Abstract
The mechanism by which gene regulatory proteins gain access to their DNA target sites is not known. In vitro, binding is inherently cooperative between arbitrary DNA binding proteins whose target sites are located within the same nucleosome. We refer to such competition-based cooperativity as collaborative competition. Here we show that arbitrarily chosen foreign DNA binding proteins, LexA and Tet repressor, cooperate with an adjacently binding endogenous activator protein, Gcn4, to coactivate expression of chromosomal reporter genes in Saccharomyces cerevisiae. Coactivation requires that the cooperating target sites be within a nucleosome-length distance; it leads to increased occupancy by Gcn4 at its binding site; and it requires both Gcn5 and Swi/Snf which, at an endogenous Gcn4-dependent promoter, act subsequent to Gcn4 binding. These results imply that collaborative competition contributes to gene regulation in vivo. They further imply that, even in the presence of the cell's full wild-type complement of chromatin remodeling factors, competition of regulatory proteins with histone octamer for access to regulatory target sites remains a quantitative determinant of gene expression levels. We speculate that initial target site recognition and binding may occur via spontaneous nucleosomal site exposure, with remodeling factor action required downstream to lock in higher levels of regulatory protein occupancy.
Eukaryotic gene regulatory regions typically encompass multiple DNA target sites for one or more regulatory proteins within a space of a few hundred base pairs or less. The proteins that bind to these sites may act cooperatively (synergistically), thereby enhancing the level of binding of the regulatory proteins over that which they could achieve acting individually, while also conferring combinatorial control over the genes.
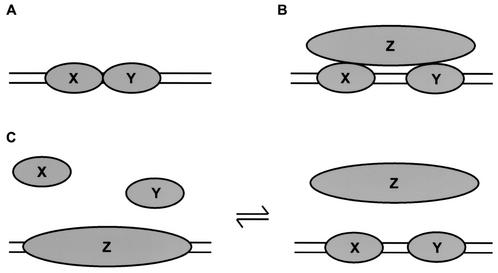
When a pair of such regulatory proteins does function cooperatively in vivo, the simplest mechanism is that the two proteins directly contact each other, with some favorable free energy of contact (Fig. 1A). A hallmark of this mechanism is that the cooperativity can be detected as the mutually cooperative binding of the two proteins to DNA in a purified in vitro system consisting only of those proteins and a DNA fragment encompassing their binding sites.
FIG. 1.
Types of protein-protein cooperativity. (A) Conventional cooperativity. Proteins X and Y touch each other with some favorable free energy. (B) Indirect cooperativity. Proteins X and Y both bind protein Z; thus, the presence of Z confers cooperativity on the binding of X and Y. Examples of such bridging occur for multiple transcription factors in the preinitiation complex (36). Alternatively, a specific Z could bend the DNA between X and Y, enabling conventional cooperative contacts (see text). The HMGI(Y) proteins in the beta interferon enhanceosome illustrate both of these (panels A and B), engaging in direct protein-protein interactions and favorably altering the DNA conformation (25). (C) Competitive cooperativity. Proteins X and Y collaborate in their competition against a common rival, Z. Z may dissociate or change conformation upon binding of X and Y. As an aside, we note that collaborative competition need not be restricted to DNA binding proteins. For example, two proteins could compete with another protein for binding sites on a fourth protein.
In other cases, studies of purified systems reveal that a pair of regulatory proteins that act cooperatively in vivo nevertheless do not bind cooperatively to naked DNA. We shall refer to such a pair of proteins as X and Y; these may be two different proteins or two molecules of the same protein. It is customary in such cases to invoke the existence of a specific additional protein, Z. If Z has the property of binding specifically to both X and Y, then the mere presence of Z in the cell automatically confers cooperativity on the binding of X and Y (Fig. 1B). (In a variant of this idea, Z may be a protein that specifically binds to and bends the intervening DNA instead, bringing the binding sites of X and Y into closer proximity, allowing favorable contacts to be made between X and Y that were inaccessible to the system with a linear DNA fragment in vitro.) In certain cases, such Z proteins have in fact been identified (e.g., references 25 and 34 to 36). In other cases (e.g., references 30, 45, and 55), no such specific Z has yet been found; many investigators simply assume that one exists and ultimately will be identified. A hallmark of this mechanism is that X and Y fail to exhibit cooperative binding to naked DNA on their own, in a purified in vitro system, but cooperativity appears once the specific Z is included in the reactions.
There appears, however, to be a third class of cooperative interactions in gene regulation in vivo which cannot be understood as either direct cooperativity (Fig. 1A) or indirect cooperativity (Fig. 1B). A customary finding is that, when constructing artificial gene activation regions, multiple binding sites placed close together along the DNA provide far higher levels of activation than do individual protein binding sites, and these effects appear to be greater than additive. The exact spacing of the binding sites, however, appears to be relatively unconstrained. These effects were investigated in detail in two studies (30, 45) which revealed that cooperativity in gene activation was due in part to cooperative interactions arising from the minimal DNA binding domains alone. Direct cooperative protein-protein interactions (Fig. 1A) were definitively ruled out, yet the available data also argued against the indirect mechanism (Fig. 1B): the diversity of origins and structures of the DNA binding domains used and the lack of constraint on their spacing along the DNA both argue against a requirement for specific protein-protein interactions with unknown protein Z. Thus, these data suggested that some other mode of cooperativity may be operative in vivo, but the data could not define what the origin or mechanism of this cooperativity might be.
Note that indirect cooperativity (Fig. 1B) can never be formally excluded, because it is impossible to prove the nonexistence of such a protein Z. However, the principle of Occam's razor asserts that, if no evidence specifically requires the postulated existence of such a protein Z and if an alternative mechanism exists that need not invoke the additional component, this alternative mechanism is to be preferred as the default explanation.
In the present study, we will show that there does exist a distinct mechanism of cooperativity, operative in eukaryotic cells in vivo, that does not require the existence of any specific interacting protein Z. Instead, a ubiquitously occurring Z confers cooperativity on arbitrary X and Y through a mechanism based on collaborative competition (Fig. 1C). X and Y bind cooperatively to DNA target sites that are otherwise occluded by the same arbitrary Z, simply as a consequence of fighting against their common competitor. Because multiple regulatory DNA target sites often occur within a nucleosome-length distance, this cooperativity is to be expected, with the ubiquitous histone octamer playing the role of Z.
Such a mechanism was suggested first from studies on purified systems in vitro (1, 32). These studies revealed that binding of arbitrary proteins to sites contained within a single nucleosome was inherently cooperative, and they provided a mechanistic framework through which this behavior could be quantitatively understood. The explanation involves a dynamic behavior intrinsic to nucleosomes which we have termed site exposure (31). Briefly, nucleosomes in vitro are in a dynamic equilibrium with transient states that have altered conformations in which the DNA has partially unwrapped (3), most likely starting from one end, such that stretches of DNA that in the time average are buried inside nucleosomes are nevertheless transiently, yet constantly, accessible even to proteins that, when bound, occlude the entire circumference of their DNA target sites. A consequence of this behavior is to confer cooperativity on the binding of arbitrarily chosen DNA binding proteins to target sites contained within the same nucleosome. Provided only that two or more proteins individually require site exposure to allow their binding to the nucleosomal DNA, then a powerful cooperativity develops between the proteins because the binding of the first protein prevents the recapture of DNA in its proximity, thereby reducing the free energy cost for the second protein to bind. We term this kind of cooperative interaction collaborative competition. Any conventional cooperativity (Fig. 1A and B) would add to this unconventional cooperativity; however, for positioned nucleosomes in vitro, the magnitude of this novel nucleosome-dependent cooperativity alone is large in comparison to most conventional cooperativities.
Subsequent studies provided evidence in support of this nucleosome-dependent cooperativity functioning in vivo (50, 51). These studies utilized plasmid-based reporter assays and investigated the effects of placing a binding site for the LexA protein (a prokaryotic repressor protein) near the binding site for an endogenous yeast activator protein in cells that constitutively expressed the LexA protein. The collaborative competition model predicts that the LexA protein in these circumstances should function as a coactivator, enhancing the activity of the endogenous gene-activating protein. In accord with this prediction, such cooperativity was observed in vivo yet was not seen on naked DNA, and in vivo the cooperativity was found to dissipate as the distance between the sites was increased over a nucleosome length.
Our new study complements and extends this work. We test the collaborative competition mechanism in vivo by using direct assays on chromosomal reporter constructs, with multiple binding partners, on both pools of cells and single cells. We vary the concentration and affinity of the binding partners. We test the key prediction that collaborative competition acts through regulatory protein binding site occupancy, and we investigate the additional effects of chromatin remodeling factors. Finally, we also extend the theoretical analysis to account for the possibility of nucleosome mobility in vivo. The previous in vivo studies are interpreted in terms of the uncoiling model of site exposure (33, 52), which presupposes that nucleosomes remain fixed in space. But in vivo, there exists the likelihood of nucleosome mobility catalyzed by protein machines containing the ISWI class of motor domains (13, 16, 20, 47, 49). In the presence of such activity, the model developed for the in vitro situation would not apply. We show below that collaborative competition will nevertheless be expected even with mobile nucleosomes, and we explain the origin of this behavior.
Our results imply that collaborative competition against nucleosomes is operative in vivo and that it provides a generally applicable mechanism for cooperativity in gene regulation. The results further provide mechanistic insight on how any protein can access its DNA binding site in vivo, and they argue against a commonly assumed role for ATP-dependent chromatin remodeling factors.
MATERIALS AND METHODS
DNA constructs and yeast manipulations.
The HIS3-derived DNA constructs (Fig. 2) were PCR amplified, gel purified, and cloned by standard techniques (24) into BamHI/XhoI sites of pRS304 (40). Modified PCR primers were used to incorporate the desired binding site and an optimized Gcn4 response element (GRE). EcoRV-linearized plasmids were integrated by one-step transformation (12) in the trp1 locus of strain W303 (a his3-11,15 trp1-1 ura3-1 leu2-112 ade2-1 can1-100; gift from Richard Gaber), strain J848 (trp1 his3 ura3 YOR290c::KanMX4), strain J3576 (trp1 his3 ura3 YGR252w::KanMX4), strain W303.1A (a W303 converted to HIS3; gift from Kevin Struhl; unpublished reagent), and strain W303.1A Gcn4-3xMyc (W303.1A with three Myc tags on the Gcn4 C terminus; gift from Kevin Struhl; unpublished reagent). J848 was created by crossing LHY298 (α trp1 his3 ura3 leu2 bar1; gift from Linda Hicke) with YOR290c (swi2Δ; EUROSCARF). J3576 was made by crossing LHY298 and YGR252w (gcn5Δ; EUROSCARF). The 154-bp spacer was constructed from two copies of a pGEM3Z polylinker and one copy each of T7 and SP6 promoters. The green fluorescent protein (GFP) gene was amplified from pEGFP-N1 (Clontech) and precisely replaced the HIS3 coding sequence. Expression plasmids for LexA (from pJWL228; gift from John Little) (39) and TetR (from pCM148; gift from Enrique Herrero) (10) were derived from pYES2 (Invitrogen). All restriction and DNA modifying enzymes were purchased from New England Biolabs and used in the recommended buffers. Strains were grown in synthetic complete media with 2% glucose or galactose, lacking uracil if needed to maintain plasmid selection. When indicated, 10 mM aminotriazole (Sigma) was added to induce histidine starvation. When designated, the inducible TetR strain was grown overnight in the presence of 10 μg of doxycycline (50% ethanol stock; Sigma)/ml.
FIG. 2.
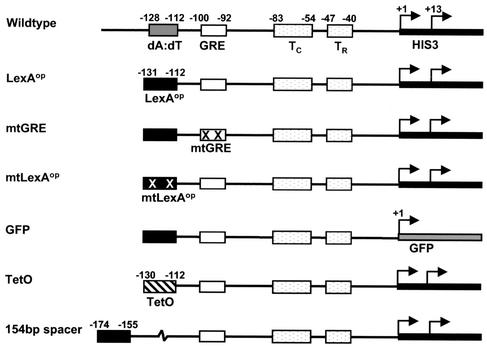
DNA constructs. Wild type indicates the natural HIS3 upstream region which contains two major transcription start sites (+1 and +13), two TATA boxes (TC and TR), a GRE, and a poly(dA-dT) element. In the LexAop construct, the poly(dA-dT) tract from the HIS3 upstream element has been replaced with the LexA operator (TACTGTATGAGCATACAGTA). The mtGRE construct is identical to LexAop except that the GRE has been mutated (ATCACTCGT; mutations shown in bold). The mtLexAop construct has two point mutations in LexAop (TACCGTATGAGCATACGGTA). In the GFP construct, the HIS3 coding sequence is replaced by that for GFP. The TetO construct replaces the poly(dA-dT) element from HIS3 with an operator for the tetracycline repressor protein (ACTCTATCAATGATAGAGT). The 154-bp spacer construct has 154 bp of nonyeast DNA between the GRE and LexAop.
High-resolution mapping.
Nuclei were prepared from 0.5- to 1.0-liter yeast cultures growing logarithmically (1.5 × 107 to 3 × 107 cells/ml) as described elsewhere (22) (except that buffer A was used in place of buffer B). Micrococcal nuclease (MNase; Sigma) reactions were performed at 0.013 U/g of nuclei at 30°C for 30 to 90 min. DNA was obtained by proteinase K (1 mg/ml) treatment, phenol-chloroform extraction, and ethanol precipitation followed by RNase A treatment. Samples were extracted and precipitated again. DNA was also purified from nuclei without MNase digestion to obtain control naked DNA. Naked DNA was digested in vitro as follows: 25 μg of naked DNA in 2.5 mM CaCl2 in a 50-μl reaction volume was treated with 0.0004 U of MNase at 30°C for 25 min. Multiple-cycle primer extension assays were performed on HindIII-digested DNA. Reaction mixtures contained 1× Taq buffer, 8 μg of digested DNA, a 0.2 mM concentration of deoxynucleoside triphosphates, 0.9 pmol of γ-32P-end-labeled primer (pRSPE; 5′GGAAACAGCTATGACCATGATTACGCCAAG), and 1 U of Taq polymerase in a 25-μl final volume. Reactions were incubated for 5 min at 94°C before initiation of 30 cycles at 94°C for 1 min, 65°C for 6 min, and 72°C for 2 min. Reactions were then incubated for 5 min at 72°C. Samples were precipitated, washed, and analyzed by 7.5% denaturing sequencing polyacrylamide gel electrophoresis (PAGE). Equivalent results were obtained using PvuII-digested DNA and a primer downstream of the area of interest (data not shown).
Indirect end labeling.
Chromatin and naked DNA were prepared from 0.5- to 1.0-liter yeast cultures grown in glucose or galactose as described above. Two micrograms of PvuII-digested DNA along with radiolabeled 100-bp and kilobase ladders (Amersham BioSciences) were run on a 1.4% agarose gel in 1× Tris-acetate-EDTA. Neutral transfer to Nytran N 0.2-μm membrane was accomplished using the Turboblotter Rapid Downward Transfer system (Schleicher & Schuell, Inc.). The DNSN probe, a PvuII-BamHI fragment from pRS304, was internally labeled (Rediprime II random labeling kit; Amersham Biosciences), purified (Nucleotide Removal Kit; Qiagen), and hybridized with UV-irradiated blots in Church & Gilbert's hybridization solution (8) at 58°C for a minimum of 6 h. Blots assayed with a probe upstream of our construct gave similar results (data not shown).
RNA protection assays.
RNA was prepared from 2 × 108 cells by using an RNeasy Midi kit (Qiagen). A260 readings were used to quantify RNA concentration. S1 nuclease assays were performed as described previously (4). No-RNA controls contained single-stranded calf thymus DNA (Sigma) in place of the RNA. Samples were hybridized with a 20- to 50-fold excess of single-stranded DED1 and HIS3 oligonucleotides (sequences in reference 7). Samples were digested with 120 U of S1 nuclease (Promega) at 37°C for 8 min. After ethanol precipitation, the products were analyzed by 8% denaturing PAGE. RNA levels were quantified using either PhosphorImager analysis (Molecular Dynamics) or Instant Imager software (ImageQuant; Molecular Dynamics), as results were determined to be equivalent (data not shown). After background correction, total HIS3 transcription levels were normalized to the DED1 control. DED1 mRNA levels are unaffected by galactose or amino acid starvation conditions (42). To correct for variations in probe labeling, we considered the values relative to those for the strain grown in glucose without aminotriazole, which should be similar for each of the isogenic strains. HIS3 mRNA levels were measured for the parental strain, W303, and were found to be negligible with the probes used (data not shown). Results shown are the average values and standard errors obtained from at least three assays.
Chromatin immunoprecipitation and quantitative PCR.
Two yeast strains were created with the integrated LexAop-containing reporter (Fig. 2): one with W303.1A (Gcn4 no tag) and one with W303.1A Gcn4-3XMyc (Gcn4 Myc tag). Both were transformed with the LexA expression plasmid and used in chromatin immunoprecipitation assays similar to those described previously (19, 41), with modifications noted. Forty milliliters of logarithmically growing yeast cells (1.2 × 107 to 2 × 107 cells/ml) were cross-linked with 1% formaldehyde for 20 min at room temperature before quenching with 360 mM glycine. Samples were incubated for 5 min and then harvested and washed twice with cold 1× TBS (20 mM Tris-HCl [pH 7.5], 150 mM NaCl). Cells were resuspended in 400 μl of FA lysis buffer (50 mM HEPES-KOH [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1%Triton X-100, 0.1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 mM phenylmethylsulfonyl fluoride). A 400-μl aliquot of zirconium beads (0.5-mm diameter) was added, and cell breakage was accomplished with 10 cycles of vortexing for 30 s followed by placing on ice for 60 s. Beads were allowed to settle, and supernatant was transferred to a new tube. A total 400 μl of FA lysis buffer was used to wash beads; the wash was combined with earlier lysate. Samples were sonicated to 200- to 800-bp fragments, with an average of 400 bp. Whole-cell extract was clarified for 20 min in a microcentrifuge. Twenty or 40% of the extract was immunoprecipitated for 3 to 4 h at room temperature with 5 μg of anti-Myc tag (06-549; Upstate) coupled to protein A-Sepharose CL-4B (Amersham Biosciences). Beads were washed for 5 min with 1 ml of FA lysis buffer twice, 1 ml of FA lysis buffer with 0.5 M NaCl twice, 1 ml of TLEND (10 mM Tris-HCl [pH 8.0], 0.25 M LiCl, 1 mM EDTA, 0.5% NP-40, 0.5% sodium deoxycholate) once, and 1 ml of TE (10 mM Tris-HCl, 1 mM EDTA) once. Immunoprecipitants were eluted in 50 mM Tris-HCl [pH 7.5], 10 mM EDTA, 1% SDS at 65°C for 20 min. Beads were removed by centrifugation before cross-linking was reversed by incubating at 65°C overnight in the presence of 0.8 mg of proteinase K/ml. DNA, in the presence of 0.4 M LiCl and 36.4 μg of glycogen/ml, was phenol-chloroform extracted, ethanol precipitated, resuspended in TE, and RNase A treated.
Quantitative PCR analysis was performed in a 50-μl volume using Pfu polymerase (Stratagene) in the presence of [α-32P]dCTP, a 1 mM concentration of primers, 0.1 mM concentration of deoxynucleoside triphosphates, and 1/50 to 1/200 immunoprecipitated DNA or 1/25,000 to 1/35,000 of total DNA. The primers used for HIS3 reactions were ChIPUP2 (5′CCAAGCTCGGAATTAACCCTCACT) and ChIPDN2 (5′GAAGATCGAGTGCTCTATCGCTAG); for reference, primers ChIPRF2 (5′AATTCGACCATTCCGACACAGACG) and ChIPRF3 (5′GATTTTATCGCTAGGTCTCCTGGC) were used; and for HIS4, ChIPHS4-3 (5′CCATCACAATCCTGACAACCAGCA) and ChIPHS4-4 (5′GCCATCCAAAAGTACCTGACCAAC) were used. The HIS3 primers used detect only the LexAop construct (Fig. 2) and not the wild-type HIS3 present (data not shown). Cycle parameters were 94°C for 4 min before 27 cycles (28 for HIS4 reactions) of 94°C for 30 s, 58°C for 30 s, 72°C for 30 s, and then 72°C for 5 min. Reactions were determined to be within linear range. Products were analyzed on 7% native PAGE. After background correction, the cross-linking efficiency was determined by taking the ratio of PCR product obtained from immunoprecipitated material over total material at a specific locus and subtracting the ratio obtained from immunoprecipitated material over total material of PCR product in the reference reactions. Immunoprecipitations were repeated twice for each strain, and PCR was performed in duplicate or in triplicate for each immunoprecipitated sample.
Flow cytometry.
GFP reporter strains with (inducible LexA) and without (LexA−) the LexA expression plasmid were switched from glucose- to galactose-containing media and allowed to grow at 30°C for 18 to 22 h (late log/early stationary). Just prior to analysis, cells were pelleted, washed, and resuspended at 2 × 106 cells/ml of water. The green fluorescence of 500,000 cells was measured using a FACSCalibur flow cytometer (Becton Dickinson) with a 530/30 band-pass filter and analyzed using CELLQuest3.1 software.
RESULTS
To test for collaborative competition in gene activation in vivo, we asked whether arbitrarily chosen DNA binding proteins would function as inducible coactivators for chromosomal reporter genes. We chose two unrelated, simple DNA binding proteins from a foreign kingdom of life (eubacteria)—LexA protein and tetracycline repressor protein from Escherichia coli—to provide a stringent test for the idea that mere occupancy of a nearby site on the DNA, not the presence of a specific gene activation domain or specific protein-protein contacts, is sufficient to enhance site occupancy of a neighboring protein. We incorporated the target sites for these DNA binding proteins in the upstream element of the Saccharomyces cerevisiae HIS3 gene near the binding site for the natural activator protein, Gcn4 (Fig. 2). Earlier studies of HIS3 gene regulation showed that binding and transcriptional activation by Gcn4 is suppressed by organization of its target site in chromatin and enhanced in the presence of a DNA element, poly(dA-dT), that can destabilize nucleosomes (14). Collaborative competition of the foreign protein and Gcn4 with histone octamer (or with higher levels of chromatin structure) should allow the foreign DNA binding protein to serve as a coactivator for HIS3 gene expression by enhancing the ability of Gcn4 to bind and to stimulate transcription.
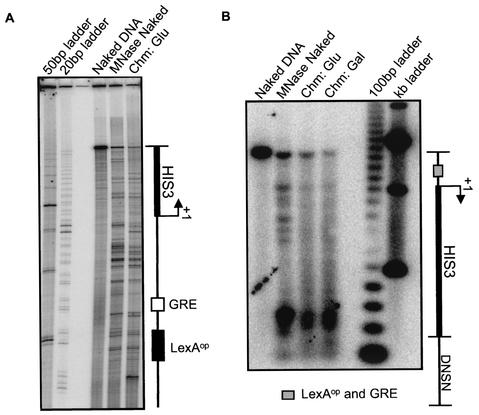
The natural yeast HIS3 locus is nucleosomal, yet the nucleosomes are not specifically positioned in the promoter region (14). Because our reporter constructs place HIS3 derivatives in a new chromosomal location, we analyzed their nucleosomal organization. High-resolution mapping of MNase accessibility by using primer extension revealed neither a single nucleosome length (147 bp) region of protection nor a single pair of strong bands representing unique nucleosome end points (Fig. 3A). Instead, differences between the native chromatin and naked DNA samples were detected along the entire length of the region probed, consistent with organization of the region in nucleosomes that are not biased to specific positions. Low-resolution mapping using indirect end labeling confirmed that the promoter region is broadly protected in chromatin, yet lacks significantly biased nucleosome positions (Fig. 3B), closely similar to results obtained for the natural HIS3 locus (14). Despite nonpositioned nucleosomes, collaborative competition could be expected when the binding sites are incorporated into the same nucleosome; the overall response of the system would represent a weighted average over many nucleosome positions, which may be equilibrating or frozen in place in each cell (see Discussion).
FIG. 3.
Nonpositioned nucleosomes on the HIS3 upstream elements. (A) Primer extension analysis of nucleosome positioning using a primer upstream of the HIS3 start site. Naked DNA, DNA treated with MNase in vitro (MNase Naked), and chromatin prepared from the LexA− strain grown in glucose (Chm: Glu) were assayed. Similar results were obtained for chromatin obtained from the inducible LexA strain grown in galactose (data not shown). The schematic indicates the positions of the binding sites for LexA (black) and Gcn4 (white). (B) An indirect end-labeling assay of nucleosome positioning was performed with a probe downstream of HIS3 gene, DNSN, on samples from the analysis shown in panel A as well as on chromatin prepared from the inducible LexA strain grown in galactose (Chm: Gal). The gray box indicates the location of the LexA and Gcn4 binding sites.
We first examined a modified HIS3 gene that incorporated a binding site for LexA (LexAop). Two yeast strains were created with the integrated LexAop-containing reporter (Fig. 2): one with a galactose-inducible source of LexA (inducible LexA) and one without (LexA−). Strains were grown in glucose (repressing condition) or galactose (inducing condition), with or without aminotriazole (a reagent that causes histidine starvation and results in elevated Gcn4 protein levels [2]). HIS3 mRNA levels were measured using an S1 nuclease protection assay.
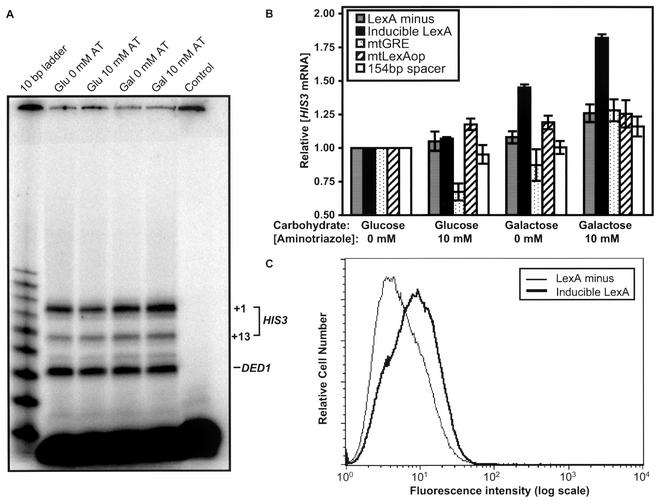
Figure 4A shows representative results for the inducible LexA strain. A LexA-dependent increase in HIS3 mRNA levels is evident in the raw data. Figure 4B summarizes the quantitative results. When no LexA expression plasmid was present (LexA− strain), no significant increase occurred in HIS3 mRNA levels under any of the conditions tested. Similar results were obtained if the LexA gene was present but repressed (inducible LexA strain; glucose). However, when the LexA gene was present and induced (inducible LexA strain; galactose), the HIS3 mRNA level increased 1.4-fold. Expression was further enhanced to 1.8-fold in the presence of aminotriazole, close to the threefold activation of the natural HIS3 locus (43) and the twofold Gcn4-dependent activation of ILV1 (26), and comparable in magnitude to physiologically important phenomena, such as dosage compensation and haploinsufficiency diseases (6, 9).
FIG. 4.
LexA coactivates gene expression in yeast. (A) Shown is an S1 assay for the inducible LexA strain grown in glucose (Glu) or galactose (Gal) with 0 or 10 mM aminotriazole (AT). The control sample is a parallel reaction lacking RNA. Total HIS3 mRNA levels from +1 and +13 initiation sites are quantified and normalized to DED1 levels. (B) The average HIS3 mRNA levels and standard errors from quantitative S1 assays are presented normalized to the glucose without aminotriazole condition for the following strains: LexA−, inducible LexA, mtGRE, mtLexAop, and 154-bp spacer. (C) Flow cytometric analysis was performed on GFP reporter strains grown in galactose. The histogram depicts the number of cells and their fluorescence intensity (log scale) for a representative run. The medians for LexA− and inducible LexA strains are 4.7 and 8.2 fluorescence units.
These results are consistent with the simple foreign DNA binding protein acting as either a direct activator or as a coactivator of HIS3 transcription in vivo. In either case, binding of LexA to its operator would be a necessary step. The mtLexAop strain, which has point mutations in the binding site for LexA that hinder the ability of LexA to bind specifically (21), suppressed the LexA-dependent activation of HIS3 (Fig. 4B). This indicates that not only must LexA protein be present in the cells, but also that LexA must be able to bind to that particular site in order to enhance transcription.
The increase in LexA-dependent expression for the inducible LexA strain in the presence of aminotriazole suggests that the response is Gcn4 dependent. In a strain carrying two point mutations in the Gcn4 binding site (mtGRE) that suppress Gcn4 binding (27), basal levels decrease in glucose with aminotriazole, as expected (38); more importantly, no LexA-dependent activation was observed (Fig. 4B). Thus, LexA acts not as a direct activator, but only as a coactivator of HIS3 expression.
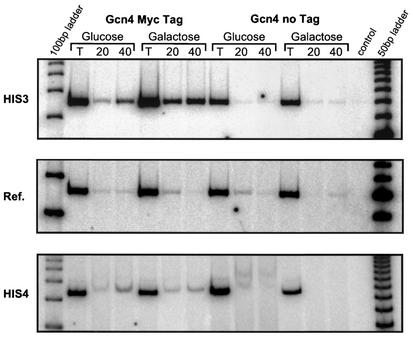
A critical prediction of the collaborative competition mechanism is that coactivation of the reporter by LexA should directly result in an increased occupancy by Gcn4 at its own binding site. Chromatin immunoprecipitated with a Myc antibody from an inducible LexA strain with a Myc-tagged Gcn4 demonstrated a 2.0- to 18-fold increase, with a geometric mean of 5.4-fold, in Gcn4 occupancy at HIS3 when LexA is expressed (Fig. 5, compare galactose versus glucose). A sister strain lacking the epitope tag (Gcn4 no tag) gave much weaker signals, consistent with background levels (Fig. 5, middle panel reference bands) and had equivalent or less immunoprecipitated material in galactose as in glucose. Furthermore, at the HIS4 locus, which has a Gcn4 binding site but no LexA binding site, cross-linking efficiency was not increased in the presence of LexA (galactose) but, rather, a slight decrease in occupancy was observed (a 0.7-fold increase for the experiment shown in Fig. 5, bottom panel). Taken together, these data confirm the prediction of the collaborative competition model that LexA binding leads to increased occupancy of the endogenous activator, Gcn4, at a closely linked binding site but has no influence on the ability of Gcn4 to bind at other loci.
FIG. 5.
LexA increases occupancy at the neighboring Gcn4 binding site. Chromatin immunoprecipitation assays were performed with anti-Myc on inducible LexA strains containing either Myc epitope-tagged Gcn4 (Gcn4 Myc tag) or wild type Gcn4 (no tag). Strains were grown in glucose or galactose. Twenty or 40% of the whole-cell extract was used in the immunoprecipitation. PCR was performed with primers to the upstream element of our modified HIS3 construct (HIS3), of HIS4, and to a reference gene (Ref.), using total DNA (T) and precipitated DNA (20 and 40). A control reaction with no template DNA was also performed.
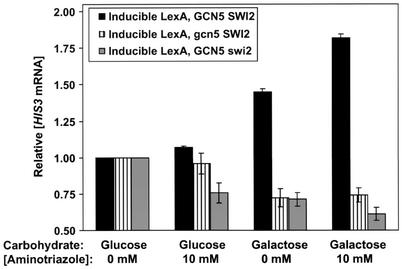
Normal transcriptional activation by Gcn4 requires the activities of both the Gcn5 histone acetyltransferase and the Swi/Snf ATP-dependent chromatin remodeling complex (11, 28, 44), which function subsequent to (downstream of) site-specific binding by Gcn4 (15, 18, 28). Thus, if LexA is coactivating via natural Gcn4 function, Gcn5 and Swi/Snf should still be required for HIS3 transcriptional activation. Consistent with this prediction, inducible LexA strains that carry either gcn5 or swi2 (the ATPase domain of Swi/Snf) deletions are unable to induce HIS3 expression under any condition (Fig. 6).
FIG. 6.
Gcn5 and Swi/Snf are required for HIS3 coactivation. Depicted are S1 assay results for the inducible LexA strain with wild-type Gcn5 and Swi/Snf (from the analysis shown in Fig. 4B), for the strain with Gcn5 deleted and for the strain with Swi2 deleted. Relative HIS3 mRNA levels were calculated as described in the legend for Fig. 4. Average values and standard errors are presented.
The lack of strongly positioned nucleosomes raised the possibility that the population of cells might include a mixture of strong responders and nonresponders. To address this question, we created variants of our LexAop-containing constructs that replaced HIS3 with a GFP reporter (Fig. 2). Quantitative measures of GFP expression in individual cells were obtained using flow cytometry for the LexA− and inducible LexA variant strains grown in galactose (Fig. 4C). We noted that comparing a strain grown in different carbohydrates was difficult because of the variations in cell physiology and GFP expression, even for the LexA− strain with the GFP reporter. We therefore compared GFP reporter strains, with or without a LexA expression plasmid, grown in galactose to minimize LexA-independent variations. Expression of LexA in this system led to a single distribution of fluorescence intensities that was shifted to higher GFP expression compared to that of the strain lacking LexA. The observed single distribution rules out the possibility that a minority of strong responder cells within the population is responsible for the entirety of the LexA-dependent gene activation, and it is consistent with the entire population of cells responding to a modest extent.
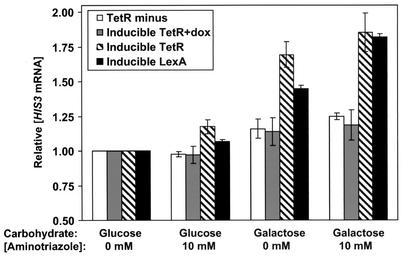
We investigated if coactivation was unique to LexA by replacing the LexA binding site with that for an unrelated protein, tetracycline repressor (TetR). The strains with the integrated TetO-containing reporter (Fig. 2) behaved similarly in glucose media, with or without aminotriazole (Fig. 7). When the strains were grown in galactose, the inducible TetR strain expressed significantly more HIS3 mRNA than the TetR− strain. Addition of aminotriazole to the galactose cultures further increased the HIS3 transcription levels for the inducible TetR strain. If the inducible TetR strain is grown in the presence of doxycycline, a tetracycline derivative that inhibits the ability of TetR to bind to TetO (10), there is no activation of HIS3 transcription in the presence of TetR. Overall, the results from the inducible TetR strain were quantitatively comparable to those from the inducible LexA strain (Fig. 7). LexA and TetR appear to affect HIS3 mRNA levels in an analogous manner.
FIG. 7.
The tetracycline repressor coactivates gene expression in yeast. S1 assay results for the TetR− strain, the inducible TetR strain with or without doxycycline (dox), and the inducible LexA strain (from the analysis shown in Fig. 4B) are shown. Relative HIS3 mRNA levels were calculated as described in the legend for Fig. 4. Average values and standard errors are shown.
Our observations that both LexA and TetR coactivate Gcn4-dependent gene expression, and that coactivation results in increased occupancy by Gcn4, are best explained by a model in which LexA or TetR are helping Gcn4 compete against a common rival. The most likely candidate for the competitor is a single histone octamer, although this need not be the case. The competitor must be large, because the foreign protein and Gcn4 binding sites are separated by 11 bp (∼40 Å) and span 40 bp (∼140 Å). Consistent with the possibility that the competitor is a histone octamer, increasing the separation of the binding sites to greater than a nucleosome length of DNA (154-bp spacer [Fig. 2]) abolished the cooperativity (Fig. 4B).
DISCUSSION
Cooperativity in gene regulation from collaborative competition with histone octamer in vivo.
The results of this study demonstrate that arbitrarily chosen foreign DNA binding proteins are able to function as coactivators of gene expression for chromosomally located eukaryotic genes in vivo. Together with the studies of Vashee et al. (50, 51), our new results establish the following. (i) Multiple arbitrarily chosen foreign DNA binding proteins coactivate, but do not directly activate, reporter gene expression. (ii) Multiple endogenous activator proteins can respond in this way. (iii) Binding of the coactivator causes increased occupancy by the endogenous activator at its own binding site. And (iv) the cooperativity decreases with increasing separation between the coactivator and activator binding sites and is eliminated when the binding sites are separated by a nucleosome length of DNA. Conventional direct or indirect cooperativities cannot plausibly account for these results, given the arbitrary and foreign nature of the coactivators. We conclude that coactivation occurs through a mechanism based on collaborative competition, with a histone octamer serving as the common competitor.
Furthermore, our laboratory showed earlier that cooperativity arising from collaborative competition with histone octamer is an inherent property of nucleosomes in vitro (32); we show below that such cooperativity is to be expected even if nucleosomes are mobile, as may occur in vivo.
We conclude from these data that there exists a mode of cooperativity that is operative in gene regulation in vivo that is distinct from both conventional direct and indirect cooperativity but acts instead through collaborative competition of diverse gene regulatory proteins against a histone octamer. This mechanism likely accounts for aspects of the behavior of artificially constructed regulatory regions (30, 45); it provides a concrete mechanism for other cases of natural promoters in which conventional direct and indirect cooperativities are considered unlikely (55); and it provides a plausible (and simpler) alternative mechanism for cases in which specific regulatory partners have been proposed yet direct evidence for the existence of such partners is lacking.
Our results are to be distinguished from studies of effects of histone loss (53), because the effects observed here are due directly to protein binding at a given regulatory locus and are operative in cells that continue living, in contrast to the artificial situation during global histone depletion.
Cooperativity arising from collaborative competition does not preclude other mechanisms of cooperativity. Collaborative competition may be superimposed upon, and additive with, conventional direct or indirect cooperativity and, thus, may contribute significantly to net regulatory behavior, even when conventional direct or indirect cooperativities are also operative.
A particularly interesting feature of the cooperativity arising from collaborative competition with nucleosomes, which distinguishes it from these other conventional modes of cooperativity, is that it arises automatically from the close juxtaposition of binding sites for arbitrarily chosen DNA binding proteins. This suggests a possible role for collaborative competition in the evolution of gene regulatory modules because it means that combinatorially regulated modules are easily assembled from randomly chosen components, with no initial requirement for coevolution. Subsequent coevolution of the chosen partners can then increase the cooperativity through conventional protein-protein contacts or bridging proteins, increasing the magnitude of the combinatorial control.
Collaborative competition with imprecisely positioned or mobile nucleosomes.
Distinct molecular mechanisms may account for collaborative competition with a histone octamer as the competitor. Our laboratory's earlier analysis applied to the case of binding of arbitrary pairs of proteins to target sites buried within the same nucleosome, with the nucleosome restricted to particular positions (32). If nucleosomes occupy a single position, this will result in a fixed amount of cooperativity which would pertain to that nucleosome in every cell in the population. Alternatively, if nucleosomes are immobile but are distributed with some probability function over many positions (in a population of cells, for example, or with nonpositioned nucleosomes in vitro), each position will result in a different cooperativity, with some positions likely resulting in no cooperativity. (For example, certain nucleosome positions may place pairs of binding sites on two different nucleosomes, or may place one or both sites in linker DNA.)
In vivo nucleosomes may be mobile, with protein binding accompanied by translocation of a competing nucleosome. Would collaborative competition still occur? The following analysis reveals that it can, with two distinct molecular origins for the resulting cooperativity.
Suppose first that a competing nucleosome is “positioned”—that is, it is energetically biased to one or more particular preferred sequence regions along the DNA—and that binding of a first regulatory protein to a target site in that nucleosome requires and is accompanied by translocation of the nucleosome off of the favored position(s) onto one or a set of less-favored positions, with a corresponding free energy cost. For simplicity, we shall take the set of less-favored positions to be equivalent and neutral, i.e., equal to random sequence DNA (a straightforward extension of these ideas relieves this assumption). The free energy diagram that describes this model is identical to that analyzed earlier (32), with the free energy cost of nucleosome translocation replacing the free energy cost of partial DNA unwrapping. Thus, it follows that this system of preferentially positioned, but mobile, nucleosomes will still allow for collaborative competition, with the magnitude of the cooperativity given by the free energy of translocating the nucleosome off the favored position(s). This free energy of translocation is equal to the difference in free energy of histone-DNA interactions, as measured in competitive nucleosome reconstitution experiments (23). Such free energies can be quite large for natural nucleosome positioning sequences—on the order of ∼2 kcal mol−1 (46), corresponding to cooperative effects on binding affinities of up to ∼30-fold.
Alternatively, even if nucleosomes (mobile or immobile) are not positioned by preferences for particular DNA sequence regions, constraints arising from the statistical positioning of nucleosomes between boundary elements with changing occupancies (17) or from higher-order chromatin structure (54) can provide comparable or even larger free energy costs for nucleosome translocation. Thus, collaborative competition would again be expected to occur.
The observed ∼2-fold increases in mRNA and protein expression levels may or may not represent the maximum that is possible with this mechanism. The absence of strongly biased nucleosome positions should reduce the cooperativity significantly below what could be obtained with a single strongly biased nucleosome position. In addition, the activation on our reporter gene constructs, which are derivatives of the natural HIS3 gene, is likely limited by other steps in the activation mechanism, since the maximal induction for the wild-type HIS3 gene is threefold (43). We chose the HIS3 gene for our initial investigations because the available evidence (14) pointed to this regulatory region as one that was poised to respond positively to collaborative competition. It will be of interest in the future to carry out analogous studies on derivatives of promoters that exhibit higher levels of inducibility and that have strongly positioned nucleosomes.
Cooperative invasion of nucleosomes by site-specific regulatory factors.
If, as appears to be the case, the common competitor in the present studies is indeed a histone octamer, this would have two important ramifications. First, it would imply that in living cells, with their full complement of chromatin remodeling factors, competition for binding by site-specific regulatory proteins with histone octamer is a quantitatively important determinant of gene expression. Second, it would mean that remodeling factors in vivo do not automatically render all chromatin sites competent for site-specific protein binding: evidently, the cell's wild-type complement of chromatin remodeling factors does not render nucleosomes “transparent” for regulatory protein binding in vivo.
What, then, would be the roles of the remodeling factors? The fact that coactivation is dependent on both Gcn5 and Swi/Snf suggests that collaborative competition and chromatin remodeling are on the same pathway for gene activation. It is possible that the initial site-specific DNA recognition and binding events themselves are under Gcn5 and/or Swi/Snf control. However, an attractive alternative possibility is that invasion of nucleosomes by gene regulatory factors may occur in stages, with initial DNA target site recognition and binding occurring independently of the remodeling factors, perhaps facilitated by dynamic properties inherent to the nucleosomes themselves. Global acetylation and deacetylation could also influence the site exposure of the binding site (15). In this initial “establishment” stage, regulatory protein binding would be sensitive to competition with histone octamer, and the cooperativity described here would be operative. Consistent with this view, Gcn4 and other transcription factors can in fact bind, site specifically, prior to targeted recruitment of the chromatin remodeling factors Gcn5 and Swi/Snf (5, 18, 28, 29, 37, 44, 48). Thus, regulatory protein binding could be followed by the recruitment and subsequent action of remodeling factors that may move the nucleosome or otherwise change its properties so as to prevent it from further competition, thereby allowing and “locking in” a higher level of occupancy of the regulatory factors.
Acknowledgments
This work was supported by an NIH grant (J.W.) and a Howard Hughes Medical Institute (HHMI) Predoctoral Fellowship (J.A.M.).
We thank Peggy Lowary for valuable scientific discussions and Eileen Chang for cloning the 154-bp spacer plasmid. We thank Paul Mason and Kevin Struhl for reagents and helpful advice in the chromatin immunoprecipitation assay and Randall Morse for the suggestion to use the HIS4 locus as a control. We gratefully acknowledge the use of the flow cytometer in the laboratory of Robert Lamb (HHMI) and instruments in the Keck Biophysics Facility at Northwestern University (http://www.biochem.northwestern.edu/Keck/keckmain.html).
REFERENCES
- 1.Adams, C. C., and J. L. Workman. 1995. Binding of disparate transcriptional activators to nucleosomal DNA is inherently cooperative. Mol. Cell. Biol. 15:1405-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht, G., H.-U. Mosch, B. Hoffmann, U. Reusser, and G. H. Braus. 1998. Monitoring the Gcn4 protein-mediated response in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 273:12696-12702. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, J. D., A. Thastrom, and J. Widom. 2002. Spontaneous access of proteins to buried nucleosomal DNA target sites occurs via a mechanism that is distinct from nucleosome translocation. Mol. Cell. Biol. 22:7147-7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1989. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York, N.Y.
- 5.Becker, P. B., and W. Horz. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71:247-273. [DOI] [PubMed] [Google Scholar]
- 6.Birchler, J. A., U. Bhadra, M. P. Bhadra, and D. L. Auger. 2001. Dosage-dependent gene regulation in multicellular eukaryotes: implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev. Biol. 234:275-288. [DOI] [PubMed] [Google Scholar]
- 7.Chen, W., S. Tabor, and K. Struhl. 1987. Distinguishing between mechanisms of eukaryotic transcriptional activation with bacteriophage T7 RNA polymerase. Cell 50:1047-1055. [DOI] [PubMed] [Google Scholar]
- 8.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Elia, A. V., G. Tell, I. Paron, L. Pellizzari, R. Lonigro, and G. Damante. 2001. Missense mutations of human homeoboxes: a review. Hum. Mutat. 18:361-374. [DOI] [PubMed] [Google Scholar]
- 10.Gari, E., L. Piedrafita, M. Aldea, and E. Herrero. 1997. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13:837-848. [DOI] [PubMed] [Google Scholar]
- 11.Georgakopoulos, T., and G. Thireos. 1992. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J. 11:4145-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito, H., Y. Fukada, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito, T., M. E. Levenstein, D. V. Fyodorov, A. K. Kutach, R. Kobayashi, and J. T. Kadonaga. 1999. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 13:1529-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iyer, V., and K. Struhl. 1995. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 14:2570-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katan-Khaykovich, Y., and K. Struhl. 2002. Dynamics of global histone acetylation and deacetylation in vivo: rapid restoration of normal histone acetylation status upon removal of activators and repressors. Genes Dev. 16:743-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kent, N. A., N. Karabetsou, P. K. Politis, and J. Mellor. 2001. In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev. 15:619-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kornberg, R. D., and L. Stryer. 1988. Statistical distributions of nucleosomes: nonrandom locations by a stochastic mechanism. Nucleic Acids Res. 16:6677-6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuo, M. H., E. vom Baur, K. Struhl, and C. D. Allis. 2000. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell 6:1309-1320. [DOI] [PubMed] [Google Scholar]
- 19.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399:609-613. [DOI] [PubMed] [Google Scholar]
- 20.Langst, G., and P. B. Becker. 2001. Nucleosome mobilization and positioning by ISWI-containing chromatin-remodeling factors. J. Cell Sci. 114:2561-2568. [DOI] [PubMed] [Google Scholar]
- 21.Lewis, L. K., G. R. Harlow, L. A. Gregg-Jolly, and D. W. Mount. 1994. Identification of high affinity binding sites for LexA which define new DNA damage-inducible genes in Escherichia coli. J. Mol. Biol. 241:507-523. [DOI] [PubMed] [Google Scholar]
- 22.Lowary, P. T., and J. Widom. 1989. Higher-order structure of Saccharomyces cerevisiae chromatin. Proc. Natl. Acad. Sci. USA 86:8266-8270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowary, P. T., and J. Widom. 1997. Nucleosome packaging and nucleosome positioning of genomic DNA. Proc. Natl. Acad. Sci. USA 94:1183-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Merika, M., and D. Thanos. 2001. Enhanceosomes. Curr. Opin. Genet. Dev. 11:205-208. [DOI] [PubMed] [Google Scholar]
- 26.Moreira, J. M., W. Horz, and S. Holmberg. 2002. Neither Reb1p nor poly(dA:dT) elements are responsible for the highly specific chromatin organization at the ILV1 promoter. J. Biol. Chem. 277:3202-3209. [DOI] [PubMed] [Google Scholar]
- 27.Mosch, H.-U., R. Graf, T. Schmidheini, and G. Braus. 1990. Three GCN4 responsive elements act synergistically as upstream and as TATA-like elements in the yeast TRP4 promoter. EMBO J. 9:2951-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Natarajan, K., B. M. Jackson, H. Zhou, F. Winston, and A. G. Hinnebusch. 1999. Transcriptional activation by Gcn4p involves independent interactions with the SWI/SNF complex and the SRB/mediator. Mol. Cell 4:657-664. [DOI] [PubMed] [Google Scholar]
- 29.Neely, K. E., A. H. Hassan, A. E. Wallberg, D. J. Steger, B. R. Cairns, A. P. Wright, and J. L. Workman. 1999. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol. Cell 4:649-655. [DOI] [PubMed] [Google Scholar]
- 30.Oliviero, S., and K. Struhl. 1991. Synergistic transcriptional enhancement does not depend on the number of acidic activation domains bound to the promoter. Proc. Natl. Acad. Sci. USA 88:224-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polach, K. J., and J. Widom. 1995. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J. Mol. Biol. 254:130-149. [DOI] [PubMed] [Google Scholar]
- 32.Polach, K. J., and J. Widom. 1996. A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. J. Mol. Biol. 258:800-812. [DOI] [PubMed] [Google Scholar]
- 33.Polach, K. J., and J. Widom. 1999. Restriction enzymes as probes of nucleosome stability and dynamics. Methods Enzymol. 304:278-298. [DOI] [PubMed] [Google Scholar]
- 34.Rabbitts, T. H. 2001. Chromosomal translocation master genes, mouse models and experimental therapeutics. Oncogene 20:5763-5777. [DOI] [PubMed] [Google Scholar]
- 35.Reeves, R., and L. Beckerbauer. 2001. HMGI/Y proteins: flexible regulators of transcription and chromatin structure. Biochim. Biophys. Acta 1519:13-29. [DOI] [PubMed] [Google Scholar]
- 36.Reinberg, D., G. Orphanides, R. Ebright, S. Akoulitchev, J. Carcamo, H. Cho, P. Cortes, R. Drapkin, O. Flores, I. Ha, J. A. Inostroza, S. Kim, T. K. Kim, P. Kumar, T. Lagrange, G. LeRoy, H. Lu, D. M. Ma, E. Maldonado, A. Merino, F. Mermelstein, I. Olave, M. Sheldon, R. Shiekhattar, L. Zawel, et al. 1998. The RNA polymerase II general transcription factors: past, present, and future. Cold Spring Harb. Symp. Quant. Biol. 63:83-103. [DOI] [PubMed] [Google Scholar]
- 37.Ryan, M. P., R. Jones, and R. H. Morse. 1998. SWI-SNF complex participation in transcriptional activation at a step subsequent to activator binding. Mol. Cell. Biol. 18:1774-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sellers, J. W., A. C. Vincent, and K. Struhl. 1990. Mutations that define the optimal half-site for binding yeast GCN4 activator protein and identify an ATF/CREB-like repressor that recognizes similar DNA sites. Mol. Cell. Biol. 10:5077-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shepley, D. P., and J. W. Little. 1996. Mutant LexA proteins with specific defects in autodigestion. Proc. Natl. Acad. Sci. USA 93:11528-11533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strahl-Bolsinger, S., A. Hecht, K. Luo, and M. Grunstein. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11:83-93. [DOI] [PubMed] [Google Scholar]
- 42.Struhl, K., W. Chen, D. E. Hill, I. A. Hope, and M. A. Oettinger. 1985. Constitutive and coordinately regulated transcription of yeast genes: promoter elements, positive and negative regulatory sites, and DNA binding proteins. Cold Spring Harb. Symp. Quant. Biol. 50:489-503. [DOI] [PubMed] [Google Scholar]
- 43.Struhl, K., and R. W. Davis. 1981. Transcription of the his3 gene region in Saccharomyces cerevisiae. J. Mol. Biol. 152:535-552. [DOI] [PubMed] [Google Scholar]
- 44.Syntichaki, P., I. Topalidou, and G. Thireos. 2000. The Gcn5 bromodomain coordinates nucleosome remodelling. Nature 404:414-417. [DOI] [PubMed] [Google Scholar]
- 45.Tanaka, M. 1996. Modulation of promoter occupancy by cooperative DNA binding and activation-domain function is a major determinant of transcriptional regulation by activators in vivo. Proc. Natl. Acad. Sci. USA 93:4311-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thastrom, A., P. T. Lowary, H. R. Widlund, H. Cao, M. Kubista, and J. Widom. 1999. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J. Mol. Biol. 288:213-229. [DOI] [PubMed] [Google Scholar]
- 47.Tsukiyama, T., C. Daniel, J. Tamkun, and C. Wu. 1995. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell 83:1021-1026. [DOI] [PubMed] [Google Scholar]
- 48.Utley, R. T., K. Ikeda, P. A. Grant, J. Cote, D. J. Steger, A. Eberharter, S. John, and J. L. Workman. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394:498-502. [DOI] [PubMed] [Google Scholar]
- 49.Varga-Weisz, P. D., M. Wilm, E. Bonte, K. Dumas, M. Mann, and P. B. Becker. 1997. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature 388:598-602. [DOI] [PubMed] [Google Scholar]
- 50.Vashee, S., K. Melchor, W. V. Ding, S. A. Johnson, and T. Kodadek. 1998. Evidence for two modes of cooperative DNA binding in vivo that do not involve direct protein-protein interactions. Curr. Biol. 8:452-458. [DOI] [PubMed] [Google Scholar]
- 51.Vashee, S., J. Willie, and T. Kodadek. 1998. Synergistic activation of transcription by physiologically unrelated transcription factors through cooperative DNA-binding. Biochem. Biophys. Res. Commun. 247:530-535. [DOI] [PubMed] [Google Scholar]
- 52.Widom, J. 2001. Role of DNA sequence in nucleosome stability and dynamics. Q. Rev. Biophys. 34:269-324. [DOI] [PubMed] [Google Scholar]
- 53.Wyrick, J. J., F. C. Holstege, E. G. Jennings, H. C. Causton, D. Shore, M. Grunstein, E. S. Lander, and R. A. Young. 1999. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402:418-421. [DOI] [PubMed] [Google Scholar]
- 54.Yao, J., P. T. Lowary, and J. Widom. 1993. Twist constraints on linker DNA in the 30-nm chromatin fiber: implications for nucleosome phasing. Proc. Natl. Acad. Sci. USA 90:9364-9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu, L., N. Sabet, A. Chambers, and R. H. Morse. 2001. The N-terminal and C-terminal domains of RAP1 are dispensable for chromatin opening and GCN4-mediated HIS4 activation in budding yeast. J. Biol. Chem. 276:33257-33264. [DOI] [PubMed] [Google Scholar]