Abstract
A human artificial chromosome (HAC) vector was constructed from a 1-Mb yeast artificial chromosome (YAC) that was selected based on its size from among several YACs identified by screening a randomly chosen subset of the Centre d’Étude du Polymorphisme Humain (CEPH) (Paris) YAC library with a degenerate alpha satellite probe. This YAC, which also included non-alpha satellite DNA, was modified to contain human telomeric DNA and a putative origin of replication from the human β-globin locus. The resultant HAC vector was introduced into human cells by lipid-mediated DNA transfection, and HACs were identified that bound the active kinetochore protein CENP-E and were mitotically stable in the absence of selection for at least 100 generations. Microdissected HACs used as fluorescence in situ hybridization probes localized to the HAC itself and not to the arms of any endogenous human chromosomes, suggesting that the HAC was not formed by telomere fragmentation. Our ability to manipulate the HAC vector by recombinant genetic methods should allow us to further define the elements necessary for mammalian chromosome function.
As the time rapidly approaches when the complete sequence of a human chromosome will be known, it is striking how little is known about how human chromosomes function. In contrast, the necessary elements for chromosomal function in yeast have been defined for several years. Three important elements appear to be required for the mitotic stability of linear chromosomes: centromeres, telomeres, and origins of replication. The ascertainment of these elements in Saccharomyces cerevisiae provided the basis for the construction of yeast artificial chromosomes (YACs), which have proven to be important tools both for the study of yeast chromosomal function and as large capacity cloning vectors (1–3).
The use of a similar strategy in human cells to produce human artificial chromosomes (HACs) might be expected to provide an important tool for the manipulation of large DNA sequences in human cells. However, of the three required chromosomal elements, only telomeres have been well defined in human cells to date. It has been demonstrated that telomeric DNA, consisting of tandem repeats of the sequence T2AG3, can seed the formation of new telomeres when reintroduced into human cells (4–6). And recently, two telomeric binding proteins, TRF1 and TRF2, have been described (7–9). The second required element, a human centromere, is thought to consist mainly of repeated DNA, specifically the alpha satellite DNA family, which is found at all normal human centromeres (10–12). However, normal human centromeres are large in size and complex in organization, and sequences lacking alpha satellite repeats also have been shown to be capable of human centromere function (13, 14). As for the third required element, the study of origins of DNA replication also has led to conflicting reports, with no apparent consensus sequence having yet been determined for the initiation of DNA synthesis in human cells (15, 16).
The production of HACs from cloned DNA sources should help to define the elements necessary for human chromosomal function and to provide an important vector suitable for the manipulation of large DNA sequences in human cells. Two approaches to generate chromosomes with the “bottom up” strategy in human cells from human elements have recently been described. Harrington et al. (17) synthesized arrays of alpha satellite DNA, which were combined in vitro with telomeres and fragmented genomic DNA, and transfected into HT1080 cells. The undefined genomic DNA component appeared to play an important role in the ability to form HACs, leaving unanswered questions as to what sequences, other than telomeres and alpha satellite DNA, were necessary for chromosome formation. Ikeno et al. (18) used two 100-kb YACs containing alpha satellite DNA from human chromosome 21 propagated in a recombination deficient strain, which necessitated transient expression of a recombination protein (Rad52) to modify the YAC with telomere sequences and selectable markers (19). Only one of the two YACs was able to form HACs in HT1080 cells, suggesting that not all alpha satellite sequences may be able to form centromeres.
Here we report construction of functional HACs from a YAC that was propagated in a recombination-proficient yeast strain and was chosen solely for its size (1 Mb) and the presence of alpha satellite DNA. This YAC contains both alpha satellite and non-alpha satellite DNA and was modified to include a putative human origin of replication and human telomeric DNA. The function and stability of HACs generated from this 1-Mb YAC in a human cell line are described.
MATERIALS AND METHODS
Plasmid Constructions.
To generate p305TGTeN, an ≈430-bp fragment of the TRP1 gene was released from plasmid pRS304 (20) by XbaI–HindIII digestion and subcloned into SalI–HindIII-digested pRS305 (20) to generate plasmid p305T. A 4.1-kb HindIII–EcoRI fragment containing the 5′ end of the β-globin gene (21) was digested from cosmid globin-1 and subcloned into the XhoI site of p305T to generate plasmid p305TG. Finally, a NotI–XhoI fragment from pNeo270 (6) containing 1.6 kb of TTAGGG repeats and a phosphoglycerate kinase driven neo gene was subcloned into the NotI–SacII sites of p305TG to generate plasmid p305TGTeN.
To generate p303UrT, a 547-bp fragment of the URA3 gene was amplified by PCR with primers 5′-GATCGCGGCCGCACAAACTTGTGTGCTTCATTGG-3′ and 5′-GATCGAATTCGTCTCCCTTGTCATCTAAACC-3′ and pRS306 (20) as the template. The PCR product was subcloned into pRS303 (20) by digestion with NotI and EcoRI and then by ligation to generate p303Ur. An SmaI–NotI fragment from pNeo270 containing 1.6-kb TTAGGG repeats was then subcloned into the NotI–SacII sites of p303Ur to generate p303UrT.
YAC Modification.
Arrayed Centre d’Étude du Polymorphisme Humain (CEPH) (Paris) YAC library clones were screened with a 32P-labeled PCR product derived by amplification of human genomic DNA with degenerate alpha satellite primers WA1 and WA2 (22). Clones with strong hybridization signals were characterized further by pulsed-field gel electrophoresis and Southern hybridization.
YAC 674E2 from the CEPH YAC library was kar-crossed to the yeast strain YPH925 as described (23). One of the resultant clones, designated 674E2–1, was transformed with NotI-linearized p305TGTeN and then with NotI-linearized p303UrT. Colonies were screened by pulsed-field gel electrophoresis and Southern hybridization for linearity and accurate modification of the YAC arms. One of the resulting clones, designated L2H2, was chosen as our HAC construct.
Cell Culture and Transfection.
HT1080 cells were obtained from the American Type Culture Collection (Rockville, MD) and maintained in DMEM plus 10% fetal bovine serum. For transfections, 2 × 105 HT1080 cells per well of a six-well plate were transfected with ≈200–400 ng of gel-purified L2H2 DNA (ref. 24; S.T.C., unpublished data) complexed with 7.5 μg of Lipofectamine (Life Technologies, Gaithersburg, MD) according to the manufacturer’s instructions. Clones were selected with 250–325 μg/ml of the neomycin analog G418. The transfection efficiency was estimated to be 4 × 10−5 colony-forming units per cell.
The presence of the HIS3 gene was determined by PCR under standard conditions on cell lysates with primers HIS3F2, 5′-CTTTCCAGAGCGGTGGTAGATCTTT-3′, and HIS3R2, 5′-ATAAGAACACCTTTGGTGGAGGGAA-3′.
Fluorescence in Situ Hybridization (FISH).
Metaphase spreads for FISH were prepared as described (25). The L2H2UN1 probe was generated by PCR amplification of gel-purified L2H2 DNA with degenerate oligonucleotide primer 6-MW (26) as described (27), followed by a second round of PCR to incorporate Spectrum Orange-conjugated dUTP (Vysis, Downer’s Grove, IL). Routine (normal stringency) hybridizations were performed in 2× standard saline citrate (SSC; 1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7)/65% formamide at 37°C. High-stringency hybridizations were in 0.5× SSC/65% formamide at 37°C. Only in the experiment represented in Fig. 3C was Cot1 DNA (20 μg) used to suppress repetitive sequences.
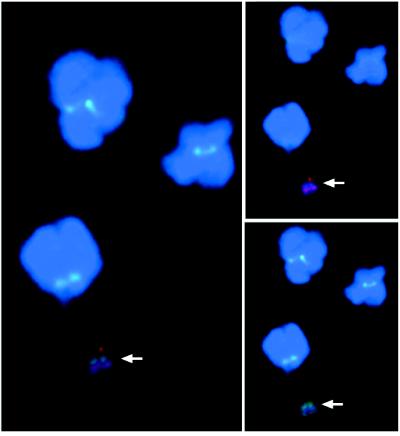
Figure 3.
HAC microdissection products used to assess the content of the HAC in 64b5. (A) HACs were microdissected from clone 64b5 after propagation in the absence of G418 selection for 78 generations. The microdissection probe was hybridized to metaphase spreads from normal human peripheral blood cells under high-stringency conditions. (B) The same microdissection probe as in A, hybridized under identical conditions to metaphase spreads from clone 64b5, gives very strong signals on the HAC. (C) HACs were microdissected from clone 64b5 after propagation in the absence of G418 selection for 72 generations. The microdissection probe was hybridized to metaphase spreads from HT1080 cells under normal stringency and blocked with Cot1 DNA. The lack of hybridization under these conditions to the arms of endogenous human chromosomes suggests that the HAC did not acquire DNA sequences by telomere fragmentation. The chromosomes were counterstained with 4′,6-diamidino-2-phenylindole.
The colocalization of the kinetochore protein CENP-E and L2H2UN1 probe was performed according to the procedure of Page et al. (28). Rabbit polyclonal antibodies to CENP-E (29) were detected with fluorescein isothiocyanate-conjugated goat anti-rabbit IgG (Kirkegaard & Perry Laboratories).
Southern Blot Hybridization.
Alpha satellite probes αXT (30) or CEP4 (kindly provided by Vysis) were hybridized overnight at 45°C in 10 ml of Hybrisol I (Oncor) and washed to a final stringency of 0.1× standard saline phosphate/EDTA (0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA)/0.1% SDS at 65°C.
RESULTS
HAC Vector Construction and Characterization.
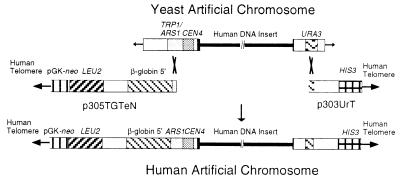
YAC 674E2, (≈1 Mb) was isolated by screening a randomly chosen subset of the CEPH YAC library (31) with a degenerate alpha satellite probe and subsequently was modified by homologous recombination to include human telomeric repeats, a putative origin of replication from the β-globin locus (16, 32), and a neomycin resistance gene (neo), which confers resistance to the neomycin analog G418 for selection in mammalian cells (Fig. 1). After the modified YAC 674E2 was analyzed for linearity, size, and the presence of the different introduced elements, one linear HAC vector clone, designated L2H2, was chosen, which contained all of the appropriate components as diagrammed in Fig. 1. However, the human insert DNA was approximately 25 kb smaller than the parental YAC, indicating that some rearrangement had taken place during the process of modification, likely within the alpha satellite array. Importantly, however, L2H2 appeared to remain stable during further propagation in yeast and was chosen for further analysis in mammalian cells.
Figure 1.
Schematic of the strategy used to modify the YAC 674E2 to generate the HAC vector, L2H2. The stepwise modification of 674E2 took place by homologous recombination in yeast, with the plasmid p305TGTeN to retrofit the left arm and then p303UrT to retrofit the right arm.
Transfection into Human HT1080 Cells.
Intact DNA from L2H2 was isolated by gel purification (24) and introduced into human HT1080 cells by lipid-mediated DNA transfection. Eighty-one G418-resistant (G418R) clones were obtained from transfection of 2.2 × 106 HT1080 cells, and PCR analysis revealed that 24 of 81 G418R clones were positive for the HIS3 gene, which lies on the opposite arm of L2H2 from neo (Fig. 1 and data not shown). In 7 of 24 of the G418R/HIS3 clones, HACs derived from L2H2 were evident in the initial screening by FISH with the probe L2H2UN1, derived by PCR with a degenerate oligonucleotide primer (26) and gel-purified L2H2 DNA as the template. The initial characterization of the 7 clonal cell lines containing HACs by FISH is summarized in Table 1. The cell line 64b5, which contained the highest proportion of metaphases with single HACs (92%), was chosen for detailed analysis.
Table 1.
Initial characterization of clonal cell lines containing HACs
| Clone | No. of HACs
|
Integration, % + HAC | |||
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | ||
| 62b7 | 0 | 68 | 28 | 4 | 0 |
| 62b10 | 0 | 40 | 40 | 10 | 10 (0) |
| 62b12 | 0 | 80 | 13 | 7 | 0 |
| 64b5 | 0 | 92 | 4 | 4 | 0 |
| 69a1 | 0 | 20 | 0 | 20 | 60 (20) |
| 69a6 | 0 | 11 | 11 | 6 | 72 (6) |
| 69a12 | 13 | 54 | 33 | 0 | 0 |
Metaphase spreads from cell lines grown in the presence of G418 were screened by FISH with the L2H2UN1 probe. Shown for each cell line is the percentage of metaphase spreads containing 0, 1, 2, or 3 HACs and the percentage of metaphase spreads containing integration events. In parentheses, the percentage of metaphase spreads displaying an integration event and an extrachromosomal element simultaneously is shown.
Cytogenetic Analysis of the HAC in Clone 64b5.
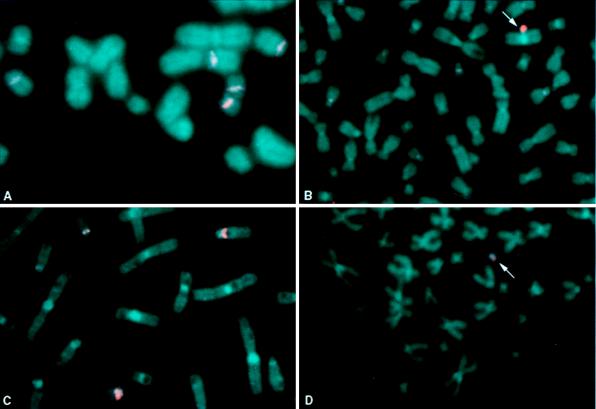
A FISH probe p305neo, derived from plasmid p305TGTeN by omitting the telomere sequences, colocalized with the L2H2UN1 probe on the HAC, providing additional evidence that the HAC in 64b5 was derived from L2H2 (data not shown).The presence of centromeric proteins was analyzed by immunofluorescence with an antibody to CENP-E (29) and FISH with the L2H2UN1 probe. The antibody to CENP-E and the L2H2UN1 probe were observed together only on the HAC (Fig. 2), suggesting that the centromeric DNA present on the HAC had characteristics of an active centromere, because the kinesin-related protein CENP-E is found at active, but not inactive, centromeres (33). The presence of telomere sequences on the HAC was confirmed by FISH with a human telomeric DNA probe (data not shown), and the size of the HAC in 64b5 was assessed by the intensity of the DNA after propidium iodine staining (18). The HACs ranged in size from 3.5 to 12.9 Mb (8.5 ± 3.3 Mb, mean ± SE), indicating that the HAC in clone 64b5 was larger than a single copy of L2H2. Multiple unsuccessful attempts were made to resolve the HAC in 64b5 by pulse-field gel electrophoresis. The large estimated size of the HAC, or alternatively the large range in estimated size (as indicated by propidium iodine staining), may account for the difficulty encountered in this procedure.
Figure 2.
Assessment of the HAC in clone 64b5 with FISH combined with immunofluorescence to demonstrate colocalization of the L2H2UN1 probe (red, rhodamine filter) and the antibody to the active kinetochore protein, centromeric binding protein E (CENP-E; green, fluorescein isothiocyanate filter) on the HAC. Chromosomes are counterstained with 4′,6-diamidino-2-phenylindole. (Left) Rhodamine and fluorescein isothiocyanate filters merged. (Upper Right) Rhodamine filter alone. (Lower Right) Fluorescein isothiocyanate filter alone. Arrow denotes the HAC.
Mitotic Stability of HACs.
To further assess the functional characteristics of the HAC, we examined its mitotic stability. The clonal cell line 64b5 was maintained in the presence or absence of G418 and analyzed at various times by FISH for retention of the HAC. The mitotic stability shown in Table 2 confirms the functional nature of the HAC centromere, which was predicted by the presence of CENP-E (Fig. 2). After 108 generations in culture without selection, 70% of the spreads retained a single HAC. During the stability assessment, occasional spreads were found that contained integrated HAC DNA (from 4% at 18 days to 20% at 108 generations in the absence of G418). The integration site remained the same over time and was observed in both the presence and absence of G418 selection, suggesting that the integration event took place early during the establishment of the clonal cell line. The mitotic stability of the HAC in clone 69a12 also was evaluated, and 56% of spreads retained a single HAC after 81 days in the absence of G418 selection.
Table 2.
Mitotic stability of the HAC in clonal cell lines 64b5 and 69a12
| Clone | Days | G418 | No. of HAC(s)
|
Integration, % + HAC | |||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | ||||
| 64b5 | 0 | + | 0 | 92 | 4 | 4 | 0 |
| 64b5 | 18 | + | 0 | 100 | 0 | 0 | 0 |
| 64b5 | 36 | + | 0 | 87 | 9 | 0 | 4 (4) |
| 64b5 | 108 | + | 0 | 89 | 3 | 0 | 8 (1) |
| 64b5 | 18 | − | 4 | 88 | 4 | 4 | 0 |
| 64b5 | 36 | − | 4 | 92 | 4 | 0 | 0 |
| 64b5 | 72 | − | 0 | 88 | 4 | 0 | 8 (2) |
| 64b5 | 108 | − | 0 | 70 | 6 | 4 | 20 (9) |
| 69a12 | 0 | + | 13 | 54 | 33 | 0 | 0 |
| 69a12 | 81 | − | 9 | 56 | 0 | 9 | 26 (9) |
Cell lines 64b5 and 69a12 were passaged 1:8 every 3 days, in the presence (+) or absence (−) of G418, for the indicated number of days. Metaphase spreads were analyzed for the presence of the HAC with probe L2H2UN1. Shown is the percentage of metaphase spreads containing 0, 1, 2, or 3 HACs and the percentage of metaphase spreads displaying an integration event. In parentheses, the percentage of metaphase spreads displaying both an integration event and an extrachromosomal element simultaneously is shown.
The HAC in Clone 64b5 Was Not Derived by Telomeric Fragmentation of Endogenous Chromosomes.
To characterize the origin of the HAC contained in clone 64b5, microdissection of the HAC was used to evaluate whether the HAC was generated by telomeric fragmentation of endogenous chromosomes. If the HAC had picked up HT1080 chromosomal DNA during its formation, FISH with the microdissection products should localize the chromosomal regions that had been acquired. Therefore, HACs were microdissected from 64b5 after 78 generations in the absence of G418 selection, and the dissected DNA was amplified essentially as described for human chromosomes (27). The microdissection probe was then hybridized under high-stringency conditions to metaphase spreads from normal human peripheral blood cells or from 64b5 (Fig. 3 A and B, respectively). On normal human metaphases, the microdissection probe hybridized to the centromeres of multiple human chromosomes, including 4, 14, 18, and 22, but not specifically to the arms of any chromosomes (Fig. 3A). On metaphase spreads from 64b5, the same microdissection probe gave signals only on the HAC itself and not on any other regions of endogenous HT1080 chromosomes (Fig. 3B). This intense hybridization signal only to the HAC is likely caused by the highly chimeric structure of L2H2 (see below) and the high affinity of the microdissection products for the HAC from which they were generated. Because the high-stringency conditions used for these analyses might preclude detection of single-copy sequences on chromosome arms, the microdissection probe was also hybridized under normal-stringency conditions with suppression of repetitive sequences to HT1080 cells, but only centromeric signals were observed (Fig. 3C). These data strongly suggested that the HAC has not acquired DNA from the arms of endogenous chromosomes. This conclusion was supported when 64b5 was evaluated by whole chromosome paints. No hybridization to the HAC was observed when each of the 24 individual chromosome paint probes (34) was hybridized to metaphase preparations from 64b5 (data not shown).
Chromosomal Origin of the Alpha Satellite DNA in L2H2.
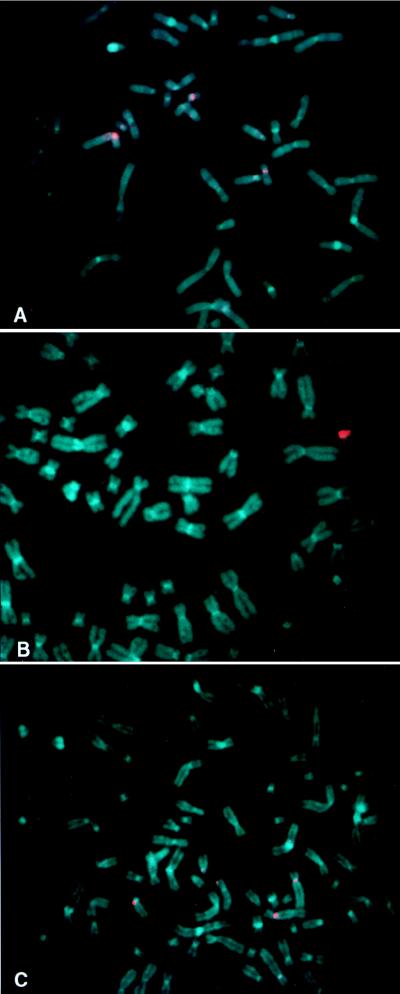
To assess the chromosomal origin of the original HAC vector DNA, L2H2, FISH was performed with the L2H2UNI probe on metaphase spreads from normal human peripheral blood cells. Signals were strongest at the centromeres of chromosomes 14 and 22 (Fig. 4A). No specific hybridization was observed to noncentromeric regions. Under the same conditions, hybridization to 64b5 showed very strong signals on the HAC, in addition to the faint centromeric signals (Fig. 4B). To increase the specificity of alpha satellite DNA hybridization, FISH also was performed under high-stringency conditions. On metaphase spreads from normal human peripheral blood lymphocytes, signals remained strongest at the centromeres of chromosomes 14 and 22, with weak signals visible at the centromeres of other chromosomes, including chromosome 18 (Fig. 4C). On metaphases from 64b5, only the HAC could be easily distinguished (Fig. 4D). Similar results were observed on normal human peripheral blood lymphocytes with biotin-labeled genomic DNA from the parental YAC (data not shown). These data suggested that sequences from one of the highly conserved centromeric regions of chromosome 14 or 22 were contained within the YAC. The centromeres of chromosomes 14 and 22 contain many identical alpha satellite sequences, including the major alpha satellite sequence, αXT (35).
Figure 4.
Assessment of the human chromosomal origin of the HAC vector L2H2 by FISH with the L2H2UN1 probe under normal-stringency (A and B) and high-stringency (C and D) conditions to assess the specific origins of both the non-alpha satellite and alpha satellite DNA sequences. (A and C) Normal human peripheral blood cells. (B and D) Clone 64b5. Arrows identify the HAC in clone 64b5.
A possible explanation for the hybridization of the HAC microdissection products to the several centromeric regions, specifically those of chromosomes 4, 14, 18, and 22, is that the centromeres of these chromosomes share homologous sequences, and any one of these sequences could be present in the HAC. Consistent with this hypothesis, the alphoid DNA repeat sequences from the centromeres of each of these four chromosomes form D type monomers, a characteristic of members of the alphoid suprachromosomal family 2 (10). Additionally, the degenerate oligonucleotide primer used to amplify the microdissection products has been shown to prime at specific sequences of the target DNA, rather than at random positions, and may therefore selectively amplify or enrich for certain sequences (26). To determine more precisely whether the input DNA, L2H2, contains alpha satellite DNA specific to the centromeres of chromosomes within suprafamily 2, Southern blot analysis and FISH studies were carried out with centromere-specific DNA probes. A probe specific for chromosomes 14 and 22, αXT (30), and the chromosome 4-specific probe CEP4 were used on Southern blots of XbaI and EcoRI digests of L2H2 and the parental YAC, 674E2. Both αXT and CEP4 hybridized to 674E2, as well as to L2H2, and comparison of the fragment sizes suggests that the rearrangement that resulted in the decreased size of L2H2 occurred within this alpha satellite-containing region (Fig. 5). Pulsed-field gel electrophoresis blots localized the αXT probe to within ≈225 kb of the p303UrT arm of L2H2 and also showed weak hybridization of CEP4 to the same region and to the alpha satellite DNA present in a human chromosome 22 monochromosomal hybrid (data not shown). L2H2 also was tested for the presence of several additional satellite sequences, including satellite 1, satellite 3, a 48-bp satellite, and a chromosome 22-specific alpha satellite sequence; however, none of these sequences were found on L2H2.
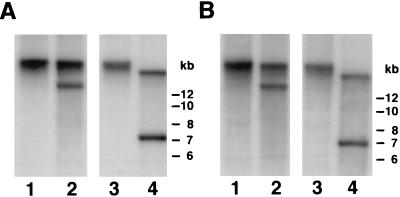
Figure 5.
Characterization of the alpha satellite DNA in L2H2. Genomic DNA from 674E2 (lanes 1 and 2) and its derivative L2H2 (lanes 3 and 4) were run in duplicate on the same gel and then hybridized with either the chromosome 14-specific/22-specific probe, αXT (A) or the chromosome 4-specific probe, CEP4, at high stringency (B). Genomic DNA was digested with XhoI (lanes 1 and 3) or EcoRI (lanes 2 and 4).
In FISH studies on the HAC in 64b5, the HAC hybridized to an alpha satellite probe from chromosomes 14 and 22 and CEP4 but not to an alpha satellite probe from chromosome 18 (data not shown). In addition, as shown in Fig. 4C, on normal human metaphase spreads that did not contain the HAC, the L2H2UN1 FISH probe hybridized to chromosomes 14 and 22 and faintly to chromosome 18.
Chromosomal Origin of the Non-Alpha Satellite DNA in L2H2.
As discussed above, microdissection products from the HAC hybridized strongly only to the HAC itself and not to any other chromosomes in clone 64b5 (Fig. 3B). Therefore, to better understand the content of the input HAC DNA, we attempted to isolate probes directly from the parental YAC 674E2. By YAC end rescue (36), novel sequences were isolated from the left end of the insert of YAC 674E2, which mapped to chromosome 21 with the National Institute of General Medical Sciences human/rodent somatic cell hybrid-mapping panel 2 (37, 38). With inter-Alu PCR (39), we isolated fragments that displayed identity with UniGene clusters Hs.31856, expressed sequence tags weakly similar to the murine hematopoietic-specific deubiquitinating enzyme, DUB-1 (40), and Hs.24994, expressed sequence tags highly similar to murine protein NGD5 (41). These fragments were found to map to human chromosomes 7 and 20q, respectively. The chromosome 20q fragment was mapped by Southern analysis to an ≈300-kb internal fragment devoid of alpha satellite sequences (data not shown). Taken together, these data indicate that YAC 674E2 and its derivative L2H2 are chimeric and contain genomic DNA derived from multiple chromosomes, including 7, 20, and 21 and alpha satellite DNA similar to that found on chromosomes 14, 22, and 4.
DISCUSSION
We have used a 1-Mb YAC containing alpha satellite DNA to construct a functional HAC vector by modification with a selectable marker, human telomeres, and a putative origin of replication. YAC 674E2 was selected for this purpose because of its size from among several YACs identified during screening of a randomly chosen subset of the CEPH YAC library with a degenerate alpha satellite probe. Other than the presence of alpha satellite DNA (14) there was no a priori reason to expect that 674E2 would generate a functional HAC. It is now apparent that L2H2 efficiently forms artificial chromosomes as indicated by the formation of HACs in ≈30% of the clonal cell lines that contained both arms of the HAC vector. Detailed analysis of one of these clonal lines, 64b5, indicates that the HAC formed by L2H2 contains human telomeres, binds CENP-E, and is mitotically stable in the absence of G418 selection for more than 100 generations.
L2H2 DNA did not always form HACs after transfection; it was sometimes observed integrated into the HT1080 genome. One possible cause for integration could be that partially degraded L2H2 DNA that lacks crucial functional elements is being formed during the transfection protocol and, consequently, becomes integrated. Other possible explanations for the integration of L2H2 could be that the length of the telomeres is insufficient to consistently seed new telomeres (5, 6). Variability in telomere length and/or function could be a consequence of the propagation of L2H2 in yeast. Yeast will modify the human telomeres on L2H2 with the addition of yeast telomeres (42, 43). It might be possible to improve the rate of seeding functional telomeres in human cells by modifying the yeast telomerase to produce human telomeres. We recently developed such a yeast strain and are currently determining whether the rate of HAC formation is improved after propagation of L2H2 in this strain (44).
The composition of L2H2 and what contributes to its functional nature are not completely understood. To address the basic structure of L2H2, several different approaches were used, many of which were confounded by the fact that L2H2 is chimeric with respect to its chromosomal origin. Even the alpha satellite DNA, with its chromosome-specific higher order repeats (45), did not provide insight into the exact origin of the centromeric function. The chimeric nature of 674E2 could reflect both its size and its DNA content. Cocloning of DNA fragments from different chromosomes has been observed during YAC library construction and has been most commonly seen among larger YACs (46). It has been suggested that YAC chimerism is facilitated by the presence of repetitive elements such as Alu sequences within the human DNA (47). In addition, because alpha satellite repeats have been shown to be unstable in YACs, it is not surprising that this 1-Mb YAC is not composed solely of alpha satellite DNA (48).
The chimerism of L2H2 and lack of precise information about its contents has made the analysis of the HAC in 64b5 quite difficult. It is evident that the mean size of the HAC in 64b5 is ≈9 times larger than the input DNA, similar to the size of the HACs observed by Harrington et al. (17) and Ikeno et al. (18). Our data do not explain the source of the extra DNA but suggest that the HAC does not contain HT1080 chromosomal DNA acquired by telomere fragmentation (4, 5, 49).
The content of L2H2 is quite different from naturally occurring, normal human centromeres. As described above, L2H2 is highly chimeric and contains Alu repeats, gene fragments, and alpha satellite sequences, all within 1 Mb of DNA. Our current data suggest that, given a minimal alpha satellite DNA content, the specific sequence of the remaining DNA may not be as important as the ultimate size of the HAC vector. Perhaps the presence of a small amount of alpha satellite DNA is all that will be necessary to seed the formation of a functional centromere (50, 51). This hypothesis could be tested by linking YACs (52) containing small amounts of alpha satellite DNA together with non-alpha satellite DNA. An array of various sized HAC vectors of known content could be generated and assessed for function and stability. YAC-based approaches such as these should broaden our understanding of how to attain human centromere function. This knowledge will be especially important when constructing HAC vectors for the purpose of stable gene transfer and expression analysis from large genomic regions.
Acknowledgments
We thank J. Trent and F. Collins for helpful discussions, P. Gregory and A. Dutra for assistance with kinetochore and cytogenetic analysis, V. Statham and E. Green for contributing to the HAC vector construction, E. Schröck and T. Reid for chromosome-specific paints and centromere probes, K. Brown for the antibody to CENP-E, B. Pike and S. Chandrasekharappa for radiation hybrid mapping, M. Bittner and Y. Xiang for help with generating the L2H2UN1 FISH probe, D. Tagle for cosmid globin-1, J. Blancoto (Georgetown University, Washington, DC) for assistance with the mitotic stability analysis, T. de Lange (Rockefeller University, New York) for pNeo270, and Vysis, Inc. (Downer’s Grove, IL) for CEP4 DNA. This work was supported in part by the Cystic Fibrosis Foundation.
ABBREVIATIONS
- YAC
yeast artificial chromosome
- HAC
human artificial chromosome
- FISH
fluorescence in situ hybridization
- CEPH
Centre d’Étude du Polymorphism Humain
References
- 1.Murray A W, Szostak J W. Nature (London) 1983;305:189–193. doi: 10.1038/305189a0. [DOI] [PubMed] [Google Scholar]
- 2.Murray A W, Szostak J W. Annu Rev Cell Biol. 1985;1:289–315. doi: 10.1146/annurev.cb.01.110185.001445. [DOI] [PubMed] [Google Scholar]
- 3.Burke D T, Carle G F, Olson M V. Science. 1987;236:806–812. doi: 10.1126/science.3033825. [DOI] [PubMed] [Google Scholar]
- 4.Farr C, Fantes J, Goodfellow P, Cooke H. Proc Natl Acad Sci USA. 1991;88:7006–7010. doi: 10.1073/pnas.88.16.7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnett M A, Buckle V J, Evans E P, Porter A C G, Rout D, Smith A G, Brown W R A. Nucleic Acids Res. 1993;21:27–36. doi: 10.1093/nar/21.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanish J P, Yanowitz J L, De Lange T. Proc Natl Acad Sci USA. 1994;91:8861–8865. doi: 10.1073/pnas.91.19.8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bilaud T, Brun C, Ancelin K, Koering C E, Laroche T, Gilson E. Nat Genet. 1997;17:236–239. doi: 10.1038/ng1097-236. [DOI] [PubMed] [Google Scholar]
- 8.Chong L, van Steensel B, Broccoli D, Erdjument H, Hanish J, Tempst P, de Lange T. Science. 1995;270:1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- 9.van Steensel B, Smogorzewska A, de Lange T. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 10.Lee C, Wevrick R, Fisher R B, Ferguson-Smith M A, Lin C C. Hum Genet. 1997;100:291–304. doi: 10.1007/s004390050508. [DOI] [PubMed] [Google Scholar]
- 11.Manuelidas L. Chromosoma. 1978;66:23–32. doi: 10.1007/BF00285813. [DOI] [PubMed] [Google Scholar]
- 12.Willard H F. Am J Hum Genet. 1985;37:524–532. [PMC free article] [PubMed] [Google Scholar]
- 13.du Sart D, Cancilla M R, Earle E, Mao J-i, Saffrey R, Tainton K M, Kalitsis P, Martyn J, Barry A E, Choo K H A. Nat Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- 14.Choo K H A. Am J Hum Genet. 1997;61:1225–1233. doi: 10.1086/301657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stillman B. J Biol Chem. 1994;269:7047–7050. [PubMed] [Google Scholar]
- 16.Aladjem M I, Rodewald L W, Kolman J L, Wahl G M. Science. 1998;281:1005–1009. doi: 10.1126/science.281.5379.1005. [DOI] [PubMed] [Google Scholar]
- 17.Harrington J J, Van Bokkelen G, Mays R W, Gustashaw K, Willard H F. Nat Genet. 1997;15:345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- 18.Ikeno M, Grimes B, Okazaki T, Nakano M, Saitoh K, Hoshino H, McGill N I, Cooke H, Masumoto H. Nat Biotechnol. 1998;16:431–439. doi: 10.1038/nbt0598-431. [DOI] [PubMed] [Google Scholar]
- 19.Willard H F. Nat Biotechnol. 1998;16:414–416. doi: 10.1038/nbt0598-415. [DOI] [PubMed] [Google Scholar]
- 20.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collins F S, Weissman S M. Prog Nucleic Acid Res Mol Biol. 1984;31:315–462. doi: 10.1016/s0079-6603(08)60382-7. [DOI] [PubMed] [Google Scholar]
- 22.Weier H-U, Kleine H-D, Gray J W. Hum Genet. 1991;87:489–494. doi: 10.1007/BF00197174. [DOI] [PubMed] [Google Scholar]
- 23.Spencer F, Ketner G, Connelly C, Hieter P. Methods. 1993;5:161–175. [Google Scholar]
- 24.Gnirke A, Huxley C, Peterson K, Olson M. Genomics. 1993;15:659–667. doi: 10.1006/geno.1993.1121. [DOI] [PubMed] [Google Scholar]
- 25.Mogayzel P J, Henning K A, Bittner M L, Novotny E A, Schwiebert E M, Guggino W B, Jiang Y, Rosenfeld M A. Hum Mol Genet. 1997;6:59–68. doi: 10.1093/hmg/6.1.59. [DOI] [PubMed] [Google Scholar]
- 26.Telenius H, Carter N P, Bebb C E, Nordenskjöld M, Ponder B A, Tunacliffe A. Genomics. 1992;13:718–725. doi: 10.1016/0888-7543(92)90147-k. [DOI] [PubMed] [Google Scholar]
- 27.Guan X-Y, Trent J M, Meltzer P S. Hum Mol Genet. 1993;2:1117–1121. doi: 10.1093/hmg/2.8.1117. [DOI] [PubMed] [Google Scholar]
- 28.Page S L, Earnshaw W C, Choo K H A, Shaffer L G. Hum Mol Genet. 1995;4:289–294. doi: 10.1093/hmg/4.2.289. [DOI] [PubMed] [Google Scholar]
- 29.Brown K D, Wood K W, Cleveland D W. J Cell Sci. 1996;109:961–969. doi: 10.1242/jcs.109.5.961. [DOI] [PubMed] [Google Scholar]
- 30.Shiels C, Coutelle C, Huxley C. Genomics. 1997;44:35–44. doi: 10.1006/geno.1997.4817. [DOI] [PubMed] [Google Scholar]
- 31.Dausset J, Ougen P, Abderrahim H, Billault A, Sambucy J L, Cohen D, Le Paslier D. Behring Inst Mitt. 1992;91:13–20. [PubMed] [Google Scholar]
- 32.Kitsberg D, Selig S, Keshet I, Cedar H. Nature (London) 1993;366:588–590. doi: 10.1038/366588a0. [DOI] [PubMed] [Google Scholar]
- 33.Sullivan B A, Schwartz S. Hum Mol Genet. 1995;4:2189–2197. doi: 10.1093/hmg/4.12.2189. [DOI] [PubMed] [Google Scholar]
- 34.Ried T, Koehler M, Padilla-Nash H, Schröck E. In: Cells: A Laboratory Manual. Spector D L, Goldman R D, Leinwand L A, editors. Vol. 3. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. [Google Scholar]
- 35.Jørgensen A L, Kølvraa S, Jones C, Bak A L. Genomics. 1988;3:100–109. doi: 10.1016/0888-7543(88)90139-5. [DOI] [PubMed] [Google Scholar]
- 36.Riley J, Butler R, Ogilvie D, Finniear R, Powell S, Anand R, Smith J C, Markham A F. Nucleic Acids Res. 1990;18:2887–2890. doi: 10.1093/nar/18.10.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drwinga H L, Toji L H, Kim C H, Greene A E, Mulivor R A. Genomics. 1993;16:311–314. doi: 10.1006/geno.1993.1190. [DOI] [PubMed] [Google Scholar]
- 38.Dubois B L, Naylor S L. Genomics. 1993;16:315–319. doi: 10.1006/geno.1993.1191. [DOI] [PubMed] [Google Scholar]
- 39.Cotter F E, Hampton G M, Nasipuri S, Bodmer W F, Young B D. Genomics. 1990;7:257–263. doi: 10.1016/0888-7543(90)90548-9. [DOI] [PubMed] [Google Scholar]
- 40.Zhu Y, Carroll M, Papa F R, Hochstrasser M, D’Andrea A D. Proc Natl Acad Sci USA. 1996;93:3275–3279. doi: 10.1073/pnas.93.8.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wick M J, Ann D K, Loh H H. Brain Res Mol Brain Res. 1995;32:171–175. doi: 10.1016/0169-328x(95)00090-f. [DOI] [PubMed] [Google Scholar]
- 42.Brown W R A, MacKinnon P J, Villasanté A, Spurr N, Buckle V J, Dobson M J. Cell. 1990;63:119–132. doi: 10.1016/0092-8674(90)90293-n. [DOI] [PubMed] [Google Scholar]
- 43.Taylor S S, Larin Z, Tyler Smith C. Hum Mol Genet. 1994;3:1383–1386. doi: 10.1093/hmg/3.8.1383. [DOI] [PubMed] [Google Scholar]
- 44.Henning K A, Moskowitz N, Ashlock M A, Liu P P. Proc Natl Acad Sci USA. 1998;95:5667–5671. doi: 10.1073/pnas.95.10.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willard H F, Waye J S. Trends Genet. 1987;3:192–198. [Google Scholar]
- 46.Nagaraja R, Kere J, MacMillan S, Masisi M W J, Johnson D, Molini B J, Halley G R, Wein K, Trusgnich M, Eble B, et al. Nucleic Acids Res. 1994;22:3406–3411. doi: 10.1093/nar/22.16.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Green E D, Riethman H C, Dutchik J E, Olson M V. Genomics. 1991;11:658–669. doi: 10.1016/0888-7543(91)90073-n. [DOI] [PubMed] [Google Scholar]
- 48.Neil D L, Villasante A, Fisher R B, Vetrie D, Cox B, Tyler-Smith C. Nucleic Acids Res. 1990;18:1421–1428. doi: 10.1093/nar/18.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown K E, Barnett M A, Burgtorf C, Shaw P, Buckle V J, Brown W R A. Hum Mol Genet. 1994;3:1227–1237. doi: 10.1093/hmg/3.8.1227. [DOI] [PubMed] [Google Scholar]
- 50.Brown W, Tyler-Smith C. Trends Genet. 1995;11:337–339. doi: 10.1016/s0168-9525(00)89100-3. [DOI] [PubMed] [Google Scholar]
- 51.Murphy T D, Karpen G H. Cell. 1998;93:317–320. doi: 10.1016/s0092-8674(00)81158-7. [DOI] [PubMed] [Google Scholar]
- 52.Larin Z, Taylor S S, Tyler-Smith C. Nucleic Acids Res. 1996;24:4192–4196. doi: 10.1093/nar/24.21.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]