Abstract
The development of the Drosophila compound eye requires the function of a set of evolutionarily conserved genes. Among these, the Drosophila Pax-6 gene eyeless (ey) plays a major role. ey has been considered a master control gene of eye development in the animal kingdom because targeted expression of ey and vertebrate as well as invertebrate homologs lead to the formation of ectopic eyes in Drosophila. We demonstrate that an intron of the ey gene contains an enhancer that regulates the eye specific expression of the gene in the eye disc primordia of embryos and in the eye imaginal discs of third instar larvae. Moreover, a 212-bp enhancer element is necessary and sufficient for the enhancer function. It is partially conserved in Drosophila hydei and contains putative Pax-6 Paired domain binding sites. We show that several binding sites are required for the eye specific expression, and, therefore, we propose a Pax-6-like molecule to be a positive transactivator for the eye specific ey expression. This transactivator recently has been identified as twin of eyeless, the second Pax-6 gene in Drosophila.
Keywords: eye development, eyeless enhancer, transcriptional regulation, Pax-6
The development of an homogeneous epithelium into patterned units is a poorly understood process. The Drosophila compound eye provides an excellent system to identify cellular and molecular mechanisms regulating these developmental steps. Our knowledge about morphogenesis and neuronal differentiation in eye development has increased considerably during the last years, but much less is known about the initial determinative events. Recent investigations of the early steps of eye development reveal that evolutionarily conserved genes are involved in determining the different eye types in the various metazoan phyla.
The eyeless (ey) gene, which belongs to the group of Pax-6 genes, was shown to be essential for compound eye development (1). In Drosophila embryos, it is expressed in the eye disc primordia and in the central nervous system (CNS). In third instar larvae, ey expression is visible in the optic lobes of the brain, in several spots of the ventral ganglion, and in the eye imaginal discs, where it is restricted to the undifferentiated cells anterior to the morphogenetic furrow (MF) (1). Strikingly, targeted expression of ey leads to induction of ectopic eyes in Drosophila, and, also, Pax-6 homologs of mouse, ascidians, and squid have the capacity to activate the program that is responsible for eye formation when misexpressed in imaginal tissues of Drosophila (2–5). Therefore, the function of ey/Pax-6 seems to be conserved in the animal kingdom, and ey/Pax-6 had been considered to be a master control gene in eye development because it has been identified in species of various phyla and is always involved in eye development (1, 3, 4, 6–9).
Recently, a second Pax-6 gene in Drosophila, twin of eyeless (toy), has been isolated (T. Czerny, G. Halder, U. Kloter, A. Souabni, W.J.G., and M. Busslinger, unpublished work). toy is also expressed in the eye anlagen and in the CNS, but its expression starts earlier as compared with ey. Like ey, it is able to induce ectopic eyes when misexpressed in various imaginal tissues of Drosophila, and the formation of ectopic eyes by toy leads to ectopic expression of ey, suggesting a role of toy as a positive regulator of ey expression (T. Czerny, G. Halder, U. Kloter, A. Souabni, W.J.G., and M. Busslinger, unpublished work).
In addition to ey and toy, the genes dachshund (dac), eyes absent (eya), and sine oculis (so), which also are involved in eye development, have shown to be able to induce ectopic eye development in Drosophila (10–13). Homologs of these genes also play a role in vertebrate eye development, and, therefore, they seem to be key regulators of eye development as well (14–17). This implies not only the conservation of a single gene, but, rather, an entire cascade of genes regulating eye development seems to be conserved from flies to vertebrates, despite the differences in structure and development of various eye types.
Investigations concerning regulatory relationships indicate that these eye specific genes are required at different steps of compound eye development (12, 13, 18). The normal expression of ey in so and eya mutants indicates a function upstream or independent of these genes and favors a linear pathway during normal eye development (18, 19). In contrast, ey, eya, and dac activate the expression of each other during ectopic eye formation and suggest, also, the existence of regulatory loops (10–13, 18). Studying the regulation of these genes should provide insight into this complex regulatory network.
In the present investigation, we show that an intron of the ey gene targets eye specific expression of the ey gene. Moreover, a 212-bp enhancer element is necessary and sufficient to direct expression in the eye disc primordia. This element contains sequence homologies to the corresponding element of Drosophila hydei. Five putative Pax-6 Paired domain binding sites are located within the enhancer element, of which two are located in the region that is conserved in D. hydei, and mutations of four binding sites destroy the eye specific expression in embryos. Our experiments provide evidence that the eye specific aspects of the ey expression depend on a Pax-6 like molecule, which is most likely toy.
MATERIALS AND METHODS
General DNA Methods.
Isolation of DNA from plasmids, restriction endonuclease digestions, gel electrophoresis of DNA, PCR reactions and subcloning of fragments were performed as described by Sambrook et al. (20). For sequence analysis, a 3.6-kilobase (kb) EcoRI fragment with the complete intron was subcloned into pBluescript KS (+) (Stratagene). Overlapping deletions of this fragment were generated by using the exonucleaseIII-S1 method as described by the supplier (Amersham Pharmacia). DNA was sequenced by the dideoxynucleotide procedure of Sanger et al. (21). Sequencing was done on both strands of the DNA with the sequenase 2.0 DNA sequencing kit from United States Biochemical. Sequences were analyzed with the husar/gcg sequence analysis software package from the University of Heidelberg.
Reporter Constructs.
All reporter constructs are insertions into the enhancer detection vector HZ50PL (22). Restriction sites in primers used for subcloning are underlined.
E36 (Fig. 1).
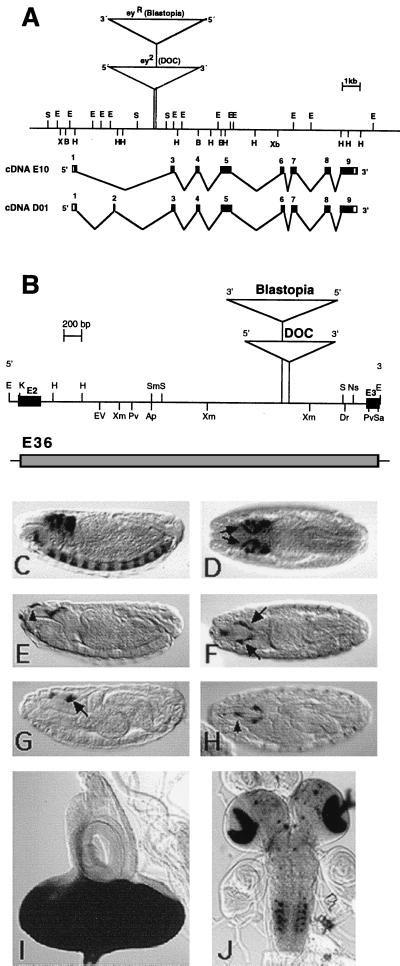
Figure 1.
(A) Genomic organization of the ey gene in D. melanogaster. The restriction map contains the insertion sites of the transposons in the alleles ey2 and eyR shown as triangles. Black boxes indicate exons of the cDNA E10 and D01, respectively. (B) Restriction map of the 3.6-kb EcoRI fragment containing the enhancer harboring intron of the ey gene. ey exon 2 (E2) and exon 3 (E3) are shown as black boxes; triangles indicate the transposon insertion sites in the alleles ey2 and eyR. Shown is expression of the eyeless+ gene in comparison with expression of reporter construct E36. (C and D) In situ hybridization of stage 15 embryos with a DIG-labeled ey probe. (C) Lateral view. (D) Dorsal view. ey is expressed in the brain, the ventral nerve cord, and the eye-antennal disc precursor cells (arrows). The 3.5-kb KpnI fragment that was used in the construct E36 is shown as a shaded box. Expression of reporter construct E36 in embryos (E–H) and larvae (I and J). Anti β-gal staining of stage 15 (E and F) and stage 17 (G and H) embryos shows expression in the eye-antennal disc precursor cells (arrows). The reporter construct shows additional expression in the region of the dorsal pouch (arrowheads). β-gal activity staining of third instar larvae shows strong signal in the eye imaginal disc (I) and a stripe in the antennal disc, in the optic lobes of the brain, and in spots in the ventral ganglion (J). In C–H, anterior is to the left. Staging is according to Campos-Ortega and Hartenstein (30). Ap, ApaI; B, BamHI; Dr, DraI; E, EcoRI; EV, EcoRV; H, HindIII; K, KpnI; Pv, PvuII; S, SalI; Sa, SacI; Sm, SmaI; Xb, XbaI; Xm, XmnI.
From the original 3.6-kb subclone, a 3.5-kb KpnI fragment (position 99–3,614 in Fig. 2) was used. The 5′ KpnI site is from the eyeless intron, and the 3′ KpnI site is from the Bluescript polylinker.
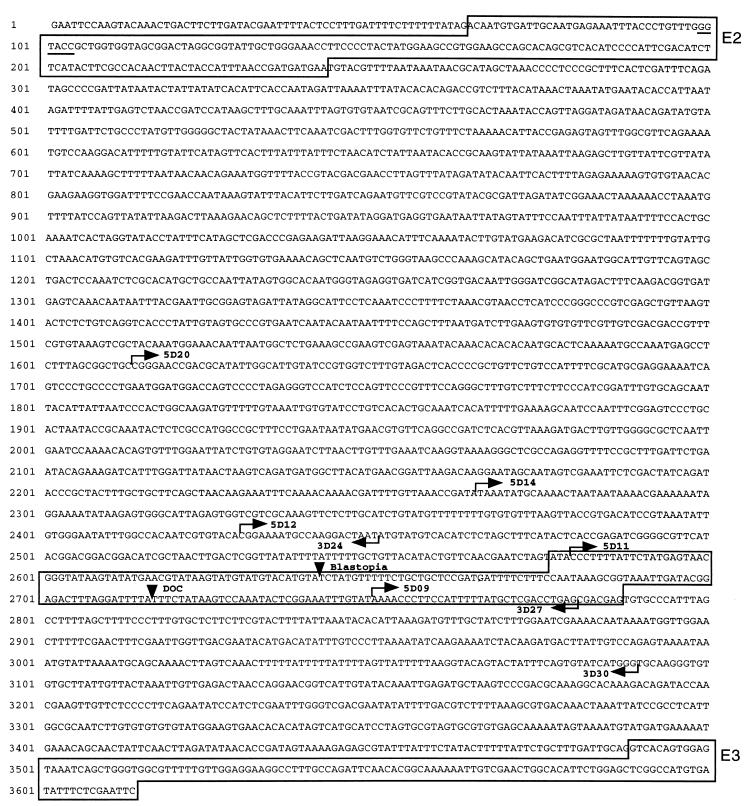
Figure 2.
Sequence of the 3.6-kb EcoRI ey enhancer fragment. Exon 2 (E2) and exon 3 (E3) are boxed in. The KpnI restriction site in exon 2 used for cloning is underlined. Insertion sites of the transposons Blastopia and DOC are indicated by arrowheads. Starting points of deletion constructs are indicated by arrows. The 212-bp fragment used in the construct D02 is boxed in.
Deletion constructs.
Various deletions of the 3.6-kb fragment were used for reporter constructs (Fig. 3 and 4). 5′ deletions were cloned into HZ50PL as NotI-KpnI fragments, 3′ deletions were cloned as KpnI fragments. For D02, the two primers Ey1 (GGCTAAATCGGTACCCTCGTCGCTCAG) and Ey2 (CAACGAATCTAGAATA CCCTTTTATTC) were used to PCR amplify a 212-bp XbaI-KpnI fragment.
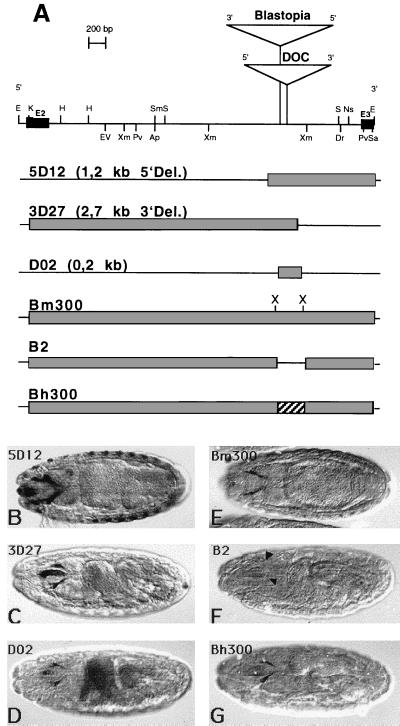
Figure 3.
Expression of different reporter constructs used to define the regulatory element. (A) Schematic presentation of the different reporter constructs and the corresponding lacZ-expression in embryos indicated by anti β-gal staining (B–G). The restriction map of the ey intron is shown at the top of A. The fragments of the intron that where used in the constructs are shown as shaded boxes. Construct Bm300 has two additional XbaI (X) sites in comparison to construct E36. In the absence of ≈300 bp, construct B2, which includes the eye specific element, expression in the eye-antennal disc precursor cells cannot be detected (F, arrowheads). Construct Bh300 contains the corresponding element of D. hydei (T. Eggert, B.H., and U.W., unpublished work) and also gives staining in the eye-antennal disc precursors (G). In all panels, anterior is to the left.
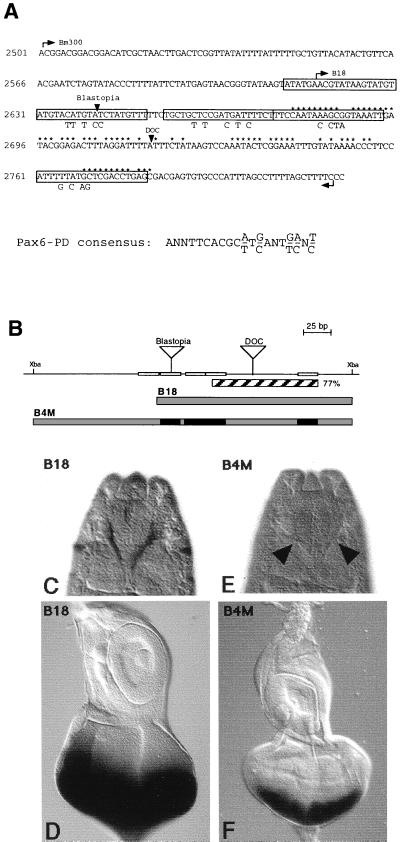
Figure 4.
Identification and mutation of putative Pax-6 Paired binding sites in the eye specific element. The sequence of the eye specific element is shown in A. Arrowheads mark the insertion sites of the transposons Blastopia and DOC. Starting points of construct B18 and Bm300 are indicated by arrows. Nucleotides that are identical to the eye specific element of D. hydei are marked by asterisks. Putative Pax-6 Paired binding sites are boxed in, and sequence changes of the putative binding sites in construct B4M are shown below the sequence. A schematic representation of the enhancer constructs is shown in B. The region with 77% homology to D. hydei is indicated as a hatched box. Putative Pax-6 Paired binding sites identified according to sequence similarities to the Pax-6 Paired consensus sequence (27) are shown as open; mutated binding sites are shown as black boxes. Triangles indicate the transposon insertions of Blastopia and DOC. Construct B18, which lacks the most 5′ binding site, still gives normal staining in embryos (C) and eye imaginal discs (D). Mutations of the other four binding sites abolish expression in embryos (arrowheads in E) and reduce the expression in eye imaginal discs to the posterior part of the eye disc (F). (C and E) Dorsal view of stage 16 embryos. In C–F, anterior of the eye-antennal disc and the embryos is to the top.
Vector cassette constructs.
The vector cassette (Fig. 3) was made by introducing two XbaI sites flanking the eye specific element (position 2,499–2,817 in Fig. 2). A 2.5-kb XbaI-KpnI fragment was amplified by using the primer Ey16 (GTCCGTCCGTTCTAGAGAACAGTCCC) and a T7 primer and a 0.8-kb XbaI-KpnI fragment was amplified by using the primer Ey12 (TCTAGATTGTGCTCTTCTTCGTTACTT) and a T3 primer. Both fragments were cloned into the KpnI site of a modified HZ50PL vector whose XbaI site had been destroyed before. The resulting construct B2 (Fig. 3) contains the ey intron without a 300-bp region, including the eye specific element. For construct Bm300 (Fig. 3), a 320-bp fragment was PCR amplified by using the primers Ey14 (AAGAGCACAATCTAGAAAGCTAAAAG) and Ey15 (GGGACTG TTTCTAGAACGGACGGAC) and was cloned into the XbaI site of the B2 construct. The point mutations in B4M (Fig. 4) were introduced one after the other by a three-step PCR amplification by using the primer couples Ey21 (CTTTCCAATACACTAGTAAATTGAT)/Ey22 (ATCAA TTTACTAGTGTATTGGAAAG), Ey23 (CCTTCCATTTGTCTAGTCGACCTGAG)/Ey24 (CTCAGGTCGACTAGACAAATGGAAGG), Ey25 (GTTTTTCTGCTGTTTCGACGTTCTT CTTTCCAA)/Ey26 (TTGGAAAGAAGAACGTCGAAACAGCAGAAAAAC), and Ey27 (GTATGTATGTATTTTTCCCTATGTTTTTCTGC)/Ey28 (GCAGAAAAACATAGGGAAAAATACATACATAC).
P-Element Mediated Transformation.
Germline transformations in cn;ry506 embryos were performed by using standard techniques (23). For each reporter construct, several independent transformant lines were generated, balanced, and analyzed.
Antibody and β-Galactosidase (β-gal) Activity Stainings.
Antibody stainings of embryos were performed according to Grossniklaus et al. (24). The mouse monoclonal anti β-gal antibody (Promega) was used at a 1:1,000 dilution. The secondary antibody was conjugated with horseradish peroxidase and was used in a 1:500 dilution (Vectastain ABC Kit, Vector Laboratories). β-gal staining of imaginal discs was done according to Bellen et al. (25).
RESULTS
An Intron of the ey Gene Contains the Enhancer for the Eye Specific Expression of the Gene.
The finding that the insertions of transposable elements in the ey mutants ey2 and eyR specifically affect gene expression in the eye disc primordia suggested that the transposable elements might disrupt an eye specific regulatory element (1). To test this hypothesis, we cloned a 3.5-kb KpnI fragment including the complete intron preceding exon 3, which marks the start of the paired domain, into the enhancer detection vector HZ50Pl (Fig. 1 A and B). The expression of a transgene from this construct E36 was compared with ey expression in wild-type embryos. ey expression starts at stage 8 in the CNS. From stage 15 on, the gene is expressed in addition to the brain and the ventral nerve cord in the eye-antennal disc precursor cells (Fig. 1 C and D). The lacZ expression of the transgene starts also at stage 15 (Fig. 1 E–H) and is restricted to the eye-antennal disc precursor cells whereas the brain and ventral nerve cord expression is missing. Additional expression in the region of the dorsal pouch and in the lateral parts of the embryo must be caused by vector sequences because ey is not expressed there (Fig. 1 E and H). In third instar larvae, the intron regulates expression in the eye disc, the optic lobes of the brain, and in several spots of the ventral ganglion (Fig. 1 I and J). Therefore, the ey intron seems to contain an enhancer for the eye specific expression of the gene in embryos and third instar larvae.
Sequence Analysis of the ey Enhancer.
To identify and characterize a minimal enhancer element, we made deletions of the 3.6-kb EcoRI fragment and used them to sequence this fragment completely and to generate shorter reporter-gene constructs to define the enhancer element in more detail. The complete sequence of the 3.6-kb fragment is shown in Fig. 2. The ey exon 2 is located at the 5′ end of the fragment, and exon 3, with part of the paired box, is located at the 3′ end. Recently isolated longer ey cDNAs indicate that the methionine in cDNA D1 (1) does not represent the initiation methionine (U.W., unpublished work). A splice site a few base pairs upstream of this ATG is used to splice exon 1 to exon 2. Therefore, the methionine of exon 1 in cDNA E10 (1) marks the 5′ end of the ORF, and exon 2 is either spliced in or skipped to generate two different transcripts and protein forms, respectively. We identified the splice form with exon 2 in embryos and larvae, and it seems to be same as the one recently identified in the adult stage (26). The insertion points of the two transposons Blastopia and DOC occur within 75 bp in the intron. Therefore, Blastopia is generating a 4-bp target sequence duplication (TGTA), and DOC is generating an 11-bp duplication (TTTCTATAAGT).
A 212-bp Enhancer Element Is Necessary and Sufficient for Expression in the Eye Disc Primordia.
The transposon insertions mentioned above suggested that the region around the insertion sites might be critical for the enhancer function, but functions of other regions cannot be excluded. To define the enhancer, we made a series of shorter reporter-gene constructs focusing on that region. The extent of all constructs is indicated in Fig. 2, and the most relevant ones for our analysis are schematically indicated in Fig. 3A. All constructs that contain a 200-bp region around the transposon insertion sites show the same spatial and temporal expression in embryos (Fig. 3 B and C) and third instar larvae (data not shown) as the original construct E36. In contrast, constructs not harboring this region (5D09 and 3D24 in Fig. 2) were negative (data not shown). Therefore, the 200-bp region is necessary for the eye specific expression. To test whether this region alone is also sufficient, we analyzed construct D02 with 212 bp of intron sequences (Fig. 3D). Also here, expression is visible, albeit a little weaker compared with constructs 3D27 and 5D12. These experiments demonstrate that the 212-bp region is not only necessary but also sufficient for the eye specific expression.
Because of the weaker expression of construct D02 as compared with the other constructs, we decided to further analyze the 212-bp element in the context of the complete intron to allow for possible enhancing effects of adjacent sequences. To do so, we generated a vector cassette by introducing two XbaI restriction sites flanking the 212-bp element a bit further away. This construct, Bm300, shows the same expression pattern as the original construct, E36, in embryos (Fig. 3E) and in larvae, demonstrating that the introduction of the restriction sites did not alter the enhancer activity. Again, a construct lacking the eye specific element but this time harboring all of the rest of the original fragment (B2) was not able to generate a pattern (Fig. 3F) except for single spots in the larval brain (data not shown).
It has been shown for several Drosophila genes that sequence conservations in regulatory regions of a gene from distantly related species could be used to identify functionally important regulatory elements. We used this approach and compared the ey intron sequence from Drosophila melanogaster with the corresponding region of the ey gene from D. hydei (T. Eggert, B.H., and U.W., unpublished work). Among other sequence conservations, we identified a region in D. hydei that is 77% homologous to the eye specific element in D. melanogaster (Fig. 4A). We were interested to know whether the homologous region from D. hydei is able to drive expression of lacZ in the same way as the D. melanogaster sequence. Therefore, we substituted the D. melanogaster eye specific element with a 300-bp fragment from D. hydei covering the homologous region. This construct, Bh300, gives also the expression pattern in the eye disc precursor cells in the embryo (Fig. 3G) and in third instar larvae (data not shown). This suggests that the important region in the 200-bp element might be located within the conserved region.
Putative Pax-6 Paired Domain Binding Sites Are Required for the ey Enhancer Function.
We next asked which gene products might be responsible for the eye specific regulation of ey. It has been shown for vertebrate Pax-6 genes that autoregulation plays an important role at least in some tissues (3); therefore, ey itself might interact with its own enhancer. Because the second Pax-6 gene in Drosophila, twin-of-eyeless (toy), is expressed earlier than ey and seems to act upstream of ey (T. Czerny, G. Halder, U. Kloter, A. Souabni, W.J.G., and M. Busslinger, unpublished work), it is also a good candidate for the enhancer activation. For this reason, we looked for potential Pax-6 Paired domain binding sites (27) within the eye specific element and identified several putative binding sites (Fig. 4A). Of the five sites identified, two are located on both sides of the DOC insertion site at the ends of the homologous region from D. hydei (Fig. 4A). The deletion of the left part of the element in construct B18, taking out the most 5′ site still gives normal staining (Fig. 4 C and D). Single mutations of the other four binding sites did not destroy the expression in the eye disc primordia of embryos and eye imaginal discs completely (data not shown). We next mutated the four putative binding sites together and, in the context of the complete intron mutations of these sites, abolished the expression in the eye disc primordia of the embryo (Fig. 4C). In eye imaginal discs, the lacZ expression is reduced to the posterior part of the eye disc (Fig. 4F) as compared with the expression in construct E36 (Fig. 1I). Therefore, the early eye specific regulation of ey in embryos seems to be mediated through a Pax-6 type protein binding to putative Pax-6 Paired domain binding sites within this ey enhancer element.
DISCUSSION
A Gene Internal Enhancer Element of 212 bp Is Necessary and Sufficient for Eye Specific ey Expression.
In this paper, we describe the identification of a regulatory element responsible for the eye specific expression of the ey gene. This element located in an intron of the ey gene includes, as expected, the transposon insertion sites, which give rise to the ey mutation ey2 and eyR. The insertion of the transposons seems not to interfere with the general transcription of the ey gene because only the expression in the eye disc promordia in embryos and in the eye imaginal discs of third instar larvae is abolished whereas ey is still normally expressed in the embryonic CNS and in the larval brain. The two alleles ey2 and eyR, therefore, can be considered as regulatory mutations that are strong hypomorphic alleles of the ey gene. Such gene internal regulatory mutations also have been reported for other genes involved in eye development like, so and orthodenticle (otd). In so1 mutant flies, a deletion in the 3′-most intron affects so expression in eye imaginal discs only whereas the expression in the optic lobe, Bolwig’s organ, and the segmental furrows is normal (28). The otd eye specific allele otduvi is also attributable to a deletion of a regulatory element affecting otd expression in photoreceptor cells (29).
A rescue experiment with a construct carrying the ey cDNA under the control of the ey enhancer resulted in a rescue of ey2 mutant eyes to almost wild-type eyes (19). This demonstrates that all of the necessary information to rescue this eye specific allele is located within the enhancer fragment. We showed that the intron drives expression of a reporter gene in the eye primordia in embryos from stage 15 on at least up to the third instar larval stage. Moreover, we could narrow down the responsible element to 212 bp, which are necessary and sufficient to regulate lacZ expression in this way, suggesting that this small element contains all of the information needed for the eye specific regulation.
In contrast to the faithful expression in the embryo at later stages in imaginal disc development, the reporter constructs are expressed posterior to the MF whereas ey is normally only expressed anterior to the MF and is down-regulated in posterior regions. Ectopic ey expression with a glass or a sevenless promoter in developing photoreceptor cells posterior to the furrow results in a rough eye phenotype, indicating that down-regulation of ey in response to the hh/dpp signaling is essential for correct photoreceptor cell formation (19). The incorrect expression we observe in late stages might be a hint for a second regulatory element that activates expression in front of the furrow and mediates down-regulation posterior to the furrow or might be necessary to maintain the initially correct expression later on.
A Pax-6-Like Protein Is Required for ey Enhancer Function.
Which genes could be considered as regulators of ey interacting with the eye specific element? ey itself would be a candidate because of the capacity for autoregulation found for other Pax-6 genes. The fact that the ey reporter construct E36 is normally expressed in an ey2 mutant background rules out this possibility (19). The best candidate as a positive regulator would be toy for several reasons. toy is expressed earlier than ey in the embryo, it is also expressed like ey in the eye primordia at stage 15 of embryogenesis, and it also has the full capacity to induce ectopic eyes. During the induction of ectopic eyes by toy, ey is activated and required for the formation of ectopic eye structures whereas in ey driven ectopic eyes toy is not expressed (T. Czerny, G. Halder, U. Kloter, A. Souabni, W.J.G., and M. Busslinger, unpublished work). These experiments put toy upstream of ey in the genetic cascade for eye development whereas genes like so, eya, and dac are considered to be downstream in this linear pathway. We therefore looked for putative Pax-6 Paired domain consensus sequences in the ey enhancer and mutated several binding sites within the enhancer element. These mutations abolished the enhancer function in embryos completely, showing that the early eye specific ey expression in embryos depends on a Pax-6 protein other than Ey.
We propose a model in which ey expression is regulated by toy, which might bind to the eye specific enhancer and thereby activate ey. This activation is likely to be part of a linear pathway because ey in turn cannot activate toy. The two genes also have nonredundant functions, as loss of ey activity is not compensated by toy. Besides this linear pathway, there is also regulatory feedback among some genes involved in early eye development. It has been shown that dac and eya activate ey expression during ectopic eye formation. In the case of eya, the reporter construct E36 was used as a marker for ey expression (11), showing that this regulatory loop acts via the ey enhancer. Because eya is not binding to DNA, this effect might be mediated by so, which can bind to DNA with its homeodomain and also forms a complex with eya. The observed ectopic expression of the reporter constructs in eye imaginal discs posterior to the MF is similar to that of so, eya, and dac. So it seems that construct E36 indeed acts as a target for the factors that are involved in the feedback loop. Whether there is a direct interaction of this complex with the ey enhancer or whether toy is activated first and in turn activates ey is not known. The fact that the mutations affect the ectopic reporter-gene expression cannot be attributable to a lack of activation by toy because toy is not expressed posterior to the MF. The introduced mutations might also have affected target sequences for the transcriptional complex involved in feedback regulation of ey. This would explain the reduced reporter gene activity in eye imaginal discs of B4M.
Eye specific expression of ey may not be regulated by toy alone because toy is also expressed in the CNS. Additional factors like otd, which confers segmental identity in the anterior head region, are likely to act in combination with toy to define eye identity, and at later stages the pathway certainly has to bifurcate. It will be interesting to reveal the complex interactions of all of these genes and to identify cofactors that are necessary for ey function during eye development.
Acknowledgments
We are grateful to Thomas Czerny and Meinrad Busslinger for sharing data before publication. We thank Wolfgang Staiber for excellent photographic work and the members of the lab for helpful discussions. This work was initiated at the Biozentrum Basel and was supported by the Kantons Basel, the Swiss National Science Foundation, and a grant from the Deutsche Forschungsgemeinschaft to U.W. (WA 556/4–1).
Footnotes
Abbreviations, ey, eyeless gene; CNS, central nervous system; MF, morphogenetic furrow; toy, twin of eyeless gene; dac, dachshund gene; eya, eyes absent gene; so, sine oculis gene; kb, kilobase; β-gal, β-galactosidase; otd, orthodenticle gene.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ131630).
References
- 1.Quiring R, Walldorf U, Kloter U, Gehring W J. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- 2.Halder G, Callaerts P, Gehring W J. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- 3.Glardon S, Callaerts P, Halder G, Gehring W J. Development (Cambridge, UK) 1997;124:817–825. doi: 10.1242/dev.124.4.817. [DOI] [PubMed] [Google Scholar]
- 4.Tomarev S I, Callaerts P, Kos L, Zinovieva R, Halder G, Gehring W, Piatigorsky J. Proc Natl Acad Sci USA. 1997;94:2421–2426. doi: 10.1073/pnas.94.6.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glardon S, Holland L Z, Gehring W J, Holland N D. Development (Cambridge, UK) 1998;125:2701–2710. doi: 10.1242/dev.125.14.2701. [DOI] [PubMed] [Google Scholar]
- 6.Ton C C, Hirvonen H, Miwa H, Weil M M, Monaghan P, Jordan T, van Heyningen V, Hastie N D, Meijers-Heijboer H, Drechsler M, et al. Cell. 1991;67:1059–1074. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- 7.Hill R E, Favor J, Hogan B L M, Ton C C T, Saunders G F, Hanson I M, Prosser J, Jordan T, Hastie N D, van Heyningen V. Nature (London) 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- 8.Hirsch N, Harris W A. J Neurobiol. 1997;32:45–61. [PubMed] [Google Scholar]
- 9.Altmann C R, Chow R L, Lang R A, Hemmati-Brivanlou A. Dev Biol. 1997;185:119–123. doi: 10.1006/dbio.1997.8573. [DOI] [PubMed] [Google Scholar]
- 10.Shen W, Mardon G. Development (Cambridge, UK) 1997;124:45–52. doi: 10.1242/dev.124.1.45. [DOI] [PubMed] [Google Scholar]
- 11.Bonini N M, Bui Q T, Gray-Board G L, Warrick J M. Development (Cambridge, UK) 1997;124:4819–4826. doi: 10.1242/dev.124.23.4819. [DOI] [PubMed] [Google Scholar]
- 12.Pignoni F, Hu B, Zavitz K H, Xiao J, Garrity P A, Zipursky S L. Cell. 1997;91:881–891. doi: 10.1016/s0092-8674(00)80480-8. [DOI] [PubMed] [Google Scholar]
- 13.Chen R, Amoui M, Zhang Z, Mardon G. Cell. 1997;91:893–903. doi: 10.1016/s0092-8674(00)80481-x. [DOI] [PubMed] [Google Scholar]
- 14.Oliver G, Mailhos A, Wehr R, Copeland N G, Jenkins N A, Gruss P. Development (Cambridge, UK) 1995;121:4045–4055. doi: 10.1242/dev.121.12.4045. [DOI] [PubMed] [Google Scholar]
- 15.Kawakami K, Ohto H, Ikeda K, Roeder R G. Nucleic Acids Res. 1996;24:303–310. doi: 10.1093/nar/24.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu X, Woo I, Her H, Beier D R, Maas R L. Development (London) 1997;124:219–231. doi: 10.1242/dev.124.1.219. [DOI] [PubMed] [Google Scholar]
- 17.Bovolenta P, Mallamaci A, Puelles L, Boncinelli E. Mech Dev. 1998;70:201–203. doi: 10.1016/s0925-4773(97)00183-4. [DOI] [PubMed] [Google Scholar]
- 18.Desplan C. Cell. 1997;91:861–864. doi: 10.1016/s0092-8674(00)80475-4. [DOI] [PubMed] [Google Scholar]
- 19.Halder G, Callaerts P, Flister S, Walldorf U, Kloter U, Gehring W J. Development (Cambridge, UK) 1998;125:2181–2191. doi: 10.1242/dev.125.12.2181. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 21.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiromi Y, Gehring W J. Cell. 1987;50:963–974. doi: 10.1016/0092-8674(87)90523-x. [DOI] [PubMed] [Google Scholar]
- 23.Rubin G M, Spradling A. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 24.Grossniklaus U, Pearson R, Gehring W J. Genes Dev. 1992;6:1030–1051. doi: 10.1101/gad.6.6.1030. [DOI] [PubMed] [Google Scholar]
- 25.Bellen H J, O’Kane C J, Wilson C, Grossniklaus U, Pearson R K, Gehring W J. Genes Dev. 1989;3:1288–1300. doi: 10.1101/gad.3.9.1288. [DOI] [PubMed] [Google Scholar]
- 26.Sheng G, Thouvenot E, Schmucker D, Wilson D S, Desplan C. Genes Dev. 1997;11:1122–1131. doi: 10.1101/gad.11.9.1122. [DOI] [PubMed] [Google Scholar]
- 27.Xu W, Rould M A, Jun S, Desplan C, Pabo C O. Cell. 1995;80:639–650. doi: 10.1016/0092-8674(95)90518-9. [DOI] [PubMed] [Google Scholar]
- 28.Cheyette B N R, Green P J, Martin K, Garren H, Hartenstein V, Zipursky S L. Neuron. 1994;12:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- 29.Vandendries E R, Johnson D, Reinke R. Dev Biol. 1996;173:243–255. doi: 10.1006/dbio.1996.0020. [DOI] [PubMed] [Google Scholar]
- 30.Campos-Ortega J A, Hartenstein V. The Embryonic Development of Drosophila melanogaster. 2nd Ed. Berlin: Springer; 1985. [Google Scholar]