were then blocked with PBS containing 5% donkey serum for 45 min at room temperature, rinsed twice with PBS, incubated in a humidified 37°C incubator for 1 to 1.5 h in PBS containing 0.5% bovine serum albumin and appropriately diluted primary antibodies, washed three times for 5 min each with PBS, and incubated in a humidified 37°C incubator for 45 min in PBS containing 0.5% bovine serum albumin and 1:200 dilutions of fluorescein isothiocyanate- and Texas Red-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories).
The cells were then washed three times for 5 min each in PBS, followed in most cases by a 5-min, room temperature incubation in PBS containing 0.5 μg of 4′,6′-diamidino-2-phenylindole (DAPI) per ml and two rinses in PBS. Slides were mounted with ProLong antifade reagent (Molecular Probes) and glass coverslips (24 by 50 mm). Conventional epifluorescence images were acquired with a Nikon Microphot FXA microscope equipped with a Quantix cooled charge-coupled device camera and pseudocolored with IPLab version 3.2 software (Scanalytics). Confocal images were obtained with the UltraVIEW (Perkin-Elmer) confocal system and a Quantix back-illuminated camera. Analysis was carried out with Openlab 3.1 software (Improvision).
RESULTS
Identification of the 32-kDa subunit of RPA as a menin-interacting protein.
A total of 1.5 × 107 transformants containing cDNAs derived from the human breast cell line HBL-100 and fused to a myristoylation signal (68) were screened for interaction with a bait protein consisting of human menin fused at its N terminus to amino acids 1 to 1067 of the human Sos protein. Seventy-nine transformants that exhibited growth at the nonpermissive temperature of 37°C upon induction of prey expression by galactose were isolated. Sequencing of the cDNAs carried by these colonies revealed 55 clones related to the Ras signaling pathway (mainly clones encoding Ras, but also two types of guanyl nucleotide-releasing factors and the Ras-related protein Ric/Rit). Previous screens with the Sos recruitment system have yielded similar types of false-positives (5, 68), reflecting the fact that overexpression of Ras or proteins that increase Ras activity can also rescue growth of the cdc25H yeast strain.
Of the remaining 24 clones, 18 reacted nonspecifically with other bait proteins and 6 exhibited menin-specific interaction. Two of these last six clones were identical and encoded amino acids 155 to 700 of the resident endoplasmic reticulum heat shock 70 protein family member GRP78 (glucose-regulated protein 78), also known as BiP; these clones may have been recovered as a result of binding to a subpopulation of misfolded Sos-menin protein present in the yeast cells at 37°C. Two other identical clones encoded a short, proline-rich, out-of-frame product that nevertheless appeared to interact specifically with menin. The final two clones contained the full-length coding region of the gene for the 32-kDa subunit of RPA, in frame with the fused myristoylation signal. These two clones also contained full-length 3′ nontranslated regions (NTRs) and some 5′ NTR sequence; however, they differed in length at the 5′ terminus by 66 nucleotides, indicating that they were derived from independent cDNA molecules. Unlike the Ras-related Ric/Rit proteins, which rescued temperature-sensitive growth at 37°C with all baits examined, the products of the two RPA2 clones interacted specifically with menin (Table 1).
TABLE 1.
Specificity of positives obtained from the Sos recruitment system screen or negative control proteins for interaction with menina
| Prey proteinb | Galactose-dependent growth at 37°C
|
|||
|---|---|---|---|---|
| Sos | Collagen | Menin | MafB | |
| Lamin C | − | − | − | − |
| MafB | ND | − | ND | +++ |
| Ric/Rit | +++ | +++ | +++ | ND |
| RPA2 (59) | − | − | ++ | ND |
| RPA2 (70) | − | − | ++ | ND |
Yeast strain cdc25H expressing the indicated bait and prey proteins was scored for growth at 37°C on CM/galactose medium lacking leucine and uracil. ND, not determined; −, no growth; ++, moderate growth; +++, robust growth. The bait proteins collagen, menin, and MafB were fused at their N termini to amino acids 1 to 1067 of human Sos. The prey proteins lamin C, MafB, Ric/Rit, and RPA2 were expressed with an N-terminal myristoylation signal. “Positives” refers to cDNA products that conferred galactose-dependent growth at 37°C; for purposes of illustration, only a subset of these are shown. MafB, a mammalian transcription factor with the ability to form homodimers (32), and the structural proteins lamin C and collagen were included as positive and negative controls, respectively.
The numbers in parentheses are identifiers for the two independent RPA2 cDNA clones obtained in the initial library screen.
Confirmation of menin-RPA2 interaction in a transcription-based yeast two-hybrid system and analysis of other RPA subunits for interaction with menin.
The interaction between menin and RPA2 was confirmed in a second yeast two-hybrid system based on Gal4-activated reporter gene transcription (Table 2). Since RPA2 is a member of a heterotrimeric complex, menin was also tested in this system for interaction with RPA1 or RPA3; however, interaction was observed only in the case of menin and RPA2 (Table 2). The specificity of the menin-RPA2 interaction was further confirmed by the observation that menin fails to interact with RPA4, an RPA2 homologue with 47% identity at the amino acid level (33). The proper folding of RPA2 in this context was also demonstrated by its ability to bind to RPA1, RPA3, and the nucleotide excision repair protein XPA but not to the Gal4 DNA-binding domain (data not shown).
TABLE 2.
Confirmation of specific menin-RPA2 interaction in a yeast two-hybrid assay based on transactivation of a Gal4-regulated lacZ reporter genea
| Proteinb | β-Galactosidase activity
|
||
|---|---|---|---|
| Gal4 DBD | Menin | p53* | |
| Gal4 AD | − | − | − |
| RPA2 (59) | − | ++ | ND |
| RPA2 (70) | − | ++ | ND |
| RPA2 (coding sequence) | − | ++ | ND |
| JunD* | − | +++ | − |
| RPA1 | − | − | ND |
| RPA3 | − | − | ND |
| RPA4 | − | − | ND |
| TAg* | − | − | ++++ |
Yeast strain Y190, which contains a genomic Gal4-regulated lacZ reporter gene, was transformed with plasmids expressing the indicated bait and prey proteins and scored for β-galactosidase activity in a colony filter lift assay after growth on CM/glucose medium lacking leucine and tryptophan at 30°C. ND, not determined; ++, +++, and ++++, moderate, high, and very high levels of β-galactosidase activity, respectively; −, no activity. The bait proteins menin, p53* (amino acids 72 to 390 of mouse p53), RPA1, RPA3, and XPA were fused at their N termini to the S. cerevisiae Gal4 DNA-binding domain (DBD). The prey proteins RPA2, JunD* (amino acids 8 to 340 of human JunD), RPA1, RPA3, RPA4, and TAg* (amino acids 87 to 708 of simian virus 40 T antigen) were fused at their N termini to the S. cerevisiae Gal4 activation domain (AD).
As indicated in Table 1, the numbers in parentheses are identifiers for the two independent RPA2 cDNA clones obtained in the initial library screen. Coding sequence refers to the cDNA sequence encoding the full-length (amino acids 1 to 271) RPA2 protein.
Effect of disease-associated MEN1 missense mutations on menin-RPA2 interaction.
Missense mutations identified in patients with multiple endocrine neoplasia type 1 were also tested for their effects on the menin-RPA2 interaction in the Gal4-based yeast two-hybrid system (Table 3). Of the 15 mutations tested, three (H139D, A160P, and A176P) abolished the interaction completely, eight (P12L, L22R, H139Y, F144V, F159C, L286P, A309P, and T344R) impaired binding to various degrees, and four (A164D, W183S, W436R, and F447S) had little or no effect. The three mutations that abolished the menin-RPA2 interaction also abolished menin-JunD binding (2) (Table 3). Three of the mutations that had no apparent effect on the menin-RPA2 interaction (A164D, W436R, and F447S) also failed to disrupt the menin-JunD interaction.
TABLE 3.
Effect of menin missense mutations on interaction with RPA2a
| Menin | β-Galactosidase activity
|
|
|---|---|---|
| RPA2 | JunD* | |
| Wild type | ++ | +++ |
| P12L | +/− | +++ |
| L22R | ++/+ | +++ |
| H139D | − | − |
| H139Y | + | − |
| F144V | +/− | +++ |
| F159C | ++/+ | +++ |
| A160P | − | − |
| A164D | ++ | +++ |
| A176P | − | − |
| W183S | ++ | − |
| L286P | + | ++ |
| A309P | +/− | ++ |
| T344R | ++/+ | + |
| W436R | ++ | +++ |
| F447S | ++ | +++ |
Bait proteins consisting of Gal4 DNA-binding domain fusions with wild-type menin or the indicated menin missense mutants were tested simultaneously for interaction with prey proteins consisting of Gal4 activation domain-RPA2 or -JunD* fusions as described in Table 2, footnote a. +, ++, and +++, low, moderate, and high levels of β-galactosidase activity, respectively; −, no activity. A level of activity that falls between two categories is indicated by a combination of two symbols separated by a slash.
Although the similarities in the effects of these MEN1 mutations on menin-RPA2 and menin-JunD binding were striking, some differences were also observed. For example, the W183S mutation, which had no noticeable effect on the menin-RPA2 interaction, abolished the menin-JunD interaction. Moreover, the P12L and F144V mutations, which had little or no effect on menin-JunD binding, strongly inhibited menin-RPA2 binding. Western blots of extracts from yeast cells expressing wild-type or mutant menin fusion proteins also ruled out the possibility that failure to interact with RPA2 was due to alterations in the expression or stability of these proteins (data not shown). However, the possibility that these mutations disrupt menin structure cannot be completely excluded.
Mapping of menin and RPA2 interaction domains.
In order to identify the domains required for the menin-RPA2 interaction, menin and RPA2 proteins with N- or C-terminal truncations were tested for their ability to bind to full-length RPA2 or menin in the Gal4-based yeast two-hybrid system (Fig. 1). Previous structural and functional studies have suggested that RPA2 consists of three major domains: an N-terminal domain that consists of the first 40 to 45 amino acids and contains multiple sites of phosphorylation; a central domain extending approximately from amino acids 43 to 171 that contains the low-affinity ssDNA-binding site; and a C-terminal region that contains a conserved domain for binding to members of various DNA repair pathways (48). Menin was therefore tested for binding to the N-terminal domain (amino acids 1 to 42), the central domain (amino acids 43 to 171), the C-terminal domain (amino acids 171 to 271), and two adjacent domains (amino acids 1 to 171 or 43 to 271) of RPA2.
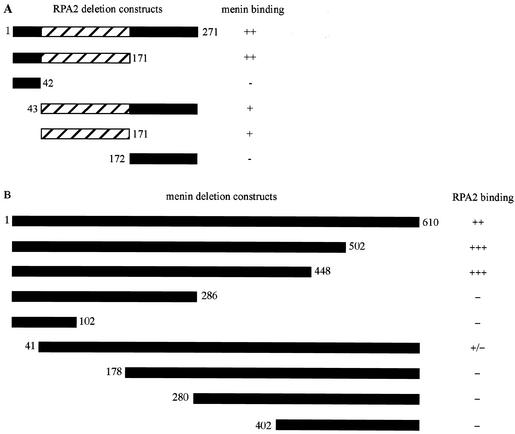
FIG. 1.
Mapping of regions involved in menin-RPA2 binding. Gal4 activation domain-RPA2 (A) and GAL4 DNA-binding domain-menin (B) fusion proteins containing the indicated regions (represented by black or hatched bars) were tested for interaction as described in Table 2, footnote a. The numbers to the left and right of the bars indicate the starting and ending amino acids, respectively, of the various menin and RPA2 constructs. The hatched area of RPA2 (amino acids 43 to 171) represents the region containing the ssDNA-binding domain.
Interaction occurred between menin and polypeptides consisting of RPA2 amino acids 1 to 171, 43 to 171, and 43 to 271, but not 1 to 42 or 172 to 271, mapping the menin-binding site(s) to the central domain of RPA2 between amino acids 43 and 171 (Fig. 1A). The N-terminal region of menin seemed to be involved in this interaction, since RPA2 bound poorly to menin lacking amino acids 1 to 40 but interacted efficiently with menin that had been C-terminally truncated at amino acid 448 (Fig. 1B). Removal of an additional 162 amino acids from the C terminus of menin abolished RPA2 binding, suggesting that residues between amino acids 286 and 448 are also important for interaction between menin and RPA2.
Demonstration of direct menin-RPA2 interaction with purified proteins.
To provide additional confirmation for the menin-RPA2 interaction and to determine whether menin binds directly to RPA2 or requires a bridging protein, we performed binding studies with recombinant menin and RPA proteins purified from E. coli (Fig. 2). Pulldown experiments with purified menin and GST or a GST-RPA2 fusion protein bound to glutathione-Sepharose showed that menin interacted specifically with RPA2 (Fig. 3A). Similarly, in reciprocal experiments, specific interaction of soluble, purified GST-RPA2 interacted specifically with FLAG-menin but not FLAG-BAP bound to anti-FLAG-agarose (Fig. 3B).
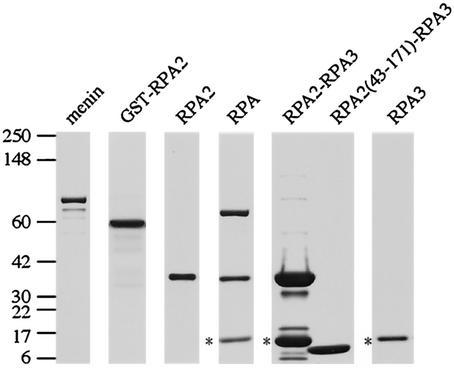
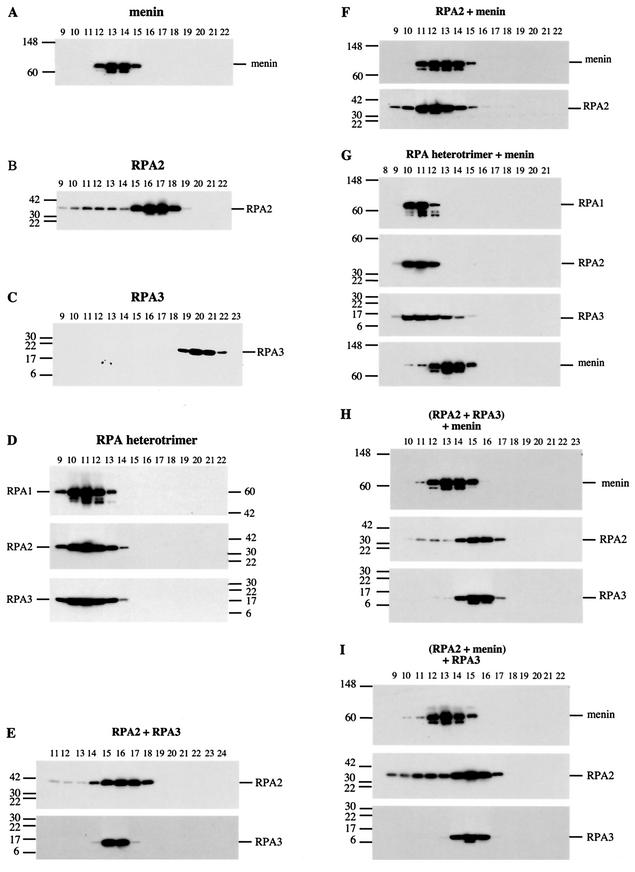
FIG.2.
Purified menin and RPA proteins used for in vitro binding studies. Purified proteins used in the experiments shown in Fig. 3 to 5 were analyzed by SDS-4 to 20% PAGE and stained with Coomassie blue. The positions of the protein standards and their corresponding molecular masses (in kilodaltons) are indicated on the left. Note that the RPA3 subunits present in the RPA heterotrimer and RPA2-RPA3 complex retained their His tags (indicated by asterisks) and therefore had a slightly reduced mobility compared to the untagged RPA3 subunit present in the truncated RPA2(43-171)-RPA3 complex. Also, the truncated RPA2(43-171) polypeptide and the RPA3 subunit were too similar in size to be resolved under these conditions, resulting in their appearance as a single band. However, the presence of both truncated RPA2 and RPA3 in the RPA(43-171)-RPA3 complex was confirmed by Western blotting (data not shown).
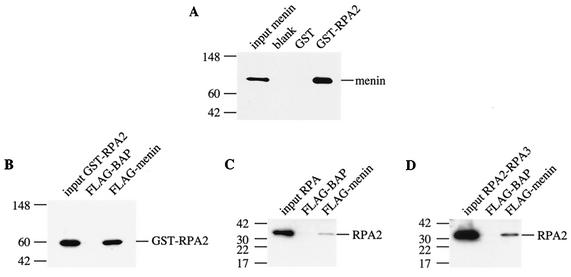
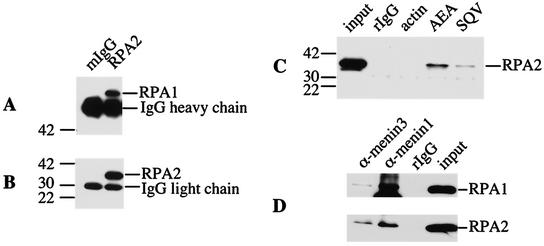
FIG.3.
Menin binds preferentially to free RPA2 in pulldown experiments with purified proteins. Binding reactions were performed by mixing GST or GST-RPA2 bound to glutathione-Sepharose with purified menin (A) or by mixing FLAG-BAP or FLAG-menin bound to anti-Flag-agarose with various purified forms of RPA2, i.e., GST-RPA2 (B), the RPA heterotrimer (C), and the RPA2-RPA3 complex (D). After incubation for 1 h at 37°C, the beads were pelleted by brief microcentrifugation and washed several times to remove nonspecifically bound proteins. Bound proteins were eluted with protein sample buffer and heating for 10 min at 70°C and analyzed by SDS-4 to 20% PAGE, followed by Western blotting with antibodies that recognize menin (SQV) or RPA2 (αSSB34A). The positions of the protein standards are indicated to the left of each panel with their corresponding molecular masses (in kilodaltons).
To determine whether binding of the other RPA subunits affects the ability of RPA2 to interact with menin, coprecipitation experiments were also performed with purified menin and trimeric RPA or an RPA2-RPA3 complex, which is thought to represent the first step in assembly of the RPA heterotrimer (26). As shown in Fig. 3C and D, menin interacted preferentially with GST-RPA2 rather than the heterotrimeric form of RPA or the RPA2-RPA3 complex, although a low level of binding to these two complexes was observed.
To study these interactions in greater detail, menin, RPA2, the RPA heterotrimer, and RPA3 were mixed together in various combinations and analyzed for comigration during gel filtration (Fig. 4). For these experiments, the GST tag was removed from RPA2 through thrombin cleavage. The migration of individual menin (Fig. 4A), RPA2 (Fig. 4B), RPA3 (Fig. 4C) and trimeric RPA (Fig. 4D) proteins is shown for purposes of comparison. A shift in the migration of RPA2 (Fig. 4F) but not the RPA (Fig. 4G) or RPA2-RPA3 (data not shown) complexes was observed in the presence of menin.
FIG.4.
Menin binds preferentially to free RPA2 in gel filtration experiments with purified proteins. Binding reactions were performed by mixing purified proteins in approximately equimolar ratios as indicated and incubating them on ice for 30 min. The binding reaction mixtures were then injected onto an analytical gel filtration column, 0.2-ml fractions were collected, and portions of these fractions were analyzed by SDS-PAGE on 4 to 20% gradient minigels, followed by Western blotting with antibodies that recognize menin (SQV), RPA1 (αSSB70C), RPA2 (αSSB34A), or RPA3 (RPA14 11.1). The positions of the protein standards and their corresponding molecular masses (in kilodaltons) are indicated to the left or right of each panel. The migration of individual menin, RPA2, and RPA3 proteins is shown in panels A to C, respectively, and the migration of the RPA heterotrimer is shown in panel group D. The migration of menin mixed with RPA2 or the RPA heterotrimer is shown in panel groups F and G, respectively. The effect of menin on a preformed RPA2-RPA3 complex and of RPA3 on a preformed menin-RPA2 complex is shown in panel groups H and I, respectively, in comparison to a mixture of RPA2 and RPA3 (group E).
The inability of menin to bind efficiently to RPA2 complexed with RPA3 or both RPA1 and RPA3 (Fig. 3 and 4) raised the possibility that binding of RPA3 to RPA2 interferes with menin's ability to interact with RPA2. This seemed especially likely because RPA3, like menin, is capable of binding to amino acids 43 to 171 of RPA2 (10). To investigate this possibility, competition experiments were performed by adding menin to a preformed RPA2-RPA3 complex (Fig. 4H) or RPA3 to a preformed menin-RPA2 complex (Fig. 4I) and monitoring their migration on an analytical gel filtration column. Comparison of the gel filtration profiles (Fig. 4E, H, and I) showed that menin was unable to disrupt a preformed RPA2-RPA3 complex, as expected based on the results in Fig. 3. In contrast, RPA3 was able to disrupt preformed menin-RPA2 complexes, suggesting that RPA3 can compete with menin for RPA2 binding and that the affinity of RPA2 for RPA3 is higher than its affinity for menin under these experimental conditions. Thus, the presence of RPA3 in the RPA2-RPA3 complexes and heterotrimeric RPA may explain the preference of menin for free RPA2 in vitro.
Inability of menin to influence DNA-binding activity of RPA in vitro.
Since the region of RPA2 that interacts with menin contains an ssDNA-binding domain, electrophoretic mobility shift assays were performed to investigate the effect of menin on RPA-ssDNA binding. During the course of the gel filtration studies described in the previous section, it became clear that menin exhibits a preference in vitro for binding to RPA2 in the absence of RPA3. However, the high affinity of the RPA heterotrimer for ssDNA (K > 109 M−1 [34], which enables RPA-ssDNA complexes to form at very low RPA concentrations) made it possible to analyze the effects of a large molar excess of menin on RPA-ssDNA binding, with the idea that this excess might shift the equilibrium of binding towards the formation of a menin-RPA complex with altered ability to bind ssDNA. RPA binding to ssDNA is also accompanied by conformational changes that stimulate phosphorylation by DNA-dependent protein kinase (8), raising the possibility that menin's ability to interact with RPA2 could be modified by RPA-ssDNA binding.
Incubation of the 32P-labeled ssDNA oligonucleotide (dC)30 with increasing amounts of purified, recombinant RPA resulted in the appearance of a single shifted complex (Fig. 5, lanes 1 to 5) that exhibited a preference for ssDNA, as evidenced by the ability of excess unlabeled ssDNA, but not double-stranded DNA (dsDNA), to compete for binding (Fig. 5, compare lanes 8 and 9). As expected, this complex contained RPA2, as indicated by a supershift in the presence of an RPA2-specific antibody (Fig. 5, lane 11). The addition of an RPA1-specific antibody resulted in partial disruption rather than a supershift of the complex (Fig. 5, lane 12), perhaps due to the location of its epitope in or around one of the RPA1 subunit's high-affinity ssDNA-binding domains. However, menin had no effect on RPA-ssDNA binding, even at a 700-fold molar excess of menin over RPA (Fig. 5, lanes 13 to 17).
As shown by the interaction domain-mapping experiments in Fig. 1, menin binds specifically to the ssDNA-binding domain located in RPA2 (amino acids 43 to 171). Although this domain is unable to bind ssDNA on its own, when complexed with RPA3 it has been shown to bind ssDNA with an affinity of approximately 1 × 105 to 5 × 105 M−1 (10). The low affinity of the RPA2 ssDNA-binding domain relative to the high-affinity domains located in RPA1 raised the possibility that the effect of menin on ssDNA binding by RPA2 could be obscured by the presence of RPA1 in the heterotrimer. To investigate this possibility, menin was tested for its effect on the ssDNA-binding activity of a complex consisting of amino acids 43 to 171 of RPA2 bound to RPA3 [RPA2(43-171)-RPA3; kindly provided by Alexey Bochkarev (9)].
Although the addition of menin inhibited RPA2(43-171)-RPA3 binding to ssDNA, this effect was nonspecific, since negative control proteins such as actin and GST as well as menin missense mutants with impaired RPA2 binding were equally adept at disrupting RPA2(43-171)-RPA3-ssDNA complex formation (data not shown). The effect of menin was also tested on RPA binding to dsDNA probes that had been damaged by treatment with UV light or cisplatin, but the results were similarly negative (data not shown). Together, these results indicate that menin does not have a significant impact on RPA's ability to bind DNA in vitro; however, the possibility cannot be excluded that menin has an effect on RPA-DNA binding in vivo, where other factors, such as posttranslational modifications or interaction with other proteins, may modify binding between menin and the RPA heterotrimer.
Coimmunoprecipitation of overexpressed menin and RPA2 in HeLa extracts.
To demonstrate that the menin-RPA2 interaction also occurs in vivo, menin was immunoprecipitated under nondenaturing conditions from HeLa cells cotransfected with menin- and RPA2-expressing plasmids, and the resulting immunoprecipitates were analyzed for the presence of associated RPA2. As a positive control, the ability of an RPA2-specific antibody to bring down both RPA1 (Fig. 6A) and RPA2 (Fig. 6B) was also demonstrated. Coprecipitation of RPA2 was observed with menin immunoprecipitated by four different antibodies: SQV, AEA, α-menin1, and α-menin3, but not normal rabbit immunoglobulin G (IgG) or an antibody against actin, indicating a specific in vivo menin-RPA2 interaction. (Fig. 6C and 6D). The detection of RPA1 as well as RPA2 in menin immunoprecipitates further suggests that menin interacts with RPA2 bound to RPA1 whether this RPA2 is part of the RPA heterotrimer or a novel complex with RPA1.
FIG. 6.
RPA2 and RPA1 are coimmunoprecipitated by menin-specific antibodies. Whole-cell (A to C) or nuclear (D) extracts prepared from HeLa cells cotransfected with menin and RPA2 were immunoprecipitated with normal mouse (mIgG) or rabbit (rIgG) IgG or antibodies that recognize RPA2 (αSSB34A), menin (AEA, SQV, α-menin1, and α-menin3), or actin (Sigma), as indicated. The immunoprecipitates were then analyzed by SDS-4 to 20% PAGE, followed by Western blotting with antibodies that recognize RPA1 (αSSB70C, A), RPA2 (αSSB34A, B and C), or a mixture of both antibodies (D). Portions of the extracts corresponding to 1/40th the amount used for each immunoprecipitation (input) were also included. The positions of the protein standards and their corresponding molecular masses (in kilodaltons) are indicated to the left of panels A, B, and C. The prominent nonspecific bands in panels A and B correspond to heavy and light chains, respectively, of the mouse antibodies present in the immunoprecipitates, which were recognized by the anti-mouse immunoglobulin-horseradish peroxidase-conjugated secondary antibodies used for Western visualization.
Similarity of in vivo menin and RPA2 subcellular localization.
To pursue our investigation of the intracellular association of menin, RPA2, and RPA1, we examined their subcellular localization in untransfected HeLa cells by immunofluorescence. As seen in previous studies (11, 52), both RPA2 and RPA1 exhibited rather diffuse nuclear signals that became distinctly punctate at the G1/S transition due to RPA entry into replication foci (see Fig. 8 and 9). RPA mobilization into foci was also observed in response to various DNA-damaging agents (see Fig. 9) (15, 22, 43, 51, 55, 58). The specificity of the α-menin3 antibody used for menin detection was demonstrated by comparison of immunofluorescence patterns from fibroblasts obtained from normal mouse embryos and from embryos with homozygous deletions in their Men1 alleles (16).
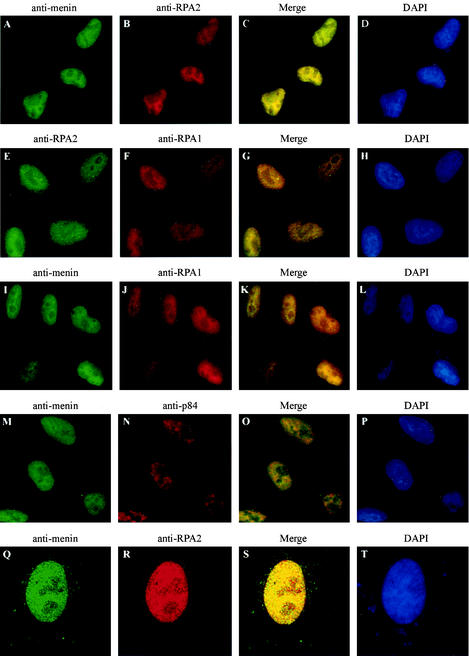
FIG.8.
The pattern of menin localization within the nucleus closely resembles that of RPA1 and RPA2. HeLa cells were analyzed by immunofluorescence with primary antibodies that recognize menin (α-menin3 [A, I, M, and Q]), RPA2 (αSSB34A [B and R] or VRQW1 [E]), RPA1 (αSSB70C [F and J]), or the nuclear antigen p84 (N5-5E10 [GeneTex] [N]) and secondary antibodies conjugated to fluorescein isothiocyanate (A, E, I, M, and Q) or Texas Red (B, F, J, N, and R), followed by DAPI staining (D, H, L, P, and T). Conventional epifluorescence (A to P) or confocal (Q to T) images were visualized through a 60× or 100× objective, respectively. Merged fluorescein isothiocyanate and Texas Red images are shown in panels C, G, K, O, and S.
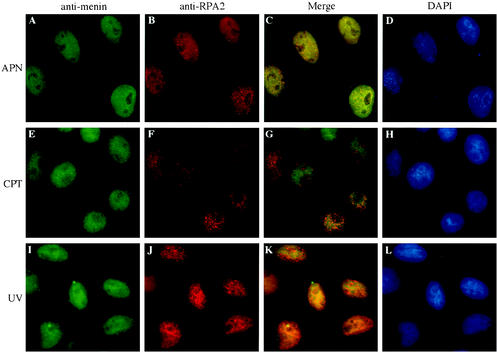
FIG. 9.
Changes in RPA2 localization that occur at the G1/S boundary or in response to DNA-damaging agents are not accompanied by detectable changes in menin localization. HeLa cells were treated with 30 μM aphidicolin for 24 h (APN), 1 μM camptothecin (CPT) for 4 h, or 60 J of UV light per m2 (UV) for 4 h, followed by conventional immunofluorescence analysis with primary antibodies that recognize menin (α-menin3 [A, E, and I]) and RPA2 (αSSB34A [B and J] or RPA34-20 [F]) and secondary antibodies conjugated to fluorescein isothiocyanate (A, E, and I) or Texas Red (B, F, and J), followed by DAPI staining (D, H, and L). Merged fluorescein isothiocyanate and Texas Red signals are shown in panels C, G, and K. All images were visualized through a 60× objective.
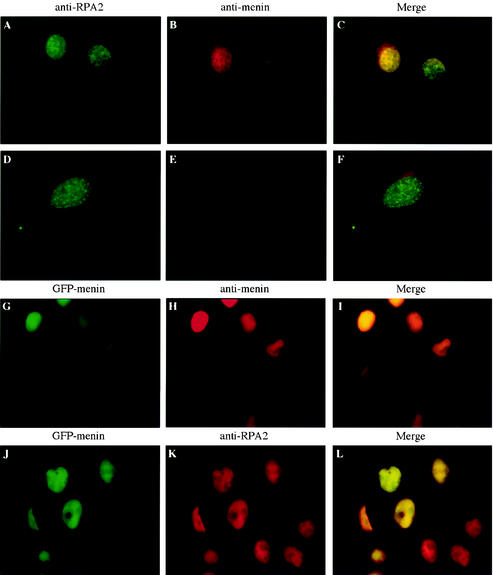
As shown in Fig. 7A to C, menin was detected almost exclusively in the nucleus of wild-type cells. As expected, menin was absent from the nucleus of menin-null cells, although some nonspecific cytoplasmic staining was observed (Fig. 7D to F). The specificity of the α-menin3 antibody was confirmed by colocalization with the fluorescent pattern generated by a GFP-menin fusion protein (Fig. 7G to I). The variable level of GFP-menin fluorescence in the nuclei of these cells correlated perfectly with the intensity of α-menin3 staining, consistent with specific recognition of the GFP-menin fusion protein by the α-menin3 antibody. In contrast, uniform levels of nuclear staining were observed for the endogenous RPA2 protein (Fig. 7J to L). The similarity of the RPA2 distributions in wild-type and menin-null cells also indicates that menin is not required for RPA2 nuclear localization.
FIG. 7.
A specific, endogenous nuclear signal is observed by immunofluorescence analysis of mammalian cells with an antibody raised against menin. Wild-type (A to C) or menin-null (D to F) mouse embryo fibroblasts or HeLa cells expressing a GFP-menin fusion protein (G to L) were analyzed by conventional immunofluorescence with primary antibodies that recognize RPA2 (αSSB34A [A, D, and K]) or menin (α-menin3 [B, E, and H]) and secondary antibodies conjugated to fluorescein isothiocyanate (A and D) or Texas Red (B, E, H, and K). Panels G and J show fluorescence from a GFP moiety fused to the N terminus of menin. Panels C, F, I, and L show merged Texas Red and fluorescein isothiocyanate or GFP-menin signals. All images were visualized through a 60× objective.
Once the specificity of the α-menin3 antibody had been demonstrated, HeLa cells were analyzed for colocalization of the menin and RPA2 proteins. As shown in Fig. 8A to C, the immunofluorescence patterns of endogenous menin and RPA2 were quite similar. Both proteins were distributed throughout the nucleus, but they were largely excluded from the nucleolus and concentrated around the periphery of the nucleoli in areas of intense DAPI staining that correlated with the presence of heterochromatin (compare Fig. 8A to C with Fig. 8D). Similar results were obtained with an independent RPA2-specific antibody (RPA34-20; data not shown). Confocal analysis also supported the interpretation that these proteins colocalized (Fig. 8Q to S).
The similarity between the menin and RPA2 signals was thus consistent with an in vivo menin-RPA2 interaction. Furthermore, the degree of similarity between the menin and RPA2 localization patterns was at least comparable to that observed between RPA1 and RPA2 (Fig. 8E to G), which are capable of forming a stable, heterotrimeric complex. As a negative control, cells were also treated with α-menin3 and an antibody specific for p84, another protein that is present in the nucleus but excluded from the nucleoli (18). As seen in Fig. 8M to O, the degree of overlap between these two proteins was less than that observed between menin and RPA2, consistent with the interpretation that menin and RPA2 colocalize as a result of a specific in vivo association. Similarities in the immunofluorescence patterns of menin and RPA1 were also observed (Fig. 8I to K), consistent with the possibility that menin interacts with trimeric RPA or some other type of RPA1-RPA2-containing complex in vivo.
However, menin was not detectable in RPA2-containing replication or repair foci (Fig. 9). Although punctate RPA2 staining indicative of early or prereplication foci was observed when cells were synchronized at the G1/S border with the DNA polymerase α inhibitor aphidicolin (29) or the ribonucleotide reductase inhibitor hydroxyurea (reviewed in reference 67) and maintained with the replication block or released into S phase, menin localization appeared to be similar to that in untreated cells (Fig. 9A to C and data not shown). Similar results were obtained when cells were treated with DNA-damaging agents such as UV light (Fig. 9I to K), which is known to induce the formation of pyrimidine dimers and lead to activation of the nucleotide excision repair pathway (reviewed in reference 6), or camptothecin (Fig. 9E to G), a topoisomerase I inhibitor that leads to the creation of dsDNA breaks and activation of dsDNA break repair pathways (reference 4 and references therein). RPA2 localization, unlike that of menin, was noticeably changed in response to UV light (Fig. 9I to K) and camptothecin (Fig. 9E to G), although the UV-induced RPA2 foci were more irregularly shaped. Consequently, the degree of overlap in the RPA2 localization patterns was, if anything, decreased in response to these DNA-damaging agents.
DISCUSSION
The role of menin in the pathology of multiple endocrine neoplasia type 1, embryonic development, and the normal regulation of cell growth and/or survival has yet to be elucidated despite its demonstrated ability to modulate the activity of transcription factors such as JunD, NF-κB, and Smad3 (2, 27, 31). The discovery of a menin-interacting protein such as RPA2, with its links to DNA replication, recombination, repair, and transcription, provides a new direction for the investigation of menin function.
The identification of multiple endocrine neoplasia type 1-associated missense mutations that disrupt binding of menin to RPA2 without affecting its binding to JunD (or vice versa), e.g., P12L, F144V, and W183S (Table 3), raises the possibility that both of these proteins are important for menin's tumor suppressor activity. However, the finding that mutants such as W436R and F447S retain the ability to interact with both RPA2 and JunD indicates that multiple endocrine neoplasia type 1 can also develop through mechanisms that do not involve perturbations in menin or JunD binding. In the case of F447S, this mechanism may be loss of the ability to repress JunD activity (2). However, the W436R mutant retains both RPA2/JunD binding and the ability to repress JunD activity; consequently, the mechanism for multiple endocrine neoplasia type 1 development in this case must lie downstream of menin-RPA2 or menin-JunD binding or involve an aspect of menin function that is unrelated to its interaction with RPA2 and JunD. Further study is needed to distinguish among these possibilities.
Similarities in the effects of N-terminal mutations such as H139D, A160P, A164D, A176P, and L286P on the interaction between menin and RPA2 or JunD are consistent with the finding that both of these interactors have binding sites within the N-terminal half of menin (Fig. 1B) (2). Furthermore, C-terminal mutations such as W436R and F447S had no effect on RPA2 binding (Table 3), consistent with gel filtration data indicating that RPA2 is capable of binding to menin C-terminally truncated at amino acid 316 (data not shown). The modest reduction in RPA2-menin binding observed as a result of the T344R mutation might be attributed to the influence of conformational changes on an upstream binding site for RPA2.
The menin-binding region of RPA2 was mapped to amino acids 43 to 171 (Fig. 1A), which also contains an ssDNA-binding domain and sequences required for interaction with RPA3 (10). This domain is believed to be important in stabilizing the interaction between the RPA heterotrimer and ssDNA and is thought to play a role in establishing RPA-ssDNA binding polarity, with RPA1 and RPA2 oriented to the 5′ and 3′ ends, respectively, of the ssDNA (17, 38). This polarity is thought to be important in positioning 5′ and 3′ endonucleases in the incision step of nucleotide excision repair (17) and for regulation of lagging-strand DNA synthesis during DNA replication (47).
Mapping the menin-binding site to the ssDNA-binding region of RPA2 raised the possibility that menin could influence RPA ssDNA-binding activity and, consequently, its functions in DNA repair, replication, or recombination. However, menin did not have a significant effect on binding of the RPA heterotrimer or RPA2(43-171)-RPA3 complex to ssDNA or undamaged or damaged dsDNA (Fig. 5 and data not shown) in vitro. This finding was consistent with the low efficiency of binding observed between menin and trimeric RPA or RPA2-RPA3 complexes relative to free RPA2 in vitro and could explain why simian virus 40 replication in vitro is not noticeably affected by the addition of purified menin (data not shown).
RPA3 was also shown to compete with menin for RPA2 binding in vitro (Fig. 4H and I), suggesting that they may have overlapping binding sites on RPA2. Although menin immunoprecipitates contained RPA1 as well as RPA2 (Fig. 6), the presence of RPA3 could not be established due to the low reactivity of available RPA3-specific antibodies and/or the low expression level of RPA3 relative to the other two subunits. Thus, it is conceivable that menin forms a complex with RPA1 and RPA2 in vivo in the absence of RPA3. Although a complex containing only these two RPA subunits has yet to be demonstrated, a significant proportion of the RPA3 population seems to be localized separately from RPA1 and RPA2 in the nucleoli of G1- or S-phase cells and in the cytoplasm during telophase (52). However, the ability of menin-specific antibodies to coprecipitate RPA1 as well as RPA2 from mammalian cell extracts (Fig. 6), together with the observed similarity in endogenous RPA1, RPA2, and menin intranuclear localization patterns (Fig. 8), raises the possibility that menin may be able to bind to the RPA heterotrimer and influence its DNA-binding activity in vivo.
Interestingly, some studies have suggested that RPA also binds to undamaged dsDNA in vivo. For instance, the bulk of chromatin-bound RPA in HeLa extracts is released by DNase I and micrococcal nuclease but not by single-strand-specific endonucleases (66), suggesting that most of this RPA is bound to dsDNA rather than ssDNA. RPA has also been identified as a repressor protein bound to the promoter of yeast metabolism and repair genes (20, 44, 61), the human metallothionein type IIA promoter (63), and a mutant endothelial nitrogen oxide synthase gene (50), raising the possibility that RPA could be involved in menin-mediated repression of JunD and NF-κB activity on certain promoters.
As alluded to previously, RPA has also been shown to undergo conformational changes upon ssDNA binding that could cause changes in its ability to interact with other proteins. For example, ssDNA binding by RPA stimulates the phosphorylation of RPA2 by DNA-dependent protein kinase, independent of nucleic acid-mediated activation of kinase activity (8). Practically nothing is known about the mechanism of potential RPA-dsDNA binding or conformational changes that might accompany this binding, but it seems likely that there are some proteins in the cell that interact preferentially with the form of RPA that binds dsDNA. Proteins that regulate transcription, such as menin, could fall into this category.
Links between RPA and transcription have also been postulated based on the observed interaction between RPA and Gal4 (25), VP16 (25, 41), p53 (19, 25, 41), Stat3 (35), and RBT1 (14), a putative transcriptional coactivator with homology to Drosophila trithorax proteins involved in developmental gene regulation and/or chromatin remodeling (12). Interestingly, Stat3 and Rbt1, like menin, have been shown to interact with the RPA2 subunit. RPA has also been identified as a component of a human RNA polymerase II complex (45). However, further investigation is required to determine the significance of these interactions.
A number of studies have suggested that menin may have a role in DNA repair or maintenance of genome stability, based on findings of elevated frequencies of spontaneous or DNA damage-induced chromosome abnormalities in peripheral blood lymphocytes from multiple endocrine neoplasia type 1-affected individuals (7, 30, 53, 59, 60, 65), raising the tantalizing possibility that menin has a role in maintaining the genomic integrity of the cell. Many proteins involved directly in repair processes, including RPA (15, 22, 43, 51, 55, 58), form foci in response to agents that cause certain types of DNA damage. Therefore, the absence of noticeable changes in menin localization in response to a wide variety of DNA-damaging agents, such as UV and camptothecin (Fig. 9) or ionizing radiation, diepoxybutane, cisplatin, ethyl methanesulfonate, and mitomycin C (data not shown), suggests that any role of menin in repair is likely to be regulatory rather than direct.
Since dsDNA break repair is thought to occur through homologous recombination and/or nonhomologous end joining, many proteins that form foci in response to dsDNA break-inducing agents and participate in DNA break repair (reviewed in reference 64) are also involved in meiotic recombination. RPA, for example, forms foci in response to dsDNA break-inducing agents such as ionizing radiation (22), camptothecin (58), and etoposide (51), has been localized to meiotic recombination nodules on mouse spermatocytes (56), and is thought to participate in the process of DNA strand exchange (reviewed in reference 28).
Although menin is ubiquitously expressed, its expression level is particularly high in the testis, consistent with a potential role in recombination (62). However, the absence of menin focus formation in response to camptothecin, ionizing radiation, or bleomycin (Fig. 9 and data not shown), suggests that any involvement of menin in recombination, as in the case of DNA repair, is likely to occur at the level of regulation rather than direct participation in the core reactions. A similar argument can be made with respect to DNA replication, based on the lack of menin mobilization into replication foci in cells synchronized at the G1/S border or released into S phase (Fig. 9). The detection of RPA2-containing foci in menin-null cells (data not shown) further indicates that menin is not essential for the assembly of DNA replication or repair complexes and that any regulation of these processes by menin is likely to occur via some other mechanism.
The requirement for RPA in DNA replication, recombination, and repair, as well as its potential role in the modulation of gene expression, makes it an attractive target for regulation by tumor suppressors or other proteins involved in the control of cell proliferation and/or apoptosis. Indeed, specific interaction between p53 and RPA is inversely correlated with the abilities of RPA to bind ssDNA and of p53 to bind transactivating sequences in the promoters of its target genes (19, 49). This interaction is disrupted by UV (1), adozelesin (42), and cisplatin (57), and a correlation between RPA-p53 binding and RPA2 phosphorylation status has been observed. RPA2 has been shown to undergo cell cycle-dependent and damage-induced phosphorylation, and although the consequences of this phosphorylation are unclear, it may influence RPA conformation, DNA-binding characteristics, and/or its interactions with other proteins, as in the case of p53.
Although treatment of cultured cells with cell-synchronizing or DNA-damaging agents and the accompanying increase in RPA2 phosphorylation does not result in detectable changes in menin localization or binding to RPA2 (Fig. 9 and data not shown), this interaction is likely to have some other important role in the control of cell growth and/or survival. The identification of RPA2 as a menin-interacting protein is thus an important step in the elucidation of menin's function as a tumor suppressor and may reveal new aspects of RPA's central role in the physiology of the cell.
Acknowledgments
We are grateful to many colleagues for their help and advice during the course of this work, especially Paul Goldsmith for purified rabbit menin- and RPA2-specific antibodies, George Poy for plasmid sequencing, John Hanover for sharing expertise in confocal microscopy, Richard Baer for providing the HBL-100 cDNA library, and Alexey Bochkarev for the gift of purified RPA2(43-171)-RPA3 for electrophoretic mobility shift assay analysis. We also thank Brian Oliver and Heinz Reiske for critical reading of the manuscript and Aniello Cerrato and Christina Heppner for helpful discussions and technical protocols.
REFERENCES
- 1.Abramova, N. A., J. Russell, M. Botchan, and R. Li. 1997. Interaction between replication protein A and p53 is disrupted after UV damage in a DNA repair-dependent manner. Proc. Natl. Acad. Sci. USA 94:7186-7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agarwal, S. K., S. C. Guru, C. Heppner, M. R. Erdos, R. M. Collins, S. Y. Park, S. Saggar, S. C. Chandrasekharappa, F. S. Collins, A. M. Spiegel, S. J. Marx, and A. L. Burns. 1999. Menin interacts with the AP1 transcription factor JunD and represses JunD-activated transcription. Cell 96:143-152. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Arnaudeau, C., C. Lundin, and T. Helleday. 2001. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 307:1235-1245. [DOI] [PubMed] [Google Scholar]
- 5.Aronheim, A., E. Zandi, H. Hennemann, S. J. Elledge, and M. Karin. 1997. Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol. Cell. Biol. 17:3094-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batty, D. P., and R. D. Wood. 2000. Damage recognition in nucleotide excision repair of DNA. Gene 241:193-204. [DOI] [PubMed] [Google Scholar]
- 7.Benson, L., K.-H. Gustavson, J. Rastad, G. Åkerström, K. Öberg, and S. Ljunghall. 1988. Cytogenetical investigations in patients with primary hyperparathyroidism and multiple endocrine neoplasia type 1. Hereditas 108:227-229. [DOI] [PubMed] [Google Scholar]
- 8.Blackwell, L. J., J. A. Borowiec, and I. A. Mastrangelo. 1996. Single-stranded-DNA binding alters human replication protein A structure and facilitates interaction with DNA-dependent protein kinase. Mol. Cell. Biol. 16:4798-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bochkarev, A., E. Bochkareva, L. Frappier, and A. M. Edwards. 1999. The crystal structure of the complex of replication protein A subunits RPA32 and RPA14 reveals a mechanism for single-stranded DNA binding. EMBO J. 18:4498-4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bochkareva, E., L. Frappier, A. M. Edwards, and A. Bochkarev. 1998. The RPA32 subunit of human replication protein A contains a single-stranded DNA-binding domain. J. Biol. Chem. 273:3932-3936. [DOI] [PubMed] [Google Scholar]
- 11.Brénot-Bosc, F., S. Gupta, R. L. Margolis, and R. Fotedar. 1995. Changes in the subcellular localization of replication initiation proteins and cell cycle proteins during G1- to S-phase transition in mammalian cells. Chromosoma 103:517-527. [DOI] [PubMed] [Google Scholar]
- 12.Calgaro, S., M. Boube, D. L. Cribbs, and H.-M. Bourbon. 2002. The Drosophila gene taranis encodes a novel trithorax group member potentially linked to the cell cycle regulatory apparatus. Genetics 160:547-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandrasekharappa, S. C., S. C. Guru, P. Manickam, S.-E. Olufemi, F. S. Collins, M. R. Emmert-Buck, L. V. Debelenko, Z. Zhuang, I. A. Lubensky, L. A. Liotta, J. S. Crabtree, Y. Wang, B. A. Roe, J. Weisemann, M. S. Boguski, S. K. Agarwal, M. B. Kester, Y. S. Kim, C. Heppner, Q. Dong, A. M. Spiegel, A. L. Burns, and S. J. Marx. 1997. Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science 276:404-407. [DOI] [PubMed] [Google Scholar]
- 14.Cho, J. M., D. J. Song, J. Bergeron, N. Benlimame, M. S. Wold, and M. A. Alaoui-Jamali. 2000. RBT1, a novel transcriptional co-activator, binds the second subunit of replication protein A. Nucleic Acids Res. 28:3478-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choudhary, S. K., and R. Li. 2002. BRCA1 modulates ionizing radiation-induced nuclear focus formation by the replication protein A p34 subunit. J. Cell. Biochem. 84:666-674. [DOI] [PubMed] [Google Scholar]
- 16.Crabtree, J. S., P. C. Scacheri, J. M. Ward, L. Garrett-Beal, M. R. Emmert-Buck, K. A. Edgemon, D. Lorang, S. K. Libutti, S. C. Chandrasekharappa, S. J. Marx, A. M. Spiegel, and F. S. Collins. 2001. A mouse model of multiple endocrine neoplasia, type 1, develops multiple endocrine tumors. Proc. Natl. Acad. Sci. USA 98:1118-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Laat, W. L., E. Appeldoorn, K. Sugasawa, E. Weterings, N. G. J. Jaspers, and J. H. J. Hoeijmakers. 1998. DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes Dev. 12:2598-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durfee, T., M. A. Mancini, D. Jones, S. J. Elledge, and W.-H. Lee. 1994. The amino-terminal region of the retinoblastoma gene product binds a novel nuclear matrix protein that co-localizes to centers for RNA processing. J. Cell Biol. 127:609-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dutta, A., J. M. Ruppert, J. C. Aster, and E. Winchester. 1993. Inhibition of DNA replication factor RPA by p53. Nature 365:79-82. [DOI] [PubMed] [Google Scholar]
- 20.Gailus-Durner, V., C. Chintamaneni, R. Wilson, S. J. Brill, and A. K. Vershon. 1997. Analysis of a meiosis-specific URS1 site: sequence requirements and involvement of replication protein A. Mol. Cell. Biol. 17:3536-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldsmith, P., P. Gierschik, G. Milligan, C. G. Unson, R. Vinitsky, H. L. Malech, and A. M. Spiegel. 1987. Antibodies directed against synthetic peptides distinguish between GTP-binding proteins in neutrophil and brain. J. Biol. Chem. 262:14683-14688. [PubMed] [Google Scholar]
- 22.Golub, E. I., R. C. Gupta, T. Haaf, M. S. Wold, and C. M. Radding. 1998. Interaction of human Rad51 recombination protein with single-stranded DNA binding protein, RPA. Nucleic Acids Res. 26:5388-5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guru, S. C., S. K. Agarwal, P. Manickam, S.-E. Olufemi, J. S. Crabtree, J. M. Weisemann, M. B. Kester, Y. S. Kim, Y. Wang, M. R. Emmert-Buck, L. A. Liotta, A. M. Spiegel, M. S. Boguski, B. A. Roe, F. S. Collins, S. J. Marx, L. Burns, and S. C. Chandrasekharappa. 1997. A transcript map for the 2.8-Mb region containing the multiple endocrine neoplasia type 1 locus. Genome Res. 7:725-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guru, S. C., P. K. Goldsmith, A. L. Burns, S. J. Marx, A. M. Spiegel, F. S. Collins, and S. C. Chandrasekharappa. 1998. Menin, the product of the MEN1 gene, is a nuclear protein. Proc. Natl. Acad. Sci. USA 95:1630-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He, Z., B. T. Brinton, J. Greenblatt, J. A. Hassell, and C. J. Ingles. 1993. The transactivator proteins VP16 and GAL4 bind replication factor A. Cell 73:1223-1232. [DOI] [PubMed] [Google Scholar]
- 26.Henricksen, L. A., C. B. Umbricht, and M. S. Wold. 1994. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 269:11121-11132. [PubMed] [Google Scholar]
- 27.Heppner, C., K. Y. Bilimoria, S. K. Agarwal, M. Kester, L. J. Whitty, S. C. Guru, S. C. Chandrasekharappa, F. S. Collins, A. M. Spiegel, S. J. Marx, and A. L. Burns. 2001. The tumor suppressor protein menin interacts with NF-κB proteins and inhibits NF-κB-mediated transactivation. Oncogene 20:4917-4925. [DOI] [PubMed] [Google Scholar]
- 28.Iftode, C., Y. Daniely, and J. A. Borowiec. 1999. Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 34:141-180. [DOI] [PubMed] [Google Scholar]
- 29.Ikegami, S., T. Taguchi, M. Ohashi, M. Oguro, H. Nagano, and Y. Mano. 1978. Aphidicolin prevents mitotic cell division by interfering with the activity of DNA polymerase-α. Nature 275:458-460. [DOI] [PubMed] [Google Scholar]
- 30.Itakura, Y., A. Sakurai, M. Katai, Y. Ikeo, and K. Hashizume. 2000. Enhanced sensitivity to alkylating agent in lymphocytes from patients with multiple endocrine neoplasia type 1. Biomed. Pharmacother. 54(Suppl. 1):187-190. [DOI] [PubMed] [Google Scholar]
- 31.Kaji, H., L. Canaff, J.-J. Lebrun, D. Goltzman, and G. N. Hendy. 2001. Inactivation of menin, a Smad3-interacting protein, blocks transforming growth factor type β signaling. Proc. Natl. Acad. Sci. USA 98:3837-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kataoka, K., K. T. Fujiwara, M. Noda, and M. Nishizawa. 1994. MafB, a new Maf family transcription activator that can associate with Maf and Fos but not with Jun. Mol. Cell. Biol. 14:7581-7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keshav, K. F., C. Chen, and A. Dutta. 1995. Rpa4, a homolog of the 34-kilodalton subunit of the replication protein A complex. Mol. Cell. Biol. 15:3119-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, C., R. O. Snyder, and M. S. Wold. 1992. Binding properties of replication protein A from human and yeast cells. Mol. Cell. Biol. 12:3050-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim, J., D. Kim, and J. Chung. 2000. Replication protein A 32 kDa subunit (RPA p32) binds the SH2 domain of Stat3 and regulates its transcriptional activity. Cell Biol. Int. 24:467-473. [DOI] [PubMed] [Google Scholar]
- 36.Kim, Y. S., A. L. Burns, P. K. Goldsmith, C. Heppner, S. Y. Park, S. C. Chandrasekharappa, F. S. Collins, A. M. Spiegel, and S. J. Marx. 1999. Stable overexpression of MEN1 suppresses tumorigenicity of RAS. Oncogene 18:5936-5942. [DOI] [PubMed] [Google Scholar]
- 37.Knapp, J. I., C. Heppner, A. B. Hickman, A. L. Burns, S. C. Chandrasekharappa, F. S. Collins, S. J. Marx, A. M. Spiegel, and S. K. Agarwal. 2000. Identification and characterization of JunD missense mutants that lack menin binding. Oncogene 19:4706-4712. [DOI] [PubMed] [Google Scholar]
- 38.Kolpashchikov, D. M., S. N. Khodyreva, D. Yu. Khlimankov, M. S. Wold, A. Favre, and O. I. Lavrik. 2001. Polarity of human replication protein A binding to DNA. Nucleic Acids Res. 29:373-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larsson, C., B. Skogseid, K. Öberg, Y. Nakamura, and M. Nordenskjold. 1988. Multiple endocrine neoplasia type 1 gene maps to chromosome 11 and is lost in insulinoma. Nature 332:85-87. [DOI] [PubMed] [Google Scholar]
- 40.Lemmens, I. H., L. Forsberg, A. A. J. Pannett, E. Meyen, F. Piehl, J. J. O. Turner, W. J. M. Van de Ven, R. V. Thakker, C. Larsson, and K. Kas. 2001. Menin interacts directly with the homeobox-containing protein Pem. Biochem. Biophys. Res. Commun. 286:426-431. [DOI] [PubMed] [Google Scholar]
- 41.Li, R., and M. R. Botchan. 1993. The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell 73:1207-1221. [DOI] [PubMed] [Google Scholar]
- 42.Liu, J.-S., S.-R. Kuo, M. M. McHugh, T. A. Beerman, and T. Melendy. 2000. Adozelesin triggers DNA damage response pathways and arrests SV40 DNA replication through replication protein A inactivation. J. Biol. Chem. 275:1391-1397. [DOI] [PubMed] [Google Scholar]
- 43.Liu, J.-S., S.-R. Kuo, X. Yin, T. A. Beerman, and T. Melendy. 2001. DNA damage by the enediyne C-1027 results in the inhibition of DNA replication by loss of replication protein A function and activation of DNA-dependent protein kinase. Biochemistry 40:14461-14668. [DOI] [PubMed] [Google Scholar]
- 44.Luche, R. M., W. C. Smart, and T. G. Cooper. 1992. Purification of the heteromeric protein binding to the URS1 transcriptional repression site in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci., USA 89:7412-7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maldonado, E., R. Shiekhattar, M. Sheldon, H. Cho, R. Drapkin, P. Rickert, E. Lees, C. W. Anderson, S. Linn, and D. Reinberg. 1996. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature 381:86-89. [DOI] [PubMed] [Google Scholar]
- 46.Marx, S. J. 2001. Multiple endocrine neoplasia type 1, p. 535-584. In J. P. Bilezikian, R. Marcus, and M. A. Levine (ed.), The parathyroids, 2nd ed. Academic Press, New York, N.Y.
- 47.Mass, G., T. Nethanel, and G. Kaufmann. 1998. The middle subunit of replication protein A contacts growing RNA-DNA primers in replicating simian virus 40 chromosomes. Mol. Cell. Biol. 18:6399-6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mer, G., A. Bochkarev, R. Gupta, E. Bochkareva, L. Frappier, C. J. Ingles, A. M. Edwards, and W. J. Chazin. 2000. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell 103:449-456. [DOI] [PubMed] [Google Scholar]
- 49.Miller, S. D., K. Moses, L. Jayaraman, and C. Prives. 1997. Complex formation between p53 and replication protein A inhibits the sequence-specific DNA binding of p53 and is regulated by single-stranded DNA. Mol. Cell. Biol. 17:2194-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miyamoto, Y., Y. Saito, M. Nakayama, Y. Shimasaki, T. Yoshimura, M. Yoshimura, M. Harada, N. Kajiyama, I. Kishimoto, K. Kuwahara, J. Hino, E. Ogawa, I. Hamanaka, S. Kamitani, N. Takahashi, R. Kawakami, K. Kangawa, H. Yasue, and K. Nakao. 2000. Replication protein A1 reduces transcription of the endothelial nitric oxide synthase gene containing a −786T->C mutation associated with coronary spastic angina. Hum. Mol. Genet. 9:2629-2637. [DOI] [PubMed] [Google Scholar]
- 51.Montecucco, A., R. Rossi, G. Ferrari, A. I. Scovassi, E. Prosperi, and G. Biamonti. 2001. Etoposide induces the dispersal of DNA ligase I from replication factories. Mol. Biol. Cell 12:2109-2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murti, K. G., D. C. He, B. R. Brinkley, R. Scott, and S.-H. Lee. 1996. Dynamics of human replication protein A subunit distribution and partitioning in the cell cycle. Exp. Cell Res. 223:279-289. [DOI] [PubMed] [Google Scholar]
- 53.Musilová, J., K. Michalová, L. Folberová, M. Neradilová, and J. Abrahamová. 1993. Chromosome sensitivity to bleomycin in patients with dominantly inherited and sporadic tumors. Neoplasma 40:93-96. [PubMed] [Google Scholar]
- 54.Ohkura, N., M. Kishi, T. Tsukada, and K. Yamaguchi. 2001. Menin, a gene product responsible for multiple endocrine neoplasia type 1, interacts with the putative tumor metastasis suppressor nm23. Biochem. Biophys. Res. Commun. 282:1206-1210. [DOI] [PubMed] [Google Scholar]
- 55.Peters, K. B., H. Wang, J. M. Brown, and G. Iliakis. 2001. Inhibition of DNA replication by tirapazamine. Cancer Res. 61:5425-5431. [PubMed] [Google Scholar]
- 56.Plug, A. W., A. H. F. M. Peters, K. S. Keegan, M. F. Hoekstra, P. de Boer, and T. Ashley. 1998. Changes in protein composition of meiotic nodules during mammalian meiosis. J. Cell Sci. 111:413-423. [DOI] [PubMed] [Google Scholar]
- 57.Riva, F., V. Zuco, A. A. Vink, R. Supino, and E. Prosperi. 2002. UV-induced DNA incision and proliferating cell nuclear antigen recruitment to repair sites occur independently of p53-replication protein A interaction in p53 wild type and mutant ovarian carcinoma cells. Carcinogenesis 22:1971-1978. [DOI] [PubMed] [Google Scholar]
- 58.Sakamoto, S., K. Nishikawa, S.-J. Heo, M. Goto, Y. Furuichi, and A. Shimamoto. 2001. Werner helicase relocates into nuclear foci in response to DNA damaging agents and co-localizes with RPA and Rad51. Genes Cells 6:421-430. [DOI] [PubMed] [Google Scholar]
- 59.Sakurai, A., M. Katai, Y. Itakura, Y. Ikeo, and K. Hashizume. 1999. Premature centromere division in patients with multiple endocrine neoplasia type 1. Cancer Genet. Cytogenet. 109:138-140. [DOI] [PubMed] [Google Scholar]
- 60.Scappaticci, S., P. Maraschio, N. del Ciotto, G. S. Fossati, A. Zonta, and M. Fraccaro. 1991. Chromosome abnormalities in lymphocytes and fibroblasts of subjects with multiple endocrine neoplasia type 1. Cancer Genet. Cytogenet. 52:85-92. [DOI] [PubMed] [Google Scholar]
- 61.Singh, K. K., and L. Samson. 1995. Replication protein A binds to regulatory elements in yeast DNA repair and DNA metabolism genes. Proc. Natl. Acad. Sci. USA 92:4907-4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stewart, C., F. Parente, F. Piehl, F. Farnebo, D. Quincey, G. Silins, L. Bergman, G. F. Carle, I. Lemmens, S. Grimmond, C. Z. Xian, S. Khodei, B. T. Teh, J. Lagercrantz, P. Siggers, A. Calendar, V. Van de Vem, K. Kas, G. Weber, N. Hayward, P. Gaudray, and C. Larsson. 1998. Characterization of the mouse Men1 gene and its expression during development. Oncogene 17:2485-2493. [DOI] [PubMed] [Google Scholar]
- 63.Tang, C.-M., A. E. Tomkinson, W. S. Lane, M. S. Wold, and E. Seto. 1996. Replication protein A is a component of a complex that binds the human metallothionein IIA gene transcription start site. J. Biol. Chem. 271:21637-21644. [DOI] [PubMed] [Google Scholar]
- 64.Thompson, L. H., and D. Schild. 2001. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat. Res. 477:131-153. [DOI] [PubMed] [Google Scholar]
- 65.Tomassetti, P., G. Cometa, E. Del Vecchio, M. Baserga, P. Faccioli, D. Bosoni, G. Paolucci, and L. Barbara. 1995. Chromosomal instability in multiple endocrine neoplasia type 1. Cancer Genet. Cytogenet. 79:123-126. [DOI] [PubMed] [Google Scholar]
- 66.Treuner, K., C. Eckerich, and R. Knippers. 1998. Chromatin association of replication protein A. J. Biol. Chem. 273:31744-31750. [DOI] [PubMed] [Google Scholar]
- 67.Yarbro, J. W. 1992. Mechanism of action of hydroxyurea. Semin. Oncol. 19(Suppl. 9):1-10. [PubMed] [Google Scholar]
- 68.Yu, X., L. C. Wu, A. M. Bowcock, A. Aronheim, and R. Baer. 1998. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J. Biol. Chem. 273:25388-25392. [DOI] [PubMed] [Google Scholar]