Abstract
In eukaryotes, mRNAs are transcribed in the nucleus and exported to the cytoplasm for translation to occur. Messenger RNAs complexed with proteins referred to as ribonucleoparticles are recognized for nuclear export in part by association with Mex67, a key Saccharomyces cerevisiae mRNA export factor and homolog of human TAP/NXF1. Mex67, along with its cofactor Mtr2, is thought to promote ribonucleoparticle translocation by interacting directly with components of the nuclear pore complex (NPC). Herein, we show that the nuclear pore-associated protein Sac3 functions in mRNA export. Using a mutant allele of MTR2 as a starting point, we have identified a mutation in SAC3 in a screen for synthetic lethal interactors. Loss of function of SAC3 causes a strong nuclear accumulation of mRNA and synthetic lethality with a number of mRNA export mutants. Furthermore, Sac3 can be coimmunoprecipitated with Mex67, Mtr2, and other factors involved in mRNA export. Immunoelectron microscopy analysis shows that Sac3 localizes exclusively to cytoplasmic fibrils of the NPC. Finally, Mex67 accumulates at the nuclear rim when SAC3 is mutated, suggesting that Sac3 functions in Mex67 translocation through the NPC.
INTRODUCTION
Nuclear export of mRNA in eukaryotes is an obligatory feature of normal gene expression. This process can be divided into two basic steps: formation of the export competent ribonucleoparticle (RNP) and translocation of the RNP through the nuclear pore complex (NPC). Beginning at transcription, mRNAs are bound and packaged by RNA binding proteins to form RNPs (reviewed in Lei and Silver, 2002). Once properly formed, the RNP is actively exported across the nuclear envelope via large aqueous channels formed by the proteinaceous NPC.
Structural integrity of the RNP is an important factor in obtaining export competence. Central to this process is the heterogeneous nuclear (hn)RNP protein Npl3, which is a highly abundant poly (A)+ RNA binding protein in Saccharomyces cerevisiae (Wilson et al., 1994). Npl3 shuttles between the nucleus and cytoplasm and is required for nuclear export of mRNA (Flach et al., 1994; Singleton et al., 1995; Lee et al., 1996). The nuclear export of Npl3 is closely tied to that of mRNA, making its localization a useful reporter for mRNA export (Krebber et al., 1999).
The overall structure of the NPC is a central framework with eightfold symmetry embedded in the membrane of the nuclear envelope. In addition, the NPC includes a nuclear-oriented basket of filaments flanked by nuclear and terminal rings as well as fibrils extending into the cytoplasm (reviewed in Stoffler et al., 1999). The NPC is composed of proteins called nucleoporins, many of which are characterized by stretches of FG (Phe-Gly) dipeptide repeats. These FG repeats are thought to serve as binding sites for soluble transport receptors during translocation (Iovine et al., 1995; Radu et al., 1995).
Translocation of RNPs through the NPC is thought to be mediated by the mRNA export factor Mex67 in S. cerevisiae or TAP/NXF1 in humans. Together as a heterodimer with its counterpart yeast Mtr2 or metazoan p15, Mex67/TAP is essential for the export of all mRNAs tested in S. cerevisiae and Drosophila (Hurt et al., 2000; Herold et al., 2001). Mex67/TAP is thought to be recruited to RNPs via interaction with the conserved mRNA export factor yeast Yra1 or human Aly/REF (Sträβer and Hurt, 2000; Stutz et al., 2000; Zenklusen et al., 2001). Once bound, Mex67/TAP may promote translocation through the NPC through serial interactions with the FG repeats of nucleoporins (Bachi et al., 2000; Sträβer et al., 2000; Strawn et al., 2001).
Mtr2 is required for proper nuclear pore targeting of Mex67. Mutation of MTR2 causes mislocalization of Mex67 from the nuclear rim to the cytoplasm, and Mtr2 itself interacts with the nucleoporin Nup85 (Santos-Rosa et al., 1998). However, there is disagreement as to whether Mtr2 promotes Mex67 interaction with nucleoporins (Sträβer et al., 2000; Strawn et al., 2001). In higher eukaryotes, the TAP cofactor p15 seems to be important for NPC targeting of TAP (Fribourg et al., 2001; Levesque et al., 2001; Wiegand et al., 2002). The function of these mRNA export receptor heterodimers is conserved because MEX67/MTR2 can be replaced by introduction of both TAP and p15 in S. cerevisiae, although Mtr2 and p15 share no sequence homology (Katahira et al., 1999). Additionally, Mtr2 has been implicated in the export of the large ribosomal subunit (Stage-Zimmermann et al., 2000; Baβler et al., 2001). Moreover, Baβler et al. (2001) described a mutant allele of MTR2 that displays a defect in ribosome but not mRNA export, suggesting that the functions of Mtr2 in both processes are distinct and separable.
An additional requirement for mRNA export is the integrity of the NPC. Three nucleoporin subcomplexes seem to be specifically required for mRNA export, the Nup84-Nup85-Nup120-Nup145-Seh1-secs13 complex, the Nsp1-Nup82-Nup159 complex, and the Nup116-Gle2 complex (Wente and Blobel, 1993; Murphy et al., 1996; Siniossoglou et al., 1996; Teixeira et al., 1997; Bailer et al., 1998; Belgareh et al., 1998; Hurwitz et al., 1998). These nucleoporins may provide docking sites for Mex67 during RNP translocation. In fact, Nup116 can be coimmunoprecipitated with Mex67 from cell lysates (Strawn et al., 2001), and Mex67 is able to bind to fragments containing the FG repeats of Nsp1, Nup116, Nup159, and Rip1 in vitro (Sträβer et al., 2000; Strawn et al., 2001). These interactions are likely to be important for Mex67-mediated RNP translocation through the NPC.
On the cytoplasmic face of the NPC exists a putative complex of proteins that is proposed to serve as the terminal docking site for RNPs. This complex consists of Nup159/Rat7, Gle1, Rip1, and Dbp5/Rat8. Nup159 localizes exclusively to the cytoplasmic fibrils of the NPC, whereas Gle1, Rip1, and Dbp5 display a wider spatial distribution, including the cytoplasmic fibrils (Kraemer et al., 1995; Schmitt et al., 1999; Strahm et al., 1999). Dbp5 is an RNA helicase that has been postulated to remodel the RNP as it is transported or to promote release of RNA binding proteins from the mRNA in the final steps of mRNA export (Snay-Hodge et al., 1998; Tseng et al., 1998). It has been suggested that a function of these nucleoporins is to position Dbp5 at the NPC (Hodge et al., 1999; Strahm et al., 1999). Alternately, these proteins may act as docking sites for RNPs as they translocate through the NPC.
The NPC-associated protein Sac3 has been implicated previously in nuclear transport. SAC3 was originally identified in a screen for suppressors of actin mutations (Novick et al., 1989) and has been subsequently shown to be required for normal mitotic progression and spindle morphology (Bauer and Kölling, 1996b). Localizing to the NPC, Sac3 associates physically with the nucleoporin Nsp1 (Jones et al., 2000). Finally, Sac3 has been ascribed a role in nuclear transport that may be related to its function in the cell cycle (Jones et al., 2000).
In this study, we have characterized the role of Sac3 in mRNA export. Mutation of SAC3 causes synthetic lethality with mutation of MEX67, MTR2, and several genes that encode cytoplasmic fibril-associated mRNA export factors. In addition, Sac3 is associated physically with Mex67, Mtr2, and other factors involved in mRNA export. By immunoelectron microscopy, Sac3 localizes exclusively to the cytoplasmic fibrils of the NPC. Finally, Mex67 accumulates at the nuclear rim in sac3− mutants, suggesting that Sac3 functions in the terminal step of Mex67 translocation through the NPC.
MATERIALS AND METHODS
Indirect Immunofluorescence and In Situ Hybridization
These procedures were performed as described previously (Krebber et al., 1999). α-Nsp1 (gift of M. Stewart; MRC LMB, Cambridge, United Kingdom) was used at a 1:1000 dilution and detected with Texas Red-conjugated donkey α-rabbit (1.5 mg/ml; Jackson Immunoresearch Laboratories, West Grove, PA) at a 1:1000 dilution. For sac3Δ139 and Δsac3-rg, in situ hybridization was performed with a Cy3-labeled oligo dT50 probe at 50 nM, and all steps between probe hybridization and the first 2× SSC wash were eliminated.
Synthetic Lethal Screen
mtr2-142 cells (PSY1720) carrying a plasmid encoding pJT10-MTR2-URA3-ADE8 (pPS1852) were subjected to ethyl methanesulfonate mutagenesis to a rate of 50% killing. We screened 15,000 colonies by the colony sectoring assay for loss of sectoring (Elledge and Davis, 1988). Candidates were selected that meet the criteria for synthetic lethality as described previously (Henry and Silver, 1996) and backcrossed to parental mtr2-142 cells (PSY1719) three times. Linkage to SAC3 was determined by crossing this strain to ACY276, which is marked with HIS3 at the SAC3 locus. A spore from this cross was further outcrossed into the S288C background by crossing twice to FY23. The resulting strain was marked with the HIS3MX6 marker downstream of sac3Δ139 by a method described previously (Longtine et al., 1998) to create PSY2555.
Immunoprecipitation
Cells (50 ml) were grown in YPD at 25–30°C to ∼2 × 107 cells/ml. All subsequent steps were performed at 4°C. Pellets were lysed in 50–100 μl of ice-cold PBSMT (2.5 mM MgCl2, 3 mM KCl, 0.5% Triton X-100 in phosphate-buffered saline) plus protease inhibitors (1 mM phenylmethylsulfonyl fluoride and 2 ng/ml each of pepstatin A, leupeptin, aprotinin, antipain, benzamidine, and chymostatin) by using glass beads in a FastPrep bead beater (6.5 m/s; Savant Instruments, Holbrook, NY). After lysis, an additional 1 ml of PBSMT was added, and lysates were clarified by centrifugation at 14,000 rpm for 10 min. Protein A-Sepharose (40 μl) was washed three times in PBSMT and added to 1 mg of lysate in a total of 1 ml of PBSMT. Then 1.5 μl of affinity-purified rabbit polyclonal α-green fluorescent protein (GFP) (0.9 mg/ml) was added and incubated overnight with agitation. Beads were washed three times with 750 μl of PBSMT and once with 750 μl of Tris-EDTA, pH 8.0. Sample buffer (20 μl) was added to samples and boiled for 5 min. Immunoprecipitates (IPs) and total lysate were resolved by 7% SDS-PAGE and transferred to nitrocellulose in 10 mM CAPS (3-[cyclohexylamino]-1 propane sulfonic acid), pH 11, 1% methanol. Blots were probed with α-GFP at 1:10,000 and α-myc (9E10, 200 μg/ml; Santa Cruz Biotechnology, Santa Cruz, CA) at 1:1000.
Immunoelectron Microscopy
Spheroplasted, Triton X-100 extracted ECFP-Sac3 cells were incubated with α-GFP antibody directly conjugated to 8-nm colloidal gold and processed for preembedding labeling as described previously (Fahrenkrog et al., 1998).
RESULTS
Sac3 Functions in mRNA Export
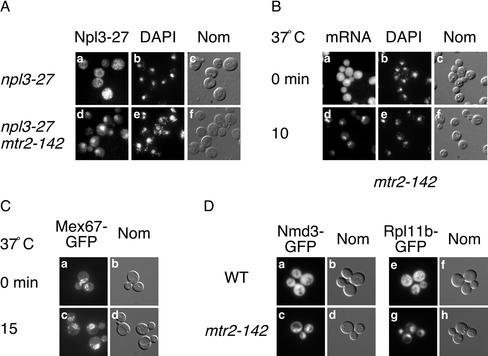
In an effort to identify genes involved in mRNA export in S. cerevisiae, we performed a screen for mutants that disrupt the export of the hnRNP protein Npl3 (Lei et al., 2001). This screen relies on a mutant form of Npl3, Npl3-27, that is slowed for nuclear import and localizes at steady state throughout the nucleus and cytoplasm of cells as determined by indirect immunofluorescence (Figure 1A, a–c) (Krebber et al., 1999). We found that mutation of the mRNA export factor MTR2 causes nuclear accumulation of Npl3-27 (Figure 1A, d–f). In addition, this mutant is temperature sensitive for growth (our unpublished data). The lesion in MTR2 maps to a single nucleotide mutation changing Gly142 to Asp, and this mutant is thus designated mtr2-142.
Figure 1.
Characterization of the mtr2-142 mutant. (A) mtr2-142 causes nuclear accumulation of Npl3-27. Localization of Npl3-27 in npl3-27 (PSY1031, a–c) and npl3-27 mtr2-142 (PSY1717, d–f) cells. Cells were shifted to 37°C for 30 min. Indirect immunofluorescence with polyclonal antibodies to Npl3 (left), DAPI (middle), and Nomarski images (right) are shown. (B) mtr2-142 cells display an mRNA export defect. Localization of poly (A)+ RNA in mtr2-142 cells (PSY1719) grown at 25°C (a–c) or shifted to 37°C for 10 min (d–f). In situ hybridization with an oligo (dT)50 probe (left), DAPI (middle), and Nomarski images (right) are shown. (C) mtr2-142 cells mislocalize Mex67 to the cytoplasm. Localization of Mex67-GFP in mtr2-142 cells (PSY2857) grown at 25°C (a and b) or shifted to 37°C for 15 min (c and d). GFP fluorescence (left) and Nomarski images (right) are shown. (D) mtr2-142 cells display a large ribosome export defect. Localization of Nmd3-GFP in wild-type (FY23, a and b) and mtr2-142 cells (PSY1719, c and d). Localization of Rpl11b-GFP in wild-type (e and f) and mtr2-142 cells (g and h). Cells were grown at 25°C.
We further characterized the mtr2-142 mutation in isolation from npl3-27 to determine its effects on nuclear export. MTR2 has been implicated in the export of mRNA as a cofactor for the export receptor Mex67 (Santos-Rosa et al., 1998). We examined the localization of mRNA in mtr2-142 cells by in situ hybridization with an oligo dT50 probe. In cells grown at permissive growth temperature, poly (A)+ RNA localizes throughout the nucleus and cytoplasm similar to wild-type (Figure 1B, a–c) but accumulates in the nuclei of cells shifted to the nonpermissive temperature for 10 min (Figure 1B, d–f). In addition, we examined the localization of Mex67-GFP in mtr2-142 cells. At permissive growth temperature, Mex67-GFP localizes to the nuclear rim as in wild type (Figure 1C, a and b). However, in cells shifted to the nonpermissive growth temperature for 15 min, Mex67-GFP mislocalizes to the cytoplasm in a punctate pattern (Figure 1C, c and d). On longer shifts to 37°C, mtr2-142 cells undergo considerable lysis (our unpublished data). The extent of Mex67 mislocalization in the mtr2-142 mutant is apparently less severe than previously published mutant alleles of mtr2 (Santos-Rosa et al., 1998). Furthermore, mtr2-142 is unable to support growth in combination with the mex67-5 mutation, indicating a synthetic lethal relationship (our unpublished data). Therefore, mtr2-142 results in a rapid accumulation of mRNA in the nucleus and mislocalization of Mex67 to the cytoplasm.
Because Mtr2 also functions in the nuclear export of large ribosomal subunits, we examined the localization of large ribosomal subunits in mtr2-142. The mtr2-33 mutant causes nuclear accumulation of the large ribosomal subunit but not of mRNA, suggesting that Mtr2 functions in discrete export pathways (Baβler et al., 2001). Export of the large ribosomal subunit can be monitored by localization of the 60S export adapter Nmd3 (Ho et al., 2000) or a large ribosomal protein such as Rpl11b (Hurt et al., 1999; Stage-Zimmermann et al., 2000). In wild-type cells, Nmd3-GFP and Rpl11b-GFP localize throughout the nucleus and cytoplasm (Figure 1D, a and b and e and f), whereas both Nmd3-GFP and Rpl11b are concentrated in the nucleus in nearly one 100% of mtr2-142 cells. These defects are apparent at the permissive growth temperature, indicating a strong inhibition of 60S export.
To determine what export factors interact genetically with Mtr2, we performed a synthetic lethal screen by the colony sectoring assay by using the mtr2-142 mutant (our unpublished data). We obtained three mutants that are likely to be complete loss of function mutations in MTR2. A fourth mutant was determined to harbor a mutation in SAC3, which encodes a nuclear pore-associated protein (Jones et al., 2000). Mapping of the genetic lesion by gap repair and sequencing revealed a mutation of a single nucleotide to cause a nonsense mutation of Trp139 to Stop thus truncating Sac3 from its wild-type length of 1301 aa. This mutant is hereby referred to as sac3Δ139.
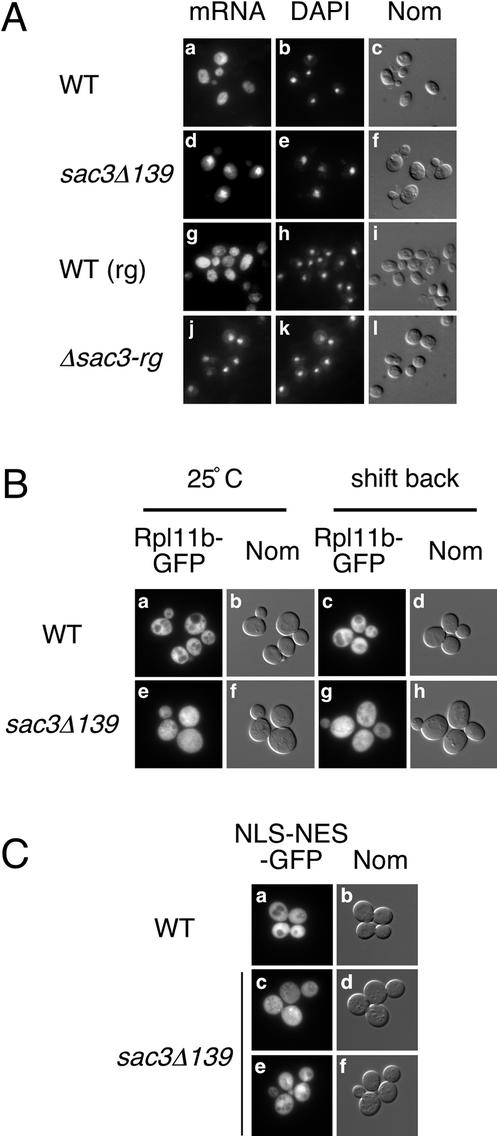
Given its interaction with mtr2-142, we examined sac3Δ139 cells for a defect in mRNA export. sac3Δ139 cells have a decreased rate of growth at all temperatures tested (our unpublished data). In wild-type cells, poly (A)+ RNA localizes throughout the nucleus and cytoplasm as monitored by in situ hybridization (Figure 2A, a–c). In sac3Δ139 cells, mRNA accumulates in the nucleus (Figure 2A, d–f). Similarly, the deletion mutant Δsac3-rg obtained from the Saccharomyces Gene Deletion Project (Winzeler et al., 1999) displays a nuclear accumulation of mRNA (Figure 2A, j–l). In the corresponding wild-type strain for Δsac3-rg mutant, poly (A)+ RNA localizes throughout the nucleus and cytoplasm (Figure 2A, g–i). The mRNA export defect of sac3Δ139 cells can be rescued by introduction of a wild-type SAC3 plasmid (our unpublished data). Furthermore, heterozygous sac3Δ139 diploid cells display a wild-type localization of mRNA, whereas diploids homozygous for sac3Δ139 accumulate mRNA in the nucleus (our unpublished data). Therefore, sac3Δ139 is a recessive loss of function mutation.
Figure 2.
Sac3 functions primarily in mRNA export. A. sac3− mutant cells display an mRNA export defect. Localization of poly (A)+ RNA in wild-type (FY23, a–c), sac3Δ139 cells (PSY2555, d–f), wild-type (PSY1930, g–i), and Δsac3-rg cells (PSY2844, j–l) grown at 30°C. In situ hybridization with an oligo (dT)50 probe (left), DAPI (middle), and Nomarski images (right) are shown. (B) sac3Δ139 cells do not display a large ribosome export defect. Localization of Rpl11b-GFP in wild-type (FY23, a–d) and sac3Δ139 cells (PSY2555, e–h) grown at 25°C (a, b, e, and f) or shifted to 37°C for 1 h and back to 25°C for 1 h (c, d, g, and h). (C) sac3Δ139 cells do not have an NES-protein export defect. Localization of NLS-NES-GFP in wild-type (FY23) and sac3Δ139 cells (PSY2555) grown at 25°C.
Because mtr2-142 cells display several defects in nuclear export, we examined sac3Δ139 cells for various nuclear export phenotypes. In wild-type cells expressing Rpl11b-GFP, the reporter localizes throughout the nucleus and cytoplasm (Figure 2B, a–b). Examination of cells after a temperature shift to 37°C to reduce the pool of ribosomes and shift back to 25°C to resume synthesis is a highly sensitive assay for large ribosome export (Hurt et al., 1999; Stage-Zimmermann et al., 2000). On a shift to 37°C and shift back to 25°C, the localization of Rpl11b-GFP is unchanged in wild-type cells (Figure 2B, c and d). In sac3Δ139 cells grown at 25°C, Rpl11b-GFP localizes throughout cells similar to wild type (Figure 2B, e and f). On a shift to high temperature and shift back, <10% of cells display a mild nuclear accumulation of the reporter (Figure 2B, g and h). This slight effect is milder than that of bona fide 60S export mutants as well as some mRNA export mutants (Stage-Zimmermann et al., 2000). Furthermore, the localization of Nmd3-GFP is unaffected in sac3Δ139 mutants (our unpublished data). The export of small ribosomal subunits is also unaffected (our unpublished data) (Moy and Silver, 1999). Finally, the ability to export an artificial reporter containing the simian virus 40 nuclear localization sequence (NLS) and the protein kinase inhibitor (PKI) nuclear export sequence (NES) fused to two GFP moieties was examined in sac3Δ139 cells. The NLS-NES-GFP reporter localizes throughout the nucleus and cytoplasm in wild-type cells (Figure 2C, a and b). In sac3Δ139 cells grown at 25°C, the NLS-NES-GFP reporter localizes throughout the nucleus and cytoplasm in most cells (Figure 2C, c and d) although in ∼15% of cells, a slight nuclear accumulation is seen (Figure 2C, e and f). When sac3Δ139 cells are grown at 30°C or shifted to 37°C for 2 h, ∼30% of cells show a slight nuclear accumulation of the NLS-NES-GFP reporter, and the same results were obtained with the Δsac3-rg strain (our unpublished data). Intense nuclear accumulation of the NLS-NES-GFP reporter in the NES-protein export receptor mutant xpo1-1 is seen in ∼100% of cells when shifted to 37°C for 15 min similar to previously reported results (our unpublished data) (Stade et al., 1997). Therefore, failure to export mRNA is the principal defect of sac3Δ139 cells.
Sac3 Interacts with mRNA Export Factors
To further test the hypothesis that Sac3 is involved in mRNA export, we tested for synthetic lethality of sac3Δ139 and various nuclear export factor mutants. This form of genetic interaction can indicate that two genes function in the same pathway or in parallel pathways. The results of these experiments are summarized in Table 1. We found that sac3Δ139 is not synthetically lethal with deletion of the NES export factor Δyrb2 or factors localized to the nuclear basket, Δmlp1 and Δmlp2. Furthermore, we found that sac3Δ139 displays synthetic lethality with a number of mRNA export factor mutants such as mex67-5, rat8-2/dbp5, rat7-1/nup159, and results in synthetic sickness with nup82-Δ108. Interestingly, all of these factors localize predominantly to the cytoplasmic fibrils of the NPC, although Mex67 and Mtr2 localize to both the nuclear and cytoplasmic faces of the pore (Kraemer et al., 1995; Hurwitz et al., 1998; Santos-Rosa et al., 1998; Strahm et al., 1999). Two mRNA export factor mutants that sac3Δ139 is not synthetically lethal with are Δnup116-5 and gle1-L356A. These results show that SAC3 interacts genetically with specific mRNA export factor genes.
Table 1.
Mutations tested for synthetic lethality with sac3Δ139
| Synthetic lethal with sac3Δ139 | Strain | Strain reference |
|---|---|---|
| Yes | mtr2-142 | This study |
| mex67-5 | Segref et al., 1997 | |
| rat8-2 (dbp5) | Snay-Hodge et al., 1998 | |
| rat7-1 (nup159) | Gorsch et al., 1995 | |
| nup82-Δ108a | Hurwitz and Blobel, 1995 | |
| No | Δmlp1/mlp1-Δ1 | Kölling et al., 1993; Strambio-de-Castillia et al., 1999 |
| Δmlp2 | Strambio-de-Castillia et al., 1999 | |
| Δyrb2 | Taura et al., 1998 | |
| Δnup116-5 | Damelin and Silver, 2003; Wente et al., 1992 | |
| gle1-L356A | Murphy and Wente, 1996 |
This mutation resulted in a synthetic sick phenotype.
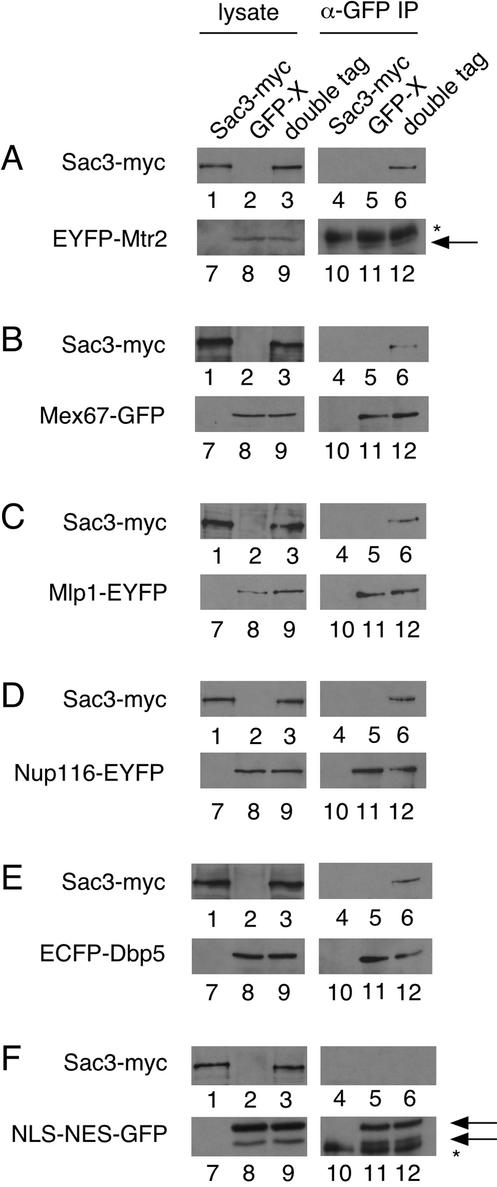
To assess whether Sac3 associates physically with mRNA export factors, we tested whether Sac3 could be coimmunoprecipitated from cell lysates with several export factors. Lysates were prepared from three different strains bearing a myc epitope-tagged Sac3, an EYFP-tagged Mtr2, or both tagged proteins. Western blotting with α-GFP to detect EYFP-Mtr2 shows expression only in the EYFP-Mtr2 and the double-tagged strains (Figure 3A, lanes 7–9). By using α-myc antibody, Sac3-myc is detectable only in the Sac3-myc and double-tagged strains (Figure 3A, lanes 1–3). Lysates from each strain were immunoprecipitated with α-GFP antibody against EYFP-Mtr2. Equal amounts of EYFP-Mtr2, which migrates just below the heavy chain of the α-GFP antibody, are immunoprecipitated from the EYFP-Mtr2 and double-tagged strains (Figure 3A, lanes 10–12). In the double-tagged strain but not single tagged strains, Sac3-myc is detectable in the immunoprecipitate (Figure 3A, lanes 4–6), indicating that Sac3 interacts physically with Mtr2. In the same manner, we examined whether Sac3 associates physically with Mex67, which interacts stably with Mtr2 (Santos-Rosa et al., 1998). Western blotting of lysates from Sac3-myc, Mex67-GFP, and double-tagged strains by using α-myc and α-GFP antibodies show even levels of expression of Sac3-myc and Mex67-GFP, respectively, only in strains with the appropriate epitope tag (Figure 3B, lanes 1–3, 7–9). When lysates from Sac3-myc, Mex67-GFP, and double-tagged strains are immunoprecipitated using α-GFP antibody, which recognizes Mex67-GFP, Mex67-GFP is efficiently immunoprecipitated in the Mex67-GFP and double-tagged strains (Figure 3B, lanes 10–12). Sac3-myc is detectable in the immunoprecipitate of the double-tagged strain only, indicating that Sac3 and Mex67 can be coimmunoprecipitated (Figure 3B, lanes 4–6). Therefore, Sac3 interacts physically with the mRNA export factors Mtr2 and Mex67.
Figure 3.
Sac3 associates physically with mRNA export factors and nuclear pore associated proteins. (A) Sac3 coimmunoprecipitates with Mtr2. Lysates (left) and α-GFP immunoprecipitates (right) from Sac3-myc (PSY2451, lanes 1, 4, 7, and 10), EYFP-Mtr2 (PSY2729, lanes 2, 5, 8 and 11), and double tagged Sac3-myc EYFPMtr2 (PSY2729, lanes 2, 5, 8 and 11), and double tagged Sac3-myc EYFP-Mtr2 (PSY2747, lanes 3, 6, 9 and 12) strains. The asterisk denotes the heavy chain of the α-GFP antibody. Arrows point to EYFP-Mtr2. Lanes 7–9 were exposed ten times longer than lanes 10–12. B. Sac3 coimmunoprecipitates with Mex67. Lysates (left) and α-GFP immunoprecipitates (right) from Sac3-myc (PSY2451, lanes 1, 4, 7 and 10), Mex67-GFP (lanes 2, 5, 8 and 11), and double tagged Sac3-myc Mex67-GFP (PSY2691, lanes 3, 6, 9 and 12) strains. C. Sac3 coimmunoprecipitates with Mlp1. Lysates (left) and α-GFP immunoprecipitates (right) from Sac3-myc (PSY2451, lanes 1, 4, 7, and 10), Mlp1-EYFP (PSY2751, lanes 2, 5, 8, and 11), and double-tagged Sac3-myc Mlp1-EYFP (PSY2752, lanes 3, 6, 9, and 12) strains. (D) Sac3 coimmunoprecipitates with Nup116. Lysates (left) and α-GFP immunoprecipitates (right) from Sac3-myc (PSY2451, lanes 1, 4, 7, and 10), Nup116-EYFP (PSY1832, lanes 2, 5, 8, and 11), and double-tagged Sac3-myc Nup116-EYFP (PSY2856, lanes 3, 6, 9, and 12) strains. (E) Sac3 coimmunoprecipitates with Dbp5. Lysates (left) and α-GFP immunoprecipitates (right) from Sac3-myc (PSY2451, lanes 1, 4, 7, and 10), ECFP-Dbp5 (PSY2726, lanes 2, 5, 8, and 11), and double-tagged Sac3-myc ECFP-Dbp5 (PSY2755, lanes 3, 6, 9, and 12) strains. (F) Sac3 does not coimmunoprecipitate with NLS-NES-GFP. Lysates (left) and α-GFP immunoprecipitates (right) from a Sac3-myc strain containing empty vector (PSY2451 transformed with pPS703; lanes 1, 4, 7, and 10), a wild-type strain expressing NLS-NES-GFP (FY23 transformed with pPS1372; lanes 2, 5, 8, and 11), and a Sac3-myc strain expressing NLS-NES-GFP (PSY2451 transformed with pPS1372; lanes 3, 6, 9, and 12). The asterisk denotes the heavy chain of the α-GFP antibody. Arrows point to two bands corresponding to NLS-NES-GFP. Samples were run on an SDS-PAGE gel. Lysate (10 μg) and 1/20th of the IP (from a total of 1 mg of lysate) were blotted with α-GFP (lanes 7–12) and 10 μg of lysate and the remainder of the IP were blotted with α-myc (9E10, lanes 1–6).
We next tested whether Sac3 interacts physically with NPC-associated proteins. It has been shown previously that Sac3 can be copurified with the nucleoporin Nsp1 from cell lysates, indicating that Sac3 associates physically with the NPC (Jones et al., 2000). We examined whether Sac3 could be coimmunoprecipiatated with the yeast homologue of mammalian Tpr, Mlp1, which has been shown to localize to the nuclear basket of the NPC (Strambio-de-Castillia et al., 1999; Kosova et al., 2000). When lysates from Sac3-myc, Mlp1-EYFP, and double-tagged strains are subjected to immunoprecipitation with α-GFP antibody against Mlp1-EYFP, Sac3-myc is visible in the immunoprecipitate of the double-tagged strain only, indicating that both proteins exist in a complex (Figure 3C). Interestingly, it has been stated as unpublished data that Mlp2, which is redundant with Mlp1, can be copurified with Mex67 and Mtr2 (Kosova et al., 2000). Furthermore, overexpression of Mlp1 causes nuclear accumulation of poly (A)+ RNA, possibly implicating Mlp1 in the mRNA export process (Kosova et al., 2000). We also examined whether Sac3 can be coimmunoprecipitated with the nucleoporin Nup116, which is essential for proper mRNA export (Wente and Blobel, 1993). In the same manner, we tested whether α-GFP immunoprecipitation against Nup116-EYFP in Sac3-myc, Nup116-EYFP, and double-tagged strains results in copurification of Sac3-myc. Western blotting shows that Sac3-myc is detectable in the immunoprecipitation from the double-tagged strain only (Figure 3D), indicating that Sac3 associates physically with Nup116.
Finally, we tested whether Sac3 could be coimmunoprecipitated with the RNA helicase Dbp5. Dbp5 is an mRNA export factor that localizes to the cytoplasmic fibrils of the NPC (Schmitt et al., 1999; Strahm et al., 1999). Sac3-myc is detectable in the α-GFP immunoprecipitate of the double-tagged but not single-tagged Sac3-myc or ECFP-Dbp5 strains (Figure 3E). To ensure that Sac3 does not simply associate with the GFP moiety of GFP-tagged proteins, we also examined whether Sac3 interacts physically with the plasmid borne NLS-NES-GFP reporter, which continuously shuttles between the nucleus and the cytoplasm through the NPC. Western blotting of lysates from Sac3-myc cells transformed with vector, wild-type cells expressing NLS-NES-GFP, and Sac3-myc cells expressing NLS-NES-GFP shows Sac3-myc expression only in Sac3-myc cells transformed with either plasmid (Figure 3F, lanes 1–3). NLS-NES-GFP expression is visible as a doublet in wild-type and Sac3-myc cells transformed with NLS-NES-GFP plasmid (Figure 3F, lanes 7–9). In cells transformed with NLS-NES-GFP, α-GFP can immunoprecipitate NLS-NES-GFP, which migrates closely to the heavy chain of the α-GFP antibody (Figure 3F, lanes 10–12). Sac3-myc is not present in the immunoprecipitates from any of the three strains (Figure 3F, lanes 4–6), indicating that Sac3-myc does not associate with all GFP-tagged proteins. In addition, we were able to detect consistently ECFP-Dbp5, EYFP-Mtr2, and Mex67-GFP in α-myc immunoprecipitations directed against Sac3-myc in the reverse experiment (our unpublished data). These results indicate that Sac3 interacts physically with the mRNA export factors Mtr2, Mex67, and Dbp5 as well as the NPC-associated proteins Mlp1 and Nup116.
Sac3 Localizes to Cytoplasmic Fibrils of NPC
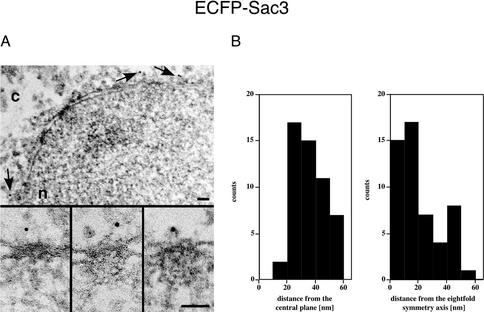
To determine the precise localization of Sac3 within the NPC, we performed immunoelectron microscopy by using epitope-tagged Sac3. Previously, it has been shown that Sac3 localizes exclusively to the nuclear rim by using a GFP-fusion (Jones et al., 2000). To localize Sac3, preembedding labeling immunoelectron microscopy by using a yeast strain expressing ECFP-Sac3 was carried out. An α-GFP antibody conjugated directly to 8-nm colloidal gold labeled only the cytoplasmic face of the NPC (Figure 4A). Quantitation of the gold particle distribution associated with the NPC with respect to the central plane of the nuclear envelope revealed that 95% of the gold particles were detected at distances from 20–60 nm (average distance 36.9 ± 10.5 nm) from the central plane (Figure 4B). With respect to the eightfold symmetry axis of the NPC, 95% of the gold particles were distributed over a broad range at distances from 0 to 50 nm (average distance 20.2 ± 15.3 nm) from this plane. From these results, we concluded that ECFP-Sac3 is associated exclusively with the cytoplasmic filaments of the NPC. We obtained similar results with C-terminal myc and GFP epitope tags on Sac3 (our unpublished data).
Figure 4.
Immunogold-localization of Sac3 in ECFP-Sac3 cells. (A) Triton X-100–extracted spheroplasts from ECFP-Sac3 cells were preimmunolabeled with a polyclonal anti-GFP antibody directly conjugated to 8-nm colloidal gold. Shown is a view along a cross-sectioned nuclear envelope stretch with labeled NPCs (arrows, top), and a gallery of selected samples of gold-labeled NPC cross sections (bottom). The anti-GFP antibody labeled exclusively the cytoplasmic periphery of the NPC. c, cytoplasm; n, nucleus. Bars, 100 nm. (B) Quantitative analysis of the gold particles associated with the NPC. Fifty-two gold particles were scored.
We also tested whether Sac3 localization is dependent on other transport factors. Dbp5 localizes to the nuclear rim and the cytoplasm in wild-type cells, but in xpo1-1 cells, Dbp5 accumulates in the nucleus, suggesting that Dbp5 shuttles through the nucleus (Hodge et al., 1999). We localized ECFP-Sac3 in xpo1-1 cells by fluorescence microscopy and found its localization to be unaffected compared with wild type (our unpublished data), indicating that it does not shuttle in a XPO1/CRM1-dependent manner. Similarly, Sac3 localization is also unaffected in the mtr2-142 mutant (our unpublished data).
Sac3 Is Required for Proper Localization of Mex67
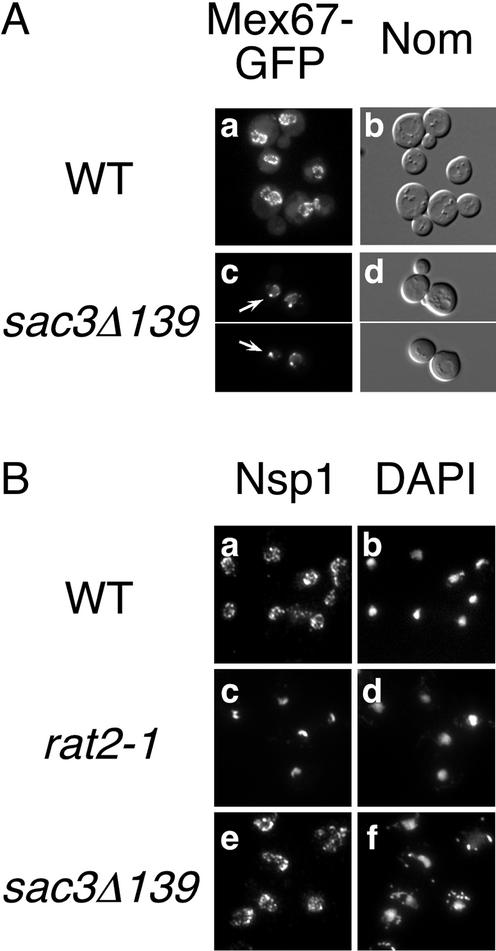
To determine whether Sac3 may function in the translocation of Mex67 through the NPC, we examined the localization of Mex67 and Mtr2 in the sac3Δ139 mutant. In wild-type cells, Mex67-GFP localizes to the nuclear rim (Figure 5A, a and b). In sac3Δ139 cells, Mex67-GFP is localized to the entire nuclear rim as in wild type; however, a single strong focus several times the intensity of the rest of the nuclear rim is visible (Figure 5A, c and d). Western blotting was performed to verify that Mex67-GFP levels are similar in wild-type and sac3Δ139 cells (our unpublished data). A similar and more profound defect of Mex67-GFP mislocalization was visible in SAC3Δ cells (ACY159) (our unpublished data). In contrast, Mtr2-GFP was unaltered in sac3− cells compared with wild type, suggesting that the defect of Mex67 localization in sac3− cells is not a result of Mtr2 mislocalization (our unpublished data). We conclude that SAC3 is required for proper localization of Mex67.
Figure 5.
Mex67 mislocalizes in sac3Δ139 cells. (A) Localization of Mex67-GFP in wild-type (MEX67-GFP, a and b) and sac3Δ139 cells (c and d, PSY2843, top and PSY2842, bottom) grown at 30°C. GFP fluorescence (left) and Nomarski images (right) are shown. Arrows point to intense foci at the nuclear rim. (B) Localization of Nsp1 in wild-type (FY23, a and b), rat2-1 (Dat4-2, c and d), and sac3Δ139 cells (e and f) grown at 25°C. Indirect immunofluorescence with polyclonal antibodies to Nsp1 (left) and 4,6-diamidino-2-phenylindole (right) are shown.
The mislocalization of Mex67 in sac3Δ139 cells is not due to pore clustering, a phenotype of some nucleoporin mutants. To examine the distribution of nuclear pores, we localized the nucleoporin Nsp1 in the sac3Δ139 mutant. By indirect immunofluorescence, Nsp1 is visible as a punctate signal at the nuclear rim in wild-type cells grown at 25°C (Figure 5B, a and b). In a pore clustering mutant rat2-1, NPCs are clustered on one side of the nucleus (Heath et al., 1995), and Nsp1 signal concentrates in a crescent shape adjacent to the DAPI signal (Figure 5B, c and d). In sac3Δ139 cells, Nsp1 localizes to the entire nuclear rim similar to wild type (Figure 5B, e and f), indicating that NPCs do not cluster in sac3Δ139 cells. The same results are obtained when cells are grown at 30°C or shifted to 37°C for 3 h (our unpublished data). Furthermore, Dbp5, which clusters in a pore clustering mutant (Snay-Hodge et al., 1998), is not mislocalized in sac3Δ139 cells (our unpublished data). Therefore, the mislocalization of Mex67 in sac3Δ139 cells is not a result of pore clustering.
DISCUSSION
Using a combination of genetic screens to identify genes involved in mRNA export, we have identified the nuclear pore-associated protein Sac3. Mutation of SAC3 causes a robust accumulation of mRNA in the nucleus but only mildly affects other nuclear export pathways. Furthermore, Sac3 interacts physically with several proteins involved in mRNA export and localizes exclusively to the cytoplasmic fibrils of the NPC. Finally, mutation of SAC3 causes mislocalization of Mex67 to a strong focus at the nuclear rim. Taken together, these results show that Sac3 is involved in mRNA export and implicate Sac3 at the step of RNP translocation through the NPC.
Earlier studies have implicated Sac3 in actin function and mitotic progression. Mutations in SAC3 were found to suppress the temperature sensitivity of the act1-1 mutant (Novick et al., 1989). A potential explanation for the suppression of act1-1 by sac3− mutants is that diminished export of the defective actin transcript decreases the deleterious effects of this actin mutation. However, this possibility is unlikely because the suppression is allele specific (Bauer and Kölling, 1996a; Novick et al., 1989), suggesting that loss of SAC3 function does not bypass the act1-1 mutation. In addition, a later study found that mutation of SAC3 causes aberrant mitosis and spindle morphology (Bauer and Kölling, 1996b). A direct role for the actin cytoskeleton has not been established in nuclear transport, but there is speculation that RNPs may travel along cellular microfilament tracks to reach their destinations. In fact, it has been suggested that Tpr and its homologues Mlp1 and 2 may form intranuclear filaments that can serve as tracks connecting the chromatin and the NPC on which RNPs travel (Cordes et al., 1997; Strambio-de-Castillia et al., 1999). It is also possible that the actin cytoskeleton may be physically attached to the cytoplasmic filaments of the NPC and necessary for proper orientation of the NPC and/or the mitotic spindle. A screen for nuclear import mutants revealed that mutation of ARP2/ACT2, an actin-related gene, causes a defect in NPC morphology (Yan et al., 1997). Currently, it is unclear as to how the nuclear transport function of Sac3 is related to its function with respect to actin.
Previously, Sac3 was ascribed a role in nuclear transport. Examination of Sac3 showed that it localizes to the nuclear rim and interacts with nucleoporins (Jones et al., 2000). Specifically, Sac3 was copurified with Nsp1 from yeast lysate and was shown to interact with Nup1 and Nup159 by yeast two hybrid. Interestingly, Nsp1 and Nup159 together with Nup82 form a nucleoporin subcomplex that functions primarily in mRNA export (Belgareh et al., 1998). Analysis of a SAC3 deletion (SAC3Δ) strain showed synthetic lethal interactions with several NES-protein export mutants and a defect in NES-protein export (Jones et al., 2000). We found that the sac3Δ139 mutant as well an independent deletion strain (Δsac3-rg) display a strong nuclear accumulation of mRNA. Consistent with our results, overexpression of SAC3 by using a galactose-inducible promoter causes an mRNA export defect (Corbett, personal communication). In addition, in sac3Δ139 cells no synthetic lethality with Δyrb2 is observed and only a mild NES-protein export defect is detected using the same reporter used in the previous study. In SAC3Δ cells, the NLS-NES-GFP reporter localizes exclusively to the nucleus in ∼30% of cells grown at 25°C and ∼50% of cells shifted to 37°C for 2 h (Corbett, personal communication). These results contrast with the mild nuclear accumulation visible in a low percentage of sac3Δ139 and Δsac3-rg cells (Figure 2C; our unpublished data). In our hands, the SAC3Δ strain seemed to revert to faster growing cells at a high rate, especially upon plasmid transformation (our unpublished data). Therefore, we speculate that a second mutation may be present in the SAC3Δ strain. Alternately, contrasting degrees of effects on NES-export in these sac3− mutants may be due to differences in strain background. SAC3 as well as a number of other genes involved in mRNA export are not essential for viability. Deletion of any of several genes such as NUP116, GLE2, and NUP133 causes profound defects in mRNA export but allows growth, although at significantly reduced rates (Wente and Blobel, 1993; Li et al., 1995; Murphy et al., 1996). Apparently, cells are able to compensate for lack of these genes because of the functional redundancy of various export factors.
Several findings support the notion that Sac3 functions at the translocation step of mRNA export. First, the sac3Δ139 mutation results in synthetic lethality when combined with mex67-5 and mtr2-142, both of which cause Mex67 to mislocalize to the cytoplasm upon a shift to nonpermissive temperature (Figure 1C; Segref et al., 1997). The sac3Δ139 mutant also mislocalizes Mex67; therefore, these combined mutations could further disrupt Mex67 localization and function and cause lethality. Additionally, Sac3 can be coimmunoprecipitated with Mex67, Mtr2, and Nup116 suggestive of interactions that may be required for RNP translocation through the NPC. To address whether SAC3 may affect the interaction of Mex67 with FG-repeat–containing nucleoporins, we performed coimmunoprecipitations of Mex67 and Nup116 as described previously (Strawn et al., 2001) in wild-type compared with sac3Δ139 cells but found the interaction to be unaffected (our unpublished data). Nevertheless, Mex67 interacts with multiple FG-repeat containing nucleoporins (Sträβer et al., 2000; Strawn et al., 2001), and Sac3 could affect any number of these or other interactions.
More specifically, Sac3 may be involved in the terminal step of export. As determined by immunoelectron microscopy, Sac3 localizes exclusively to the cytoplasmic fibrils of the NPC, suggesting that it acts at a late stage of mRNA export. Sac3 may be a static NPC-associated protein because Sac3 localizes exclusively to the nuclear rim and remains properly localized in the xpo1-1 mutant (Jones et al., 2000; our unpublished data). However, we also found that Sac3 coimmunoprecipitates with Mlp1, which localizes to the nuclear basket by immunoelectron microscopy (Strambio-de-Castillia et al., 1999; Kosova et al., 2000). It is possible that either Mlp1 or Sac3 or both proteins are mobile factors despite being preferentially localized to opposite sides of the pore at steady state. Interestingly, Sac3 was not identified in a large-scale analysis of NPC proteins despite that it meets the criteria defined in this study (Rout et al., 2000). The absence of Sac3 in this NPC purification may be a result of its sensitivity to proteolysis during the preparation or its transient interaction with the NPC. Our genetic analysis revealed specificity of interactions between sac3Δ139 and mutations in genes encoding transport factors, perhaps providing a better indication of Sac3 function than our coimmunoprecipitation experiments. All of the synthetic lethal interactors identified in our analysis correspond to mRNA export factors that localize at least partially to the cytoplasmic fibrils of the NPC.
Understanding of the release of proteins from the mRNA remains elusive. A candidate for the mediator of this process is the RNA helicase Dbp5, which localizes to the cytoplasmic fibrils and the cytoplasm (Snay-Hodge et al., 1998; Tseng et al., 1998; Strahm et al., 1999). Dbp5 can be coimmunoprecipitated with Sac3 from cell lysates. Furthermore, sac3Δ139 is synthetically lethal with a mutant of DBP5, rat8-2, and with a mutant allele of NUP159, which is required for Dbp5 localization to the nuclear rim (Hodge et al., 1999; Schmitt et al., 1999). In addition, sac3Δ139 combined with a mutant allele of NUP82 results in synthetic sickness. A phenotype of nup82-Δ108 cells is that Nup159 localization to the nuclear pore is disrupted (Hurwitz et al., 1998). Each of these proteins seems to be important for proper localization of other cytoplasmic fibril-associated proteins involved in mRNA export and may function as terminal docking sites for RNPs during NPC translocation. Further studies should elucidate the mechanism of this complex process.
Table 2.
S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| FY23 | MATa ura3-52 leu2Δ1 trp1Δ63 | Winston et al., 1995 |
| ACY159 | MATα Δsac3∷HIS3 ura3-52 leu2Δ1 his3Δ200 lys2 | Jones et al., 2000 |
| ACY276 | MATa SAC3∷SAC3-GFP-HIS3 ura3-52 leu2Δ1 his3Δ200 trp1Δ63 | This study |
| Dat4-2 | MATa rat2-1 ura3-52 leu2Δ1 trp1Δ63 | Heath et al., 1995 |
| MEX67-GFP | MATa Δmex67∷HIS3 ura3 leu2 his3 trp1 ade2 〈pUN100 Mex67-GFP LEU2〉 | Santos-Rosa et al., 1998 |
| PSY1031 | MATα np13-27 ura3-52 leu2-3,112 his3 lys1-1 trp1-1 ade2-1 ade8 can1-100 | Krebber et al., 1999 |
| PSY1717 | MATa np13-27 mtr2-142 ura3-52 leu2-3,112 his3 lys1-1 ade2-1 ade8 can1-100 | This study |
| PSY1719 | MATa mtr2-142 ura3-52 leu2-3,112 his3 lys1-1 ade2-1 ade8 can1-100 | This study |
| PSY1720 | MATα mtr2-142 ura3-52 leu2-3,112 his3 lys1-1 ade2-1 ade8 can1-100 | This study |
| PSY1832 | MATa NUP116∷NUP116-EYFP-URA3 ura3-52 leu2Δ1 trp1Δ63 | Damelin and Silver, 2000 |
| PSY2451 | MATa SAC3∷SAC3-13myc-kanMX6 ura3-52 leu2Δ1 trp1Δ63 | This study |
| PSY2555 | MATa sac3Δ139∷HIS3MX6 ura3 leu2 his3 | This study |
| PSY2691 | MATa SAC3∷SAC3-13myc-kanMX6 Δmex67∷HIS3 ura3 leu2 his3 trp1 ade2 〈pUN100 Mex67-GFP LEU2〉 | This study |
| PSY2726 | MATa DBP5∷NOP1pro-ECFP-DBP5-TRP1 ura3-52 leu2Δ1 trp1Δ63 | This study |
| PSY2729 | MATa MTR2∷NOP1pro-EYFP-MTR2-TRP1 ura3-52 leu2Δ1 trp1Δ63 | This study |
| PSY2747 | MATa SAC3∷SAC3-13myc-kanMX6 MTR2∷NOP1pro-EYFP-MTR2-URA3 leu2Δ1 trp1Δ63 | This study |
| PSY2751 | MATa MLP1∷MLP1-EYFP-URA3 ura3-52 leu2Δ1 trp1Δ63 | This study |
| PSY2752 | MATa MLP1∷MLP1-EYFP-URA3 SAC3∷SAC3-13myc-kanMX6 ura3-52 leu2Δ1 trp1Δ63 | This study |
| PSY2755 | MATa SAC3∷SAC3-13myc-kanMX6 DBP5∷NOP1pro-ECFP-DBP5-TRP1 ura3-52 leu2Δ1 trp1Δ63 | This study |
| PSY2842 | MATa sac3Δ139∷HIS3MX6 Δmex67∷HIS3 ura3 leu2 his3 trp1 〈pUN100 Mex67-GFP LEU2〉 | This study |
| PSY2843 | MATα sac3Δ139∷HIS3MX6 Δmex67∷HIS3 ura3 leu2 his3 trp1 〈pUN100 Mex67-GFP LEU2〉 | This study |
| PSY2844 | MATa Δsac3∷kanMX4 ura3Δ0 leu2Δ0 his3Δ1 met15Δ0 | Winzeler et al., 1999 |
| PSY2856 | MATa SAC3∷SAC3-13myc-kanMX6 NUP116∷NUP116-EYFP-URA3 ura3-52 leu2Δ1 trp1Δ63 | This study |
| PSY2857 | MATα mtr2-142 Δmex67∷HIS3 ura3 leu2 his3 lys1 trp1 ade2 〈pUN100 Mex67-GFP LEU2〉 | This study |
ACKNOWLEDGMENTS
We thank G. Blobel, D. Botstein, C. Cole, A. Corbett, E. Hurt, M. Rout, and S. Wente for sharing strains and plasmids. We are grateful to M. Stewart for α-Nsp1 antibodies. We thank A. Corbett for communicating unpublished data, M. Damelin for critical comments on the manuscript, and members of the Silver laboratory for thoughtful discussions. E.P.L. and T.M. were supported by grants from the Ryan Foundation and the National Cancer Institute. C.S. was supported by grants from the National Institutes of Health and American Cancer Society. B.F. was supported by a grant from the M.E. Müller Foundation. H.K. was supported by a grant from the Deutsche Forschungsgemeinshaft. This work was supported by grants from the National Institutes of Health and Human Frontiers Science Project (to P.A.S. and U.A.).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–08–0520. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-08-0520.
REFERENCES
- Bachi A, et al. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA. 2000;6:136–158. doi: 10.1017/s1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailer SM, Siniossoglou S, Podtelejnikov A, Hellwig A, Mann M, Hurt E. Nup116p and nup100p are interchangeable through a conserved motif which constitutes a docking site for the mRNA transport factor gle2p. EMBO J. 1998;17:1107–1119. doi: 10.1093/emboj/17.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baβler J, Grandi P, Gadal O, Lessmann T, Petfalski E, Tollervey D, Lechner J, Hurt E. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol Cell. 2001;8:517–529. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- Bauer A, Kölling R. Characterization of the SAC3 gene of Saccharomyces cerevisiae. Yeast. 1996a;12:965–975. doi: 10.1002/(SICI)1097-0061(199608)12:10%3C965::AID-YEA999%3E3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Bauer A, Kölling R. The SAC3 gene encodes a nuclear protein required for normal progression of mitosis. J Cell Sci. 1996b;109:1575–1583. doi: 10.1242/jcs.109.6.1575. [DOI] [PubMed] [Google Scholar]
- Belgareh N, Snay-Hodge C, Pasteau F, Dagher S, Cole CN, Doye V. Functional characterization of a Nup159p-containing nuclear pore subcomplex. Mol Biol Cell. 1998;9:3475–3492. doi: 10.1091/mbc.9.12.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes VC, Reidenbach S, Rackwitz HR, Franke WW. Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J Cell Biol. 1997;136:515–529. doi: 10.1083/jcb.136.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damelin M, Silver PA. Mapping interactions between nuclear transport factors in living cells reveals pathways through the nuclear pore complex. Mol Cell. 2000;5:133–140. doi: 10.1016/s1097-2765(00)80409-8. [DOI] [PubMed] [Google Scholar]
- Damelin M, Silver PA. In situ analysis of spatial relationships between proteins of the nuclear pore complex. Biophys J. 2002;83:3626–3636. doi: 10.1016/S0006-3495(02)75363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge SJ, Davis RW. A family of versatile centromeric vectors designed for use in the sectoring-shuffle mutagenesis assay in Saccharomyces cerevisiae. Gene. 1988;70:303–312. doi: 10.1016/0378-1119(88)90202-8. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog B, Hurt EC, Aebi U, Pante N. Molecular architecture of the yeast nuclear pore complex: localization of Nsp1p subcomplexes. J Cell Biol. 1998;143:577–588. doi: 10.1083/jcb.143.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flach J, Bossie M, Vogel J, Corbett A, Jinks T, Willins DA, Silver PA. A yeast RNA-binding protein shuttles between the nucleus and the cytoplasm. Mol Cell Biol. 1994;14:8399–8407. doi: 10.1128/mcb.14.12.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fribourg S, Braun IC, Izaurralde E, Conti E. Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol Cell. 2001;8:645–656. doi: 10.1016/s1097-2765(01)00348-3. [DOI] [PubMed] [Google Scholar]
- Gorsch LC, Dockendorff TC, Cole CN. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J Cell Biol. 1995;129:939–955. doi: 10.1083/jcb.129.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath CV, Copeland CS, Amberg DC, Del Priore V, Snyder M, Cole CN. Nuclear pore complex clustering and nuclear accumulation of poly(A)+ RNA associated with mutation of the Saccharomyces cerevisiae RAT2/NUP120 gene. J Cell Biol. 1995;131:1677–1697. doi: 10.1083/jcb.131.6.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry MF, Silver PA. A novel methyltransferase (Hmt1p) modifies poly(A)+-RNA-binding proteins. Mol Cell Biol. 1996;16:3668–3678. doi: 10.1128/mcb.16.7.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold A, Klymenko T, Izaurralde E. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA. 2001;7:1768–1780. [PMC free article] [PubMed] [Google Scholar]
- Ho JH, Kallstrom G, Johnson AW. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J Cell Biol. 2000;151:1057–1066. doi: 10.1083/jcb.151.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge CA, Colot HV, Stafford P, Cole CN. Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1-1 cells. EMBO J. 1999;18:5778–5788. doi: 10.1093/emboj/18.20.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt E, Hannus S, Schmelzl B, Lau D, Tollervey D, Simos G. A novel in vivo assay reveals inhibition of ribosomal nuclear export in ran-cycle and nucleoporin mutants. J Cell Biol. 1999;144:389–401. doi: 10.1083/jcb.144.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt E, Sträβer K, Segref A, Bailer S, Schlaich N, Presutti C, Tollervey D, Jansen R. Mex67p mediates nuclear export of a variety of RNA polymerase II transcripts. J Biol Chem. 2000;275:8361–8368. doi: 10.1074/jbc.275.12.8361. [DOI] [PubMed] [Google Scholar]
- Hurwitz ME, Blobel G. NUP82 is an essential yeast nucleoporin required for poly(A)+ RNA export. J Cell Biol. 1995;130:1275–1281. doi: 10.1083/jcb.130.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz ME, Strambio-de-Castillia C, Blobel G. Two yeast nuclear pore complex proteins involved in mRNA export form a cytoplasmically oriented subcomplex. Proc Natl Acad Sci USA. 1998;95:11241–11245. doi: 10.1073/pnas.95.19.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine MK, Watkins JL, Wente SR. The GLFG repetitive region of the nucleoporin Nup116p interacts with Kap95p, an essential yeast nuclear import factor. J Cell Biol. 1995;131:1699–1713. doi: 10.1083/jcb.131.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AL, Quimby BB, Hood JK, Ferrigno P, Keshava PH, Silver PA, Corbett AH. SAC3 may link nuclear protein export to cell cycle progression. Proc Natl Acad Sci USA. 2000;97:3224–3229. doi: 10.1073/pnas.050432997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J, Sträβer K, Podtelejnikov A, Mann M, Jung JU, Hurt E. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kölling R, Nguyen T, Chen EY, Botstein D. A new yeast gene with a myosin-like heptad repeat structure. Mol Gen Genet. 1993;237:359–369. doi: 10.1007/BF00279439. [DOI] [PubMed] [Google Scholar]
- Kosova B, Panté N, Rollenhagen C, Podtelejnikov A, Mann M, Aebi U, Hurt E. Mlp2p, a component of nuclear pore attached intranuclear filaments, associates with Nic96p. J Biol Chem. 2000;275:343–350. doi: 10.1074/jbc.275.1.343. [DOI] [PubMed] [Google Scholar]
- Kraemer DM, Strambio-de-Castillia C, Blobel G, Rout MP. The essential yeast nucleoporin NUP159 is located on the cytoplasmic side of the nuclear pore complex and serves in karyopherin-mediated binding of transport substrate. J Biol Chem. 1995;270:19017–19021. doi: 10.1074/jbc.270.32.19017. [DOI] [PubMed] [Google Scholar]
- Krebber H, Taura T, Lee MS, Silver PA. Uncoupling of the hnRNP Npl3p from mRNAs during the stress-induced block in mRNA export. Genes Dev. 1999;13:1994–2004. doi: 10.1101/gad.13.15.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Henry M, Silver PA. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- Lei EP, Krebber H, Silver PA. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev. 2001;15:1771–1782. doi: 10.1101/gad.892401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei EP, Silver PA. Protein, and RNA export from the nucleus. Dev Cell. 2002;2:261–272. doi: 10.1016/s1534-5807(02)00134-x. [DOI] [PubMed] [Google Scholar]
- Levesque L, Guzik B, Guan T, Coyle J, Black BE, Rekosh D, Hammarskjold ML, Paschal BM. RNA export mediated by tap Involves NXT1-dependent interactions with the nuclear pore complex. J Biol Chem. 2001;28:28. doi: 10.1074/jbc.M106558200. [DOI] [PubMed] [Google Scholar]
- Li O, Heath CV, Amberg DC, Dockendorff TC, Copeland CS, Snyder M, Cole CN. Mutation or deletion of the Saccharomyces cerevisiae RAT3/NUP133 gene causes temperature-dependent nuclear accumulation of poly(A)+ RNA and constitutive clustering of nuclear pore complexes. Mol Biol Cell. 1995;6:401–417. doi: 10.1091/mbc.6.4.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Moy TI, Silver PA. Nuclear export of the small ribosomal subunit requires the ran-GTPase cycle and certain nucleoporins. Genes Dev. 1999;13:2118–2133. doi: 10.1101/gad.13.16.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R, Watkins JL, Wente SR. GLE2, a Saccharomyces cerevisiae homologue of the Schizosaccharomyces pombe export factor RAE1, is required for nuclear pore complex structure and function. Mol Biol Cell. 1996;7:1921–1937. doi: 10.1091/mbc.7.12.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R, Wente SR. An RNA-export mediator with an essential nuclear export signal. Nature. 1996;383:357–360. doi: 10.1038/383357a0. [DOI] [PubMed] [Google Scholar]
- Novick P, Osmond BC, Botstein D. Suppressors of yeast actin mutations. Genetics. 1989;121:659–674. doi: 10.1093/genetics/121.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Moreno H, Simos G, Segref A, Fahrenkrog B, Panté N, Hurt E. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol Cell Biol. 1998;18:6826–6838. doi: 10.1128/mcb.18.11.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt C, et al. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J. 1999;18:4332–4347. doi: 10.1093/emboj/18.15.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton DR, Chen S, Hitomi M, Kumagai C, Tartakoff AM. A yeast protein that bidirectionally affects nucleocytoplasmic transport. J Cell Sci. 1995;108:265–272. doi: 10.1242/jcs.108.1.265. [DOI] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Snay-Hodge CA, Colot HV, Goldstein AL, Cole CN. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 1998;17:2663–2676. doi: 10.1093/emboj/17.9.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Stage-Zimmermann T, Schmidt U, Silver PA. Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol Biol Cell. 2000;11:3777–3789. doi: 10.1091/mbc.11.11.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffler D, Fahrenkrog B, Aebi U. The nuclear pore complex: from molecular architecture to functional dynamics. Curr Opin Cell Biol. 1999;11:391–401. doi: 10.1016/S0955-0674(99)80055-6. [DOI] [PubMed] [Google Scholar]
- Sträβer K, Baβler J, Hurt E. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J Cell Biol. 2000;150:695–706. doi: 10.1083/jcb.150.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sträβer K, Hurt E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahm Y, Fahrenkrog B, Zenklusen D, Rychner E, Kantor J, Rosbash M, Stutz F. The RNA export factor Gle1p is located on the cytoplasmic fibrils of the NPC and physically interacts with the FG-nucleoporin Rip1p, the DEAD-box protein Rat8p/Dbp5p and a new protein Ymr255p. EMBO J. 1999;18:5761–5777. doi: 10.1093/emboj/18.20.5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-de-Castillia C, Blobel G, Rout MP. Proteins connecting the nuclear pore complex with the nuclear interior. J Cell Biol. 1999;144:839–855. doi: 10.1083/jcb.144.5.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn LA, Shen T, Wente SR. The GLFG regions of Nup116p and Nup100p serve as binding sites for both Kap95p and Mex67p at the nuclear pore complex. J Biol Chem. 2001;276:6445–6452. doi: 10.1074/jbc.M008311200. [DOI] [PubMed] [Google Scholar]
- Stutz F, Bachi A, Doerks T, Braun IC, Seraphin B, Wilm M, Bork P, Izaurralde E. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA. 2000;6:638–650. doi: 10.1017/s1355838200000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taura T, Krebber H, Silver PA. A member of the Ran-binding protein family, Yrb2p, is involved in nuclear protein export. Proc Natl Acad Sci USA. 1998;95:7427–7432. doi: 10.1073/pnas.95.13.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MT, Siniossoglou S, Podtelejnikov S, Benichou JC, Mann M, Dujon B, Hurt E, Fabre E. Two functionally distinct domains generated by in vivo cleavage of Nup145p: a novel biogenesis pathway for nucleoporins. EMBO J. 1997;16:5086–5097. doi: 10.1093/emboj/16.16.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng SS, Weaver PL, Liu Y, Hitomi M, Tartakoff AM, Chang TH. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J. 1998;17:2651–2662. doi: 10.1093/emboj/17.9.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Blobel G. A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J Cell Biol. 1993;123:275–284. doi: 10.1083/jcb.123.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Rout MP, Blobel G. A new family of yeast nuclear pore complex proteins. J Cell Biol. 1992;119:705–23. doi: 10.1083/jcb.119.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand HL, Coburn GA, Zeng Y, Kang Y, Bogerd HP, Cullen BR. Formation of Tap/NXT1 heterodimers activates Tap-dependent nuclear mRNA export by enhancing recruitment to nuclear pore complexes. Mol Cell Biol. 2002;22:245–256. doi: 10.1128/MCB.22.1.245-256.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SM, Datar KV, Paddy MR, Swedlow JR, Swanson MS. Characterization of nuclear polyadenylated RNA-binding proteins in Saccharomyces cerevisiae. J Cell Biol. 1994;127:1173–84. doi: 10.1083/jcb.127.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Yan C, Leibowitz N, Mélèse T. A role for the divergent actin gene, ACT2, in nuclear pore structure and function. EMBO J. 1997;16:3572–3586. doi: 10.1093/emboj/16.12.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Vinciguerra P, Strahm Y, Stutz F. The yeast hnRNP-Like proteins Yra1p and Yra2p participate in mRNA export through interaction with Mex67p. Mol Cell Biol. 2001;21:4219–4232. doi: 10.1128/MCB.21.13.4219-4232.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]