Abstract
The down-regulation of the high-molecular-weight isoforms of tropomyosin (TM) is considered to be an essential event in cellular transformation. In ras-transformed fibroblasts, the suppression of TM is dependent on the activity of the Raf-1 kinase; however, the requirement for other downstream effectors of Ras, such as MEK and ERK, is less clear. In this study, we have utilized the mitogen-activated protein kinase scaffolding protein Kinase Suppressor of Ras (KSR) to further investigate the regulation of TM and to clarify the importance of MEK/ERK signaling in this process. Here, we report that overexpression of wild-type KSR1 in ras-transformed fibroblasts restores TM expression and induces cell flattening and stress fiber formation. Moreover, we find that the transcriptional activity of a TM-α promoter is decreased in ras-transformed cells and that the restoration of TM by KSR1 coincides with increased transcription from this promoter. Although ERK activity was suppressed in cells overexpressing KSR1, ERK inhibition alone was insufficient to upregulate TM expression. The KSR1-mediated effects on stress fiber formation and TM transcription required the activity of the ROCK kinase, because these effects could be suppressed by the ROCK inhibitor, Y27632. Overexpression of KSR1 did not directly regulate ROCK activity, but did permit the recoupling of ROCK to the actin polymerization machinery. Finally, all of the KSR1-induced effects were mediated by the C-terminal domain of KSR1 and were dependent on the KSR-MEK interaction.
Malignant transformation results in the down-regulation of the high-molecular-weight isoforms of the actin-binding protein tropomyosin (TM-1, -2, and -3) (23, 39). This loss in TM expression is not an artifact due to prolonged cell culture or deliberate transformation, given that TM levels are also reduced in freshly isolated human tumor tissues (1, 2, 7, 13, 14). Several lines of evidence indicate that the suppression of TM is essential for transformation. In particular, down-regulation of TM-1 by the expression of a TM-1 antisense construct is sufficient to induce anchorage independence in immortal cells (8). Moreover, the reintroduction of TM-1 or TM-2 into ras- or src-transformed cells reduces or eliminates anchorage-independent growth and tumorigenicity (15, 22, 44, 45, 60). Consequently, the TM isoforms TM-1 and -2 can be classified as class II tumor suppressors (27).
Despite the apparent importance of TM down-modulation in tumorigenesis, the precise mechanisms and proteins involved in regulating TM expression have not been fully elucidated. For example, although TM mRNA levels are reduced in tumor cells (11, 19), it is not known whether this reduction is due to decreased transcriptional activity or mRNA instability. In addition, studies have suggested a role for the Ras-Raf-MEK-mitogen-activated protein kinase (MAPK) cascade in this process; however, the exact contribution of the individual pathway components is not clear. Activated alleles of both Ras and Raf can induce the down-regulation of high-molecular-weight TM proteins in NIH 3T3 cells, and the Raf-1 kinase is required for TM suppression in ras-transformed fibroblasts (23). Surprisingly, however, pharmocological inhibition of MEK, the well-characterized effector of Raf-1, is insufficient to restore TM expression in either ras- or raf-transformed fibroblasts (23). Likewise, MEK inhibition does not restore the expression of human TM-1 in transformed MDA-MB-231 breast cancer cells (51). While these findings would suggest that MEK activity is not required for TM down-modulation, a role for MEK has been inferred from studies examining other transformed cell types. In c-jun-transformed FR3T3 cells, TM levels are down-regulated through an autocrine pathway that is disrupted by MEK inhibitor treatment (31). MEK inhibition has also been shown to induce the re-expression of one or more TM isoforms in ras-transformed RIE and NRK cells (40, 53). Adding further confusion regarding the involvement of MEK in TM regulation is the finding that in ras-transformed NIH 3T3 fibroblasts, treatment with low concentrations of radicicol, the HSP90-binding antibiotic, is able to fully restore TM-2 expression without inhibiting MEK/ERK signaling (25). In addition, although pharmocological inhibition of MEK does not upregulate TM levels in ras-transformed fibroblasts, overexpression of a dominant-negative form of MEK1 does restore TM expression (23). Given that the dominant-negative MEK protein is not as efficient an inhibitor of ERK phosphorylation as the pharmocological MEK inhibitors, the effect of the dominant-negative mutant cannot be attributed simply to a block in ERK activity (23). Thus, while MEK may play a role in TM regulation, its contribution to this process may be more complex than originally anticipated.
In addition to Ras, Raf, MEK, and MAPK, another component of this important signaling cascade is the protein Kinase Suppressor of Ras (KSR). KSR1 was initially discovered to be a regulator of Ras-mediated signaling by genetic screens performed in Drosophila melanogaster and Caenorhabditis elegans (59, 61). More recently, KSR1 has been shown to function as a scaffolding protein that enables Raf activation and facilitates signal transmission from Raf-1 to MEK and ERK (4, 35, 37, 38, 48, 55). As would be expected for a scaffolding protein, the effect of KSR1 on Ras-mediated signaling varies significantly with the level of KSR1 protein expressed. At low levels of expression, KSR1 enhances Ras signaling, while at high levels of expression, KSR1 suppresses many Ras-mediated events, including focus formation in mammalian fibroblasts, germinal vesicle breakdown in Xenopus oocytes, and R7 photoreceptor formation in the Drosophila eye (10, 12, 21, 24). This biphasic response is consistent with the theoretical model for a scaffolding protein, which predicts that the scaffold must be in stoichiometric equilibrium with its binding partners (30). When all components of the signaling complex are expressed at equivalent levels, a scaffold will enhance the efficiency and specificity of signaling. However, high overexpression of the scaffold will lead to a separation of the individual components, thus preventing their interaction and signal transmission.
Among the proteins with which KSR1 has been shown to interact are Raf-1, MEK, and MAPK, as well as 14-3-3 proteins, G protein-βγ, heat shock protein 70 (Hsp70), Hsp90, cdc37, and C-TAK1 (6, 10, 37, 56, 63). In particular, the interaction between KSR1 and MEK appears to be crucial for KSR1 function. MEK constitutively associates with the C-terminal region of KSR1, and all genetically identified loss-of-function mutations mapping to the KSR1 C-terminal domain have been found to disrupt MEK binding (36, 48, 56). At least one important consequence of the KSR-MEK interaction is the ability of KSR1 to transport MEK from the cytoplasm to the plasma membrane, thus localizing MEK with its upstream activator Raf-1 and downstream effector ERK (37). The translocation of the KSR1 complex to the cell surface occurs in response to signaling events and is mediated by the KSR1 cysteine-rich C1 domain (66). Interestingly, KSR1 has also been shown to shuttle through the nucleus in a manner that is dependent on its interaction with MEK (9). Whether KSR1 performs any function in the nucleus and whether this is another critical aspect of the KSR-MEK interaction are currently unknown. Moreover, the effects of KSR1 on gene expression and other cellular properties have not been previously addressed.
In this report, we have utilized the MAPK scaffold KSR1 to gain further insight into the mechanisms regulating TM expression in ras-transformed cells. Here, we find that overexpression of KSR1 reverts the ras-transformed phenotype and restores TM expression. All the effects of KSR1 are dependent on the binding of MEK to KSR1 and require the activity of the ROCK kinase.
MATERIALS AND METHODS
Reagents.
TM311, a monoclonal antibody that recognizes a common epitope in the first exon of TM-1, TM-2, TM-3, and TM-6 (41, 54) and anti-myosin light chain (MLC) antibody (clone MY-21) were purchased from Sigma (St. Louis, Mo.). Anti-phospho-Erk and anti-phospho-MEK antibodies and the MEK inhibitor PD98059 were obtained from Cell Signaling Technology (Beverly, Mass.). Anti-Erk-1 antibody (clone K-23) was purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.), anti-phospho-MLC antibody (pp2b) was a generous gift from F. Matsumura (Rutgers University, New Brunswick, N.J.) (34), and Y27632 was obtained from either Biomol (Plymouth Meeting, Pa.) or Calbiochem (La Jolla, Calif.).
Cell culture.
NIH 3T3 and v-Ki-ras-transformed NIH 3T3 cells (Larry Feig, Tufts University, Boston, Mass.) were cultured at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (DMEM; Mediatech, Herndon, Va.) supplemented with 10% donor calf serum (DCS; BioWhittaker, Walkersville, Md.) and 1 μg of gentamicin per ml (InVitrogen, Carlsbad, Calif.).
Generation of recombinant adenoviruses.
The generation of adenoviruses expressing the various KSR1 constructs has been described previously (36). All KSR1 clones were constructed to contain two copies of a polyomavirus-derived epitope tag (Pyo; amino acids MEYMPME) at the amino terminus. DNA sequences were inserted into the pAdTrack-CMV (cytomegalovirus) vector coexpressing the green fluorescent protein (GFP) for the generation of recombinant adenoviruses according to the procedures of He et al. (17). Viruses were purified over CsCl2 gradients and dialyzed against a phosphate-buffered saline (PBS)-glycerol solution. Viral titers were determined by serial dilution, and ras-transformed cells were infected with the minimal virus titer required to achieve ∼100% infectivity as determined by GFP expression.
Infection of cells.
Cells were seeded in 24-well plates at a density of 5 × 104 cells per well in DMEM with 10% DCS. The following day, the medium was aspirated, and cells were incubated with 200 μl of Optimem (InVitrogen) containing 0.4 μl of Lipofectamine (InVitrogen) and 0.2 to 0.5 μl of adenovirus. After 4 h of incubation, the infection medium was removed, and 1 ml of DMEM with 10% DCS was added. For experiments performed in six-well plates, all parameters were multiplied by 5. Cells were photographed with a Nikon Coolpix 990 digital camera mounted on a Nikon Eclipse TS100 inverted microscope with an epifluorescence attachment.
Immunofluorescence.
Cells were grown in 24-well plates or chamber slides. At the appropriate time after adenoviral infection, cells were fixed for 10 min with a solution of 4% paraformaldehyde in PBS (pH 7.4). The cells were then permeabilized with 0.2% Triton X-100 in PBS for 5 min and subsequently blocked with 0.5% bovine serum albumin in PBS for 1 h. To identify actin stress fibers, samples were incubated with rhodamine-conjugated phalloidin (Sigma).
Cell lysis and Western blotting.
For preparation of whole-cell extracts, cells were washed twice in cold PBS and then scraped into sodium dodecyl sulfate (SDS) cell lysis buffer (62.5 mM Tris [pH 6.8], 1% SDS) containing protease and phosphatase inhibitors (10 μg of leupeptin per ml, 0.1 U of aprotinin per ml, 2 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, 10 mM sodium fluoride). The samples were passed three to five times through a 25-gauge needle to reduce viscosity. Protein concentrations were determined by the bicinchoninic acid assay (Pierce, Rockford, Ill.). Equal amounts of protein were separated by electrophoresis on Nupage Bis-Tris-polyacrylamide gels (InVitrogen) and subsequently blotted onto nitrocellulose membranes. Equal loading of the samples was assessed by staining the blots with a 0.1% Ponceau S-5% acetic acid solution (Sigma). The membranes were probed with antibodies directed against the relevant proteins followed by goat anti-mouse or goat anti-rabbit antibodies conjugated to horseradish peroxidase (Pierce). Proteins were visualized by chemiluminescence with SuperSignal as substrate (Pierce). Blots were exposed to Kodak X-Omat autoradiography film.
Reporter constructs.
The partial sequence of the rat TM-α promoter was a generous gift from N. Ruiz-Opazo (Boston University, Boston, Mass.) (20). A stretch of 1,286 bp of the promoter plus the 5′ untranslated region of the TM-α gene (GenBank accession no. J05467; bases 344 to 1629) was subcloned into pGL3 (Promega) upstream of the gene encoding firefly luciferase. pFA2-Elk1 transactivator plasmid (Stratagene, La Jolla, Calif.) contains the yeast GAL4 DNA binding domain (DBD; residues 1 to 147) fused with the DNA activation domain of Elk-1 (residues 307 to 427). pFR-Luc reporter plasmid (Stratagene) contains a synthetic promoter with five tandem repeats of the yeast GAL4 binding elements that control expression of the firefly luciferase gene. The SRE reporter (Stratagene) contains five copies of the sequence AGGATGTCCATATTAGG ACATCT upstream of a basic TATA element controlling the firefly luciferase reporter gene. pRL-null (Promega, Madison, Wis.), which contains the renilla luciferase gene without enhancer elements, was used in the reporter assays as an internal control for transfection efficiency and protein levels (5).
Elk-1 trans-reporter assays.
To determine Elk-1 activity, a trans-reporter assay was performed with the pFA2-Elk1 transactivator plasmid and the pFR-Luc reporter plasmid (see above). Transcription from the pFR-Luc reporter plasmid is dependent on the activation status of the GAL4 DBD-Elk-1 fusion protein, and the activity levels of firefly luciferase reflect the activation status of Elk-1. Dual-luciferase reporter assays were performed according to the manufacturers' protocols (Stratagene and Promega). Briefly, 5 × 104 cells per well were seeded into 24-well plates. After 24 h, cells were transfected by adding 250 μl of Optimem containing 4 μl of PLUS reagent, 1 μl of Lipofectamine, 400 ng of pFR-Luc reporter plasmid, 20 ng of pFA2-Elk1 transactivator plasmid, and 2 ng of pRL-null. As a negative control, the pFA2-Elk1 transactivator plasmid was replaced with the pFC2-dbd plasmid containing only the GAL4 DBD. Four hours after transfection, 1 ml of DMEM with 10% DCS was added to the wells. The following day, cells were washed once with cold PBS and lysed in 100 μl of passive lysis buffer (Promega) containing protease and phosphatase inhibitors (10 μg of leupeptin per ml, 0.1 U of aprotinin per ml, 2 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, 10 mM sodium fluoride). Dual-luciferase-reporter assays were performed on a LB96V Microlumat luminometer (Perkin-Elmer, Gaithersburg, Md.) with 20 μl of lysate per assay. Reporter activities were normalized for pRL-null activity as previously described (5).
TM-α and SRE cis-reporter assays.
To determine SRE-dependent transcriptional activity and transcription from the TM-α promoter, dual-luciferase reporter assays were performed with the pSRE-Luc construct (Stratagene) and the pGL3-TM-α luciferase construct, respectively, in combination with pRL-null (Promega). As negative controls, the SRE and TM-α reporter constructs were replaced with pMCS-Luc (Stratagene) and pGL3 (Promega) respectively, both of which contain the luciferase gene without any enhancer elements. Cells were transfected by adding 200 μl of Optimem containing 1 μl of Lipofectamine, 4 μl of PLUS reagent, 0.4 μg of the reporter plasmid or control plasmid, and 2 ng of pRL-null. Cell extracts were prepared and analyzed as described above.
RESULTS
KSR1 overexpression induces cell flattening and stress fiber formation in ras-transformed fibroblasts.
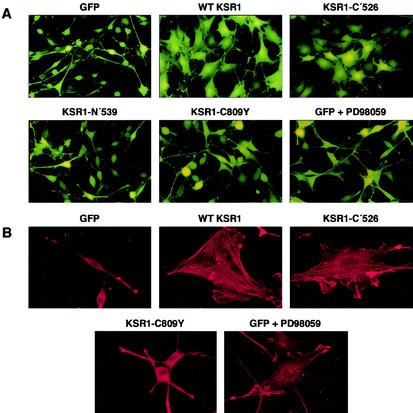
Unlike the flat phenotype of nontransformed fibroblasts, cells expressing activated Ras proteins are rounded and display a spindle-shaped morphology. To examine whether KSR1 overexpression alters this morphology, ras-transformed fibroblasts were infected with an adenovirus expressing GFP in combination with wild-type (WT) KSR1. While expression of GFP alone did not alter the ras-transformed phenotype, overexpression of KSR1 (10- to 20-fold over endogenous levels) resulted in a dramatic flattening of the cells (Fig. 1A). To determine which region of KSR1 was responsible for this effect and to evaluate the significance of the KSR1-MEK interaction in mediating this change, cells were infected with adenoviruses encoding GFP and either the amino-terminal domain (N′539), the C-terminal domain (C′526), or a mutant of KSR1 deficient in MEK binding (C809Y). As shown in Fig. 1A, overexpression of KSR1-C′526 also induced a marked flattening of the ras-transformed cells, while overexpression of KSR1-N′539 or the MEK-binding mutant KSR1-C809Y did not (Fig. 1A).
FIG. 1.
Overexpression of WT KSR1 or KSR1-C′526 induces morphological changes and stress fiber formation in ras-transformed cells. (A) ras-transformed cells were infected with adenoviral constructs expressing GFP alone or GFP in combination with either WT KSR1, the carboxy-terminal region KSR1-C′526, the amino-terminal region KSR1-N′539, or the MEK-binding-deficient mutant KSR1-C809Y. Cells were photographed 24 h after infection. ras-transformed cells overexpressing GFP alone are spindle shaped. Overexpression of WT-KSR1 or KSR1-C′526 induces flattening of these cells. Overexpression of KSR1-C809Y or KSR1-N′539 has no effect. Treatment of ras-transformed cells overexpressing GFP with PD98059 induced some cell flattening. (B) The ras-transformed cells described in panel A were stained for stress fibers with rhodamine-conjugated phalloidin. Overexpression of WT KSR1 and KSR1-C′526 induces stress fiber formation. Overexpression of KSR1-C809Y has no effect on the actin cytoskeleton. Although the MEK inhibitor PD98059 induces some cell flattening, the stress fibers in these cells are much thinner than those observed in cells overexpressing WT KSR1 or KSR1-C′526.
In addition to the morphological changes observed in fibroblasts, ras-induced transformation leads to a loss of actin stress fibers. To examine the consequences of KSR1 expression on stress fiber formation, cells infected with various KSR1-expressing adenoviruses were stained with phalloidin-rhodamine conjugate. In contrast to control ras-transformed fibroblasts, which were spindle shaped and contained no stress fibers, cells overexpressing either WT KSR1 or KSR1-C′526 were flat and had well-defined stress fibers (Fig. 1B). Overexpression of either KSR1-N′539 or KSR1-C809Y failed to mediate these effects (Fig. 1B) (data not shown). Treatment of ras-transformed cells with the MEK inhibitor PD98059 induced partial cell flattening, but the stress fibers observed in these cells were very thin in comparison to those found in KSR1-expressing cells (Fig. 1B). Taken together, these results demonstrate that overexpression of KSR1 induces morphological changes and stress fiber formation in ras-transformed fibroblasts. Moreover, our findings indicate that these effects are mediated by the C-terminal domain of KSR1 and are dependent on the ability of KSR1 to bind MEK.
KSR1 overexpression restores TM expression in ras-transformed fibroblasts.
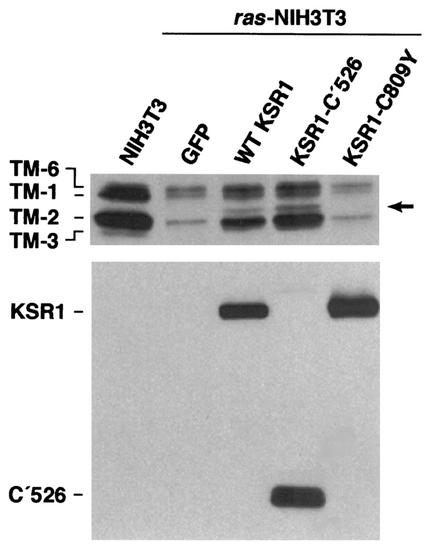
Transformation of fibroblasts with the ras oncogene results in a dramatic down-regulation of the high-molecular-weight isoforms (TM-1, -2, and -3) of TM (15, 22, 23, 44) (Fig. 2). Although the Ras-mediated suppression of TM requires Raf activity, the contribution of its downstream target, MEK, is less clear. Pharmacological inhibition of MEK has minimal effects on TM levels, and yet, expression of a dominant-inhibitory form of MEK1 does restore TM expression in ras-transformed fibroblasts (23). Therefore, while MEK enzymatic activity may not be essential for TM regulation, MEK may have other functions that are required for this process. KSR1 is another component of the Ras-Raf-MEK-MAPK signaling cascade that functions, at least in part, by facilitating signal transmission from Raf-1 to MEK. Therefore, to determine whether overexpression of KSR1 has an effect on TM regulation, we examined TM protein levels in ras-transformed fibroblasts infected with the various KSR1-expressing adenoviruses. Western blot analysis of whole-cell extracts revealed that overexpression of WT KSR1 resulted in a significant increase in the levels of TM-1 or -6 and TM-2 (Fig. 2). Overexpression of the MEK-binding mutant KSR1-C809Y or KSR-N′539 was unable to restore TM levels, but overexpression of KSR1-C′526 resulted in an even higher level of TM expression than was observed in WT KSR1-expressing cells (Fig. 2). In addition, a putative novel isoform of TM was detected in the WT-KSR1- and KSR1-C′526-expressing cells (Fig. 2, arrow). Thus, the morphological changes and stress fiber formation induced by KSR1 in ras-transformed fibroblasts correlate well with the restoration of TM protein levels. Furthermore, like the KSR1-induced effects on cell morphology and stress fiber formation, the restoration of TM expression was mediated by the KSR1 C-terminal domain and required the KSR1-MEK interaction.
FIG. 2.
Overexpression of WT KSR1 or KSR1-C′526 restores TM expression in ras-transformed cells. Whole-cell extracts of untransformed NIH 3T3 or ras-transformed cells infected with adenoviral constructs expressing GFP alone or GFP in combination with either WT KSR1, KSR1-C′526, or KSR1-C809Y were fractionated on 10% Bis-Tris gels. Western blots of the gels were probed with the TM311 monoclonal antibody to detect the high-molecular-weight isoforms of TM (top panel) and with anti-Pyo antibody to detect Pyo-tagged KSR1 proteins (lower panel). ras-transformed cells express significantly lower levels of TM than nontransformed NIH 3T3 cells. Overexpression of either WT KSR1 or KSR1-C′526 induces a strong increase in expression of TM-1, TM-6, and TM-2 and the expression of a seemingly novel TM isoform (arrow). Overexpression of KSR1-C809Y has no effect on TM levels.
The ras oncogene suppresses transcription from the TM-α promoter.
mRNA levels for the high-molecular-weight isoforms of TM are reduced in ras-transformed cells (7, 11, 33). To date, however, it is unclear whether this reduction is due to destabilization of TM mRNA and/or decreased transcription of the TM genes. To gain further insight into the mechanism of TM mRNA reduction, we examined the transcriptional activity of the TM-α gene promoter, which regulates the transcription of, among others, the high-molecular-weight TM isoforms TM-2, -3, and -6 (16, 18, 28, 29, 43). Using a luciferase reporter construct containing a partial TM-α promoter, we found that in comparison to untransformed NIH 3T3 cells, transcription was decreased threefold in cells stably transformed by the Ki-rasV12 oncogene (Fig. 3A). In addition, transient expression of H-rasV12 together with the TM-α reporter construct resulted in a twofold decrease in transcription compared to the level in cells cotransfected with a control vector and the reporter construct (Fig. 3A). These findings indicate that the loss of TM mRNA in ras-transformed cells is due, at least in part, to decreased TM transcription.
FIG. 3.
Analysis of TM-α promoter activity in ras-transformed cells. (A) The ras oncogene suppresses transcription from the TM-α promoter. Nontransformed NIH 3T3 cells and cells stably transformed with v-Ki-ras were transiently transfected with a TM-α reporter construct in combination with pRL-null (left panel). In a separate experiment, NIH 3T3 cells were transiently cotransfected with the TM-α reporter construct and either pSRα expressing the H-rasV12 oncogene or the vector pSRα (right panel). In both assays, dual-luciferase reporter assays were performed, and transcription from the TM-α promoter was normalized according to the activity of pRL-null. The normalized activity in nontransformed NIH 3T3 cells was set at 1. Both stable and transient expression of the ras oncogene results in decreased TM-α promoter activity. (B) Overexpression of KSR1 enhances TM-α promoter activity in v-Ki-ras-transformed cells. Ras-transformed cells were infected with adenoviruses expressing GFP alone or GFP in combination with the indicated KSR variants. The following day, cells were transfected with a TM-α-luciferase reporter construct in combination with pRL-null. To assess the effects of the MEK inhibitors PD98059 and UO126 on TM-α transcription, cells expressing GFP alone were treated with the inhibitor prior to and after transfection with the TM-α reporter construct. Dual-luciferase reporter assays were performed, and transcription from the TM-α promoter was normalized according to the activity of pRL-null. The normalized reporter activity in cells expressing GFP alone was set at 1. Both WT KSR1 overexpresssion and KSR1-C′526 overexpression enhance TM-α transcriptional activity. Neither treatment of ras-transformed cells with MEK inhibitors nor overexpression of KSR1-C809Y had any effect on TM-α transcriptional activity. None of the adenovirus constructs nor the MEK inhibitors had any significant effect on the control reporter construct pGL3, which lacks the TM-α promoter (data not shown).
KSR1 overexpression enhances transcription from the TM-α promoter in ras-transformed fibroblasts.
To investigate whether the KSR1-induced restoration of TM protein levels in ras-transformed fibroblasts might be due to enhanced transcription of TM genes, we examined the effects of KSR1 overexpression on the transcriptional activity of the TM-α promoter. ras-transformed cells infected with the various KSR1-expressing adenoviruses were transiently transfected with the TM-α reporter construct, and transcriptional activity was measured 24 h later. In addition, since we have previously found that treatment with the MEK inhibitor PD98059 fails to restore TM expression in ras-transformed fibroblasts (23), we also tested the effects of PD98059 and the unrelated MEK inhibitor UO126 on the transcriptional activity of the TM promoter. For these assays, cells were treated with 50 μM PD98059 or 10 μM UO126 1 day prior to transfection of the reporter construct and were again treated with 50 μM PD98059 or 10 μM UO126, respectively, 4 h after transfection (similar results were obtained with cells that were not pretreated with the MEK inhibitors prior to transfection). As depicted in Fig. 3B, TM-α promoter activity was two- to threefold higher in ras-transformed fibroblasts overexpressing WT KSR1 than in cells expressing GFP alone. A five- to eightfold increase in TM promoter activity was detected in cells expressing KSR1-C′526, while no change in activity was observed in cells expressing the MEK-binding deficient mutant KSR1-C809Y. Thus, a strong correlation is observed between the effect of the KSR1 proteins on TM protein expression and their effect on TM promoter activity. Moreover, in agreement with our previous finding that the MEK inhibitor PD98059 does not restore TM protein levels in ras-transformed fibroblasts, pharmacological inhibition of MEK had no effect on TM transcription.
Effect of KSR1 overexpression on MEK and ERK phosphorylation and ERK activity in ras-transformed fibroblasts.
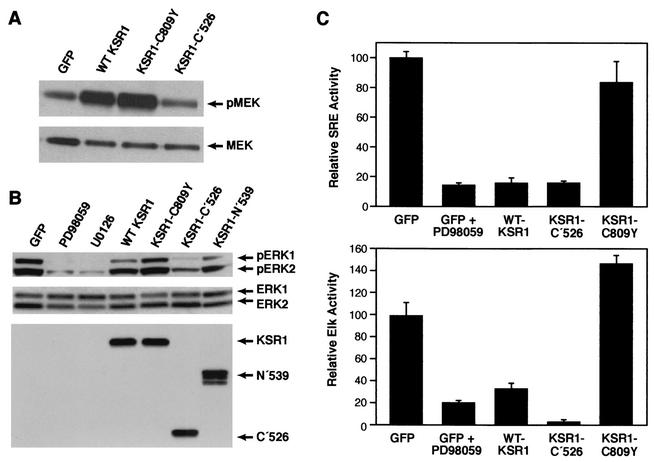
Overexpression of KSR1 has been shown to interfere with mitogen-induced ERK activity in nontransformed cells (24, 65). To determine whether KSR1 overexpression alters MEK/ERK signaling in ras-transformed fibroblasts and to evaluate the potential importance of MEK/ERK signaling to the KSR1-mediated effects, we examined MEK phosphorylation, ERK phosphorylation, and ERK activity in cells overexpressing the various KSR1 proteins. Using antibodies that specifically recognize MEK phosphorylated at Ser 217/219, we found that MEK was constitutively phosphorylated on these activating sites in ras-transformed cells. Overexpression of WT KSR1 or KSR1-C′526 actually enhanced the levels of phospho-MEK detected (Fig. 4A), while overexpression of KSR1-C809Y had no effect, suggesting that the KSR1-MEK interaction facilitates MEK phosphorylation in these cells. ERK-1 and ERK-2 were also constitutively phosphorylated on activating sites in ras-transformed cells. These high levels of phospho-ERK could be significantly reduced by treatment of cells with the MEK inhibitors PD98059 and UO126 (Fig. 4B). Notably, the effects of KSR1 overexpression on ERK phosphorylation were not necessarily those predicted based on the MEK phosphorylation data. As expected, overexpression of KSR1-N′539 or KSR1-C809Y had no effect on ERK phosphorylation (Fig. 4B). However, KSR1-C′526, which enhanced MEK phosphorylation, suppressed phospho-ERK levels, as did WT KSR1, albeit to a far lesser extent. The fact that WT KSR1 had such a modest effect on ERK phosphorylation is consistent with the notion that the inhibition of MEK activity is not the predominant mechanism by which KSR1 affects cell morphology and gene expression in ras-transformed fibroblasts.
FIG. 4.
Effect of KSR1 overexpression on MEK and ERK phosphorylation. Whole-cell extracts were made from ras-transformed cells infected with adenoviral constructs expressing GFP alone or GFP in combination with the indicated KSR1 protein. Equal amounts of protein were fractionated on Bis-Tris gels, and the phosphorylation status of MEK and ERK was determined by Western blot analysis with anti-phospho-MEK (A, top panel) and anti-phospho-ERK antibodies (B, top panel). Total MEK and ERK levels were assessed by probing blots with anti-MEK (A, bottom panel) and anti-ERK (B, middle panel) antibodies. Western blot analysis with the anti-Pyo antibody was also performed to demonstrate the equivalent expression of the Pyo-tagged KSR1 variants (B, bottom panel). Overexpression of WT KSR1 and KSR1-C′526 facilitates MEK phosphorylation. In contrast, KSR1-C′526 markedly suppressed phospho-ERK levels, as did WT KSR1, albeit to a far lesser extent. PD98059 and UO126 suppressed ERK phosphorylation to the strongest extent. KSR1-N′539 and KSR1-C809Y had no effect on the phosphorylation levels of MEK and ERK. (C) KSR1 overexpression inhibits signaling downstream of ERK. ras-transformed cells were infected with adenoviral constructs expressing GFP alone or GFP and the indicated KSR1 protein. Eighteen hours following infection, the cells were transfected with the SRE reporter (top panel) or Elk reporter (bottom panel) constructs in combination with pRL-null. To assess the effects of the MEK inhibitor PD98059 on transcriptional activities, cells expressing GFP alone were treated with the inhibitor prior to and after transfection with the reporter constructs. Dual-luciferase reporter assays were performed, and transcriptional activities were normalized according to the activity of pRL-null. The normalized reporter activity in cells expressing GFP alone was set at 100. Treatment with the MEK inhibitor PD98059 and overexpression of WT KSR1 or KSR1-C′526 suppressed the activities of both the SRE and Elk-1 reporters. In contrast, overexpression of KSR1-C809Y did not suppress those activities. Neither the constructs nor the inhibitor had a significant effect on the control reporter constructs pFA2-dbd and pMCS-Luc, which lack the response elements for Elk and SRE, respectively (data not shown).
To investigate the effects of the KSR1 proteins on ERK activity in vivo, we measured the transcriptional activities of SRE- and Elk-1-dependent reporter constructs. Although phosphorylation by ERK is the primary mechanism controlling Elk-1 activity, SRE activity is regulated in a more complex manner and is only partially dependent on ERK-mediated phosphorylation of the Ets family members Elk-1, SAP-1, and SAP-2. In SRE-dependent cis-reporter assays, we found that PD98059, WT KSR1, and KSR1-C′526 all inhibited SRE-dependent transcription to approximately the same extent (Fig. 4C). The KSR1-mediated inhibition was dependent on MEK binding, because expression of the KSR1-C809Y mutant had no significant effect on SRE activity. The suppressive effect of WT KSR1 on SRE reporter activity exceeded that which was expected based on the modest ability of this protein to inhibit ERK phosphorylation. In Elk-1-dependent trans-reporter assays, Elk-1 transcription was reduced by either PD98059 treatment or by WT KSR1 overexpression (five- and threefold inhibitions, respectively). The KSR1-C′526 protein caused a much greater reduction in Elk-1-dependent transcription (>20-fold reduction), correlating with its strong inhibitory effect on ERK phosphorylation (Fig. 4C). Consistent with the results from the SRE assay, overexpression of the KSR1-C809Y mutant had no suppressive effect. Together, these findings demonstrate that overexpression of KSR1 inhibits ERK activity in ras-transformed fibroblasts and that this inhibition is dependent on the KSR1-MEK interaction. Moreover, the ability of KSR1 to block ERK activity (as determined by the activation of nuclear substrates such as Elk-1) does not necessarily correlate with its effect on ERK phosphorylation levels.
MEK-binding-independent events are not implicated in KSR1-mediated TM regulation.
The results presented above indicate that the KSR1-MEK interaction is critical for the upregulation of TM as well as for the inhibition of ERK activity that is observed in KSR1-expressing ras-transformed cells. While the ability of KSR1 to restore TM levels does in fact correlate with its ability to inhibit ERK, ERK inhibition cannot be the sole mechanism by which KSR1 mediates its effects, given that suppression of ERK signaling by pharmacological inhibitors does not restore TM levels in these cells (23). Based on these results, we hypothesize that KSR-induced effects rely either on MEK-binding-dependent events other than ERK inhibition or on inhibition of ERK activity (which is MEK-binding dependent) plus additional events, which may or may not depend on MEK binding. If the KSR1-mediated effects do indeed require MEK-binding-independent effects as well as MEK-binding-dependent ERK inhibition, we might expect that the MEK-binding-deficient mutant of KSR1 (C809Y) could restore TM levels in cells where ERK activity is inhibited. To address this possibility, ras-transformed cells overexpressing KSR1-C809Y were treated with PD98059 prior to transfection with the TM reporter, and transcriptional activity was measured 24 h after transfection. To control for PD98059 activity, a parallel set of cells were transfected with the SRE reporter. As shown in Fig. 5, the addition of PD98059 to KSR1-C809Y-overexpressing cells completely suppressed SRE activity, indicating that the drug was functional. Under these conditions of ERK inhibition, however, KSR1-C809Y was still unable to induce TM promoter activity in ras-transformed cells. Thus, while the role of ERK inhibition in TM regulation remains uncertain, it is unlikely that KSR1 utilizes a MEK-binding-independent pathway to upregulate TM transcription.
FIG. 5.
PD98059 does not enhance TM transcription in KSR1-C809Y-overexpressing cells. (A) ras-transformed cells overexpressing KSR1-C809Y were treated with PD98059 prior to and after transfection of the TM-α reporter (gray bars). As a control, cells were treated with vehicle only (dimethyl sulfoxide [DMS]; open bars). Dual-luciferase reporter assays were performed, and transcription from the TM-α promoter was normalized according to the activity of pRL-null. The normalized reporter activity in dimethyl sulfoxide-treated cells was set at 1. No effect of PD98059 treatment on TM-α reporter activity was observed in ras-transformed cells overexpressing KSR1-C809Y. (B) As a control for the effectiveness of PD98059 treatment, KSR1-C809Y-overexpressing cells treated as described in panel A were transiently transfected with an SRE reporter, and SRE-dependent transcription was determined. SRE activity was normalized according to the activity of pRL-null, and the activity in dimethyl sulfoxide-treated cells was set at 100. PD98059 treatment resulted in a 10- to 20-fold reduction in SRE activity, indicating that the drug was effective in KSR1-C809Y-overexpressing cells.
ROCK activity is required for the KSR1-induced effects observed in ras-transformed fibroblasts.
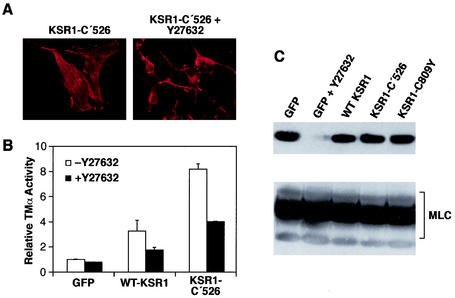
Activities and/or events other than the inhibition of ERK nuclear target signaling appear to be required for the KSR1-induced upregulation of TM. As shown in Fig. 1B, both WT KSR1 and KSR1-C′526 are able to promote stress fiber formation in ras-transformed fibroblasts. Since stress fiber formation in fibroblasts is dependent on the activity of Rho and its downstream effector, ROCK (46, 47, 49, 58, 62), we next examined whether the KSR1-induced phenotypic changes in ras-transformed cells require ROCK activity (Fig. 6). For these assays, cells infected with the KSR1-C′526-expressing adenoviruses were treated with the ROCK inhibitor Y27632 (10 μM for 16 h—conditions that effectively block ROCK enzymatic activity) (Fig. 6C), after which they were stained with rhodamine-conjugated phalloidin. Upon Y27632 treatment, cells overexpressing KSR1-C′526 became rounded and lost their stress fibers, demonstrating that ROCK activity is needed for the KSR1-induced stress fiber formation and cell flattening (Fig. 6A). The same results were observed for ras-transformed cells overexpressing WT KSR1 (data not shown). To examine whether the enhanced transcription from the TM promoter also requires ROCK activity, cells overexpressing WT KSR1 or the KSR1-C′526 mutant were treated with Y27632, and the transcriptional activity of the TM-α reporter was determined. As shown in Fig. 6B, inhibition of ROCK activity resulted in a 50% decrease in TM-α promoter activity in ras-transformed cells overexpressing either WT KSR1 or KSR1-C′526. It should be noted that TM-α promoter activity in the Y27632-treated KSR1-overexpressing cells is still higher than that observed in untreated control cells (∼1.7-fold increase for WT KSR1 and ∼4-fold increase for KSR1-C′526), indicating that ROCK-independent pathways also contribute to the KSR1-mediated effects on TM expression.
FIG. 6.
ROCK activity is required for KSR1-induced effects in ras-transformed cells. ras-transformed cells overexpressing GFP alone or in combination with KSR1-C′526 were treated with the ROCK inhibitor Y27632 (10 μM) and analyzed for stress fiber expression (A) and TM-α reporter activity (B). Treatment of cells overexpressing KSR1-C′526 with Y27632 led to a loss of stress fibers and rounding of cells (A). Treatment of ras-transformed cells overexpressing KSR1-C′526 and WT KSR1 resulted in a decrease in TM-α promoter activity (B). Dual-luciferase reporter assays were performed, and transcription from the TM-α promoter was normalized according to the activity of pRL-null. The normalized reporter activity in cells expressing GFP alone was set at 1. (C) KSR1 overexpression does not alter ROCK activity. Whole-cell extracts of ras-transformed cells infected with adenoviral constructs expressing GFP alone or GFP in combination with either WT KSR1, KSR1-C′526, or KSR1-C809Y were fractionated on Bis-Tris gels, and Western blots were probed with anti-phospho-MLC antibody pp2b (top panel). As a control, extracts from ras-transformed cells expressing GFP alone and treated with Y27632 were also analyzed. Blots were stripped and reprobed with anti-MLC antibody to determine total levels of MLC (bottom panel).
KSR1 does not regulate ROCK activity.
The above results demonstrate that the effects of KSR1 are at least partially dependent on ROCK activity, suggesting that KSR1 may act either upstream of or parallel to ROCK-induced events. To determine whether KSR1 functions upstream of ROCK, we investigated the effects of KSR1 overexpression on ROCK activity in ras-transformed cells. Because ROCK has been shown to phosphorylate MLC on serine 19 (62), we measured ROCK activity by assessing the in vivo phosphorylation status of MLC. Using an antibody specific for phospo-Ser19-MLC (Fig. 6C), we found that MLC was constitutively phosphorylated on Ser19 in ras-transformed cells and that this phosphorylation could be completely blocked by Y27632 treatment, demonstrating that ROCK is responsible for the majority of MLC phosphorylation in ras-transformed cells. As shown in Fig. 6C, overexpression of WT KSR1, KSR1-C′526, or KSR1-C809Y did not alter the phosphorylation status of MLC. Thus, while the KSR1-mediated effects observed in ras-transformed cells require ROCK enzymatic activity, KSR1 does not act upstream to directly regulate ROCK activity. In addition, these findings, taken together with those presented in Fig. 1 to 3, suggest that overexpression of KSR1 proteins competent to bind MEK provides a function, such as the restored expression of TM, that allows ROCK to be recoupled to the actin polymerization machinery in ras-transformed fibroblasts.
DISCUSSION
In this report, we have further explored the mechanism by which TM is down-regulated in ras-transformed fibroblasts and have analyzed the effects of KSR1 on TM expression. KSR1 is a scaffold protein that plays an important role in signal transmission downstream of Ras; however, little is known regarding the mechanisms employed and to what extent KSR1 affects cellular behavior and gene expression. In the present study, we show that overexpression of KSR1 upregulates TM expression and dramatically alters the cellular morphology and cytoskeletal architecture of ras-transformed cells. These effects were achieved without any discernible increase in the activity of ROCK, the Rho effector that regulates stress fiber formation. We and others have shown previously that the forced expression of TM-1 or TM-2 is sufficient to mediate the morphological flattening of transformed cells and to restore stress fiber formation (15, 22, 44, 60). Thus, the multiple effects induced by KSR1 expression may be entirely attributable to its ability to normalize TM protein levels.
The decrease in TM expression in ras-transformed cells is associated with a comparable reduction in TM mRNA levels (7, 11, 33). In previous studies, we have been unable to detect any significant change in the stability of TM-2 transcripts in ras-transformed fibroblasts, suggesting that the low mRNA levels observed in these cells may be due to a decrease in transcriptional activity (G. M. Fuhler, R. A. J. Janssen, and J. W. Mier, unpublished observations). Our findings presented in Fig. 3 support this model in that the reporter activity from a TM-α promoter construct is diminished in cells either stably or transiently expressing activated ras alleles. Moreover, we find that the restoration of TM levels mediated by KSR1 is likely to be due to increased TM transcription, because overexpression of either the WT or C-terminal domain of KSR1 resulted in increased TM-α reporter activity in ras-transformed cells (Fig. 3). Treatment with the MEK inhibitor PD98059, however, had no effect on the transcriptional activity of the TM-α promoter, further supporting our earlier report that MEK inhibition is not sufficient to restore TM protein expression in ras-transformed NIH 3T3 cells (23).
High overexpression of KSR1 has previously been shown to suppress Ras-dependent signaling, yet it is unclear exactly where in the Raf-MEK-ERK cascade KSR1 prevents signal transmission. In regard to MEK activation, we report here that overexpression of KSR1 in ras-transformed cells does not block, but instead, enhances the phosphorylation of MEK on activating sites. This increase was found to be dependent on the KSR1-MEK interaction in that it was not observed in cells expressing the KSR1-C809Y mutant defective in MEK binding. In agreement with our results, it has recently been reported that KSR1 facilitates the phosphorylation of MEK by Raf and that this process requires the interaction of KSR1 with Raf and MEK (37, 38, 48).
In regard to ERK, the effect of KSR1 overexpression is more complex. We find that overexpression of either WT or the C-terminal domain of KSR1 inhibits ERK activity, whereas only the C-terminal domain induces a significant reduction in ERK phosphorylation. Because the C-terminal region of KSR1 contains the binding site for MEK but not ERK, overexpression of this protein presumably segregates MEK from ERK, thus preventing ERK phosphorylation. As for the full-length protein, our findings are similar to those reported by Sugimoto et al. (57), where overexpression of KSR1 in mitogen-treated cells inhibited ERK activity towards Elk-1 but not ERK phosphorylation. Because of KSR1's scaffolding function, it has been thought that some of the suppressive effects of high KSR1 overexpression are due to the disruption of the Raf-MEK-ERK signaling complex. However, this does not appear to be the case in our studies, since overexpression of full-length WT KSR1 did not prevent MEK activation or significantly block the phosphorylation of ERK by MEK. Thus, it is unlikely that the upregulation of TM by KSR1 is due to a disruption of the Raf-MEK-ERK signaling complex, given that both WT and the C-terminal domain of KSR1 restore TM levels, yet only the C-terminal domain appears to disrupt the interactions required for signal transmission from Raf to ERK.
It is unclear how WT KSR1 inhibits Elk- and SRE-dependent transcription without an equivalent inhibition in MEK and ERK phosphorylation. In the work by Sugimoto et al., it was suggested that that KSR1 might activate PP2B, which dephosphorylates Elk-1 (57). Alternatively, KSR1 overexpression may inhibit Elk-1 phosphorylation by altering the localization of phosphorylated ERK or MEK-ERK complexes. In support of this hypothesis, Stewart et al. have found that overexpression of KSR1 leads to the formation of a high-molecular-weight complex at the cell membrane containing KSR1 and functionally active MEK and ERK (56). In addition, it has recently been reported that high levels of KSR1 can interfere with the nuclear cycling of MEK (9). Thus, KSR1 may serve as an anchor preventing the localization of active ERK or MEK-ERK complexes to the nucleus.
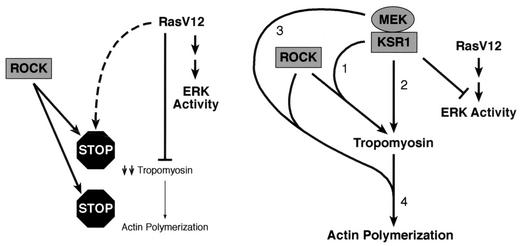
The restoration of TM by KSR1 correlates with a decrease in Elk-1 and SRE-dependent transcriptional activity. Yet, a block in ERK nuclear signaling cannot be the sole mechanism by which KSR1 restores TM expression, because treatment of ras-transformed cells with MEK inhibitors efficiently inhibits ERK nuclear signaling, but does not restore TM levels or enhance transcription from the TM-α promoter. Moreover, the difference between the effects of KSR1 and PD98059 on TM expression is not due to a difference in the degree of ERK inhibition, since both KSR1 and PD98059 suppressed SRE and Elk-1 activities to the same extent. Therefore, it is likely that KSR1 affects cellular events in addition to ERK nuclear signaling (Fig. 7).
FIG. 7.
Summary of findings in this study. In ras-transformed fibroblasts, the level of ERK activity is high, ROCK is active but uncoupled from the actin polymerization machinery (stop sign), TM levels are down-regulated in an ERK-independent way, and no stress fibers are formed. Overexpression of KSR1 suppresses ERK activity and induces the upregulation of TM via a ROCK-dependent pathway (numbered 1) and a ROCK-independent pathway (numbered 2). The interaction of KSR with MEK is required for these effects. The KSR-MEK complex may also recouple ROCK activity towards the actin-polymerization machinery (numbered 3). Tropomyosin and ROCK appear to mutually depend on each other for the induction of actin polymerization (numbered 4).
Based on the fact that KSR1-C809Y is unable to induce the KSR1-mediated changes, we conclude that the interaction with MEK is required. Although we cannot exclude the possibility that the C809Y mutation alters other properties of KSR1 (such as localization, or interactions with unknown components of the KSR1 scaffolding complex), this mutation has clearly been shown to abolish MEK binding (9, 36, 56). In light of these results, it is interesting to speculate that the interaction with KSR1 may modulate MEK's ability to interact with proteins or targets other than ERK. Previously, we have shown that overexpression of a dominant-negative form of MEK, MEK1-S218A, S222A, also induces restored expression of TM in ras-transformed fibroblasts. As was observed for WT KSR1, this mutant did not suppress the constitutively high levels of phosphorylated MEK and only moderately inhibited the phosphorylation of ERK (23). Consequently, the dominant-negative form of MEK1, like KSR1, may modulate the effects of MEK on effectors other than ERK. In support of the model that MEK may have functions in addition to ERK regulation, activated MEK1 has recently been found to regulate the transcriptional activity of MyoD through interactions with the MyoD transcriptional complex (42).
Further studies will be needed to fully define the MEK-dependent events that are required for the KSR1-mediated regulation of TM. In particular, it is unclear whether MEK enzymatic activity is needed for these effects. In the studies presented here, we find that MEK is phosphorylated on activating sites in cells overexpressing KSR1, indicating that KSR1 does not disrupt MEK activation. However, we have also observed that MEK inhibitor treatment does not abolish the effect of either WT KSR1 or KSR1-C′526 on TM promoter activity (unpublished observations). While this result would suggest that MEK enzymatic activity might not be essential for TM regulation, we also find that MEK activity is not completely suppressed in these cells, and we have preliminary evidence that the KSR1-MEK interaction may protect MEK from inhibitor binding (unpublished observations). Therefore, it is difficult to interpret the results of these experiments. Nonetheless, in cells expressing the MEK-binding deficient mutant KSR1-C809Y, MEK activity and ERK activation are fully inhibited by PD98059 treatment (Fig. 5).
Overexpression of KSR1 not only restores TM expression but also induces cell flattening and stress fiber formation. The exact mechanism by which KSR1 brings about these morphological changes is unknown. Under normal conditions, stress fiber formation is mediated by signaling downstream of the Rho effector ROCK. ROCK regulates cytoskeletal architecture by the phosphorylation of MLC phosphatase, MLC, and LIM kinase (3, 26, 32, 58, 62). In ras-transformed cells, we found that MLC is constitutively phosphorylated due to the activity of ROCK, because treatment of cells with the ROCK inhibitor Y27632 blocked MLC phosphorylation (Fig. 6C). Stress fiber formation and increased TM transcription induced by KSR1 overexpression also required ROCK activity and could be suppressed by Y27632 treatment. Y27632, however, did not completely abrogate the effects of KSR1 on TM transcription, indicating that KSR1 may also act via ROCK-independent pathways. Thus, while ras transformation suppresses TM expression and uncouples ROCK activity from the actin polymerization machinery (40, 50), overexpression of KSR1 restores TM levels and allows the connection between ROCK and the cytoskeleton to be reestablished. KSR1 does not directly regulate ROCK enzymatic activity, and we have also found that it does not alter the subcellular localization of ROCK (unpublished observations). Therefore, it is possible that KSR1 may act as a direct link between MLC activity and stress fiber formation or that it may affect other factors that contribute to stress fiber formation, such as LIMK and cofilin (58, 64). Interestingly, stress fiber formation induced by the overexpression of TM-1 in ras-transformed cells is also dependent on ROCK activity (52), suggesting that the limiting factor for stress fiber formation in ras-transformed cells may be the low level of TM expression. As a result, the elevated levels of TM observed in these cells overexpressing KSR1 may complement the constitutive ROCK activity to promote stress fiber formation (Fig. 7).
In summary, our results provide new insights into TM regulation in ras-transformed fibroblasts and into the role played by the KSR1 scaffold in gene expression, cytoskeletal architecture, and signaling through the MAPK cascade. Here, we demonstrate for the first time that the loss of TM expression in ras-transformed cells and the restoration of TM levels by KSR1 overexpression are due, at least in part, to altered transcription. In addition, our findings further support the model that the interaction with MEK is critical for KSR1 function. All KSR1-mediated effects observed in this study were dependent on MEK binding and required the activity of the ROCK kinase. Finally this study provides support for the existence of alternative functions or substrates for the Raf-MEK-ERK signaling cascade that govern the expression of genes associated with the maintenance of the actin cytoskeleton.
Acknowledgments
We thank D. Ritt (National Cancer Institute—Frederick, Frederick, Md.), G. M. Fuhler (Rijksuniversiteit Groningen, Groningen, The Netherlands), and B. C. Mosterman and K. G. Veenstra (Beth Israel Deaconess Medical Center, Boston, Mass.) for technical assistance; F. N. van Leeuwen (Nijmegen Center for Molecular Life Sciences, Nijmegan, The Netherlands) and W. C. Weinberg (Center for Biologics Evaluation and Research, FDA, Bethesda, Md.) for critically reading the manuscript; and E. Bonvini (Center for Biologics Evaluation and Research) for valuable help and advice.
This research was made possible in part by funds from the Netherlands Organization for Scientific Research (grant 901-10-092); a grant from the Alexander and Margaret Stewart Trust, Washington, D.C., to R.A.J.J.; NCI grant CA74401 to J.W.M.; and funds from DHHS/NIH/NCI to D.K.M.
REFERENCES
- 1.Alaiya, A. A., B. Franzen, K. Fujioka, B. Moberger, K. Schedvins, C. Silfversvard, S. Linder, and G. Auer. 1997. Phenotypic analysis of ovarian carcinoma: polypeptide expression in benign, borderline and malignant tumors. Int. J. Cancer 73:678-683. [DOI] [PubMed] [Google Scholar]
- 2.Alaiya, A. A., M. Oppermann, J. Langridge, U. Roblick, L. Egevad, S. Brindstedt, M. Hellstrom, S. Linder, T. Bergman, H. Jornvall, and G. Auer. 2001. Identification of proteins in human prostate tumor material by two-dimensional gel electrophoresis and mass spectrometry. Cell. Mol. Life Sci. 58:307-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amano, M., M. Ito, K. Kimura, Y. Fukata, K. Chihara, T. Nakano, Y. Matsuura, and K. Kaibuchi. 1996. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase). J. Biol. Chem. 271:20246-20249. [DOI] [PubMed] [Google Scholar]
- 4.Anselmo, A. N., R. Bumeister, J. M. Thomas, and M. A. White. 2002. Critical contribution of linker proteins to raf kinase activation. J. Biol. Chem. 277:5940-5943. [DOI] [PubMed] [Google Scholar]
- 5.Behre, G., L. T. Smith, and D. G. Tenen. 1999. Use of a promoterless Renilla luciferase vector as an internal control plasmid for transient co-transfection assays of Ras-mediated transcription activation. BioTechniques 26:24-26, 28. [DOI] [PubMed] [Google Scholar]
- 6.Bell, B., H. Xing, K. Yan, N. Gautam, and A. J. Muslin. 1999. KSR-1 binds to G-protein beta gamma subunits and inhibits beta gamma-induced mitogen-activated protein kinase activation. J. Biol. Chem. 274:7982-7986. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharya, B., G. L. Prasad, E. M. Valverius, D. S. Salomon, and H. L. Cooper. 1990. Tropomyosins of human mammary epithelial cells: consistent defects of expression in mammary carcinoma cell lines. Cancer Res. 50:2105-2112. [PubMed] [Google Scholar]
- 8.Boyd, J., J. I. Risinger, R. W. Wiseman, B. A. Merrick, J. K. Selkirk, and J. C. Barrett. 1996. Regulation of microfilament organization and anchorage-independent growth by tropomyosin 1. Proc. Natl. Acad. Sci. USA 92:11534-11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brennan, J. A., D. J. Volle, O. V. Chaika, and R. E. Lewis. 2002. Phosphorylation regulates the nucleocytoplasmic distribution of kinase suppressor of Ras. J. Biol. Chem. 277:5369-5377. [DOI] [PubMed] [Google Scholar]
- 10.Cacace, A. M., N. R. Michaud, M. Therrien, K. Mathes, T. Copeland, G. M. Rubin, and D. K. Morrison. 1999. Identification of constitutive and Ras-inducible phosphorylation sites of KSR: implications for 14-3-3 binding, mitogen-activated protein kinase binding, and KSR overexpression. Mol. Cell. Biol. 19:229-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper, H. L., B. Bhattacharya, R. H. Bassin, and D. S. Salomon. 1987. Suppression of synthesis and utilization of tropomyosin in mouse and rat fibroblasts by transforming growth factor alpha: a pathway in oncogene action. Cancer Res. 47:4493-4500. [PubMed] [Google Scholar]
- 12.Denouel-Galy, A., E. M. Douville, P. H. Warne, C. Papin, D. Laugier, G. Calothy, J. Downward, and A. Eychene. 1998. Murine Ksr interacts with MEK and inhibits Ras-induced transformation. Curr. Biol. 8:46-55. [DOI] [PubMed] [Google Scholar]
- 13.Franzen, B., S. Linder, A. A. Alaiya, E. Eriksson, K. Fujioka, A. C. Bergman, H. Jornvall, and G. Auer. 1997. Analysis of polypeptide expression in benign and malignant human breast lesions. Electrophoresis 18:582-587. [DOI] [PubMed] [Google Scholar]
- 14.Franzen, B., S. Linder, K. Uryu, A. A. Alaiya, T. Hirano, H. Kato, and G. Auer. 1996. Expression of tropomyosin isoforms in benign and malignant human breast lesions. Br. J. Cancer 73:909-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gimona, M., J. A. Kazzaz, and D. M. Helfman. 1996. Forced expression of tropomyosin 2 or 3 in v-Ki-ras-transformed fibroblasts results in distinct phenotypic effects. Proc. Natl. Acad. Sci. USA 93:9618-9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodwin, L. O., J. P. Lees-Miller, M. A. Leonard, S. B. Cheley, and D. M. Helfman. 1991. Four fibroblast tropomyosin isoforms are expressed from the rat alpha-tropomyosin gene via alternative RNA splicing and the use of two promoters. J. Biol. Chem. 266:8408-8415. [PubMed] [Google Scholar]
- 17.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helfman, D. M., S. Cheley, E. Kuismanen, L. A. Finn, and Y. Yamawaki-Kataoka. 1986. Nonmuscle and muscle tropomyosin isoforms are expressed from a single gene by alternative RNA splicing and polyadenylation. Mol. Cell. Biol. 6:3582-3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hendricks, M., and H. Weintraub. 1981. Tropomyosin is decreased in transformed cells. Proc. Natl. Acad. Sci. USA 78:5633-5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrera, V. L., and N. Ruiz-Opazo. 1990. Regulation of alpha-tropomyosin and N5 genes by a shared enhancer. Modular structure and hierarchical organization. J. Biol. Chem. 265:9555-9562. [PubMed] [Google Scholar]
- 21.Huang, A. M., and G. M. Rubin. 2000. A misexpression screen identifies genes that can modulate RAS1 pathway signaling in Drosophila melanogaster. Genetics 156:1219-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janssen, R. A., and J. W. Mier. 1997. Tropomyosin-2 cDNA lacking the 3′ untranslated region riboregulator induces growth inhibition of v-Ki-ras-transformed fibroblasts. Mol. Biol. Cell 8:897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janssen, R. A., K. G. Veenstra, P. Jonasch, E. Jonasch, and J. W. Mier. 1998. Ras- and Raf-induced down-modulation of non-muscle tropomyosin are MEK-independent. J. Biol. Chem. 273:32182-32186. [DOI] [PubMed] [Google Scholar]
- 24.Joneson, T., J. A. Fulton, D. J. Volle, O. V. Chaika, D. Bar-Sagi, and R. E. Lewis. 1998. Kinase suppressor of Ras inhibits the activation of extracellular ligand-regulated (ERK) mitogen-activated protein (MAP) kinase by growth factors, activated Ras, and Ras effectors. J. Biol. Chem. 273:7743-7748. [DOI] [PubMed] [Google Scholar]
- 25.Kim, P. N., E. Jonasch, B. C. Mosterman, J. W. Mier, and R. A. J. Janssen. 2001. Radicicol suppresses transformation and restores tropomyosin-2 expression in both ras- and MEK-transformed cells without inhibiting the Raf/MEK/ERK signaling cascade. Cell Growth Differ. 12:543-550. [PubMed] [Google Scholar]
- 26.Kimura, K., M. Ito, M. Amano, K. Chihara, Y. Fukata, M. Nakafuku, B. Yamamori, J. Feng, T. Nakano, K. Okawa, A. Iwamatsu, and K. Kaibuchi. 1996. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science 273:245-248. [DOI] [PubMed] [Google Scholar]
- 27.Lee, S. W., C. Tomasetto, and R. Sager. 1991. Positive selection of candidate tumor-suppressor genes by subtractive hybridization. Proc. Natl. Acad. Sci. USA 88:2825-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lees-Miller, J. P., L. O. Goodwin, and D. M. Helfman. 1990. Three novel brain tropomyosin isoforms are expressed from the rat α-tropomyosin gene through the use of alternative promoters and alternative RNA processing. Mol. Cell. Biol. 10:1729-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lees-Miller, J. P., and D. M. Helfman. 1991. The molecular basis for tropomyosin isoform diversity. BioEssays 13:429-437. [DOI] [PubMed] [Google Scholar]
- 30.Levchenko, A., J. Bruck, and P. W. Sternberg. 2000. Scaffold proteins may biphasically affect the levels of mitogen-activated protein kinase signaling and reduce its threshold properties. Proc. Natl. Acad. Sci. USA 97:5818-5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ljungdahl, S., S. Linder, B. Franzen, B. Binetruy, G. Auer, and M. C. Shoshan. 1998. Down-regulation of tropomyosin-2 expression in c-Jun-transformed rat fibroblasts involves induction of a MEK1-dependent autocrine loop. Cell Growth Differ. 9:565-573. [PubMed] [Google Scholar]
- 32.Maekawa, M., T. Ishizaki, S. Boku, N. Watanabe, A. Fujita, A. Iwamatsu, T. Obinata, K. Ohashi, K. Mizuno, and S. Narumiya. 1999. Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science 285:895-898. [DOI] [PubMed] [Google Scholar]
- 33.Matsumura, F., J. J. Lin, S. Yamashiro-Matsumura, G. P. Thomas, and W. C. Topp. 1983. Differential expression of tropomyosin forms in the microfilaments isolated from normal and transformed rat cultured cells. J. Biol. Chem. 258:13954-13964. [PubMed] [Google Scholar]
- 34.Matsumura, F., S. Ono, Y. Yamakita, G. Totsukawa, and S. Yamashiro. 1998. Specific localization of serine 19 phosphorylated myosin II during cell locomotion and mitosis of cultured cells. J. Cell Biol. 140:119-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morrison, D. K. 2001. KSR: a MAPK scaffold of the Ras pathway? J. Cell Sci. 114:1609-1612. [DOI] [PubMed] [Google Scholar]
- 36.Müller, J., A. M. Cacace, W. E. Lyons, C. B. McGill, and D. K. Morrison. 2000. Identification of B-KSR1, a novel brain-specific isoform of KSR1 that functions in neuronal signaling. Mol. Cell. Biol. 20:5529-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Müller, J., S. Ory, T. Copeland, H. Piwnica-Worms, and D. K. Morrison. 2001. C-TAK1 regulates Ras signaling by phosphorylating the MAPK scaffold, KSR1. Mol. Cell 8:983-993. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen, A., W. R. Burack, J. L. Stock, R. Kortum, O. V. Chaika, M. Afkarian, W. J. Muller, K. M. Murphy, D. K. Morrison, R. E. Lewis, J. McNeish, and A. S. Shaw. 2002. Kinase suppressor of Ras (KSR) is a scaffold which facilitates mitogen-activated protein kinase activation in vivo. Mol. Cell. Biol. 22:3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pawlak, G., and D. M. Helfman. 2001. Cytoskeletal changes in cell transformation and tumorigenesis. Curr. Opin. Genet. Dev. 11:41-47. [DOI] [PubMed] [Google Scholar]
- 40.Pawlak, G., and D. M. Helfman. 2002. Post-transcriptional down-regulation of ROCKI/Rho-kinase through an MEK-dependent pathway leads to cytoskeleton disruption in Ras-transformed fibroblasts. Mol. Biol. Cell 13:336-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Percival, J. M., G. Thomas, T. A. Cock, E. M. Gardiner, P. L. Jeffrey, J. J. Lin, R. P. Weinberger, and P. Gunning. 2000. Sorting of tropomyosin isoforms in synchronised NIH 3T3 fibroblasts: evidence for distinct microfilament populations. Cell Motil. Cytoskel. 47:189-208. [DOI] [PubMed] [Google Scholar]
- 42.Perry, R. L., M. H. Parker, and M. A. Rudnicki. 2001. Activated MEK1 binds the nuclear MyoD transcriptional complex to repress transactivation. Mol. Cell 8:291-301. [DOI] [PubMed] [Google Scholar]
- 43.Pittenger, M. F., J. A. Kazzaz, and D. M. Helfman. 1994. Functional properties of non-muscle tropomyosin isoforms. Curr. Opin. Cell Biol. 6:96-104. [DOI] [PubMed] [Google Scholar]
- 44.Prasad, G. L., R. A. Fuldner, and H. L. Cooper. 1993. Expression of transduced tropomyosin 1 cDNA suppresses neoplastic growth of cells transformed by the ras oncogene. Proc. Natl. Acad. Sci. USA 90:7039-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prasad, G. L., L. Masuelli, M. H. Raj, and N. Harindranath. 1999. Suppression of src-induced transformed phenotype by expression of tropomyosin-1. Oncogene 18:2027-2031. [DOI] [PubMed] [Google Scholar]
- 46.Ridley, A. J., and A. Hall. 1992. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 70:389-399. [DOI] [PubMed] [Google Scholar]
- 47.Ridley, A. J., and A. Hall. 1994. Signal transduction pathways regulating Rho-mediated stress fibre formation: requirement for a tyrosine kinase. EMBO J. 13:2600-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roy, F., G. Laberge, M. Douziech, D. Ferland-McCollough, and M. Therrien. 2002. KSR is a scaffold required for activation of the ERK/MAPK module. Genes Dev. 16:427-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sahai, E., A. S. Alberts, and R. Treisman. 1998. RhoA effector mutants reveal distinct effector pathways for cytoskeletal reorganization, SRF activation and transformation. EMBO J. 17:1350-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sahai, E., M. F. Olson, and C. J. Marshall. 2001. Cross-talk between Ras and Rho signalling pathways in transformation favours proliferation and increased motility. EMBO J. 20:755-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seddighzadeh, M., S. Linder, M. C. Shoshan, G. Auer, and A. A. Alaiya. 2000. Inhibition of extracellular signal-regulated kinase 1/2 activity of the breast cancer cell line MDA-MB-231 leads to major alterations in the pattern of protein expression. Electrophoresis 21:2737-2743. [DOI] [PubMed] [Google Scholar]
- 52.Shah, V., S. Bharadwaj, K. Kaibuchi, and G. L. Prasad. 2001. Cytoskeletal organization in tropomyosin-mediated reversion of ras-transformation: evidence for Rho kinase pathway. Oncogene 20:2112-2121. [DOI] [PubMed] [Google Scholar]
- 53.Shields, J. M., H. Mehta, K. Pruitt, and C. J. Der. 2002. Opposing roles of the extracellular signal-regulated kinase and p38 mitogen-activated protein kinase cascades in Ras-mediated downregulation of tropomyosin. Mol. Cell. Biol. 22:2304-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stamm, S., D. Casper, J. P. Lees-Miller, and D. M. Helfman. 1993. Brain-specific tropomyosins TMBr-1 and TMBr-3 have distinct patterns of expression during development and in adult brain. Proc. Natl. Acad. Sci. USA 90:9857-9861.7694294 [Google Scholar]
- 55.Sternberg, P. W., and J. Alberola-Ila. 1998. Conspiracy theory: RAS and RAF do not act alone. Cell 95:447-450. [DOI] [PubMed] [Google Scholar]
- 56.Stewart, S., M. Sundaram, Y. Zhang, J. Lee, M. Han, and K.-L. Guan. 1999. Kinase suppressor of Ras forms a multiprotein signaling complex and modulates MEK localization. Mol. Cell. Biol. 19:5523-5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sugimoto, T., S. Stewart, M. Han, and K. L. Guan. 1998. The kinase suppressor of Ras (KSR) modulates growth factor and Ras signaling by uncoupling Elk-1 phosphorylation from MAP kinase activation. EMBO J. 17:1717-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sumi, T., K. Matsumoto, and T. Nakamura. 2000. Specific activation of LIM kinase 2 via phosphorylation of threonine 505 by ROCK, a Rho-dependent protein kinase. J. Biol. Chem. 276:670-676. [DOI] [PubMed] [Google Scholar]
- 59.Sundaram, M., and M. Han. 1995. The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell 83:889-901. [DOI] [PubMed] [Google Scholar]
- 60.Takenaga, K., and A. Masuda. 1994. Restoration of microfilament bundle organization in v-raf-transformed NRK cells after transduction with tropomyosin 2 cDNA. Cancer Lett. 87:47-53. [DOI] [PubMed] [Google Scholar]
- 61.Therrien, M., H. C. Chang, N. M. Solomon, F. D. Karim, D. A. Wassarman, and G. M. Rubin. 1995. KSR, a novel protein kinase required for RAS signal transduction. Cell 83:879-888. [DOI] [PubMed] [Google Scholar]
- 62.Totsukawa, G., Y. Yamakita, S. Yamashiro, D. J. Hartshorne, Y. Sasaki, and F. Matsumura. 2000. Distinct roles of ROCK (Rho-kinase) and MLCK in spatial regulation of MLC phosphorylation for assembly of stress fibers and focal adhesions in 3T3 fibroblasts. J. Cell Biol. 150:797-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xing, H., K. Kornfeld, and A. J. Muslin. 1997. The protein kinase KSR interacts with 14-3-3 protein and Raf. Curr. Biol. 7:294-300. [DOI] [PubMed] [Google Scholar]
- 64.Yang, N., O. Higuchi, K. Ohashi, K. Nagata, A. Wada, K. Kangawa, E. Nishida, and K. Mizuno. 1998. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature 393:809-812. [DOI] [PubMed] [Google Scholar]
- 65.Yu, W., W. J. Fantl, G. Harrowe, and L. T. Williams. 1998. Regulation of the MAP kinase pathway by mammalian Ksr through direct interaction with MEK and ERK. Curr. Biol. 8:56-64. [DOI] [PubMed] [Google Scholar]
- 66.Zhou, M., D. A. Horita, D. S. Waugh, R. A. Byrd, and D. K. Morrison. 2002. Solution structure and functional analysis of the cysteine-rich C1 domain of kinase suppressor of Ras (KSR). J. Mol. Biol. 315:435-446. [DOI] [PubMed] [Google Scholar]