Abstract
The Pax-6 gene encodes a transcription factor containing both a paired and a homeodomain and is highly conserved among Metazoa. In both vertebrates and invertebrates, Pax-6 is required for eye morphogenesis, development of parts of the central nervous system, and, in some phyla, for the development of olfactory sense organs. Ectopic expression of Pax-6 from insects, mammals, cephalopods, and ascidians induces ectopic eyes in Drosophila, suggesting that Pax-6 may be a universal master control gene for eye morphogenesis. Platyhelminthes are an ancient phylum, originating from the base of spiralian protostomes, that bear primitive eyes, consisting of a group of rhabdomeric photoreceptor cells enclosed in a cup of pigment cells. The analysis of Pax-6 and its expression pattern should provide insights into the ancestral function of Pax-6 in eye morphogenesis. We have identified the Pax-6 gene of the planarian Dugesia(G)tigrina (Platyhelminthes; Turbellaria; Tricladida). This gene shares significant sequence identity and conserved genomic organization with Pax-6 proteins from other phyla. Phylogenetic analysis indicates that it clusters with the other Pax-6 genes, but in the most basal position. DtPax-6 is expressed as a single transcript in both regenerating and fully grown eyes, and electron microscopy studies show strong expression in the perykarion of both photoreceptor and pigment cells. Very low levels of expression also are detectable in other body regions. Because a bona fide Pax-6 homolog so far has not been detected in diploblastic animals, we speculate that Pax-6 may be typical for triploblasts and that the appearance of additional Pax genes may have coincided with increasingly complex body plans.
Keywords: homeobox, paired box, eye evolution, regeneration
Developmental mechanisms in species from various animal phyla display profound similarities, suggesting that they were already in place very early in the evolutionary history of metazoans (1). These observations have led to the description of a common molecular ancestor (2). However, the different body plans observed today are the likely product of alterations in developmental pathways accumulated during evolution. It is therefore equally important to elucidate the differences in developmental pathways. Some of the best-studied cases of conservation relate to transcription factors. A special group of developmental regulators is the Pax gene family of transcription factors. Characteristic for this family is the presence of the 128-aa-long, conserved DNA-binding motif, the paired domain (3). Pax genes originally were described in Drosophila and since have been identified in a large variety of triploblastic animals (4) and, more recently, in the diploblastic phylum Cnidaria (5, 6).
In addition to the paired domain, some Pax genes encode a conserved octapeptide C-terminal to the paired domain and/or a partial or complete homeodomain (7). Pax-6 is one of the Pax genes that has a complete homeodomain. It was identified originally in humans and mouse (8, 9). Since then, Pax-6 was identified in a number of vertebrates and in a variety of invertebrates, including Drosophila (10), the ascidian Phallusia mammillata (11), the sea urchin Paracentrotus lividus (12), the cephalopod Loligo opalescens (13), the nemertean Lineus sanguineus (14), the nematode Caenorhabditis elegans (15, 16), and the cephalochordate Branchiostoma floridae (17). Sequence comparison of all these genes with the vertebrate genes shows striking conservation in their DNA-binding domains. Their genomic organization, domain sequences, and function are highly conserved in the Pax-6 homologs analyzed so far in the animal kingdom (4). Expression studies in animals of different phyla reveal conserved expression in the central nervous system, the developing eye, and in other cephalic organs. The induction of supernumerary eyes in Drosophila by ectopic expression in imaginal discs of Pax-6 genes of Drosophila, mouse, ascidian, and squid indicate that the Pax-6 dependent eye developmental pathway is well conserved and may have a common evolutionary (monophyletic) origin (18–21).
Very recently, Pax genes also were identified in several Cnidarians, including the sea nettle Chrysaora quinquecirrha and the hydra Hydra littoralis (5), the coral Acropora millepora (6), and the hydromedusa Podocoryne carnea (H. Gröger, P.C., W.J.G., and V. Schmid, unpublished observations). Although Pax genes with an octapeptide and complete homeodomain were identified (5), and some relationship with Pax-6 was demonstrated, no unambiguous Pax-6 gene was identified. This observation raises the possibility that authentic Pax-6 genes are part of the evolutionary history of triploblastic animals and that the appearance of additional Pax genes in triploblastic animals may have contributed to the increase in body plan complexity.
In this study we mainly address the following questions. Do animals at the base of the triploblastic phylogenetic tree have real Pax-6 genes or rather do they have a gene more like Pax-B (5)? Is this putative Pax-6 gene expressed in the eye, and, if so, is the expression pattern restricted to accessory cells or is it also seen in photoreceptor cells?
Planarians originally were considered the stereotypical representatives of the simplest organism in the tree of life, possessing three tissue layers (triploblastic), bilateral symmetry, cephalization, and complex organ systems. However, more recent data, including molecular analyses of 18S ribosomal RNA and homeobox genes, resulted in the inclusion of planarians in the lophotrochozoa protostomia clade (22–25). Planarians can be considered simplified organisms by progenesis (26). Nevertheless, planarians are at the base of the lophotrochozoa clade and, as such, are a suitable model with which to study evolutionary questions. Planarians have been studied in great detail for their regenerative abilities, but their development is extremely modified (27) or absent, as in the species of this study, Dugesia(G)tigrina, which reproduces exclusively by asexual fission. The eye spots of planarians consist of two cell types—a bipolar nerve cell with a rhabdomere as a photoreceptive structure and a cup-shaped structure composed of pigmented cells (28)—and represent one of the most ancestral and simple types of visual systems. The turnover of the eyes is supported by the differentiation of undifferentiated and totipotent cells (neoblasts) present in the adult. In the early stages of regeneration the primitive pigmented cells originate from the blastema (regenerative mesenchymal tissue) close to the primitive visual cells derived from nerve cells from the regenerated cephalic ganglia. This mixture of cells then is rearranged, each cell moves to its respective position, and a normal eye spot is formed (29).
We report the identification of a Pax-6 gene from the planarian Dugesia(G)tigrina (DtPax-6). The high degree of amino acid sequence identity in the paired and homeodomain and the conservation of the genomic organization suggest that DtPax-6 is orthologous to known vertebrate and invertebrate Pax-6 genes. The expression of DtPax-6 in intact and regenerating planarians suggests a putative role in development and maintenance of the eye. Furthermore, the positioning of the planarians at the base of the protostomes may also indicate that the ancestors of protostomes and deuterostomes already possessed a Pax-6 gene active in eye development.
MATERIALS AND METHODS
Species.
The planarians used in this study belong to an asexual race (class A; ref. 30) of the species Dugesia (G) tigrina (Girard), and specimens were collected near Barcelona. They were maintained in spring water. Two-week-starved organisms were used in all experiments. Planarians, 9–10 mm long, were cut postpharyngeally according to Saló and Baguñà (31) and left regenerating in Petri dishes with spring water in the dark at 17°C.
Isolation of the DtPax-6 Gene.
A Pax-6 fragment was amplified by PCR from planarian genomic DNA with two degenerate primers corresponding to the conserved residues 48–52 (GCVSK) and 99–103 (WEIRD) of the paired domain as described in ref. 14. A nonamplified planarian genomic library in λ FixII was screened with the 162-bp amplified Pax-6 fragment based on ref. 32. Two phages of 17 kb were isolated, both containing the 5′ and partial coding region of the planarian Pax-6 gene. The cDNA DtPax-6 ends were amplified by rapid amplification of cDNA ends by PCR (RACE-PCR) using the Amplifinder TM RACE (CLONTECH) kit with RNA obtained as described in ref. 33. Amplified fragments were cloned in pBluescript (Stratagene), and the sequence was determined by dideoxy sequencing with the ABI Prism kit (Perkin–Elmer).
Phylogenetic Analysis of the Pax-6 Paired Domains and Homeodomains.
The phylogenetic trees of Pax paired and homeodomain sequences were inferred by using the mega software (34). The Poisson-correction distance of protein sequences was used for the evolutionary distances, and the neighbor-joining method was used for tree construction. The paired-domain sequences and homeodomain sequences were as used in ref. 5 except for the sea urchin sequences Su-Pax 258, Su-Pax B, Su-Pax B1, and C. elegans 258 (35).
Reverse Transcription–PCR (RT-PCR) and Northern Blot.
One microgram of total RNA was denatured in the presence of random hexamers and incubated with Moloney murine leukemia virus-reverse transcriptase (Promega) at 42°C for 1 h. Of the cDNA reaction mixture, 1/25 was used for the PCR with specific primers. HB-F and HB-R primers, respectively, covered cDNA positions 1226–1250 and 1363–1387 (residues 5–12 and 50–58 of the homeodomain). PCR cycles were performed as follows: 5 min at 94°C, 30 cycles (30 sec at 94°C; 30 sec at 60°C; 30 sec at 72°C), and 2 min at 72°C. The presence of a 45-bp intron in the amplified sequence allowed us to distinguish cDNA from genomic amplification. As an internal control we coamplified Dth-2 transcripts, which are homogeneously distributed along the planarian anteroposterior axis (32). Dth-2 primers covered cDNA positions 376–400 (exon 1) and 817–841 (exon 3), respectively.
Northern blot analyses were performed by standard procedures (36).
In Situ Hybridization.
In situ hybridizations were carried out in frozen or paraffin sections with 35S riboprobes basically according to ref. 32. Electron microscopy in situ hybridization was performed over ultrathin sections from Lowicryl K4M embedded samples (37). Immunocytochemical detection of digoxigenin was performed with antidigoxigenin antibody conjugated to gold particles that were 10 nm in diameter (Boehringer Mannheim).
RESULTS
Isolation and Sequence Comparison of DtPax-6 with Other Pax Genes.
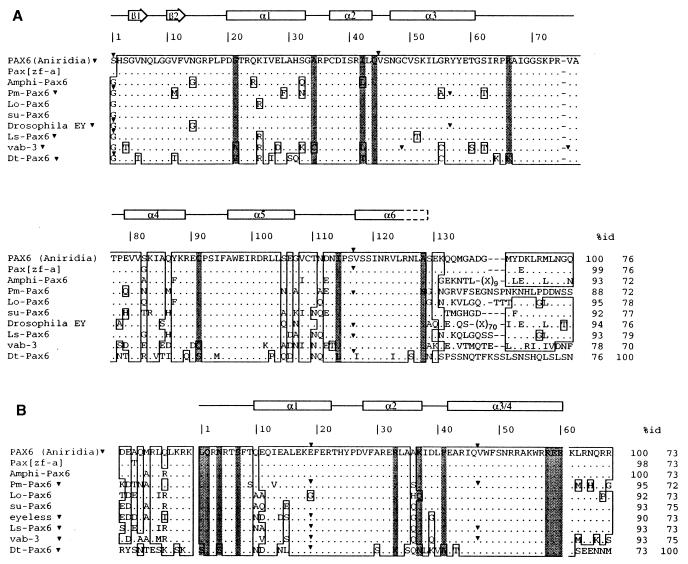
Initial isolation and partial characterization of a planarian Pax-6 homolog was achieved with PCR amplification of genomic DNA with two sets of degenerate oligonucleotides (11) and subsequent screening of a genomic library. A complete cDNA of 2124 bp was identified by RACE-PCR of both ends. The ORF encodes 577 aa, with two regions of high sequence conservation in the deduced protein, the paired domain, and the homeodomain. Comparison with Pax-6 sequences of other species (Fig. 1) shows the lowest identity described to date with values between 72–79% and 72–75% in the paired domain and homeodomain, respectively. However, some of the characteristic residues of family VI and the conserved residues that define their DNA-binding specificity positions 42, 44, and 47 of the paired domain are present in DtPax-6. No other region of similarity was observed, even in the linker region, where the presence of a conserved motif of 11 aa was described (4). The flanking regions of the homeodomain show the lowest similarity. The C-terminal region comprises 193 aa rich in serine (13%), asparagine (12%), proline (7%), and threonine (5.5%). Thirty percent of these serines and other less frequent residues scattered throughout the sequence also are present in the same position of the Drosophila Pax-6 homolog eyeless.
Figure 1.
Alignments of the amino acid sequences of the paired domains (A) and homeodomains (B) of Pax-6 homologs of some vertebrates and invertebrates. The secondary structure of the domains are shown along the top. The consensus sequence for all genes of the paired family are compared with human PAX6 (Aniridia) and zebrafish as representatives for vertebrate sequences, Amphioxus Branchiostoma floridae (AmphiPax-6) (17), Ascidian Phallusia mammillata (PmPax-6) (11), cephalopod Loligo opalescens (LoPax-6) (13), sea urchin Paracentrotus lividus (PlPax-6) (12), Drosophila (Ey) (10), nemertean Lineus sangineus (LsPax-6) (14), and the nematode C. elegans (vab-3) (15, 16). Solid arrowheads indicate the position of intron–exon boundaries. Shaded bars indicate the Pax-6 specific amino acids (38). Percentages of sequence identity of the paired domains and homeodomains were determined by comparison with human PAX6 and planarian Pax-6, respectively, and are indicated after the sequences.
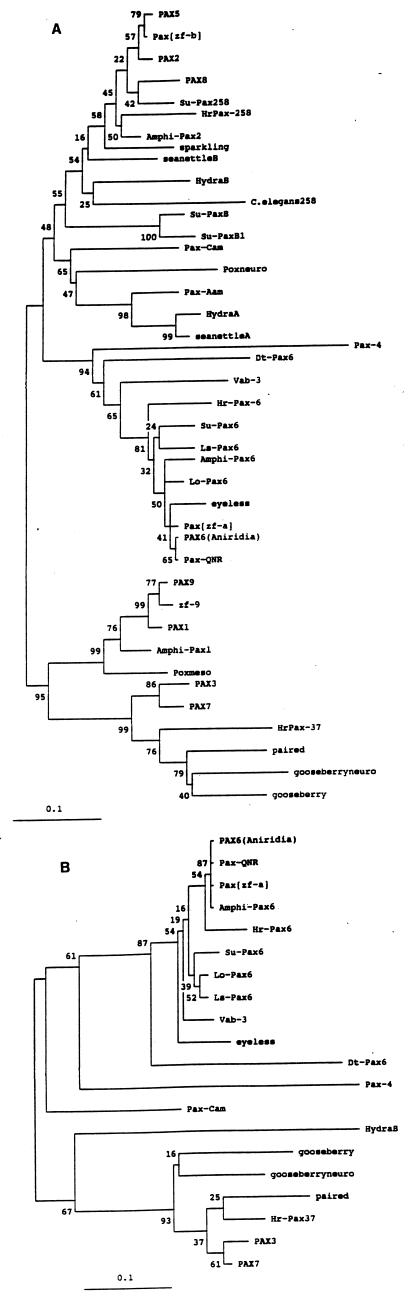
Phylogenetic trees were constructed for paired and homeodomain sequences. The clustering of DtPax-6 with the other Pax-6 genes is supported by a high bootstrap value. Similar results were obtained for paired domain and homeodomain sequences (Fig. 2).
Figure 2.
Phylogenetic trees of Pax gene paired domains (A) and homeodomains (B). The planarian DtPax-6 paired domains and homeodomains clearly cluster with other Pax-6 genes. Human Pax genes: PAX1, 2, 3, 5, 6 (Aniridia), 7, 8, 9; mouse Pax gene: Pax-4; quail Pax gene: Pax-QNR; zebrafish Pax genes: Pax[zf-a], [zf-b], zf-9; sea urchin (Paracentrotus lividus) Pax genes: Su-Pax B, B1, 6, 258; Halocynthia roretzi Pax genes: HrPax-37, 6, 258; Amphioxus Pax genes: Amphi-Pax1, 2, 6; Loligo opalescens Pax gene: Lo-Pax6; Drosophila Pax genes: sparkling, eyeless, poxneuro, poxmeso, paired, gooseberry neuro, gooseberry; C. elegans Pax genes: C. elegans 258, vab-3; Lineus sanguineus Pax gene: Ls-Pax6; Dugesia(G)tigrina Pax gene: Dt-Pax6; seanettle Pax genes: seanettle A, B; Hydra Pax genes: Hydra A, B; Acropora millepora Pax genes: Pax-Aam, Pax-Cam.
DtPax-6 Genomic Organization.
The genomic organization was determined by partial sequencing of a genomic 2.6-kb XbaI fragment from the λ FixII phages and genomic PCRs with specific oligos close to the putative splicing sites. The paired box has two introns located in the first codon and between codons 116 and 117, as is the case in several vertebrates and invertebrates. The DtPax-6 paired box does not contain additional introns. Additional introns have been described for vertebrates (a third intron position between codons 44 and 45), for Drosophila and Phallusia (a specific third intron located at codon 56), and for C. elegans with two new intron positions (Fig. 1A). In the DtPax-6 homeodomain we observed a single, small (45-nt) intron position at codon 19, though a second intron at position 47, which is present in most metazoa analyzed except eyeless of Drosophila (10), is absent in planarian (Fig. 1B). The three conserved splice sites and the phylogenetic tree analysis identify DtPax-6 as a true Pax-6 homolog.
DtPax-6 Expression in Intact and Regenerating Adults.
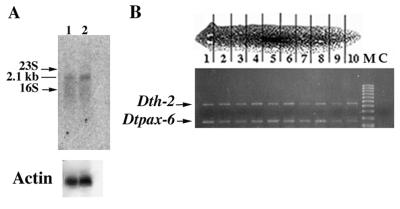
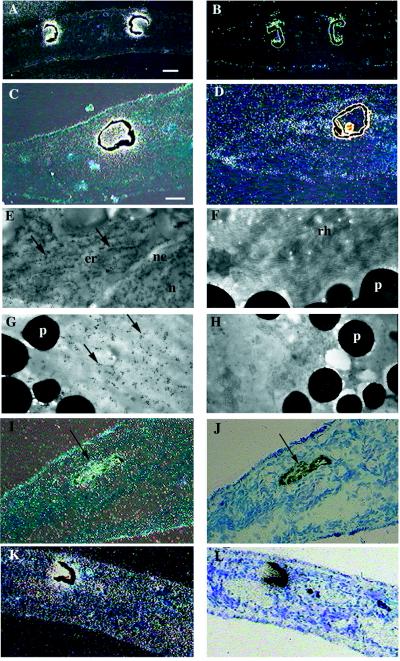
Pax-6 expression in intact and regenerating planarians was analyzed by Northern blotting and in situ hybridization. Northern blot analysis shows a unique 2.1-kb transcript in adults and in regenerative stages without any evidence for differential splicing (Fig. 3A). Regional RT-PCR with specific primers of DtPax-6 that surround one intron position to distinguish DNA from RNA amplification demonstrates the presence of DtPax-6 transcripts along the entire planarian body axis at low concentrations (Fig. 3B). DtPax-6 spatial expression was determined on sagittal and transversal paraffin sections of adult intact and regenerating planarians. In adults, DtPax-6 was expressed continuously and uniformly all over the eye (Fig. 4 A–D). In situ electron microscopy showed the distribution of DtPax-6 transcript in the perykarion of pigment cells and the endoplasmic reticulum of photoreceptor cells (Fig. 4 E–H), while the rhabdomeric region of the photoreceptor cells is negative (Fig. 4F). No expression was detected in other organs such as cephalic ganglia or lateral sense organs (auricula), although it is possible that weak DtPax-6 expression in these regions would not be detected by in situ hybridization. During the early stages of head regeneration, DtPax-6 expression was detected in a group of pigmented cells close to the dorsal epidermis (Fig. 4 I and J), which constitutes the earliest visible sign of eye regeneration. This early expression in the eye primordia was maintained throughout regeneration (Fig. 4 K and L).
Figure 3.
DtPax-6 expression in Dugesia(G)tigrina. (A) Northern blot: 10 μg of poly(A)+ from intact adults (lane 1) and 7-day-regenerating (lane 2) planarians hybridized at high stringency conditions with 0.55-kb DtPax-6 3′ cDNA fragment. A single band of 2.1 kb was observed in both lanes. Blots were rehybridized with a Drosophila 5C actin gene probe (39) to control for levels of RNA loaded. The transcript size and the positions of 23S and 16S RNAs are indicated. (B) Adult planarian DtPax-6 gene expression at different anteroposterior (1–10) regions deduced by RT-PCR amplification using specific oligonucleotides from exons 3 and 4 from the DtPax-6 cDNA sequence (positions 1226–1250 and 1363–1387, respectively) that produce a fragment of 162 nt. As an internal control, we used in the same amplification two new sets of specific oligonucleotides from the Dth-2 cDNA sequence [positions 376–400 and 817–841 (32), respectively] that produce a control fragment of 465 nt. Lane M, 100-bp ladder (Pharmacia) molecular size marker. Lane C, non-retro-transcribed planarian total RNA as a control.
Figure 4.
DtPax-6 expression in intact and regenerating planarian adults by in situ hybridization. (A–H) Intact organisms. (I and J) Seven days of regeneration. (K and L) Fourteen days of regeneration. In situ hybridizations were performed in paraffin sections with 35S-labeled RNA antisense (A, C, I–L) and sense (B and D) or by electron microscopy with digoxigenin-labeled RNA antisense (E–G) and sense (H). A and B are transversal sections. C, D, and I–L are sagittal sections; dorsal is in the top and ventral is in the bottom; anterior is to the left and posterior is to the right. A–D, I, and K are dark-field views; J and L are bright-field views. (A and C) Dark-field view of transversal and sagittal paraffin sections at the eye level of an intact planarian. The dorsal position of the eyes is easily observable by the presence of pigmented cells. Clear homogeneous expression can be observed exclusively in the whole eye spot. (B and D) Similar head sections hybridized with a sense probe. Note the absence of silver grains in the eye and the artifactual grains surrounding the pigment cells because of the pigment granules that are refractile in dark-field. (E) Electron microscopy in situ hybridization of the perikaryon of a photoreceptor cell with gold particles (arrows), indicating the presence of Dt-Pax6 transcript in the endoplasmic reticulum. (F) No staining is visible in the rhabdomeric region of the photoreceptor cell. (G) Electron microscopy in situ hybridization of the eye pigmented cell with positive gold particles (arrows), indicating the presence of DtPax-6 transcript in the cytoplasmic region in between the pigment granules. (H) No staining is visible with the sense probe in the eye pigmented cell cytoplasm. (I and J) Dark- and bright-field sagittal section of 7-day head-regenerating planarians. DtPax-6 expression already was observed when the first sign of eye pigment cell differentiation appears at 7 days and is maintained throughout regeneration. (K and L) Fourteen days of regeneration. n, nucleus; nm, nuclear envelope; er, endoplasmic reticulum; p, pigment granules; rh, rhabdomeres.
DISCUSSION
The planarian Dugesia(G)tigrina has a bona fide Pax-6 gene. The phylogenetic analysis shows clustering of DtPax-6 with the other Pax-6 genes with a very high probability. This notion is corroborated further by the conservation of specific residues and several intron positions, two in the paired and one in the homeodomain, allowing it to be considered as a Pax-6 ortholog. Nevertheless, this gene has the lowest degree of sequence identity when compared with the other Pax-6 sequences. Sequence identity essentially is confined to the DNA-binding domains where most of the Pax-6 specific residues are conserved. No additional sequence homology was found in the linker region between the two domains and in the carboxyl-terminal region. Only a short stretch of amino acids with sequence similarity can be observed N-terminal to the homeodomain. This low degree of sequence conservation may well correlate with the basal position of planarians in the lophotrochozoa (spiralian) protostome clade. The observation that a true Pax-6 homolog is present in planarians but has not been identified in diploblastic animals suggests that Pax-6 might be typical for triploblasts, and the appearance of additional Pax genes may have coincided with the appearance of increasingly complex body plans.
The second question we wanted to address in this study relates to the role of Pax-6 in eye development. Planarian DtPax-6 is expressed continuously in the adult eyes. Initial expression also coincides with the first signs of eye differentiation during cephalic regeneration. The expression of developmental regulatory genes in adults is usual in Dugesia(G)tigrina (25, 26, 32, 40), since planarians show great morphological plasticity because of the presence throughout their life of an undifferentiated totipotent cell type, the neoblast. Regulatory genes are expressed continuously in association with cell turnover and change of proportions in the continuous growth and degrowth or regeneration processes. This is also the case for DtPax-6 as an important regulator in eye development and regeneration in Dugesia. A similar association of continued Pax-6 expression with regeneration capacity has been described for the urodele eye (41). Urodele Pax-6 is expressed in all stages of lens regeneration, while in the intact eye Pax-6 expression is still detected in the lens, neural retina, and pigment epithelium. In amphibians that lose the capacity to regenerate lens, such as the axolotl, Pax-6 is expressed during development, declines in larval stages, and is repressed in adults (41). Therefore, our results suggest that Pax-6 expression in adults could be related to the capacity to regenerate the eyes. The RT-PCR amplification experiments show a distribution of DtPax-6 along the anteroposterior axis, indicating that, as in other organisms, DtPax-6 may be used for developmental processes in structures other than the eye.
Our attempts to induce ectopic eyes by expressing DtPax-6 in Drosophila have been unsuccessful. This is a negative result and, therefore, difficult to interpret. However, the Pax-6 genes from mouse, squid, and sea squirt, all of which show considerably less sequence divergence, have been shown by the same method to be capable of inducing ectopic eyes in Drosophila. Therefore, we assume that the planarian gene has diverged too much to be able to substitute functionally for the Drosophila gene. Judging from the evolutionary analysis of 18S RNA (23), it appears that Triclads evolve faster than other branches of the Platyhelminthes. Therefore, the more slowly evolving marine Polyclads may have a Pax-6 gene that is less diverged from that of higher metazoa than the gene from freshwater planarians.
The observation that DtPax-6 is expressed both in the pigment cells and in the photoreceptors provides further evidence for an early and basic function of Pax-6 in eye evolution. In contrast to higher animals whose Pax-6 is not expressed in differentiated photoreceptor cells (42–44), we demonstrate that DtPax-6 is also expressed in differentiated photoreceptors, suggesting that it is also involved in the maintenance of their differentiated state and in the regulation of opsin genes that are essential for vision. Other genes such as the Otx-related gene, Crx, have been shown to regulate rhodopsin expression in mouse (45). This observation gives a hint as to how increasingly complex eyes may have been built during evolution. The insertion of other genes, including regulatory genes, in the developmental cascade can lead to alterations in structure and function of the eye with changes advantageous to the organism being retained and disadvantageous alterations being selected against.
Acknowledgments
We thank Dr. M. Iglesias for helping in the cryosections, C. Lopez for performing electron microscopic in situ hybridization, and Dr. Daniel Martinez for making software available to P.C. This work was supported by grants from the Swiss National Science Foundation and the Kantons of Basel to W.G., Texas Advanced Research Program Grant 011618-061 to W.-H.L., a grant from the Dirección General de Investigación Cientifica y Técnica to E.S. (Ministerio de Educación y Ciencia, Spain, PB92-0551 and PB95-0579), and Formación de Personal Investigador fellowship to A.M.M.-M. and E.C. from Ministerio de Educación y Ciencia, España, Comisio Interdepartamental de Recerca i Innovacio Tecnologica (Generalitat de Catalunya), and AECI-Mutis, Ministerio de Asuntos Exteriores, respectively.
ABBREVIATIONS
- RT-PCR
reverse transcription–PCR
- RACE-PCR
rapid amplification of cDNA ends by PCR
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ010498).
References
- 1.Valentine J W, Erwin D H, Jablonski D. Developmental Biology. 1996;173:373–381. doi: 10.1006/dbio.1996.0033. [DOI] [PubMed] [Google Scholar]
- 2.Slack J M W, Holland P W H, Graham C F. Nature (London) 1993;361:490–492. doi: 10.1038/361490a0. [DOI] [PubMed] [Google Scholar]
- 3.Bopp D, Burri M, Baumgartner S, Frigerio G, Noll M. Cell. 1986;47:1033–1040. doi: 10.1016/0092-8674(86)90818-4. [DOI] [PubMed] [Google Scholar]
- 4.Callaerts P, Halder G, Gehring W J. Annu Rev Neurosci. 1997;20:483–532. doi: 10.1146/annurev.neuro.20.1.483. [DOI] [PubMed] [Google Scholar]
- 5.Sun H, Rodin A, Zhou Y, Dickinson D P, Harper D E, Hewett-Emmett D, Li W-H. Proc Natl Acad Sci USA. 1997;94:5156–5161. doi: 10.1073/pnas.94.10.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Catmull J, Hayward D C, McIntyre N E, Reece-Hoyes J S, Mastro R, Callaerts P, Ball E E, Miller D J. Dev Genes Evol. 1998;208:352–356. doi: 10.1007/s004270050191. [DOI] [PubMed] [Google Scholar]
- 7.Noll M. Curr Opin Genet Dev. 1993;3:595–605. doi: 10.1016/0959-437x(93)90095-7. [DOI] [PubMed] [Google Scholar]
- 8.Ton C C, Hirvonen H, Miwa H, Weil M M, Monaghan P, Jordan T, Van Heyningen V, Hastie N D, Meijers-Heijboer H, Drechsler M. Cell. 1991;67:1059–1074. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- 9.Walther C, Guénet J L, Simon D, Deutsch U, Jostes B, Goulding M, Plachov D, Balling R, Gruss P. Genomics. 1991;11:424–434. doi: 10.1016/0888-7543(91)90151-4. [DOI] [PubMed] [Google Scholar]
- 10.Quiring R, Walldorf U, Kloter U, Gehring W J. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- 11.Glardon S, Callaerts P, Halder G, Gehring W J. Development. 1997;124:817–825. doi: 10.1242/dev.124.4.817. [DOI] [PubMed] [Google Scholar]
- 12.Czerny T, Busslinger M. Mol Cell Biol. 1995;15:2858–2871. doi: 10.1128/mcb.15.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomarev S I, Callaerts P, Kos L, Zinovieva R, Halder G, Gehring W, Piatigorsky J. Proc Natl Acad Sci USA. 1997;94:2421–2426. doi: 10.1073/pnas.94.6.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loosli F, Kmita-Cunisse M, Gehring W J. Proc Natl Acad Sci USA. 1996;93:2658–2663. doi: 10.1073/pnas.93.7.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chisholm A D, Horvitz H R. Nature (London) 1995;377:52–55. doi: 10.1038/377052a0. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Emmons S W. Nature (London) 1995;377:55–59. doi: 10.1038/377055a0. [DOI] [PubMed] [Google Scholar]
- 17.Glardon S, Holland L Z, Gehring W J, Holland N D. Development. 1998;125:2701–2710. doi: 10.1242/dev.125.14.2701. [DOI] [PubMed] [Google Scholar]
- 18.Halder G, Callaerts P, Gehring W J. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- 19.Halder G, Callaerts P, Gehring W J. Curr Opin Genet Dev. 1995;5:602–609. doi: 10.1016/0959-437x(95)80029-8. [DOI] [PubMed] [Google Scholar]
- 20.Gehring W J. Science. 1996;272:468–469. doi: 10.1126/science.272.5261.468. [DOI] [PubMed] [Google Scholar]
- 21.Gehring W J. Genes Cells. 1996;1:11–15. doi: 10.1046/j.1365-2443.1996.11011.x. [DOI] [PubMed] [Google Scholar]
- 22.Aguinaldo A, Turbeville J, Linford L, Rivera M, Garey J, Raff R, Lake J. Nature (London) 1997;387:489–493. doi: 10.1038/387489a0. [DOI] [PubMed] [Google Scholar]
- 23.Carranza S, Baguñà J, Riutort M. Mol Biol Evol. 1997;14:485–497. doi: 10.1093/oxfordjournals.molbev.a025785. [DOI] [PubMed] [Google Scholar]
- 24.Balavoine G. C R Acad Sci Paris/Life Sciences. 1997;320:83–94. doi: 10.1016/s0764-4469(99)80090-0. [DOI] [PubMed] [Google Scholar]
- 25.Bayascas J R, Castillo E, Muñoz-Mármol A M, Saló E. Development. 1997;124:141–148. doi: 10.1242/dev.124.1.141. [DOI] [PubMed] [Google Scholar]
- 26.Bayascas J R, Castillo E, Saló E. Dev Genes Evol. 1998;208:467–473. doi: 10.1007/s004270050204. [DOI] [PubMed] [Google Scholar]
- 27.Le Moigne A. Bull Soc Zool Fr. 1963;88:403–422. [Google Scholar]
- 28.Kishida Y. Sci Rep Kanazawa Univ. 1967;12:75–110. [Google Scholar]
- 29.Kishida Y. Sci Rep Kanazawa Univ. 1967;12:111–142. [Google Scholar]
- 30.Ribas M, Riutort M, Baguñà J. J Zool (London) 1989;218:609–626. [Google Scholar]
- 31.Saló E, Baguñà J. J Embryol Exp Morph. 1984;83:63–80. [PubMed] [Google Scholar]
- 32.Garcia-Fernàndez J, Baguñà J, Saló E. Development. 1993;118:241–253. doi: 10.1242/dev.118.1.241. [DOI] [PubMed] [Google Scholar]
- 33.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 34.Kumar S, Tamura K, Nei M. mega: Molecular Evolutionary Genetics Analysis. University Park: Pennsylvania State Univ.; 1993. , Version 1.01. [Google Scholar]
- 35.Czerny T, Bouchard M, Kozmik Z, Busslinger M. Mech Dev. 1997;67:179–192. doi: 10.1016/s0925-4773(97)00119-6. [DOI] [PubMed] [Google Scholar]
- 36.Garcia-Fernàndez J, Baguñà J, Saló E. Proc Natl Acad Sci USA. 1991;88:7338–7342. doi: 10.1073/pnas.88.16.7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puvion-Dutilleul F, Puvion E. Eur J Cell Biol. 1989;49:99–109. [PubMed] [Google Scholar]
- 38.Loosli F. Ph.D. thesis. Basel: Basel Univ.; 1995. [Google Scholar]
- 39.Fyrberg E A, Bond B J, Hershey N D, Mixter K S, Davidson N. Cell. 1981;24:107–116. doi: 10.1016/0092-8674(81)90506-7. [DOI] [PubMed] [Google Scholar]
- 40.Muñoz-Màrmol A M, Casali A, Castillo E, Bayascas J R, Saló E. Dev Genet Evol. 1997;207:296–305. doi: 10.1007/s004270050117. [DOI] [PubMed] [Google Scholar]
- 41.Del Rio-Tsonis K, Washabaugh C H, Tsonis P A. Proc Natl Acad Sci USA. 1995;92:5092–5096. doi: 10.1073/pnas.92.11.5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hitchcock P F, Macdonald R E, VanDeRyt J T, Wilson S W. J Neurobiol. 1996;29:399–413. doi: 10.1002/(SICI)1097-4695(199603)29:3<399::AID-NEU10>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 43.Belecky-Adams T, Tomarev S, Li H S, Ploder L, McInnes R R, Sundin O, Adlere R. Invest Ophthalmol Vis Sci. 1997;38:1293–1303. [PubMed] [Google Scholar]
- 44.Hirsch N, Harris W A. J Neurobiol. 1997;32:45–61. [PubMed] [Google Scholar]
- 45.Chen R, Wang Q, Nie Z, Sun H, Lennon G, Copeland N, Gilbert D, Jenkins N, Zack D. Neuron. 1997;19:1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]