Abstract
The factors that organize the internal membranes of cells are still poorly understood. We have been addressing this question using striated muscle cells, which have regular arrays of membranes that associate with the contractile apparatus in stereotypic patterns. Here we examine links between contractile structures and the sarcoplasmic reticulum (SR) established by small ankyrin 1 (sAnk1), a ∼17.5-kDa integral protein of network SR. We used yeast two-hybrid to identify obscurin, a giant Rho-GEF protein, as the major cytoplasmic ligand for sAnk1. The binding of obscurin to the cytoplasmic sequence of sAnk1 is mediated by a sequence of obscurin that is C-terminal to its last Ig-like domain. Binding was confirmed in two in vitro assays. In one, GST-obscurin, bound to glutathione-matrix, specifically adsorbed native sAnk1 from muscle homogenates. In the second, MBP-obscurin bound recombinant GST-sAnk1 in nitrocellulose blots. Kinetic studies using surface plasmon resonance yielded a KD = 130 nM. On subcellular fractionation, obscurin was concentrated in the myofibrillar fraction, consistent with its identification as sarcomeric protein. Nevertheless, obscurin, like sAnk1, concentrated around Z-disks and M-lines of striated muscle. Our findings suggest that obscurin binds sAnk1, and are the first to document a specific and direct interaction between proteins of the sarcomere and the SR.
INTRODUCTION
The mechanisms that organize the internal membrane systems in eukaryotic cells are poorly understood. We are studying this question in striated muscle, which has two highly organized internal membranes, the transverse (t-) tubules and the sarcoplasmic reticulum (SR; Franzini-Armstrong, 1994; Flucher and Franzini-Armstrong, 1996; Leong and MacLennan, 1998).
The SR of striated muscle is an intricate web of intracellular membranes that, with the sarcolemma and t-tubules, modulate cytosolic Ca2+ during contraction and relaxation (Franzini-Armstrong and Peachey, 1981; Flucher, 1992; Franzini-Armstrong, 1999). The SR closely surrounds and interacts with the myofibrils, but the molecules that organize and stabilize these connections are not known. We recently characterized a small form of ankyrin in the SR that may play an important role in these processes (Zhou et al., 1997).
Ankyrins are multifunctional adaptor proteins of the plasma membrane and intracellular membranes (Bennett and Chen, 2001). They contribute to the formation and cytoskeletal anchoring of specialized membrane domains. They also participate in signal transduction and protein sorting through their binding to ion pumps, Ca2+ release channels, and cell adhesion molecules (De Matteis and Morrow, 1998; Rubstov and Lopina, 2000).
Mammals have three distinct ankyrin genes, ANK1, ANK2, and ANK3, that produce a variety of tissue-specific and developmentally regulated products (Sedgwick and Smerdon, 1999; Bennett and Chen, 2001). Canonical ankyrins, encoded by all three genes, have molecular masses of ∼190–210-kDa and share three highly conserved structural domains: an NH2-terminal, ∼89-kDa membrane-binding domain, a central ∼62-kDa spectrin-binding region, and a ∼55-kDa COOH-terminal domain (Mohler et al., 2002). However, giant proteins encoded by ANK2 and ANK3 that carry additional structural motifs (Otto et al., 1991; Kordeli and Bennett, 1991; Kunimoto, 1995; Zhang and Bennett, 1998), and smaller forms encoded by ANK1 and ANK3 that lack domains of the canonical ankyrins (Peters et al., 1995; Devarajan et al., 1996; Hoock et al., 1997; Zhou et al., 1997; Birkenmeier et al., 1998; Gallagher and Forget, 1998), have also been identified.
Ankyrin 2 (or ankyrin B) is mainly expressed in brain and cardiac muscle (Otto et al., 1991; Tuvia et al., 1999), whereas ankyrin 3 (or ankyrin G) products are present in epithelial cells, the nervous system, striated muscle and intracellular organelles including the Golgi apparatus and lysosomes (Kordeli et al., 1995; Peters et al., 1995; Devarajan et al., 1996; Hoock et al., 1997; Gagelin et al., 2002). Isoforms of ankyrin 1 (or ankyrin R) are present in red blood cells, neurons and striated muscle (Lambert et al., 1990; Lux et al., 1990; White et al., 1992; Birkenmeier et al., 1993).
The ANK1 products in striated muscle include large, canonical (∼210 kDa) and small (∼17–19-kDa) ankyrins (Gallagher et al., 1997; Zhou et al., 1997; Birkenmeier et al., 1998). The large ankyrin isoform is concentrated at the sarcolemma of skeletal myofibers, whereas sAnk1 is targeted to the SR, and specifically to the network SR, which is primarily responsible for Ca2+ uptake. Consistent with its location, sAnk1 is present in a reticulum around the Z-disks and M-lines of the contractile apparatus. sAnk1 lacks both membrane and spectrin-bindiing sequences and retains only a short sequence from the COOH-terminus of large ankyrin 1. The NH2-terminal portion of sAnk1 contains a unique 73-amino acid segment, the first 29 residues of which are hydrophobic and anchor the protein to the SR membrane. The remaining 126 amino acids of sAnk1 face the myoplasm (our unpublished studies). In short, sAnk1 seems to be organized in the network SR to enable it to interact with myofibrillar proteins.
We tested this idea by identifying potential binding partners of sAnk1. Using the yeast two-hybrid screen, we show that obscurin (Young et al., 2001), a giant (∼800 kDa) sarcomeric protein, is a major cytoplasmic ligand for sAnk1. The binding to sAnk1 is mediated by the region of obscurin immediately following its last Ig-like domain. We use in vitro assays, kinetic analysis by surface plasmon resonance, and immunofluorescence techniques to demonstrate that this binding is specific, direct and likely to be physiological.
MATERIALS AND METHODS
Yeast Two-hybrid Screening and β-Galactosidase Assay
The Matchmaker LexA two-hybrid system was used as prescribed by the manufacturer (Clontech, Palo Alto, CA). A fragment encoding the COOH-terminal tail of sAnk1aa29–155 (Birkenmeier et al., 1998) was inserted into the pLexA bait vector at EcoRI/XhoI sites after PCR amplification with primers (i): 5′ ACTGGAATTCGTCAAGGGTTCCCTGTGC 3′ (sense) and (ii): 5′ ACTGCTCGAGCTGCTTGCCCCTTTT 3′ (antisense). A cDNA library was constructed from skeletal muscle of adult rats in the pB42AD prey vector (a kind gift from Dr. T. Gustafson, Metabolex, Hayward, CA). Saccharomyces cerevisiae strain EGY48 was sequentially transformed with reporter p8op-lacZ, bait and prey plasmids. True transformants were selected by plating on induction media (i.e., SD Gal/Raf lacking Ura, His, Trp, and Leu) in the presence of 80 mg/L X-gal. Positive pB42AD plasmids were recovered by electroporation in bacterial strain DH5α and sequenced.
For domain mapping, deletion constructs of the COOH-terminal sequence of sAnk129–155 and obscurin clone F were generated by PCR amplification and introduced into the EcoRI/XhoI sites of the pLexA bait and pB42AD prey vectors, respectively. The following sets of primers were used for amplification of the sAnk1 deletion constructs; for sAnk1-A, the sense primer (i) was used in combination with the antisense primer (iii): 5′ ACTGCTCGAGTTGTTCCTCTGTCAC 3′, for generation of sAnk1-B the sense primer (iv): 5′ ACTGGAATTCTTCACAGACGAACAG 3′ was used along with the antisense primer (ii) and for sAnk1-C, the sense primer (v): 5′ ACTGGAATCATCTCCACCAGGGTG 3′ was used in conjunction with the antisense primer (vi): 5′ ACTGCTCGAGTCCACTCCCTCTTAG 3′. Similarly, for generation of the obscurin clone F deletion constructs, the following primer sets were used. For amplification of obscurin clone F1, the sense primer (vii): 5′ ACTGGAATTCTCCCGTTCACCCTCC 3′ was used along with the antisense primer (viii): 5′ ACGTCTCGAGCCAGTCCACATTGCC 3′, for obscurin clone F2, the sense primer (ix): 5′ ACTGGAATTCTGGGCAGCTGCTGGC 3′ was used with the antisense primer (x): 5′ ACGTCTCGAGGCCCTGGGAGGGGCC 3′, and for generation of obscurin clone F3, the sense primer (xi): 5′ ACTGGAATTCACTCCAGCCTCAGAGCCC 3′ was used in combination with the antisense primer (xii): 5′ ACGTCTCGAGACTGCCTCCTTCCTCCTT 3′. All sense primers carry an EcoRI recognition sequence, whereas all the antisense primers contain an XhoI site for insertion into the bait and prey vectors. The authenticity of each of these constructs was verified by sequence analysis. Subsequently, different combinations of bait and prey plasmids (see RESULTS) were sequentially transformed into EGY48 S. cerevisiae yeast cells and transformants were processed as described above.
Liquid β-galactosidase assays were performed as described in the Clontech Yeast Protocols Handbook, using chlorophenol red-β-d-galactopyranoside as substrate. For each interaction tested, five independent colonies were assayed and each experiment was repeated twice. Results represent average values.
Generation and Purification of Glutathione-S-transferase and Maltose-Binding Protein (MBP) Fusion Proteins
Obscurin clones B, F, and F3 were isolated by restriction digestion of pB42AD with EcoRI/XhoI and inserted into pGEX4T-1 at EcoRI/XhoI sites (Amersham Pharmacia Biotech, Piscataway, NJ) to generate glutathione-S-transferase (GST)-fusion proteins. Obscurin clones F and F3 were also introduced into pMAL-c2X vector at EcoRI/SalI sites (New England Biolabs, Beverly, MA; XhoI and SalI have compatible ends) to produce MBP-fusion proteins. A PCR-amplified fragment of sAnk1aa29–155 was inserted into pGEX4T-1 at EcoRI/XhoI sites and pMAL-c2X at EcoRI/SalI sites. Recombinant polypeptides were expressed by induction with 0.3 mM isopropyl β-d-thioglucopyranoside (IPTG) for 3 h and purified by affinity chromatography on glutathione-agarose (GST-fusion proteins) or amylose resin (MBP-fusion proteins) columns, following the manufacturers' instructions.
GST “Pull Down” Assay
Homogenates of quadriceps muscle of adult, Sprague Dawley rats (Zivic-Miller Laboratories, Zelienople, PA) were prepared at RT for 2–3 min with a Brinkman Polytron homogenizer at setting 3 (VWR, West Chester, PA) in 10 mM NaPO4, pH 7.2, 2 mM EDTA, 10 mM NaN3, 120 mM NaCl, 0.5% deoxycholate, 0.5% NP-40, supplemented with protease inhibitors (Roche, Indianapolis, IN). Equal amounts of recombinant GST and GST-obscurin clone F proteins were bound to glutathione-Sepharose and mixed with 0.5 mg of quadriceps muscle homogenate at 4°C for 16 h. Beads were washed in the cold with 10 mM NaPO4, pH 7.2, 120 mM NaCl, 10 mM NaN3, 0.1% Tween-20, and heated for 5 min at 90°C in 2× SDS Laemmli sample buffer. The soluble fraction was analyzed by SDS-PAGE, transferred to nitrocellulose, and probed with antibodies to sAnk1.
Blot Overlay
The blot overlay assays were performed as previously described, with some modifications (Kontrogianni-Konstantopoulos et al., 2000). In brief, aliquots (∼2.5 μg) of bacterially expressed, affinity-purified MBP and MBP-obscurin clone F proteins were separated by 10% SDS-PAGE and transferred to nitrocellulose. Blots were first incubated in buffer A (50 mM Tris, pH 7.2, 120 mM NaCl, 3% BSA, 2 mM dithiothreitol, 0.5% NP-40, 0.1% Tween-20) plus protease inhibitors for 3 h at 25°C and then with 3 μg/ml GST or GST-sAnk1 polypeptides in buffer A for 16 h at 4°C. Blots were washed extensively with buffer A and once with buffer B (PBS, pH 7.2, 10 mM NaN3, 0.1% Tween-20). Subsequently they were blocked in buffer C (PBS, pH 7.2, 10 mM NaN3, 0.1% Tween-20, 3% dry milk) and probed with antibodies to sAnk1 or GST.
Kinetic Analysis of sAnk1 Binding to Obscurin with a Surface Plasmon Resonance Biosensor
Real-time evaluation of the kinetics of binding of sAnk1 to obscurin was performed with a BIACORE 3000 instrument (Biacore, Uppsala, Sweden). The association (ka, M−1 s−1) and dissociation (kd, s−1) rate constants as well as the association (KA, M−1) and dissociation (KD, M) equilibrium constants were determined. Studies were performed at 25°C on a carboxymethyl-dextran sensor chip (CM5) activated in the presence of a 1:1 NHS:EDC (N-hydroxy-succinimide:1-ethyl-3,3-dimethylaminopropyl-carbodiimide) mixture, according to the manufacturer's instructions (Amine Coupling Kit; Biacore). Approximately 10,000 Resonance Units (RU) of affinity-purified goat anti-GST antibody were immobilized on flow cells 1 and 2 (FC1 and FC2) of the activated CM5 chip through standard amine coupling, using the GST kit from Biacore.
Control GST-protein (∼25 kDa), GST-sAnk1 (∼39 kDa), GST-obscurin-F3 (∼38.5 kDa), MBP-sAnk1 (∼57 kDa), and MBP-obscurin-F3 (∼66 kDa) were produced and affinity-purified, as described above. Two sets of kinetics studies were performed. In the first, GST-obscurin-F3 was immobilized on FC2, via its interaction with the immobilized GST-antibody, and MBP-sAnk1 was used as analyte. In the second, GST-sAnk1 was used as ligand and captured on FC2, and MBP-obscurin-F3 served as analyte. In both experiments, control GST-protein was bound to GST-antibody, immobilized on FC1, to correct for bulk effects and nonspecific binding. Each experimental set was repeated three times and yielded highly reproducible results.
All samples were diluted in freshly degassed, filter-sterilized HBS-EP buffer (0.01 M HEPES, pH 7.4, 0.15 M NaCl, 3 mM EDTA, 0005% Surfactant P20) that contained 10 mM maltose, to eliminate nonspecific binding of the MBP fusion proteins to the dextran matrix of the sensor chip. The same solution was also used as the running buffer during the kinetics assays.
Approximately 300–350 RU of control GST-protein and ligand (i.e., GST-sAnk1 or GST-obsurin-F3) were bound on FC1 and FC2, respectively. The concentrations of the analytes used, MBP-sAnk1 or MBP-obscurin-F3, were 300, 250, 200, 150, 100, 50, 25 and 0 nM. The flow rate for analyte injection was 20 μl/min. For each analyte concentration, association was measured for 180 s and dissociation was measured over a further 180 s. After dissociation of the analyte, the surface was regenerated with a 10-μl injection of 10 mM glycine, pH 2.2, at a flow rate of 10 μl/min. Data were evaluated with the 1:1 Langmuir model and the Heterogeneous Ligand model (BIAevaluation Software 3.1, Biacore).
Preparation of Soluble and Insoluble Muscle Fractions
Quadriceps muscle from adult rats was homogenized as above in lysis buffer (10 mM NaPO4, pH 7.2, 120 mM NaCl, 2% Triton-100) in the presence of a cocktail of protease inhibitors (Roche) and incubated on ice for 1 h with occasional mixing. The homogenate was centrifuged at 14,000 × g for 30 min at 4°C to yield a supernatant (the detergent soluble, or DS, fraction) and pellet. The pellet was dissolved in 2% SDS, 1% β-mercaptoethanol (β-ME), 120 mM NaCl, plus protease inhibitors, incubated on ice for 30 min with occasional mixing and spun at 14,000 × g for 30 min at 4°C. The resultant supernatant was designated as the detergent insoluble (DI) fraction. Muscle was also homogenized in the presence of 10 mM NaPO4, pH 7.2, 2 mM EDTA, 10 mM NaN3, 120 mM NaCl, 1% NP-40, 0.5% deoxycholate plus protease inhibitors, incubated on ice for 1 h with occasional mixing, and spun, as above. Protein was measured with Bradford reagent (Bio-Rad, Hercules, CA), and ∼50 μg from each sample was solubilized in 2× SDS Laemmli sample buffer at 42°C for 30 min. (This treatment is required for obscurin to penetrate the gel). Samples were fractionated on 4–10% gradient or 12% SDS-PAGE, transferred to nitrocellulose (30 V, 16 h, 4°C), blocked with buffer C and incubated with antibodies to sAnk1 or obscurin, at a concentration of 100 ng/ml, in the same solution. Immunoreactive bands were visualized with a chemiluminescence detection kit (Tropix, Bedford, MA).
Immunofluorescence Labeling of Adult Skeletal and Cardiac Muscle
GST-obscurin clone B fusion protein was used to generate rabbit antibodies. Antiserum was affinity-purified over GST and GST-obscurin clone B columns to prepare antibodies specific for obscurin. Affinity-purified rabbit antibodies to sAnk1 have been described (Zhou et al., 1997).
Frozen longitudinal and cross sections of adult rat myocardium and quadriceps muscle were prepared as described (Williams et al., 2001). Sections were blocked with PBS containing 1 mg/ml BSA, 10 mM NaN3, for 1–2 h at 25°C. Primary antibodies, including antiobscurin (3 μg/ml), anti-sAnk1 (3 μg/ml), or ChromaPure rabbit IgG (3 μg/ml; Jackson ImmunoResearch Laboratories Inc., West Grove, PA), together with a mAb to α-actinin (1:500; Sigma) were incubated with the sections for 12 h at 4°C. Samples were counterstained with either goat anti-rabbit Alexa568 or goat anti-rabbit Alexa568 and goat anti-mouse Alexa488 (Molecular Probes, Eugene, OR) at 1:100 dilution, for 1 h at 25°C. In immunodepletion experiments, antiobscurin was preabsorbed with 100 μg of GST-obscurin clone B antigen per milliliter of diluted, affinity-purified serum for 8–12 h at 4°C before labeling. Sections were washed extensively with PBS, 1 mg/ml BSA, 10 mM NaN3, mounted with Vectashield (Vector Laboratories, Burlingame, CA), and analyzed with a Zeiss 410 confocal laser scanning microscope (Carl Zeiss, Inc., Tarrytown, NY) equipped with a 63×, NA 1.4 objective.
Materials
Unless otherwise noted, all reagents were from Sigma Chemical Co. (St. Louis, MO) and were the highest grade available.
RESULTS
sAnk1 is an integral protein of the network SR, positioned with its COOH-terminal hydrophilic tail facing the myoplasm (Zhou et al., 1997; our unpublished results). To identify potential binding partners of sAnk1 in skeletal muscle, we used its COOH-terminal tailaa29–155 as “bait” to screen a cDNA library from adult rat skeletal muscle (Figure 1A). Approximately 5 × 106 transformants were screened, of which 12 “prey” clones met the stringent criteria for positive interactions. These were further characterized by DNA sequencing. Notably, 6 of the 12 positive clones carried overlapping fragments of the COOH-terminal region of obscurin (Figure 1B; Young et al., 2001). No other “prey” sequence was present in more than 1 of the 12 clones examined.
Figure 1.
Yeast two-hybrid analysis identified obscurin as the major cytoplasmic ligand for sAnk1. (A) The COOH-terminal hydrophilic tail of sAnk129–155 was used as bait to screen a rat skeletal muscle cDNA library. (B) Six of the 12 positive preys encoded overlapping fragments of the 2.7-kb, COOH-terminal sequence of obscurin.
The longest obscurin clone (A, ∼2.7 kb) contains a Rho-guanine nucleotide exchange factor (Rho-GEF) domain next to a pleckstrin homology (PH) region, followed by two immunoglobulin-like (Ig) domains and a long, nonmodular COOH-terminus of 413 amino acids (Figure 2A). The 3′ end of the 6 obscurin “preys” all contained an in-frame stop codon, followed by a 3′UTR of 525 base pairs that carries stop codons in all three reading frames (Figure 2B). Unlike the 3′UTR of human obscurin (Young et al., 2001), the rat sequence contains a consensus polyadenylation signal upstream of the poly-A tail (Figure 2B, underlined). The smallest positive clone (F) encodes the most C-terminal 353 amino acids of obscurin as well as the 3′UTR, but it lacks the structural motifs present in the larger clone A. Thus, the sAnk1-binding site should lie within the COOH-terminal 353 amino acid residues of rat obscurin.
Figure 2.
Sequence of the C-terminal region of rat obscurin. (A) Sequence comparison between the COOH-terminal sequence of the human and rat obscurin orthologues. The first line represents amino acids5816–6620 of the human protein (Young et al., 2001), the third line shows the corresponding rat sequence, and the middle line shows shared residues. Overall homology and similarity are ∼77% and ∼83%, respectively. The Ig-like, PH and (partial) Rho-GEF domains are in boxes. The positions of obscurin clones A–F are denoted by arrows. (B) A 525 base pairs 3′UTR is carried by all six obscurin clones; the putative polyadenylation signal is underlined.
Comparison of the partial sequence of rat obscurin with its human orthologue (Young et al., 2001) showed extensive homology, especially within the signaling and structural domains. In particular, the Rho-GEF, PH, and two Ig-like domains shared 98, 95, 94, and 80% identity, respectively (Figure 2A). The COOH-terminal sequence showed lower identity (68%). Overall, clone A was 83% similar and 77% identical to its human orthologue.
We used the yeast two-hybrid screen to identify the region of the cytoplasmic sequence of sAnk1 that binds obscurin and to define more precisely the sequence of obscurin that binds sAnk1. For sAnk1, bait constructs encoding amino acids 29–89, 61–130, and 90–155, were coexpressed in yeast with the prey vector encoding obscurin clone F (Figure 3A). Only sAnk1aa61–130 elicited expression of β-galactosidase, suggesting that this 70 amino acid sequence, but not flanking regions of sAnk1, contains the binding site for obscurin. In a similar experiment, we expressed three fragments of obscurin clone F in the prey vector and assayed their ability to interact with the full cytoplasmic sequence of sAnk1, expressed in the bait vector. In this case, two fragments of obscurin clone F, encoding amino acid residues 447–596 and 501–621, equivalent to amino acid sequences 6258–6407 and 6312–6432 of human obscurin (Young et al., 2001), gave positive responses. Clone F1aa447–596 gave only half the response of clone F3aa501–621 and of full length clone F (Figure 3B), suggesting that it contains only part of the binding site for sAnk1. It therefore seems likely that the binding site on obscurin for sAnk1 is contained in clone F3aa501–621, equivalent to amino acids 6312–6432 of the human obscurin orthologue.
Figure 3.
Regions of sAnk1 and obscurin involved in binding. To identify the minimal sequences of sAnk1 and obscurin required for their association, a series of deletion constructs were generated and expressed in the pLexA-bait and pB42AD-prey vectors respectively. Yeast two-hybrid analysis followed by a β-galactosidase assay in solution indicated that these were confined to a 70 amino acid sequence in sAnk1 (sAnk1aa61–130) and a 120-amino acid segment in obscuring (obscurinaa501–627) that corresponds to amino acid sequences 6312–6432 of the human obscurin homologue.
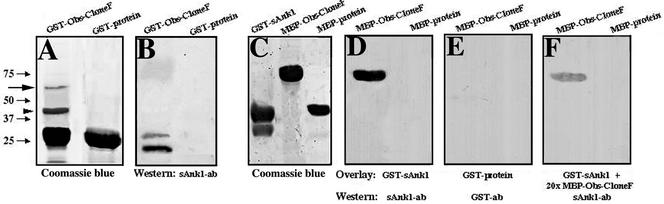
We performed a GST pull down assay to confirm that sAnk1 binds obscurin. For these experiments, we chose to use the original clones identified in the yeast two-hybrid screen, because they were more readily separated from GST alone. When we expressed obscurin clone F as a GST-fusion protein, the mass of the resultant polypeptide was only ∼45 kDa, instead of the expected ∼64 kDa (Figure 4A, arrow), probably because of endogenous bacterial proteases. The truncated band of ∼45 kDa presumably contains GST (25 kDa) and the NH2-terminal ∼20 kDa encoded by clone F (Figure 4A, arrowhead). The truncated, ∼45-kDa GST-obscurin F fusion protein includes the entire sequence that we identified above as containing the minimal binding site for sAnk1 (Figure 3B). When skeletal muscle lysates were incubated with GST-obscurin clone F bound to glutathione matrix, native sAnk1 was efficiently and specifically adsorbed, as detected by Western blotting with antibodies to sAnk1 (Figure 4B). This interaction required obscurin clone F, because GST alone failed to adsorb sAnk1 (Figure 4B).
Figure 4.
Direct, specific binding of sAnk1 to obscurin in vitro. (A) Equivalent amounts of GST-obscurin clone F fusion protein and GST were subjected to SDS-PAGE and stained with Coomassie blue. The GST-obscurin clone F fusion protein yielded two products, the intact protein at ∼64 kDa (arrow) and a major breakdown product at ∼45 kDa (arrowhead). GST-protein is ∼25 kDa. (B) After binding to glutathione matrices, recombinant GST-obscurin clone F, but not GST, adsorbed native sAnk1 from homogenates of quadriceps muscle, as shown by SDS-PAGE and immunoblotting with antibodies to sAnk1. (C) Bacterially expressed GST-sAnk1 (∼39 kDa), MBP-obscurin clone F (∼80 kDa), and MBP-protein (∼42 kDa) were separated by SDS-PAGE and stained with Coomassie blue. (D) Purified GST-sAnk129–155 binds to immobilized MBP-obscurin clone F fusion protein but not to MBP, as shown by blotting with antibodies to sAnk1. (E) No binding to MBP-obscurin or MBP was detected in blots similar to those in D incubated with GST alone and probed with antibodies to GST. (F) The specific binding seen in D is significantly reduced by adding a 20-fold excess of MBP-obscurin clone F fusion protein to the overlay buffer along with recombinant GST-sAnk129–155.
We also performed an overlay assay with sAnk1aa29–155 and obscurin clone F, which were expressed in bacteria as GST- and MBP-fusion proteins (Figure 4C), respectively, to learn if sAnk1 binds obscurin directly. Equivalent amounts of MBP-obscurin clone F and MBP-protein were separated by SDS-PAGE, transferred to nitrocellulose, and incubated with GST-sAnk1 or GST-protein. Recombinant sAnk1 specifically bound to MBP-obscurin-F peptide, but not to MBP-protein, as shown by immunoblotting with antibodies to sAnk1 (Figure 4D). No specific binding was detected when an identical blot was overlaid with GST-protein and subsequently probed with antibodies to GST (Figure 4E). Furthermore, addition of a 20-fold excess of MBP-obscurin-F fusion protein to the overlay buffer significantly reduced binding of GST-sAnk1 (Figure 4F). These results showed that sAnk1 and obscurin bind to each other directly.
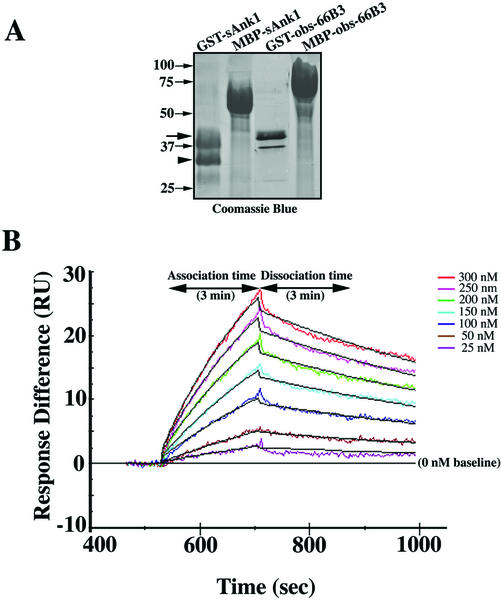
We used a Biacore 3000 surface plasmon resonance biosensor (for review, see Myszka, 1997) to perform kinetic studies of the binding of sAnk1 to obscurin. Recombinant GST-sAnk1aa29–155 (∼39 kDa), MBP-sAnk1aa29–155 (∼57 kDa), GST-obscurin-F3aa501–621 (∼38.5 kDa), and MBP-obscurin-F3aa501–621 (∼66 kDa) were generated (Figure 5A). Two sets of kinetic studies were performed. In the first, GST-obscurin-F3 was used as ligand, with GST alone as control (see MATERIALS AND METHODS). Different concentrations of MBP-sAnk1, ranging from 0 to 300 nM, were flowed over immobilized GST-obscurin-F3 and GST. Real-time binding of MBP-sAnk1 to GST-obscurin-F3 was evaluated after subtraction of nonspecific binding to control GST. The sensograms for the different analyte concentrations were overlaid and aligned to the baseline, established in the absence of MBP-sAnk1 (Figure 5B). An increasing amount of MBP-sAnk1 bound to GST-obscurin-F3 during the injection period (3 min), and binding reversed relatively slowly during the dissociation phase (3 min). The sensogram data were quantified by performing kinetics analysis using BIA evaluation 3.1 software and fitted with the 1:1 Langmuir binding model. Three independent kinetic experiments were performed that yielded similar results (Table 1). The kinetic constants for the “on” (ka) and “off” (kDa) reactions were 1.42 × 104 M−1 s−1 and 1.79 × 10−3 s−1, respectively. An average binding affinity, KD, of 130 nM was determined. χ2 values calculated for these experiments were very low, suggesting that these values provided an excellent fit to the data.
Figure 5.
Real time kinetic analysis of the binding of sAnk1 to obscurin by surface plasmon resonance. (A) Bacterially expressed, affinity purified GST-sAnk1 (∼39 kDa), MBP-sAnk1 (∼57 kDa), GST-obscurin-F3 (∼38.5 kDa), and MBP-obscurin-F3 (∼66 kDa) were fractionated on SDS-PAGE followed by Coomassie blue staining. Note the heterogeneity of the GST-sAnk1 construct, which contains a major breakdown product at ∼32 kDa (arrowhead) along with the intact protein (39 kDa, arrow). (B) A series of binding curves were generated in real time (experiment 1 of Table 1) in which GST-obscurin-F3 was used as ligand and MBP-sAnk1, at concentrations from 0 to 300 nM, as analyte. In this experiment, immobilized GST-obscurin-F3 was present in FC2, and GST in FC1. After correction for nonspecific binding and alignment of the sensograms (see RESULTS), curves of the data, shown in different colors, were fitted with the 1:1 Langmuir binding model, shown in black. The y-axis indicates the resonance units (RU) generated by the binding of MBP-sAnk1 to surface-bound GST-obscurin-F3, with each curve representing a different concentration of analyte. The results are consistent with 1:1 binding of the sAnk1 and obscurin fusion proteins, with an affinity of 130 nM (Table 1).
Table 1.
Kinetics analysis of sAnk1 binding to obscurin by surface plasmon resonance
| Experiment | ka (1/Ms) | kd (1/s) | KA (1/M) | KD (M) | χ2 |
|---|---|---|---|---|---|
| 1 | 1.16e4 | 1.53e-3 | 7.58e6 | 1.32e-7 | 0.15 |
| 2 | 1.13e4 | 1.68e-3 | 6.72e6 | 1.49e-7 | 0.28 |
| 3 | 1.97e4 | 2.16e-3 | 9.16e6 | 1.09e-7 | 0.16 |
| Mean | 1.42e4 | 1.79e-3 | 7.82e6 | 1.3e-7 | |
| 4 | 1.26e4 | 2.37e-3 | 4.95e6 | 2.0e-7 | 0.28 |
In experiments 1–3, we used recombinant GST-obscurin-F3 as ligand and MBP-sAnk1 as analyte. The mean values reported are only for experiments 1–3. In experiment 4, we used GST-sAnk1 as ligand and MBP-obscurin-F3 as analyte. We analyzed all our results with the 1:1 Langmuir binding model to calculate the kinetic and binding constants reported. The χ2 value for each experiment indicates that this model provides an excellent fit to the data.
We performed a similar set of kinetic studies using GST-sAnk1 as ligand and MBP-obscurin-F3 as analyte. Our results (unpublished data) could be fit with the 1:1 Langmuir binding model with a KD of 200 nM (Table 1). Although we obtained a better fit of these data with the Heterogeneous Ligand model, consistent with the presence of two subpopulations of recombinant GST-sAnk1 in our fusion protein preparation (Figure 5A, lane 1), the low χ2 value obtained with the 1:1 Langmuir analysis suggests that most of the GST-sAnk1 remains intact and active during the biosensor assay. Thus, these studies collectively demonstrated a dissociation equilibrium constant (KD) for the binding of sAnk1 to obscurin in the middle nM range.
We used the GST-obscurin clone B fusion protein as an antigen to generate polyclonal antibodies in rabbits. Specific antibodies were affinity-purified and used to characterize obscurin in muscle homogenates. Quadriceps skeletal muscle was homogenized with nondenaturing detergents and the soluble and insoluble material was analyzed by SDS-PAGE and immunoblotting. Small amounts of obscurin were present in the DS fraction, but an ∼800-kDa immunoreactive band was readily and specifically detected in the DI pellet (Figure 6A). Unlike obscurin, sAnk1 was enriched in the DS fraction, with only trace amounts in the DI fraction (Figure 6B). These results are consistent with a sarcomeric location for obscurin as well as with the known concentration of sAnk1 in the network SR (Zhou et al., 1997).
Figure 6.
Obscurin is present in the detergent insoluble fraction of muscle lysates. (A) Western blotting with antibodies to obscurin clone B shows that obscurin is primarily in the detergent insoluble (DI) pellet of adult skeletal muscle lysates (arrow). Smaller amounts of obscurin are detected in the detergent soluble (DS) fractions. (B) Unlike obscurin, sAnk1 (arrowheads) is enriched in the DS fraction.
The insolubility of obscurin in muscle homogenates made studies of its association with sAnk1 in solution difficult. We therefore used immunofluorescence combined with confocal microscopy to compare the subcellular distributions of obscurin and sAnk1. Frozen sections of adult quadriceps were labeled with antibodies to obscurin together with antibodies to α-actinin, to mark Z-lines. Obscurin was present in striations in register with Z-lines as well as midway in between at M-lines (Figure 7, A–C). In cross sections, obscurin assumed a reticular distribution (Figure 7D). Both patterns were identical to those earlier reported for sAnk1 (Zhou et al., 1997).
Figure 7.
The subcellular distribution of obscurin in skeletal and cardiac myofibers. (A–C) Obscurin (A) is present in striations in longitudinal sections of quadriceps myofibers. Lines correspond to Z-disks (arrows) and M-lines (arrowheads), as shown by double immunostaining with antibodies to the Z-disk marker α-actinin (B). In the color overlay (C), obscurin is in red, α-actinin in green, and areas with both proteins in yellow. (D) Labeling for obscurin shows a reticular pattern in cross sections of quadriceps skeletal muscle. (E) Prominent staining of obscurin occurs at M-lines (arrowhead) in adult myocardium, with weaker labeling at Z-disks (arrow), marked by anti–α-actinin. Colors are as in panel C. (F) Like obscurin, sAnk1 in cardiac muscle concentrates over M-lines (arrowhead) and to a lesser extent around Z-disks (arrow), stained with anti–α-actinin. In (F), sAnk1 is shown in red and α-actinin in green. (G) No labeling was detected when nonimmune rabbit IgG was used. H. Preabsorption of the obscurin antibody with its antigen eliminated the labeling observed in panel A.
We also examined the subcellular localization of obscurin and sAnk1 in adult myocardium. In ventricular muscle, both obscurin and sAnk1 are primarily at M-lines and to a lesser extent at Z-disks (Figure 7, E and F). These observations are consistent with the distribution of the network SR in cardiac muscle, which, unlike skeletal muscle, is more prevalent at the level of the M-line. No labeling was detected when primary antibodies were either replaced by nonimmune rabbit IgG (Figure 7G) or preadsorbed with their respective antigens (Figure 7H).
DISCUSSION
As part of an effort to understand how the SR becomes aligned with the myofibrils of striated muscle, we used the yeast two-hybrid screen to identify ligands of sAnk1. sAnk1 is enriched in the network SR, to which it is anchored by its NH2-terminal, hydrophobic head (Zhou et al., 1997; our unpublished studies). Its COOH-terminal sequence extends into the cytoplasm, where it may interact with other proteins. Here we show that the major ligand of the cytoplasmic domain of sAnk1 is the COOH-terminal region of obscurin, a giant protein of striated muscle that concentrates at the level of the Z-disks and M-lines (Young et al., 2001). Studies in vitro indicate that the binding of sAnk1 to obscurin is direct and specific. Kinetic analysis of the interaction of sAnk1 to obscurin in real time yielded a binding affinity of 130 nM, consistent with strong but reversible binding between the hydrophilic tail of sAnk1 and the COOH-terminus of obscurin. Furthermore, obscurin and sAnk1 have similar distributions in striated muscle, consistent with an association of these two proteins in vivo. Notably, in skeletal muscle both sAnk1 and obscurin are present at the level of the Z-disk and the M-line, consistent with the distribution of the network SR (Franzini-Armstrong, 1994). Our results are the first to demonstrate an interaction between proteins of the SR and the contractile apparatus. They are also unique in other respects, as they demonstrate previously unknown properties of both the ankyrin and the titin superfamilies of proteins.
The C-terminal portion of the large, canonical forms of ankyrin (see Introduction) has been referred to as the “regulatory domain” of the protein, because its phosphorylation alters the affinity of the spectrin- and membrane-binding domains of ankyrin for their respective ligands (Bennett, 1992). Although it is clear that this region must bind at least transiently to kinases and phosphatases, a ligand for the C-terminal sequence of ankyrin has not yet been identified. Here we show that the C-terminal region of sAnk1, between amino acids 61 and 130, has binding activity for obscurin. This binding site contains sequence that is shared with the C-terminal region of the large, canonical ankyrin 1 (sAnk1aa74–130) as well as sequence that is unique to sAnk1 (sAnk1aa61–73). It remains to be determined if the binding of sAnk1 to obscurin is mediated by the sequence shared with the large canonical ankyrin 1 or by the unique sequence, or if both must be present to form the binding site.
Obscurin resembles two other giant proteins of striated muscle, titin and nebulin (Wang, 1996; Gregorio et al., 1999). Like those proteins, obscurin contains multiple Ig domains and it is tightly associated with the contractile apparatus, as indicated by its limited solubility in nondenaturing detergents (Isaacs et al., 1989; Granzier and Wang, 1993). It therefore seems probable that, like titin and nebulin, obscurin binds to one or more proteins of the contractile apparatus with high affinity. Consistent with this, obscurin binds titin, though in a region of that protein that does not precisely coincide with the Z-disk or M-line (Young et al., 2001). In addition to its ability to bind titin, our results clearly show that obscurin also binds sAnk1. It is the first member of the titin superfamily to be shown to bind to an integral membrane protein.
The antibodies to obscurin that we have generated to date are to its C-terminal, sAnk1-binding region. This raises some questions about the isoforms of obscurin we have studied and also about its possible location and orientation in the sarcomere. The C-terminal sequence is present in the ∼800-kDa product of the obscurin gene and in some of its smaller, alternatively spliced products, but not in those products that contain COOH-terminal ser/thr kinase domains (Russel et al., 2002). Although our antibodies recognize some smaller proteins in adult skeletal muscle (our unpublished observations), most of the obscurin we detect is ∼800 kDa. Thus, obscurin at Z-disks and M-lines is likely to be ∼800 kDa in mass.
As this giant form of obscurin is predicted to be 208 nm in length (Young et al., 2001), its positioning within the sarcomere may be difficult to determine from immunolabeling with antibodies to its C terminus alone. For example, obscurin also may be present in the interior of the sarcomere, where its COOH-terminal epitopes could be inaccessible. Alternatively, obscurin may be concentrated at many points along the surface of the sarcomere in a longitudinal orientation, with its COOH-terminal region exposed and accessible to antibodies only at the peripheries of the Z-disk and M-line. Finally, obscurin may be completely restricted to the Z-disk and M-line, either with its NH2-terminal region buried in the interior of those structures and its COOH-terminal sequence exposed at the surface and accessible to sAnk1, or with both NH2- and COOH-terminal regions at the periphery. The latter possibility, which is consistent with results from other laboratories that used antibodies to more N-terminal epitopes to label obscurin at Z-lines and M-lines (Young et al., 2001; Bang et al., 2001), suggests that obscurin surrounds the myofibril at the ends and in the middle of each sarcomere.
If this is correct, obscurin also differs from titin and nebulin in its positioning with respect to the contractile apparatus. Titin and nebulin are both found in the interior of the sarcomere, where they extend longitudinally, from the Z-disk to the M-line and along the thin filaments, respectively (Ojima et al., 2000; Trombitas et al., 2000). Titin and nebulin play important roles in determining the longitudinal dimensions of these sarcomeric elements (Nwe et al., 1999; Nwe and Shimada, 2000; McElhinny et al., 2001; Sutko et al., 2001; Granzier and Labeit, 2002; Watanabe et al., 2002), but a similar “molecular ruler” to determine the diameter of the sarcomere has not yet been identified. As the first member of the titin superfamily to associate with the sarcomere only at its periphery, obscurin may serve this function.
Our results demonstrate that sAnk1, a protein of the SR, binds to obscurin, a protein that associates preferentially with the contractile apparatus of striated muscle. This binding interaction is probably limited to muscle, however. Although sAnk1 is also expressed in the brain (White et al., 1992), the 800-kDa form of obscurin is expressed only in skeletal and cardiac tissue (Young et al., 2001) and so is unlikely to be involved in linking the endoplasmic reticulum (ER) to the cytoskeleton in other cells. Studies from several in vitro systems have documented that the cytoskeleton contributes to dynamic changes in the ER but is not necessary for its organization or maintenance (Baumann and Walz, 2001; Voeltz et al., 2002). Indeed, the ER in nonmuscle cells appears to be only weakly anchored to the cytoskeleton, as it continuously changes its shape and subcellular location (Koch and Booth, 1988; Yang et al., 1997; Voeltz et al., 2002).
The links between the SR and the contractile apparatus in muscle cells are likely to be much firmer than the links between the ER and the cytoskeleton in nonmuscle cells, because the orientation of the SR with respect to the contractile elements is invariant in each type of striated fiber. Our results suggest that the binding of sAnk1 to obscurin in vivo is in part responsible for this orientation (Figure 8). For example, the C-terminal region of obscurin is sited appropriately to bind to sAnk1 in the network SR. Similarly, sAnk1 is located and oriented in the SR membrane to permit its C-terminal domain to interact with obscurin. These two molecules bind to each other with high affinity and are enriched in the same regions of skeletal and cardiac muscle. Given their close association, we propose that the binding of sAnk1 to obscurin contributes to the regular alignment of the network SR with the Z-disks and M-lines of striated muscle. Experiments to test this model are now in progress.
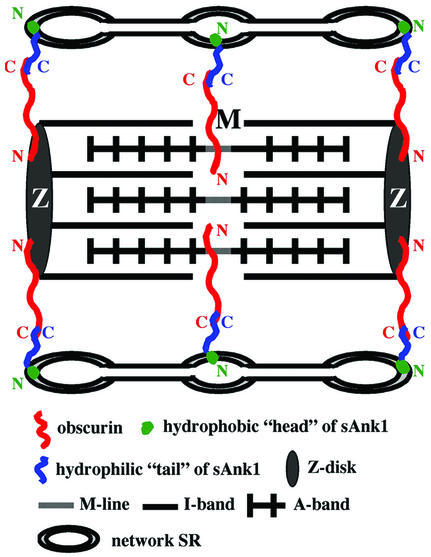
Figure 8.
Model of sAnk1 association with obscurin. sAnk1 is anchored to the network SR, around Z-disks and M-lines, through its hydrophobic NH2-terminus, with its hydrophilic COOH-terminal portion facing the myoplasm. Obscurin appears to surround the myofibrils at Z-disks and M-lines, with its COOH-terminus exposed to allow it to bind to sAnk1. Thus, sAnk1 binding to obscurin may contribute to the sarcomeric alignment of the network SR.
ACKNOWLEDGMENTS
We thank D.C. Catino, W.G. Resneck, and A. O'Neill for their expert assistance. This work was supported by a grant from the National Institutes of Health to R.J.B. (RO1 HL 64304) and by fellowships to A.K.-K. and E.J. from T32 AR 07293 (Dr. M. Schneider, P.I.).
Abbreviations used:
- β-ME
β-mercaptoethanol
- DI
detergent-insoluble
- DS
detergent-soluble
- ER
endoplasmic reticulum
- FC
flow cell
- GST
glutathione-S-transferase
- MBP
maltose binding protein
- sAnk1
small ankyrin 1
- RU
resonance units
- SR
sarcoplasmic reticulum
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–07–0411. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–07–0411.
The accession number for the partial rat obscurin sequence is AY167411.
REFERENCES
- Bang ML, et al. The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ Res. 2001;89:1065–1072. doi: 10.1161/hh2301.100981. [DOI] [PubMed] [Google Scholar]
- Baumann O, Walz B. Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int Rev Cytol. 2001;205:149–214. doi: 10.1016/s0074-7696(01)05004-5. [DOI] [PubMed] [Google Scholar]
- Bennett V. Ankyrins. Adaptors between diverse plasma membrane proteins and the cytoplasm. J Biol Chem. 1992;267:8703–8706. [PubMed] [Google Scholar]
- Bennett V, Chen L. Ankyrins and cellular targeting of diverse membrane proteins to physiological sites. Curr Opin Cell Biol. 2001;13:61–67. doi: 10.1016/s0955-0674(00)00175-7. [DOI] [PubMed] [Google Scholar]
- Birkenmeier CS, White RA, Peters LL, Hall EJ, Lux SE, Barker JE. Complex patterns of sequence variation and multiple 5′ and 3′ ends are found among transcripts of the erythroid ankyrin gene. J Biol Chem. 1993;81:2144–2149. [PubMed] [Google Scholar]
- Birkenmeier CS, Sharp JJ, Gifford EJ, Deveau SA, Barker JE. An alternative first exon in the distal end of the erythroid ankyrin gene leads to production of a small isoform containing an NH2-terminal membrane anchor. Genomics. 1998;50:79–88. doi: 10.1006/geno.1998.5305. [DOI] [PubMed] [Google Scholar]
- De Matteis MA, Morrow JS. The role of ankyrin and spectrin in membrane transport and domain formation. Curr Opin Cell Biol. 1998;10:542–549. doi: 10.1016/s0955-0674(98)80071-9. [DOI] [PubMed] [Google Scholar]
- Devarajan P, Stabach PR, Mann AS, Ardito T, Kashgarian M, Morrow JS. Identification of a small cytoplasmic ankyrin (AnkG119) in the kidney and muscle that binds beta I sigma spectrin and associates with the Golgi apparatus. J Cell Biol. 1996;133:819–830. doi: 10.1083/jcb.133.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flucher BE. Structural analysis of muscle development: transverse tubules, sarcoplasmic reticulum, and the triad. Dev Biol. 1992;154:245–260. doi: 10.1016/0012-1606(92)90065-o. [DOI] [PubMed] [Google Scholar]
- Flucher BE, Franzini-Armstrong C. Formation of junctions involved in excitation-contraction coupling in skeletal and cardiac muscle. Proc Natl Acad Sci USA. 1996;93:8101–8106. doi: 10.1073/pnas.93.15.8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzini-Armstrong C, Peachey LD. Striated muscle-contractile and control mechanisms. J Cell Biol. 1981;91:166–186. doi: 10.1083/jcb.91.3.166s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzini-Armstrong C. The sarcoplasmic reticulum and the transverse tubules. In: Engel AG, Franzini-Armstrong C, editors. Myology. New York: McGraw-Hill, Inc.; 1994. pp. 176–199. [Google Scholar]
- Franzini-Armstrong C. The sarcoplasmic reticulum and the control of muscle contraction. FASEB J. 1999;13(Suppl.2):S266–S270. doi: 10.1096/fasebj.13.9002.s266. [DOI] [PubMed] [Google Scholar]
- Gagelin C, Bruno C, Deprette C, Ludosky MA, Recouvreur M, Cartaud J, Cognard J, Raymond G, Kordeli E. Identification of Ank(G107), a muscle-specific ankyrin-G isoform. J Biol Chem. 2002;277:12978–12987. doi: 10.1074/jbc.M111299200. [DOI] [PubMed] [Google Scholar]
- Gallagher PG, Tse WT, Scarpa AL, Lux SE, Forget BG. Structure and organization of the human ankyrin-1 gene. Basis for complexity of pre-mRNA processing. J Biol Chem. 1997;272:19220–19228. doi: 10.1074/jbc.272.31.19220. [DOI] [PubMed] [Google Scholar]
- Gallagher PG, Forget BG. An alternate promoter directs expression of a truncated, muscle-specific isoform of the human ankyrin 1 gene. J Biol Chem. 1998;273:1339–1348. doi: 10.1074/jbc.273.3.1339. [DOI] [PubMed] [Google Scholar]
- Granzier HL, Wang K. Gel electrophoresis of giant proteins: solubilization and silver-staining of titin and nebulin from single muscle fiber segments. Electrophoresis. 1993;14:56–64. doi: 10.1002/elps.1150140110. [DOI] [PubMed] [Google Scholar]
- Granzier H, Labeit S. Cardiac titin: an adjustable multi-functional spring. J Physiol. 2002;541(Pt 2):335–342. doi: 10.1113/jphysiol.2001.014381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorio CC, Granzier HL, Sorimachi H, Labeit S. Muscle assembly: a titanic achievement? Curr Opin Cell Biol. 1999;11:18–25. doi: 10.1016/s0955-0674(99)80003-9. [DOI] [PubMed] [Google Scholar]
- Hoock TC, Peters LL, Lux SE. Isoforms of ankyrin-3 that lack the NH2-terminal repeats associate with mouse macrophage lysosomes. J Cell Biol. 1997;136:1059–1070. doi: 10.1083/jcb.136.5.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs WB, Kim IS, Struve A, Fulton AB. Biosynthesis of titin in cultured skeletal muscle cells. J Cell Biol. 1989;109:2189–2195. doi: 10.1083/jcb.109.5.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch GLE, Booth C. Dissociation and re-assembly of the endoplasmic reticulum in live cells. J Cell Sci. 1988;91:511–522. doi: 10.1242/jcs.91.4.511. [DOI] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A, Huang S-C, Benz EJ., Jr A nonerythroid isoform of protein 4.1R interacts with components of the contractile apparatus in skeletal myofibers. Mol Biol Cell. 2000;11:3805–3817. doi: 10.1091/mbc.11.11.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordeli E, Bennett V. Distinct ankyrin isoforms at neuron cell bodies and nodes of Ranvier resolved using erythrocyte ankyrin-deficient mice. J Cell Biol. 1991;114:1243–1259. doi: 10.1083/jcb.114.6.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordeli E, Lambert S, Bennett V. AnkyrinG. A new ankyrin gene with neural-specific isoforms localized at the axonal initial segment and node of Ranvier. J Biol Chem. 1995;270:2352–2359. doi: 10.1074/jbc.270.5.2352. [DOI] [PubMed] [Google Scholar]
- Kunimoto M. A neuron-specific isoform of brain ankyrin, 440-kD ankyrinB, is targeted to the axons of rat cerebellar neurons. J Cell Biol. 1995;131:1821–1829. doi: 10.1083/jcb.131.6.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S, Yu H, Prchal JT, Lawler J, Ruff P, Speicher D, Cheung MC, Kan YW, Palek J. cDNA sequence for human erythrocyte ankyrin. Proc Natl Acad Sci USA. 1990;87:1730–1734. doi: 10.1073/pnas.87.5.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong P, MacLennan DH. Complex interactions between skeletal muscle ryanodine receptor and dihydropyridine receptor proteins. Biochem Cell Biol. 1998;76:681–694. doi: 10.1139/bcb-76-5-681. [DOI] [PubMed] [Google Scholar]
- Lux SE, John KM, Bennett V. Analysis of cDNA for human erythrocyte ankyrin indicates a repeated structure with homology to tissue-differentiation and cell-cycle control proteins. Nature. 1990;344:36–42. doi: 10.1038/344036a0. [DOI] [PubMed] [Google Scholar]
- Myszka DG. Kinetic analysis of macromolecular interactions using surface plasmon resonance biosensors. Curr Opin Biotechnol. 1997;8:50–57. doi: 10.1016/s0958-1669(97)80157-7. [DOI] [PubMed] [Google Scholar]
- McElhinny AS, Kolmerer B, Fowler VM, Labeit S, Gregorio CC. The N-terminal end of nebulin interacts with tropomodulin at the pointed ends of the thin filaments. J Biol Chem. 2001;276:583–592. doi: 10.1074/jbc.M005693200. [DOI] [PubMed] [Google Scholar]
- Mohler PJ, Gramolini AO, Bennett V. Ankyrins. J Cell Sci. 2002;115:1565–1566. doi: 10.1242/jcs.115.8.1565. [DOI] [PubMed] [Google Scholar]
- Nwe TM, Maruyama K, Shimada Y. Relation of nebulin and connectin (titin) to dynamics of actin in nascent myofibrils of cultured skeletal muscle cells. Exp Cell Res. 1999;252:33–40. doi: 10.1006/excr.1999.4611. [DOI] [PubMed] [Google Scholar]
- Nwe TM, Shimada Y. Inhibition of nebulin and connectin (titin) for assembly of actin filaments during myofibrillogenesis. Tissue Cell. 2000;32:223–227. doi: 10.1054/tice.2000.0109. [DOI] [PubMed] [Google Scholar]
- Ojima K, et al. Identification, tissue expression and chromosomal localization of human Obscurin-MLCK, a member of the titin and Dbl families of myosin light chain kinases. Gene. 2002;282:237–46. doi: 10.1016/s0378-1119(01)00795-8. [DOI] [PubMed] [Google Scholar]
- Otto E, Kunimoto M, McLaughlin T, Bennett V. Isolation and characterization of cDNAs encoding human brain ankyrins reveal a family of alternatively spliced genes. J Cell Biol. 1991;114:241–253. doi: 10.1083/jcb.114.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters LL, John KM, Lu FM, Eicher EM, Higgins A, Yialamas M, Turtzo LC, Otsuka AJ, Lux SE. Ank3 (epithelial ankyrin), a widely distributed new member of the ankyrin gene family and the major ankyrin in kidney, is expressed in alternatively spliced forms, including forms that lack the repeat domain. J Cell Biol. 1995;130:313–330. doi: 10.1083/jcb.130.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov AM, Lopina OD. Ankyrins. FEBS Lett. 2000;482:1–5. doi: 10.1016/s0014-5793(00)01924-4. [DOI] [PubMed] [Google Scholar]
- Russell MW, Raeker MO, Korytkowski KA, Sonneman KI. Identification, tissue expression, and chromosomal localization of human obscurin-MLCK, a member of the titin and Dbl families of myosin light-chain kinases. Gene. 2002;282:237–246. doi: 10.1016/s0378-1119(01)00795-8. [DOI] [PubMed] [Google Scholar]
- Sedgwick SG, Smerdon SJ. The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem Sci. 1999;24:311–316. doi: 10.1016/s0968-0004(99)01426-7. [DOI] [PubMed] [Google Scholar]
- Sutko JL, Publicover NG, Moss RL. Titin: an elastic link between length and active force production in myocardium. Circulation. 2001;104:1639–1645. [PubMed] [Google Scholar]
- Trombitas K, Freiburg A, Greaser M, Labeit S, Granzier HL. From connecting filaments to co-expression of titin isoforms. Adv Exp Med Biol. 2000;481:405–418. doi: 10.1007/978-1-4615-4267-4_24. [DOI] [PubMed] [Google Scholar]
- Tuvia S, Buhusi M, Davis L, Reedy M, Bennett V. Ankyrin-B is required for intracellular sorting of structurally diverse Ca2+ homeostasis proteins. J Cell Biol. 1999;147:995–1008. doi: 10.1083/jcb.147.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Rolls MM, Rapoport TA. Structural organization of the endoplasmic reticulum. EMBO Rep. 2002;3:944–950. doi: 10.1093/embo-reports/kvf202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. Titin/connectin and nebulin: giant protein rulers of muscle structure and function. Adv Biophys. 1996;33:123–134. [PubMed] [Google Scholar]
- Watanabe K, Nair P, Labeit D, Kellermayer MS, Greaser M, Labeit S, Granzier H. Molecular mechanics of cardiac titin's PEVK and N2B spring elements. J Biol Chem. 2002;277:11549–11558. doi: 10.1074/jbc.M200356200. [DOI] [PubMed] [Google Scholar]
- White RA, Birkenmeier CS, Peters LL, Barker JE, Lux SE. Murine erythrocyte ankyrin cDNA: highly conserved regions of the regulatory domain. Mammal Genome. 1992;3:281–285. doi: 10.1007/BF00292156. [DOI] [PubMed] [Google Scholar]
- Williams MW, Resneck WG, Kaysser T, Ursitti JA, Birkenmeier CS, Barker JE, Bloch RJ. Na,K-ATPase in skeletal muscle: two populations of beta-spectrin control localization in the sarcolemma but not partitioning between the sarcolemma and the transverse tubules. J Cell Sci. 2001;114:751–762. doi: 10.1242/jcs.114.4.751. [DOI] [PubMed] [Google Scholar]
- Yang L, Guan T, Gerace L. Integral membrane proteins of the nuclear envelope are dispersed throughout the endoplasmic reticulum during mitosis. J Cell Biol. 1997;137:1199–1210. doi: 10.1083/jcb.137.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P, Ehler E, Gautel M. Obscurin, a giant sarcomeric Rho guanine nucleotide exchange factor protein involved in sarcomere assembly. J Cell Biol. 2001;154:123–136. doi: 10.1083/jcb.200102110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bennett V. Restriction of 480/270-kD Ankyrin G to axon proximal segments requires multiple ankyrin G-specific domains. J Cell Biol. 1998;142:1571–1581. doi: 10.1083/jcb.142.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Birkenmeier CS, Williams MW, Sharp JJ, Barker JE, Bloch RJ. Small, membrane-bound, alternatively spliced forms of ankyrin 1 associated with the sarcoplasmic reticulum of mammalian skeletal muscle. J Cell Biol. 1997;136:621–631. doi: 10.1083/jcb.136.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]