Abstract
Cdc6p has an essential function in the mechanism and regulation of the initiation of DNA replication. Budding yeast Cdc6p binds to chromatin near autonomously replicating sequence elements in late M to early G1 phase through an interaction with Origin Recognition Complex or another origin-associated factor. It then facilitates the subsequent loading of the Mcm family of proteins near autonomously replicating sequence elements by an unknown mechanism. All Cdc6p homologues contain a bipartite Walker ATP-binding motif that suggests that ATP binding or hydrolysis may regulate Cdc6p activity. To determine whether these motifs are important for Cdc6p activity, mutations were made in conserved residues of the Walker A and B motifs. Substitution of lysine 114 to alanine (K114A) in the Walker A motif results in a temperature-sensitive phenotype in yeast and slower progression into S phase at the permissive temperature. A K114E mutation is lethal. The Cdc6K114E protein binds to chromatin but fails to promote loading of the Mcm proteins, suggesting that ATP binding is essential for this activity. The mutant arrests with a G1 DNA content but retains the ability to restrain mitosis in the absence of DNA replication, unlike depletion of Cdc6p. In contrast, Cdc6p containing a double alanine mutation in the Walker B motif, DE(223, 224)AA, is functional, and the mutant exhibits an apparently normal S phase. These results suggest that Cdc6p nucleotide binding is important for establishing the prereplicative complex at origins of DNA replication and that the amino terminus of Cdc6p is required for blocking entry into mitosis.
The initiation of DNA replication in eukaryotes occurs at multiple cis-acting sequences, termed replicators, ensuring that DNA replication occurs only during the S phase of the cell division cycle. Origin Recognition Complex (ORC) is a multisubunit protein complex that binds to replicators (1), is required for initiation (2–4) and so fulfills the classical definition of an initiator protein. However, ORC itself does not appear to have an origin unwinding activity (5), and it is known that many additional proteins are required for initiation (6). Because the genome must be precisely duplicated, each active replicator is normally allowed to initiate replication only once per cell cycle, and numerous studies have revealed that control of initiation mediates this precision (2, 7, 8).
The initiation phase of DNA replication has been most studied in the unicellular eukaryote Saccharomyces cerevisiae. Simple DNA sequences, termed autonomously replicating sequence (ARS) elements, have been shown to function as replicators and to contain origins of replication, the actual physical sites where DNA synthesis begins (9–12). These replicators exist in at least three states during the cell cycle, reflecting the recruitment of replication factors in an ordered fashion before the initiation of bidirectional replication (13–21). In order of assembly, these are the post-replicative complex (post-RC) that is detected after initiation in S phase until M phase; the pre-replicative complex (pre-RC) that exists from late mitosis to G1 phase; and the pre-initiation complex that is present in late G1. The precise protein composition at ARS elements during each of these states remains unknown; however, ORC, Cdc6p, Cdc45p, and the Mcm proteins are known components (16, 20).
ORC is bound to chromatin and to specific replicators at all times during the cell cycle (16, 18, 20). In vivo genomic footprints of ARS1 after initiation produce a pattern of DNase I cleavage that is very similar to that obtained with ORC and naked ARS1 DNA, suggesting that ORC alone may be bound to origins in the post-RC state (13). The pre-RC complex detected by genomic footprinting requires at least the presence of ORC, Cdc6p, and the Mcm proteins (14, 15, 17). Cdc6p is required to load the Mcm proteins onto chromatin and this can occur only from late in mitosis to late in G1 phase, when the M-phase and S-phase cyclin-dependent kinases are inactive (15, 17, 18, 20, 22, 23). Similar observations have been made in other species including Xenopus and Schizosaccharomyces pombe (7, 24, 25). This assembly is inhibited by active cyclin-dependent kinases that are in turn inhibited by Sic1p (20, 26). After activation of the Clb-cyclin-dependent kinases in late G1 by degradation of Sic1p, Cdc45p binds to chromatin as part of a large complex called the preinitiation complex (21).
It is clear that Cdc6p levels in the cell control the frequency of initiation throughout the genome (10) and gain-of-function mutations in CDC6 can cause reinitiation in a single cell cycle (18). To investigate whether the ATP-binding motifs present in Cdc6p are important for either chromatin binding or Mcmp loading, we have made mutations in conserved regions of the Walker A and B motifs required for stable binding and hydrolysis of ATP (27). This analysis reveals that Cdc6p binds to chromatin regardless of mutations in the ATP-binding sites, at least one of which would be predicted to severely impair ATP binding. Although Cdc6K114Ep cannot load Mcm proteins or promote initiation, the cells terminally arrest with a G1 content and do not undergo the reductional mitosis that occurs with depletion of Cdc6p (28). These observations strongly suggest that the presence of Cdc6p is sufficient to inhibit mitosis in the absence of ongoing DNA synthesis. However, Cdc6K114Ep containing a deletion of the amino terminus is no longer able to inhibit this reductional division. The K114A mutation confers a temperature-sensitive phenotype, and this mutant exhibits a slower progression into S phase. Interestingly, although the adjacent DE residues contained in the Walker B motif are conserved in all Cdc6 homologues reported so far, a DE(223, 224)AA mutation results in no obvious growth phenotype in S. cerevisiae. In addition, the timing of chromatin binding for Cdc6p and all six Mcm proteins was observed in a complete cell cycle.
MATERIALS AND METHODS
Strains and Plasmids.
Genetic manipulation and transformation of yeast was done by using standard techniques (29, 30). All strains of S. cerevisiae are derivatives of W303 (31) and are listed in Table 1. pMW71 contains the 2.4-kb MluI to NheI genomic CDC6 sequence in pRS415. All mutations in CDC6 were introduced by using site-directed mutagenesis of single-stranded uracil-containing phagemid DNA. After DNA sequencing, restriction fragments containing only the desired mutation were exchanged into pMW71. These are pMW95 (K114E), pMW125 (K114A), pMW126 (K114R), pMW302 (ΔN) that deletes amino acids 2–48, pMW369 (DE(223, 224)AA), and pMW389 (ΔN, K114E). After testing complementation by plasmid shuffling, wild-type CDC6 and selected mutants were cloned into pRS405 and then integrated into the LEU2 locus of K4055 after linearization with HpaI. Genomic Southern blots confirmed single-copy integrations and the sequence of the integrated CDC6 was determined after PCR to confirm the presence of the desired mutation. K4055 and derivatives were always cultured in synthetic complex (SC) medium (29) lacking methionine and also without leucine for the CDC6 integrants. Methionine (2 mM) was added to the medium or plates to repress the wild-type copy of CDC6 under control of the MET3 promoter, as in Fig. 2B. YB511 was created to detect all six Mcm proteins with specific antibodies. Details of construction are available on request.
Table 1.
Strains
| Strain | Genotype | Source |
|---|---|---|
| W303-1A | Mata leu2-3,112 trp1-1 ura3-1 his3-11,15 can1-100 ade2-1 | R. Rothstein |
| K4055 | W303 Mata cdc6Δ∷hisG pMET3-CDC6∷TRP1 | K. Nasmyth |
| K6210 | W303 Mata MCM7(Myc7)-URA3 | " |
| YB209 | W303 Mata cdc6Δ/pRS426-CDC6 | (10) |
| YB495 | W303 Matα cdc6Δ/pRS426-CDC6 | This study |
| YB496 | YB209/pMW71 | " |
| YB497 | YB209/pMW126 | " |
| YB502 | K4055 CDC6∷LEU2 | " |
| YB503 | K4055 cdc6K114A∷LEU2 | " |
| YB504 | K4055 cdc6K114E∷LEU2 | " |
| YB505 | K4055 cdc6DE(223,224)AA∷LEU2 | " |
| YB506 | K4055 cdc6ΔN,K114E∷LEU2 | " |
| YB507 | YB209 CDC6∷LEU2 | " |
| YB508 | YB209 cdc6K114A∷LEU2 | " |
| YB510 | YB209 cdc6DE(223,224)AA∷LEU2 | " |
| YB511 | W303 Mata dbf2-1 MCM4(T7)-HIS3 MCM6(HA3)-LEU2 MCM7(Myc7)-URA3 | " |
Figure 2.
Growth characteristics of CDC6 ATP-binding site mutants. (A) A schematic of Cdc6p showing the regions of homology to RFC and the AAA+ superfamily of proteins (44, 45). Below is an alignment (from top to bottom) of the human (50), Xenopus (7), S. pombe (51), and S. cerevisiae (40) Cdc6p Walker A and B motifs (underlined). The amino acid changes in Cdc6p are indicated by the arrows at lysine 114 and the aspartic and glutamic acids 223 and 224. (B) Strains YB502, YB503, YB504, and YB505 were streaked onto SC plates lacking methionine (−Met), expressing pMET3-CDC6WT, or containing 2 mM methionine (+Met), allowing expression of only the indicated CDC6 allele, and photographed after 2 days at 30°C and 37°C.
Fluorescence-Activated Cell Sorter (FACS) Analysis.
For the methionine shutoff experiments in Fig. 3, all cultures were grown at 30°C to logarithmic phase. Methionine was added to 2 mM and cell aliquots were processed as described (18). To synchronize cultures in G1 (Fig. 4), cells were grown to logarithmic phase in YPD (1% yeast extract, 2% bactopeptone, 2% glucose) at 25°C and α-factor was added to 10 μg/ml. After 1 hr, additional α-factor was added to 5 μg/ml, and the cultures were incubated 1.5 hr further. Time points were taken after collecting the cells, resuspending in an equivalent volume of YPD lacking α-factor, and culturing at 25°C.
Figure 3.
cdc6K114E fails to initiate S phase but inhibits mitosis. Asynchronous cultures of K4055, YB504, and YB506 were grown to exponential phase in SC medium lacking methionine. Methionine was added to 2 mM to repress the wild-type pMET3-CDC6 present in all strains, and time points were processed for FACS analysis. From left to right, strains encode no CDC6, or either cdc6K114E or cdc6ΔN, K114E integrated at the LEU2 locus and under the control of the CDC6 promoter.
Figure 4.
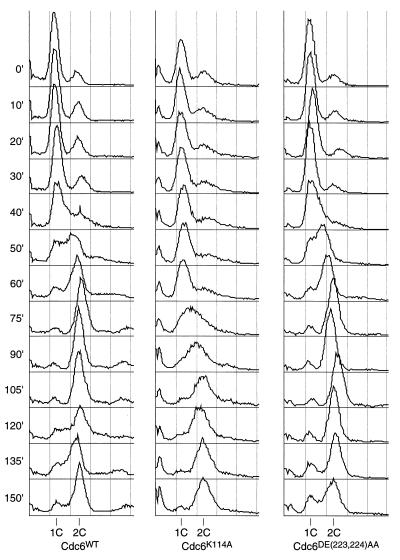
The cdc6K114A mutant moves more slowly into S phase. YB507, YB508, and YB510 were grown to exponential phase in YPD, arrested in G1 with α-factor, released from the block and monitored by FACS for entry into S phase.
Chromatin-Binding Analysis.
For analyzing Cdc6p and Mcm protein chromatin binding through the cell cycle (Fig. 1), YB511 was grown to exponential phase in YPD at 25°C and shifted to 37°C for 3 hr. The culture was returned to 25°C and cell aliquots were taken and processed as below. For the experiments with conditional expression of CDC6, K4055, and YB502–505 were grown at 30°C to logarithmic phase. Cultures were synchronized in G2/M after two additions of nocodazole to 20 μg/ml at 0 and 60 min. At 120 min, 2 mM methionine was added to the culture (to repress the wild-type pMET3-CDC6) and incubated an additional 30 min. Cultures were then released into SC medium without nocodazole but containing 10 μg/ml α-factor with or without 2 mM methionine. Time points were taken at 0, 30, 60, and 90 min after nocodazole release. All samples were processed into soluble and insoluble (chromatin) fractions after Nonidet P-40 lysis of yeast spheroplasts as described (18). Equivalent amounts of these fractions were loaded onto SDS-10% polyacrylamide gels, transferred to nitrocellulose, and probed by using monoclonal antibodies against Mcm2p (Mcm2–18, 1:5,000), Mcm3p (Mcm3–28, 1:5,000), Mcm4p-T7 [T7 (Novagen) 1:5,000], Mcm5p (Mcm5–32, 1:5,000), Mcm6p-HA3 (12CA5, 1:5,000), Mcm7p-Myc7 (9E10, 1:1,000), Orc3p (SB3, 1:50,000), Orc6p (SB49, 1:1,000), and Cdc6p (9H8/5, 1:1,000) (17). The monoclonal antibodies against the Mcm proteins will be described elsewhere (M. Akiyama, M.W., and B.S., manuscript in preparation).
Figure 1.
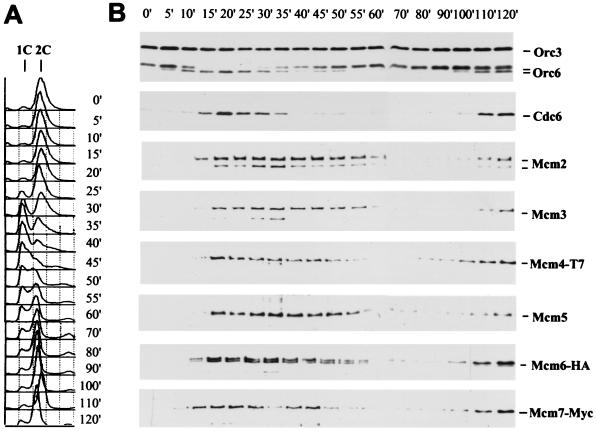
Chromatin binding analysis of Cdc6p and Mcm proteins in the cell cycle. (A) FACS profile of YB511 after a dbf2–1 arrest (37°C) and release at 25°C. (B) Chromatin pellets of YB511 were probed for Orc3p, Orc6p, Cdc6p, and the six Mcm proteins by using monoclonal antibodies specific for each protein.
RESULTS
Timing of Cdc6p and Mcm Protein Chromatin Association.
ORC is bound to chromatin at ARS elements during all stages of the cell division cycle (16, 18) and recruits other proteins that are required for the initiation of DNA replication. Cdc6p associates with chromatin late in mitosis and promotes the binding of the Mcm proteins near ARS elements (17, 20); however, it is not known whether all Mcm proteins become associated with chromatin at the same time relative to Cdc6p binding. We examined the timing of Cdc6p chromatin binding at 5-min intervals after release from a dbf2–1 block that arrests cells in late mitosis and compared this to the chromatin association of all six Mcm proteins. Soluble and chromatin-enriched fractions were isolated (18), and proteins present in the chromatin fraction are shown in Fig. 1B. Orc3p and Orc6p chromatin binding are shown because ORC is chromatin bound throughout the cell cycle. Note that Orc6p becomes phosphorylated at the G1-S transition and dephosphorylated at the passage through mitosis (18). Thus, along with DNA content analysis (Fig. 1A), this is an excellent marker for cell-cycle stage.
Cdc6p and all six Mcm proteins load onto chromatin as cells exit mitosis, with Cdc6p loading first at 15 min after release from the dbf2–1 block (Fig. 1B). All proteins remain on chromatin throughout G1, but the majority of Cdc6p is unloaded as cells prepare to enter S phase. Thus it is likely that Cdc6p is removed from all origins before entry into S phase. In contrast, the Mcm proteins are gradually released, perhaps reflecting their release from both early and late origins and as cells replicate DNA (16). Cdc6p and the Mcm proteins then rebind to chromatin for the next chromosome replication cycle at 110 min when the cells still have a G2/M DNA content.
Mutations in the CDC6 Walker A Box Impair Its Function.
S. cerevisiae Cdc6p contains a bipartite Walker ATP-binding motif (27) that is conserved in numerous Cdc6p homologues (Fig. 2A). To test their importance for Cdc6p activity, we made changes in the conserved Walker A and B motifs and expressed the mutant genes from the endogenous CDC6 promoter. Lysine 114 of the Walker A motif is expected to be critical for correct orientation of the β-γ phosphates of ATP (32). A conservative mutation to arginine, cdc6K114R, had no effect on cell growth or in S-phase progression as determined by plasmid complementation and flow cytometry (data not shown). However, an alanine mutation, cdc6K114A, resulted in a temperature-sensitive phenotype and a mutation to glutamate, cdc6K114E, was lethal (Fig. 2B). The glutamate substitution would create an ionic repulsion with the phosphates of ATP in the binding pocket and probably hinder ATP binding (33). Both the cdc6K114A and cdc6K114E mutants are recessive to the wild type in diploid yeast (data not shown).
The adjacent aspartic and glutamic acid residues in the Walker B motif are thought to be important for proper positioning of ATP through coordination of a Mg2+ ion and have been proposed to play a role in ATP hydrolysis (34–36). Surprisingly, the double mutation cdc6DE(223, 224)AA had little effect on cell growth either at 30°C or 37°C (Fig. 2B), even though these residues are absolutely conserved in the human, Xenopus, and S. pombe CDC6 homologues. This observation suggests that an intact Walker B motif is not essential for Cdc6p activity.
Cdc6K114Ep Is Capable of Inhibiting Mitosis in the Absence of Initiation.
S. cerevisiae depleted of wild-type Cdc6p fail to initiate DNA synthesis and eventually undergo an aberrant reductional anaphase in which cells randomly segregate unreplicated chromosomes, which is a lethal event (28). A similar phenotype was observed when the S. pombe CDC6 homologue, cdc18+ is deleted (37), demonstrating a conserved role in coupling the initiation and possibly completion of DNA synthesis to mitosis. To test whether an intact ATP-binding motif was required for this activity, cells containing either no additional CDC6 or CDC6 derivatives integrated at the LEU2 locus under control of the CDC6 promoter were examined for DNA content after shutoff of the methionine-dependent pMET3-CDC6 at an unlinked locus.
As previously demonstrated, cells depleted of Cdc6p initially accumulate with a 1C DNA content because they are incapable of initiating DNA synthesis. Beginning at 180 min after shutoff of pMET3-CDC6, these cells undergo a premature mitosis and accumulate with less than 1C DNA content (ref. 28; Fig. 3). The cdc6K114E mutant also accumulates with a 1C DNA content because it fails to enter S phase. However, these cells remain arrested in G1 and fail to undergo this reductional division. This observation demonstrates that the Walker A motif of Cdc6p is required for the initiation of DNA replication, but it is not necessary to prevent G1 cells from undergoing reductional mitosis. If the amino terminus of cdc6K114E is deleted, cells also accumulate with a 1C DNA content but now undergo a reductional division beginning at 180 min (Cdc6ΔN, K114E, Fig. 3). Thus, although the amino terminus is dispensable for the essential role of Cdc6p in initiation (38), it is required to inhibit premature entry into mitosis.
cdc6K114A Is Slow to Initiate S Phase.
To test whether the nonlethal mutations affected S-phase progression, strains containing only the wild-type or mutant CDC6 alleles were arrested in G1 by using α-factor, released from the arrest and then monitored by flow cytometry. The wild-type CDC6 and cdc6DE(223, 224)AA strains initiated S phase with very similar kinetics, approximately 40 min after the α-factor release (Fig. 4). The cdc6K114A strain, however, initiated S phase much later, between 60 and 75 min after release from the G1 block (Fig. 4). This observation could indicate an inefficient establishment of pre-RCs throughout the genome, as occurs with the original cdc6–1 allele at the permissive temperature (10) and is consistent with a defect in the initiation function of Cdc6p. The cdc6DE(223, 224)AA mutant does exhibit a phenotype if this experiment is performed in SC media, since the cells enter S phase more slowly relative to the wild type, although S-phase progression is not as slow as for the cdc6K114A mutant (data not shown).
Cdc6K114Ep Binds to Chromatin but Fails to Load Mcm Proteins.
One essential function of Cdc6p in initiation is to promote the binding of Mcm proteins to chromatin at or near ARS elements. Studies using both chromatin binding (17, 18) and chromatin coimmunoprecipitation of ARS sequences with anti-Cdc6p or Mcm antibodies (16, 20) revealed that Cdc6p binds to ARS sequences and that Cdc6p is absolutely required for the subsequent association of the Mcm proteins. However, it is not known whether the binding of Cdc6p to chromatin and its ability to load the Mcm proteins are dependent on ATP binding or hydrolysis.
To test these possibilities, we determined whether the CDC6 ATP-binding site mutants allow chromatin binding of either Cdc6p itself or the Mcm proteins. Because the cdc6K114E allele does not allow growth of yeast, strains were grown by using a wild-type CDC6 gene under control of the MET3 promoter, arrested in G2/M with nocodazole and then released to a G1 (α-factor) block expressing only the wild-type and mutant alleles of CDC6 from the natural promoter. Cells were arrested in α-factor to provide a uniform arrest point in the cell cycle. Thus the experiment determines whether Cdc6p or Mcmp can be loaded onto chromatin and also measures the stability of the proteins in the α-factor arrested state. Soluble and chromatin-enriched fractions were isolated and the chromatin binding of Cdc6p, ORC, and Mcm proteins was examined as cells entered G1 phase (18).
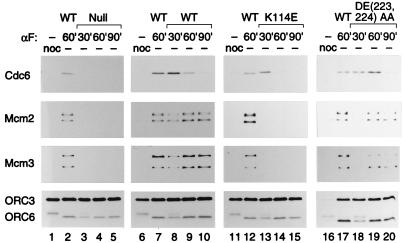
Wild-type and mutant Cdc6 proteins are not bound to chromatin in cells arrested at G2/M by nocodazole (Fig. 5, lanes 1, 6, 11, and 16) but do bind to chromatin within 30 min of release from the nocodazole block to the α-factor block (lanes 8, 13, and 18). Chromatin samples of cells released from the nocodazole block expressing wild-type pMET3-CDC6 are shown as a control (lanes 2, 7, 12, and 17). After a prolonged α-factor arrest, the amounts of soluble (not shown) and chromatin-bound Cdc6p diminished (e.g., lanes 8–10), probably reflecting the rapid turnover of this protein (28). Cdc6K114Ep was less stable in the α-factor arrested cells than the wild type or Cdc6DE(223, 224)AAp (lanes 13–15 vs. lanes 8–10; 18–20) in this experiment. Nevertheless, Cdc6K114Ep did bind to chromatin (Fig. 5, lane 13). As a control, no chromatin-bound Cdc6p was detected when the MET3 promoter was shut off and no additional copy of CDC6 was present (lanes 3–5), indicating that the pMET3-CDC6 copy was completely repressed.
Figure 5.
Cdc6 proteins bind to chromatin but Cdc6K114Ep fails to promote loading of Mcm proteins. Western blots of chromatin fractions from cells (K4055, YB502, YB504, YB505) blocked in G2/M with nocodazole and released to an α-factor (G1) block for 30′, 60′, or 90′ expressing the indicated CDC6 alleles. Lanes 1, 6, 11, and 16 are the chromatin fractions at the nocodazole arrest. Lanes 2, 7, 12, and 17 are the chromatin fractions released to α-factor for 60′ in cells expressing wild-type CDC6 from the MET3 promoter (i.e., no methionine). The next three lanes of each set (e.g., 3–5) represent chromatin fractions released to an α-factor block for 30′, 60′, and 90′ expressing, from left to right, no CDC6, CDC6WT, cdc6K114E, or cdc6DE(223, 224)AA. Blots were probed for Cdc6p, Mcm2p, Mcm3p, and Orc3p plus Orc6p.
The chromatin binding of both Mcm2p and Mcm3p was also determined. At the nocodazole block, the Mcm proteins were not bound to chromatin but were present in the soluble fraction (not shown), as previously reported (17, 18). On release from the G2/M block, the Mcm proteins were loaded onto chromatin in G1 when the wild-type Cdc6p (Fig. 5, lanes 8–10), Cdc6DE(223, 224)AAp (lanes 18–20), or Cdc6K114Ap (not shown) proteins were expressed. In contrast, the Mcm proteins were not loaded when no Cdc6p was expressed (lanes 3–5) or only the Cdc6K114Ep mutant was expressed (lanes 13–15), suggesting that ATP binding was required for Cdc6 to load the Mcm proteins.
As a control, Orc3p and Orc6p were bound to chromatin at all times during this experiment. Interestingly, Orc6p went from a phosphorylated to a nonphosphorylated form of greater mobility between the G2/M block and release into the G1 arrest (Fig. 5, compare lanes 1 and 2). Dephosphorylation after release from nocodazole probably reflects inactivation of the mitotic form of the Cdc28p kinase and an unknown phosphatase. Dephosphorylation of Orc6p served as a marker for progression into G1. At 30 min after release from nocodazole, only half of the cells have a G1 DNA content (not shown), and Orc6p was still partially phosphorylated (Fig. 5, lanes 3, 8, 13, and 18). By 60 min, all of the cells had a G1 DNA content and Orc6p was completely dephosphorylated (Fig. 5, lanes 4, 9, 14, and 19).
DISCUSSION
Cdc6p is required at a very early stage leading to the initiation of DNA replication. It is synthesized in late M phase (28, 39), binds to chromatin at ARS elements (20) and is required for the subsequent loading of the Mcm proteins (17). We show that Cdc6p is unloaded from chromatin before all of the Mcm proteins, perhaps being unloaded simultaneously from origins that fire both early and late in S phase. In contrast, the Mcm proteins exit chromatin more slowly, maybe in a replicon-specific manner (Fig. 1; 16, 20). In addition, Cdc6p somehow acts to inhibit mitosis in S. cerevisiae. Overproduction of Cdc6p causes a transient delay in mitosis (40), and depletion of Cdc6p results in an initial G1 arrest, followed by a reductional mitosis resulting in haploid cells with less than 1C DNA content (28).
All Cdc6p homologues contain a conserved bipartite ATP-binding motif, strongly suggesting that ATP binding and perhaps hydrolysis are important aspects of Cdc6p function. We have shown that mutations in the Walker A motif inactivate Cdc6p. A K114A mutation results in a ts phenotype, and a K114E mutation is lethal. Both of these proteins bind to chromatin with apparently normal kinetics after release from a mitotic block. Cdc6K114Ep, however, is unable to load Mcm2p and Mcm3p, strongly suggesting that Cdc6p requires ATP binding to promote loading of the Mcm proteins.
An earlier report (41) has suggested that GST-Cdc6p expressed in Escherichia coli is an ATPase; however, all of our attempts to detect a specific ATP-binding or ATPase activity for Cdc6p have been unsuccessful (J. Mendez, M.W., and B.S., unpublished work). It is possible that ATP binding and hydrolysis are regulated through interaction with other proteins or with DNA. It is noteworthy that the related Orc1p has conserved Walker A and B motifs and origin DNA modulates its ATPase activity (5).
Cdc6K114Ep binds to chromatin, and so presumably its interaction near ARS elements is maintained. This mutant protein is also able to inhibit mitosis in the absence of the initiation of DNA replication, since we do not observe a reductional division in the absence of S phase. Because the ability to inhibit mitosis does not require the Mcm-loading activity of Cdc6p, by this criterion the mitotic function is separable from this essential initiation function. These results demonstrate that it is not merely the absence of initiation that triggers a reductional mitosis, but that some active function of Cdc6p is required to inhibit mitosis. Deletion of the amino terminus of Cdc6p abrogates this effect. Thus the inhibition of mitosis may be caused by the ability of Cdc6p to bind and inhibit Cdc28p kinase (42, 43) or to interact with the Cdc4p ubiquitin-dependent proteolytic pathway (38), since both of these interactions require the amino terminus of Cdc6p.
Two recent reports have shown a similarity of Cdc6p to a broad family of ATPases (44, 45), including RFC, a structure-specific DNA-binding protein that loads the ring-shaped proliferating cell nuclear antigen molecule onto primed DNA (46). This family is involved in diverse cellular functions, but all somehow couple ATP binding to structural changes in themselves or other proteins. The similarity extends over eight regions corresponding to seven of the eight RFC homology boxes (47) and the sensor I motif (33), a region of the protein predicted to be important for coordinating ATP binding to structural changes or activities of this class of proteins. Inclusion of Cdc6p in this family suggests that Cdc6p may couple ATP binding or hydrolysis to remodeling or loading proteins directly at replicators. This suggestion is in excellent agreement with the known requirement of Cdc6p for promoting assembly of the Mcm proteins at origins. Furthermore, we recently reported a cdc6 mutant in which Mcm proteins remained bound to chromatin after S phase, causing reinitiation of DNA replication (18). One of the mutations in cdc6–3 maps to the conserved sensor I motif, raising the possibility that this mutation may alter the conformation of Cdc6p to mimic an activated ATP-bound state.
Walker’s original designation of the B motif contained a stretch of hydrophobic residues ending in an aspartic acid residue (27), because a number of ATP-binding proteins, kinases, helicases, and ATP synthases have been reported to contain signature motifs that are variations of the “B motif” (34, 48, 49). Cdc6p homologues conform to this designation in having five hydrophobic residues followed by DEMD (Fig. 2A). This is similar to the B motif of the “DEAD/H box” family of helicases (34). By analogy to ATP-binding proteins of known structure (35, 36), these acidic residues of Cdc6p should be important for coordinating the Mg2+ ion in the bound Mg2+ATP and also for hydrolysis of ATP. Surprisingly, mutation of both DE residues to alanine in CDC6 has little effect on the viability of yeast or progression into S phase. This observation suggests that if Cdc6p is an ATPase, hydrolysis of ATP may not be required for its activity. ATP hydrolysis might instead be linked to the inactivation of Cdc6p.
A recent report also examined mutations in the Cdc6p Walker A and B motifs (44). It was found that an E224G mutation in the Walker B motif of CDC6 is lethal when the protein is overproduced, blocking cells in late G1 to early S phase and this protein is unable to load Mcm5p onto chromatin. Although we agree that the Walker A-box is essential for Cdc6p activity, we suggest that the integrity of the B motif is not essential for Cdc6p function, since we see little effect of the DE(223, 224)AA mutation or by overproducing this mutant Cdc6p with or without the amino terminus (not shown). In addition, cdc6E224G can complement a CDC6 deletion strain by using a plasmid shuffle assay, albeit with very slow growth (our unpublished results). Perhaps the E224G mutation additionally alters the conformation of Cdc6p outside of the B motif, resulting in a protein that interferes with initiation when overproduced.
We suggest a model for Cdc6p activity based on the available evidence. Cdc6p is synthesized in late M phase (28, 39), and then likely associates with ORC bound to replicators (Fig. 1; refs. 10, 20). Soon after binding to chromatin, Cdc6p promotes loading of the Mcm proteins in a step that requires ATP binding but not ATP hydrolysis. This proposal is exactly analogous to RFC, which requires ATP binding but not ATP hydrolysis to load proliferating cell nuclear antigen onto DNA (46). Hydrolysis of ATP may instead lock Cdc6p to an inactive ADP-bound state that cannot be recycled until a subsequent round of cell division or perhaps de novo synthesis of Cdc6p. Because purified Cdc6p does not alter the ORC footprint at ARS1 (M.W. and B.S., unpublished data) and Cdc6p chromatin binding during late M and G1 phase does not correlate precisely with the presence of the pre-RC footprint (C.L. and B.S., unpublished data), we suggest that another factor(s) is responsible for the loss of the hypersensitive site and extended footprint seen in the post-RC to pre-RC transition.
Clearly, Cdc6p has other functions in the chromosome replication cycle. Cdc6p bound to replicators may be essential for inhibiting mitosis until initiation of DNA replication has begun. The unreplicated chromatin in G1 cells may resemble newly synthesized chromatin in G2 cells, and the cell division machinery may not be able to distinguish these two states. The presence of Cdc6p on chromatin precisely during G1 (Fig. 1) may signal that the chromatin is in a G1 state. When the initiation of DNA replication is blocked because of the absence of Cdc6p but other mitotic functions proceed normally, such as the activation of the Cdc28/Clb2p kinase, the unreplicated chromosomes become attached to spindles and then undergo an abnormal reductional division. When Cdc6p is present and able to bind to ATP, it promotes the efficient loading of the Mcm proteins that stages the origin for the subsequent steps leading to initiation, e.g., binding of Cdc45p (21) and action of the Cdc7/Dfb4p kinase (2–4).
Acknowledgments
We thank J. Chong and A. Verreault for comments on the manuscript, J. Duffy for preparation of figures, K. Nasmyth for strains, J. Diffley for the Cdc6 antibody, M. Coronesi for FACS analysis, and P. Wendel for expert technical help. This work was supported by a grant from the National Institutes of Health (GM45436 to B.S.). M.W. and C.L. were supported by fellowships from the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation (DRG-1241 and DRG-1308). M.W. is also a Special Fellow of the Leukemia Society.
ABBREVIATIONS
- SC
synthetic complex
- ARS
autonomously replicating sequence
- post-RC
post-replicative complex
- pre-RC
pre-replicative complex
- FACS
fluorescence-activated cell sorter
- ORC
Origin Recognition Complex
References
- 1.Bell S P, Stillman B. Nature (London) 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 2.Diffley J F X. Genes Dev. 1996;10:2819–2830. doi: 10.1101/gad.10.22.2819. [DOI] [PubMed] [Google Scholar]
- 3.Stillman B. Science. 1996;274:1659–1663. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- 4.Piatti S. Prog Cell Cycle Res. 1997;3:143–156. doi: 10.1007/978-1-4615-5371-7_12. [DOI] [PubMed] [Google Scholar]
- 5.Klemm R D, Austin R J, Bell S P. Cell. 1997;88:493–502. doi: 10.1016/s0092-8674(00)81889-9. [DOI] [PubMed] [Google Scholar]
- 6.Dutta A, Bell S P. Annu Rev Cell Dev Biol. 1997;13:293–332. doi: 10.1146/annurev.cellbio.13.1.293. [DOI] [PubMed] [Google Scholar]
- 7.Coleman T R, Carpenter P B, Dunphy W G. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- 8.Newlon C S. Cell. 1997;91:717–720. doi: 10.1016/s0092-8674(00)80459-6. [DOI] [PubMed] [Google Scholar]
- 9.Loo S, Fox C A, Rine J, Kobayashi R, Stillman B, Bell S. Mol Biol Cell. 1995;6:741–756. doi: 10.1091/mbc.6.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang C, Weinreich M, Stillman B. Cell. 1995;81:667–676. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- 11.Newlon C S, Theis J F. Curr Opin Genet Dev. 1993;3:752–758. doi: 10.1016/s0959-437x(05)80094-2. [DOI] [PubMed] [Google Scholar]
- 12.Bielinsky A-K, Gerbi S A. Science. 1998;279:95–98. doi: 10.1126/science.279.5347.95. [DOI] [PubMed] [Google Scholar]
- 13.Diffley J F X, Cocker J H, Dowell S J, Rowley A. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- 14.Cocker J H, Piatti S, Santocanale C, Nasmyth K, Diffley J F X. Nature (London) 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- 15.Santocanale C, Diffley J F X. EMBO J. 1996;15:6671–6679. [PMC free article] [PubMed] [Google Scholar]
- 16.Aparicio O M, Weinstein D M, Bell S P. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 17.Donovan S, Harwood J, Drury L S, Diffley J F X. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang C, Stillman B. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Owens J C, Detweiler C S, Li J J. Proc Natl Acad Sci USA. 1997;94:12521–12526. doi: 10.1073/pnas.94.23.12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka T, Knapp D, Nasmyth K. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- 21.Zou L, Stillman B. Science. 1998;280:593–596. doi: 10.1126/science.280.5363.593. [DOI] [PubMed] [Google Scholar]
- 22.Piatti S, Böhm T, Cocker J H, Diffley J F X, Nasmyth K. Genes Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- 23.Detweiler C S, Li J J. J Cell Sci. 1997;110:753–763. doi: 10.1242/jcs.110.6.753. [DOI] [PubMed] [Google Scholar]
- 24.Jallepalli P, Kelly T J. Curr Opin Cell Biol. 1997;9:358–363. doi: 10.1016/s0955-0674(97)80008-7. [DOI] [PubMed] [Google Scholar]
- 25.Nishitani H, Nurse P. Prog Cell Cycle Res. 1997;3:135–142. doi: 10.1007/978-1-4615-5371-7_11. [DOI] [PubMed] [Google Scholar]
- 26.Dahmann C, Diffley J F X, Nasmyth K A. Curr Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- 27.Walker J, Saraste M, Runswick M J, Gay N J. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piatti S, Lengauer C, Nasmyth K. EMBO J. 1995;14:3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rose M, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 30.Guthrie, C. & Fink, G. R. (1991) Methods Enzymol. 194. [DOI] [PubMed]
- 31.Thomas B J, Rothstein R. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 32.Saraste M, Sibbald P R, Wittinghofer A. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 33.Guenther B, Onrust R, Sali A, O’Donnell M, Kuriyan J. Cell. 1997;91:335–345. doi: 10.1016/s0092-8674(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 34.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Nucleic Acids Res. 1989;17:4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Story R M, Steitz T A. Nature (London) 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- 36.Subramanya H S, Bird L E, Brannigan J A, Wigley D B. Nature (London) 1996;384:379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- 37.Muzi-Falconi M, Brown G W, Kelly T J. Proc Natl Acad Sci USA. 1996;93:1566–1570. doi: 10.1073/pnas.93.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drury L S, Perkins G, Diffley J F X. EMBO J. 1997;16:5966–5976. doi: 10.1093/emboj/16.19.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McInerny C J, Partridge J F, Mikesell G E, Creemer D P, Breeden L L. Genes Dev. 1997;11:1277–1288. doi: 10.1101/gad.11.10.1277. [DOI] [PubMed] [Google Scholar]
- 40.Bueno A, Russell P. EMBO J. 1992;11:2167–2176. doi: 10.1002/j.1460-2075.1992.tb05276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zwerschke W, Rottjakob H-W, Kuntzel H. J Biol Chem. 1994;269:23351–23356. [PubMed] [Google Scholar]
- 42.Elsasser S, Lou F, Wang B, Campbell J L, Jong A. Mol Biol Cell. 1996;7:1723–1735. doi: 10.1091/mbc.7.11.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown G W, Jallepalli P V, Huneycutt B J, Kelly T J. Proc Natl Acad Sci USA. 1997;94:6142–6147. doi: 10.1073/pnas.94.12.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perkins G, Diffley J F X. Mol Cell. 1998;2:23–32. doi: 10.1016/s1097-2765(00)80110-0. [DOI] [PubMed] [Google Scholar]
- 45.Neuwald, A. F., Aravind, L., Spouge, J. L. & Koonin, E. V. (1999) Genome Res., in press. [PubMed]
- 46.Tsurimoto T, Stillman B. J Biol Chem. 1991;266:1950–1960. [PubMed] [Google Scholar]
- 47.Cullmann G, Fien K, Kobayashi R, Stillman B. Mol Cell Biol. 1995;15:4661–4671. doi: 10.1128/mcb.15.9.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hodgman T C. Nature (London) 1988;333:22–23. doi: 10.1038/333022b0. [DOI] [PubMed] [Google Scholar]
- 49.Linder P, Lasko P F, Leroy P, Nielsen P J, Nishi K, Schnier J, Slonimski P P. Nature (London) 1989;337:121–122. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- 50.Williams R S, Shohet R V, Stillman B. Proc Natl Acad Sci USA. 1997;94:142–147. doi: 10.1073/pnas.94.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kelly T J, Martin G S, Forsburg S L, Stephen R J, Russo A, Nurse P. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]