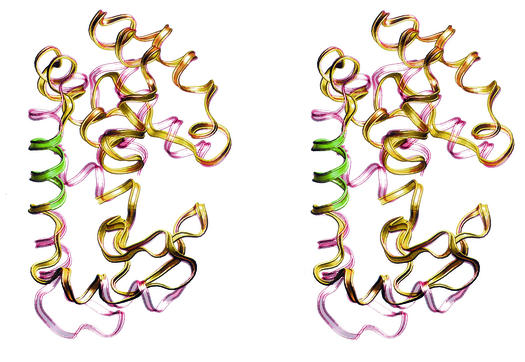Abstract
Helicobacter pylori is an important pathogen of the gastric system. The clinical outcome of infection is thought to be correlated with some genetic features of the bacterium. However, due to the extreme genetic variability of this organism, it is hard to draw definitive conclusions concerning its virulence factors. Here we describe a novel H. pylori gene which expresses an autolytic enzyme that is also capable of degrading the cell walls of both gram-positive and gram-negative bacteria. We designated this gene lys. We found this gene and observed its expression in a number of unrelated clinical strains, a fact that suggests that it is well conserved in the species. A comparison of the nucleotide sequences of lys and the hypothetical gene HP0339 from H. pylori strain ATCC 26695 revealed almost total identity, except for the presence of an insertion consisting of 24 nucleotides in the lys sequence. The coding sequences of lys and HP0339 show a high degree of homology with the coding sequence of bacteriophage T4 lysozyme. Because of this similarity, it was possible to model the three-dimensional structures of both the lys and HP0339 products.
The past few years have been marked by successful completion of numerous genome sequencing projects, ranging from projects involving the short genomes of prokaryotes to projects involving the human genome (9). The availability of the complete genome sequences of different organisms should have a profound impact on the development of new therapeutic strategies and on the identification of new genetic diseases.
Direct comparisons of genes whose functions have already been well characterized with newly sequenced genomes might allow workers to identify highly homologous predicted open reading frames. However, the hypothetical functions of the genes would be inferred solely on the basis of sequence homology with known genes, and they may not be the true functions in vivo. In addition, a gene may have a new function that is different from that deduced for its counterparts in other organisms.
For this reason, it is important to verify the actual function of any putative gene. By combining database searches with standard molecular biology techniques, it is possible to provide a complete description of the role of a gene.
The present work arose from our interest in using glycosidases in transglycosylation reactions for the synthesis of biologically relevant oligosaccharides (54). In order to identify new members belonging to such families, we focused on T4 lysozyme as a reference enzyme in our study. In fact, it has been demonstrated that bacteriophage T4 lysozyme, a common β(1-4)-N-acetylmuramidase, can acquire transglycolytic activity if a key residue is mutated (28).
In this study we investigated the presence in recently sequenced genomes of a gene whose predicted product exhibits a high level of homology with T4 lysozyme.
Screening for a lysozyme-like candidate revealed a putative gene in the genome of Helicobacter pylori (strain ATCC 26695), an important pathogen of the digestive system. Further analysis confirmed that the most important features of the product of this putative gene should match those of the class of proteins to which the reference lysozyme belongs. A reverse transcription-PCR (RT-PCR) assay demonstrated that the gene is expressed in vivo, and a zymogram analysis of crude protein extracts of H. pylori strains showed that a lytic activity is present on H. pylori and both gram-positive and gram-negative bacterial cell walls. Cloning and expression of the gene obtained from one of our bacterial strains confirmed the hydrolytic action of the protein. Finally, theoretical modeling of the structure of H. pylori lysozyme revealed a very high degree of structural similarity with T4 lysozyme.
MATERIALS AND METHODS
Database search and multiple alignment.
The complete nucleotide sequence of the lysozyme gene of bacteriophage T4 (GenBank accession number AF158101) was submitted to a FASTA3 server (http://www.cmbi.kun.nl/bioinf/tools/fasta.shtml) (40) for a nucleotide-versus-protein database search.
A multiple alignment was constructed by using a CLUSTALW server (http://www.cmbi.kun.nl/bioinf/tools/clustalw.shtml) (52) with the sequence of the product of the H. pylori putative gene (HP0339) and five other lysozyme sequences, including those of enterobacterial phage P1, bacteriophage P21, bacteriophage PA2, enterobacterial phage P22, and bacteriophage T4. The amino acid sequences of the proteins can be obtained through the National Center for Biotechnology Information protein database by using the following accession numbers: HP0339, C64562; P1, CAA61013; P21, P51771; PA2, WMBPP2; P22, NP_059622; and T4, 230620.
Bacterial strains and culture conditions.
H. pylori strains were isolated by gastric biopsy from 38 patients with gastritis and were maintained at −70°C in brucella broth containing 20% glycerol.
Bacteria were cultured at 37°C under microaerobic conditions in an anaerobic jar with a gas-generating kit (Oxoid, Basingstoke, England) on 7% horse blood agar plates supplemented with H. pylori selective medium (DENT) or in liquid cultures containing brucella broth (Difco), 10% calf fetal serum, and a selective antibiotic cocktail (DENT).
Escherichia coli XL1 blue and BL21 were used as hosts for plasmid cloning and protein expression, respectively. They were grown in Luria-Bertani broth (Sigma) supplemented with ampicillin (100 μg/ml) for recombinant clone selection.
Protein extraction.
H. pylori cells grown in 50-ml liquid cultures were collected by centrifugation and were washed rapidly twice with 5 ml of ice-cold phosphate-buffered saline (PBS) (8.1 mM Na2HPO4, 1.5 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl; pH 7.4). Cell pellets were resuspended in 100 μl of PBS containing 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol (DTT), 0.5 μg of pepsatinin per ml, and 0.6 μg of leupeptin per ml and were lysed by sonication at 150 W for 5 min on ice.
DNase and RNase were each added to a final concentration of 40 μg/ml, and the extracts were kept on ice for 1 h before centrifugation at 8,000 × g for 10 min to separate the supernatants (crude extracts) from the remaining particulate matter. The total protein concentration was determined with the bicinchoninic acid protein assay reagent (Pierce).
Genomic DNA extraction.
The H. pylori genomic DNA used for PCR was prepared by resuspending bacterial cells grown on agar plates in 50 μl of sterile water and boiling them for 10 min. After centrifugation for 15 min at 10,000 × g, 10 μl of supernatant was used for each PCR analysis.
The H. pylori DNA used for randomly amplified polymorphic DNA (RAPD) analysis was extracted from 50 ml of liquid culture by the cetyltrimethylammonium bromide method (6).
PCR.
Each PCR was carried out by using genomic DNA or cDNA as the template in a 100-μl mixture containing 0.5 U of Taq DNA polymerase (Sigma), 1× standard PCR buffer (Sigma), each deoxynucleoside triphosphate (dNTP) at a concentration of 0.1 mM, and 50 pmol of each primer with the following cycling conditions: 94°C for 3 min, 55°C for 1 min, and 72°C for 1 min for 35 cycles in a Progene DNA thermal cycler.
The following oligonucleotide primers were designed by using the previously published sequence of the putative lysozyme gene of H. pylori strain ATCC 26695: forward primer 5′-GATTCAGAGGGGTTTTCG-3′ and reverse primer 5′-AAATTTCCATATTCCTTGACG-3′. These primers amplify the complete coding sequence.
vacA has been used as a control gene for RT-PCR and PCR. For detection of vacA, forward primer 5′-ATGGAAATACAACAAACACAC-3 and reverse primer 5′-CCTGAGACCGTTCCTACAGC-3′ were used.
RT.
Total RNA was isolated by lysing the bacterial cells in 50 ml of a liquid culture in guanidinium thiocyanate-phenol buffer (TRIZOL; Gibco) and by using DNase I treatment to remove contaminating genomic DNA (amplification grade DNase I kit; Sigma) as recommended by the manufacturer. One microgram of total RNA that was denatured at 70°C for 10 min was mixed with an RT mixture (50 mM Tris-HCl [pH 8.3], 75 mM KCl, 3 mM MgCl2, 10 mM DTT, 200 U of Moloney murine leukemia virus reverse transcriptase [GIBCO-BRL], 30 U of RNase-OUT [GIBCO-BRL], 50 pmol of random exaprimers, each dNTP at a concentration of 0.5 mM) and incubated for 1 h at 37°C. After cDNA synthesis, the reaction was blocked by heating the mixture at 95°C for 5 min. To confirm the absence of contamination by genomic DNA, samples were prepared without reverse transcriptase treatment.
RAPD analyses.
For each strain, RAPD analyses were carried out by using one of the following primers (2): 1247 (5′-AAGAGCCCCGT-3′), 1254 (5′-CCGCAGCCAA-3′), 1282 (5′-AACGCGCAAC-3′), or 1283 (5′-GCGATCCCC-3′).
RAPD analyses were performed in 50-μl mixtures containing 10 ng of genomic DNA, 3.5 mM MgCl2, 50 pmol of one of the four primers, 0.5 U of Taq DNA polymerase (Sigma), and each dNTP at a concentration of 100 μM in the appropriate reaction buffer (Sigma). A Progene thermal cycler was used for 40 cycles of amplification (94°C for 30 s, 28°C for 1 min, and 72°C for 2 min). After PCR, 20-μl aliquots were subjected to electrophoresis on a 2% agarose gel in 1× Tris-acetate buffer.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Protein analyses were performed by using a denaturing 12% polyacrylamide gel (acrylamide/bisacrylamide ratio, 29:1) and the Laemmli method (30). After electrophoresis, proteins bands were visualized by Coomassie blue staining.
Preparation of E. coli and H. pylori murein sacculi.
E. coli DH5α was grown in 1 liter of Luria-Bertani broth at 30°C overnight. Murein sacculi were prepared by boiling bacterial cells in 4% SDS for 4 h as previously described (21) and by washing the sacculi with 2 M NaCl to remove peptidoglycan-associated proteins.
H. pylori peptidoglycan was isolated from bacterial cells scraped from agar plates and resuspended in 10 mM HEPES (pH 7.5). The bacterial suspension was slowly dropped onto an equal volume of a boiling solution of 8% SDS and processed as described above for E. coli sacculi.
Murein sacculi were kept at −20°C in Milli-Q water until they were required and, before a zymogram gel was cast and a lyso-plate assay was performed, were homogenized by sonication at 220 W for 3 min.
Spot assay and zymogram analysis.
For a spot assay with Micrococcus cells, 20 μg of total proteins from an H. pylori crude extract was placed on a 1% agar plate containing dried cells of Micrococcus lysodeikticus (0.05%, wt/vol) in 20 mM phosphate buffer (pH 7.0)-10 mM MgCl2. After incubation overnight at 37°C, the plate was checked for areas of lysis.
An agarose plate containing H. pylori sacculi was prepared adding 10 mg of peptidoglycan to 200 μl of 1% molten agarose in 20 mM phosphate buffer (pH 7.0)-10 mM MgCl2 and pouring the preparation into the wells of a 24-well plate (Falcon). After the agarose solidified, recombinant enzyme was spotted, the plate was incubated overnight at 37°C, and lytic zones were visualized by staining the gel with 0.1% methylene blue in 0.01% KOH.
A zymogram analysis of peptidoglycan hydrolysis was performed as previously described (50). Protein extracts and/or recombinant purified enzymes were subjected to electrophoresis in SDS-PAGE gels containing heat-inactivated M. lysodeikticus cells (0.05%, wt/vol) or E. coli murein sacculi (0.01%, wt/vol) as a substrate.
Following electrophoresis, each gel was soaked for 30 min in Milli-Q water at room temperature and then incubated in renaturation buffer (20 mM sodium phosphate buffer [pH 7.0], 0.1% Triton X-100, 10 mM MgCl2) at 37°C for 18 h with gentle agitation. Bands with lytic activity were visualized by staining the gel with 0.1% methylene blue in 0.01% KOH.
Molecular masses were determined by comparison with prestained molecular weight standards that were electrophoresed on the same gel.
Cloning, expression, and purification of recombinant protein.
The coding sequences of the H. pylori lys and HP0339 genes were obtained by PCR performed by using genomic DNA from one of our clinical strains (designated HPTS142) and vector construct GHPAE79 obtained from the American Type Culture Collection (ATCC) as templates. For cloning, the following primers were used: forward primer 5′-AAGAATTCGATTCAGAGGGGTTTTCG-3′ (EcoRI site underlined) and reverse primer 5′-AACTCGAGAAATTTCCATATTCCTTGACG-3′ (XhoI site underlined).
The genes were cloned by using EcoRI and XhoI restriction sites in the pGEX-4T-1 expression vector (Pharmacia Biotech) and standard protocols (45).
Recombinant proteins were overexpressed in E. coli BL21; bacteria harboring the pGEX-lys and pGEX-HP0339 plasmids were incubated in Luria-Bertani medium containing ampicillin (100 μg/ml) at 30°C until the optical density at 600 nm was 1.0. Then 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added to induce expression of the recombinant protein, and the preparation was incubated for 4 h at 30°C to minimize the presence of inclusion bodies. The cells were harvested by centrifugation at 10,000 × g for 15 min, and the pelleted bacteria were resuspended and incubated in 10 ml of TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) per g containing lysozyme (100 μg/ml) for 1 h at 37°C. Following incubation, a solution containing 0.1% sodium deoxycholate was added, the preparation was sonicated at 220 W for 2 min on ice, and the insoluble fraction was separated by centrifugation (10,000 × g, 15 min, 4°C).
Inclusion bodies were solubilized in 8 M urea-5 mM DTT, and protein refolding was obtained by dialyzing the solution against 50 volumes of TE buffer containing 1 mM DTT and 0.4 M arginine at 4°C overnight, followed by dialysis against 50 volumes of PBS containing 1 mM DTT at 4°C for 8 h.
Recombinant protein purification was performed by affinity chromatography by using glutathione-Sepharose 4B resin (Pharmacia-Biotech) according to manufacturer's instructions.
The glutathione S-transferase (GST) tail was removed from fusion protein bound to the resin matrix by incubating the resin in cleavage buffer (20 mM Tris-HCl [pH 8.4], 150 mM NaCl, 2.5 mM CaCl2) containing 0.5 U of thrombin (Novagen) per μg of fusion protein.
Molecular modeling by homology.
Templates were identified by submitting the complete amino acid sequence of HP0339 to the 3D-pssm protein fold recognition server (http://www.bmm.icnet.uk/servers/3dpssm) (24, 25). The Swiss-Pdb Viewer (http://www.expasy.ch/spdbv) (19) was used to align and compare the amino acid sequence of the target with the sequences and structures of the templates.
The resulting structure was modeled by submitting its complete amino acid sequence to the Swiss Model automated protein modeling server (http://www.expasy.ch/swissmod/SWISS-MODEL.html) (47), using the same set of templates. The resulting model was minimized with Swiss-Pdb Viewer by 20 steps of in vacuo steepest descent minimization. Parts of the model exhibiting poor structural conformation were remodeled by using the Swiss-Pdb Viewer loop library. The final model was minimized as described above and was checked for quality by submitting the structure to a WHAT IF web server (http://www.cmbi.kun.nl:1100/WIWWWI/) (20, 55). The same protocol was used for modeling the Lys protein.
Nucleotide sequence accession number.
The nucleotide sequence of the lys gene from clinical strain HPTS142 has been deposited in the GenBank database under accession number AY054410.
RESULTS
Data bank searches.
We investigated the presence of genes likely to express lysozyme-like proteins in recently sequenced genomes. We approached the problem by searching the genomic databases for bacteriophage T4 lysozyme homologues, since this gene-enzyme system has been widely investigated in specific studies of the effect of mutagenesis on enzyme activity (3, 29, 39). The availability of many T4 mutants and our knowledge of both the amino acid sequence (22) and the three-dimensional structure of the protein (42) make the T4 lysozyme a model that is particularly suitable for comparative studies.
A similarity search performed with the FASTA3 server from the CMBI website of Nijmegen University and the Protein Information Resource database (http://pir.georgetown.edu) (7) resulted in identification of four homologues of the T4 lysozyme; two of them were phage lysozymes, and the others were defined as hypothetical proteins (i.e., proteins whose nucleotide coding sequences are known but whose expression and function have not been tested yet).
Among the hypothetical proteins, the one that showed the highest homology with T4 lysozyme was the product of the putative HP0339 gene of H. pylori strain ATCC 26695, whose genome has recently been completely sequenced (53).
The predicted HP0339 gene codes for a hypothetical protein consisting of 116 residues and having a molecular mass of 13,562 Da. As shown in Fig. 1, 32 (27%) of the 116 residues of the product of the HP0339 gene are identical to residues of T4 lysozyme. In addition, many other amino acid substitutions are conservative, further increasing the overall similarity.
FIG. 1.
CLUSTALW alignment. The T4 lysozyme amino acid sequence was used as a reference for alignment of the HP0339 amino acid sequence. Matching residues are indicated by asterisks, whereas conserved and semiconserved residues are indicated by colons and dots, respectively. The residues forming the catalytic triad are highlighted.
Moreover, the alignment procedure confirmed that T4 lysozyme regions important for its enzymatic activity are conserved in the product of the HP0339 gene. In fact, it has been reported that the catalytic activity of T4, like the catalytic activities of most lysozymes, is largely due to three amino acids. In the case of the T4 lysozyme, catalysis takes place due to the concerted action of Glu11, Asp20, and Thr26 with the substrate (29). These three amino acids are present in the product of the HP0339 gene and are located at the same relative distance; they are separated by six and seven residues.
The homology at the amino acid level found between the T4 lysozyme and the product of the H. pylori gene prompted us to investigate whether the HP0339 gene codes for a protein with lysozyme-like activity against bacterial cell walls.
H. pylori strain screening.
H. pylori is characterized by high DNA polymorphism in the population, and there are differences in gene content among strains (23). For this reason, in order to obtain a high level of confidence, we collected evidence of the presence of the gene from a nonnegligible number of H. pylori strains. We examined bacteria isolated by endoscopy from 38 patients with H. pylori infections. All of the clinical strains analyzed had the phenotype CagA+ VacA+.
RAPD analysis was used for genetic typing of the clinical isolates. Stable band patterns with a high discriminatory capacity were obtained by using four different primers (2), and a comparison of the patterns showed that the strains isolated could be grouped into at least 30 distinct RAPD types having different genotypes and exhibiting various degrees of DNA polymorphism (data not shown).
Screening for the lysozyme-like gene in clinical strains was performed by PCR by using genomic DNA extracted by boiling water suspensions of bacteria. Specific primers were designed for the HP0339 nucleotide sequence (GenBank accession no. AE000551) to amplify a 348-bp fragment. Since all of the strains tested were VacA S1 or S2, two primers that amplify an internal region of the H. pylori vacA gene were used as controls for PCRs.
Interestingly, PCR genomic screening identified 21 unrelated strains that were positive for the HP0339 gene, but the gene was not detected in the other nine unrelated strains.
To verify the HP0339 nucleotide sequence reported in the GenBank database, we purchased from the ATCC the vector construct GHPAE79 (ATCC microbial special collection no. 627346). This construct contains a genomic insert corresponding to nucleotides 350692 to 351039 of the AE000511 locus of H. pylori strain ATCC 26695, including the complete coding sequence for the hypothetical HP0339 protein.
Lysozyme-like genes amplified by PCR from six unrelated clinical strains and from the ATCC construct were then isolated and sequenced. To ensure accuracy, the entire sequence was determined for each sample at least twice on one strand and once on the other strand. A comparison between the DNA sequence of the gene from our clinically isolated strains and that of the HP0339 gene showed that there was 96% identity in the complete sequence, except that only the clinical strains contained an insertion consisting of 24 consecutive nucleotides located between nucleotide positions 138 and 162 of the HP0339 coding sequence. The other differences observed in the nucleotide sequences were single substitutions, and most of them corresponded to consensus mutations.
Lysozyme-like gene expression in vivo.
In order to test whether the H. pylori HP0339 lysozyme-like gene was actually transcribed in vivo into an mRNA, total RNA was extracted from H. pylori cells, and an RT-PCR assay was carried out with a set of primers that amplify the complete coding sequence. vacA primers were used as a control to ensure the quality and integrity of the bacterial RNA.
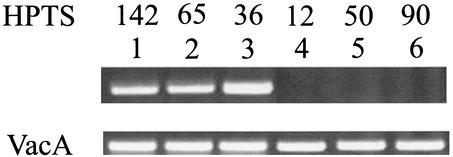
Strains HPTS142, HPTS65, HPTS36, HPTS12, HPTS50, and HPTS90 were tested for gene transcription by RT-PCR (Fig. 2). Products of the appropriate size could be amplified only from the first three strains, indicating that lysozyme-like gene expression was not present in HPTS12, HPTS50, and HPTS90 (Fig. 2). To confirm that the gene expressed in strains HPTS142, HPTS65, and HPTS36 was indeed the gene studied, fragments from two strains were sequenced (data not shown).
FIG. 2.
RT-PCR analysis of lysozyme-like gene expression in clinically isolated H. pylori strains. By using the vacA gene as an internal standard, lysozyme-positive strains HPTS142, HPTS65, and HPTS36 (lanes 1 to 3) and lysozyme-negative strains HPTS12, HPTS50, and HPTS90 (lanes 4 to 6) were analyzed for gene expression. Additional details are described in Materials and Methods.
Cell wall lytic activity in H. pylori protein extracts.
To demonstrate the presence of cell wall hydrolytic activity in isolated H. pylori strains, crude protein extracts were spotted on agar plates prepared with 20 mM phosphate buffer (pH 7.0)-10 mM MgCl2 containing M. lysodeikticus cells as a substrate. After overnight incubation at 37°C, the protein extracts from our clinical strains of H. pylori that express lysozyme-like enzyme induced the lysis of M. lysodeikticus cells. No hydrolytic activity was found in the extracts from H. pylori strains that do not have the lysozyme-like enzyme (data not shown).
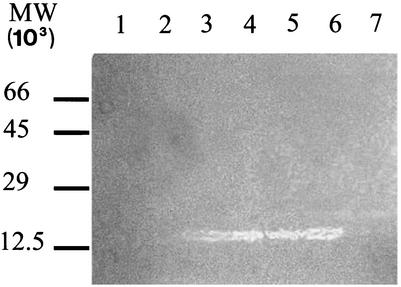
Zymogram gel analysis of peptidoglycan hydrolysis was used to better characterize the activity tested by the spot assay. Crude H. pylori protein extracts were subjected to SDS-PAGE in a gel containing 0.05% M. lysodeikticus cells. Following gel treatment with renaturation buffer (Fig. 3), only protein extracts from lysozyme-positive strains were able to solubilize the peptidoglycan substrate and produce a clear band in the gel after staining with methylene blue. Bovine serum albumin and ovalbumin were electrophoresed in parallel on the same zymogram gel as negative controls (Fig. 3, lane 7)
FIG. 3.
Zymogram analysis of H. pylori protein extracts. Portions (10 μg) of total proteins extracted from H. pylori strains were loaded on an SDS-12% polyacrylamide gel containing 0.05% (wt/vol) M. lysodeikticus cells. After electrophoresis, the gel was incubated in renaturation buffer overnight and stained with 0.1% methylene blue. Lanes 1 and 2, crude protein extracts from lysozyme-negative strains HPTS12 and HPTS90, respectively; lanes 3, 4, 5, and 6, crude protein extracts from lysozyme-positive strains HPTS142, HPTS65, HPTS61, and HPTS36, respectively; lane 7, bovine serum albumin and ovalbumin (2 μg). The positions of molecular weight (MW) markers are indicated on the left.
To eliminate the possibility that the clear lytic zones on the zymogram were due to an endoprotease instead of a lysozyme, protein extracts were treated with three protease inhibitors (leupeptin, aprotinin, and pepstatin; 0.1 μg/ml) before the extracts were loaded on the gel. In these conditions, the activity of the lytic enzyme was not affected, demonstrating that no protease was responsible for lysis.
The enzyme has an apparent molecular mass of 13.5 kDa, as determined by comparison with calibrated SDS-PAGE standards, which corresponds to the theoretical molecular mass estimated for the lysozyme-like gene product. Despite the apparent strong activity revealed in the zymogram, a gel electrophoresed in parallel and stained with either Coomassie blue or silver nitrate did not show any relevant band in that region (data not shown), indicating that the Lys protein is not a major protein in the bacteria.
Cloning, overexpression, and purification of the Lys protein.
To characterize the protein encoded by the gene and to verify that it was the cause of the lytic activity detected by spot assay and zymogram analyses, a PCR fragment coding for the protein, obtained from clinical strain H. pylori HPTS142, was cloned into the pGEX-4T-1 vector. The pGEX-4T vector is designed for inducible high-level intracellular expression of genes as fusion products with Schistosoma japonicum GST. A thrombin cleavage sequence, located immediately upstream of the multiple cloning site on the plasmid, enables subsequent removal of the GST tail. The E. coli host XL1 blue was transformed with recombinant plasmid pGEX-lys for cloning, and the E. coli host BL21 was transformed with positive recombinants for expression.
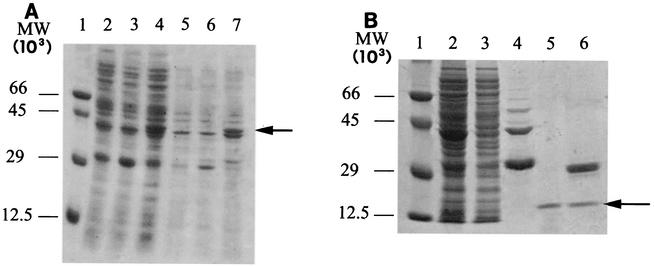
In order to extract the recombinant protein, whole bacterial cells were induced with IPTG and then sonicated after incubation in the presence of hen egg lysozyme and ionic detergent. In these conditions, a recombinant fusion protein was efficiently expressed and appeared in SDS-PAGE gels as a predominant band corresponding to a molecular mass of 44 kDa; this protein was absent in the BL21 host transformed with the pGEX-4T vector plasmid and in nontransformed BL21 cells (Fig. 4A). As estimated by SDS-PAGE, the expression level of the soluble fraction of the recombinant protein was about 20 μg/100 ml of bacterial culture. Moreover, a large percentage of the fusion protein was expressed as inclusion bodies (Fig. 4A, lane 7). Efficient extraction with 8 M urea and proper refolding by sequential dialysis in physiological buffer allowed us to retrieve the fusion protein from the inclusion bodies.
FIG. 4.
SDS-PAGE analysis of GST-Lys protein expression (A) and purification (B). (A) E. coli BL21 cells containing the pGEX-lys plasmid were grown at 30°C to an optical density at 600 nm of 1.0 and then induced with 0.1 mM IPTG for 4 h. Lane 1 contained molecular weight (MW) standards; the values on the left indicate molecular weights. Lane 2, crude soluble extract from nontransformed BL21 host cells; lane 3, crude soluble extract from BL21 cells containing nonrecombinant plasmid pGEX; lane 4, crude soluble extract from BL21 host cells containing the pGEX-lys recombinant plasmid; lane 5, solubilized pellet from nontransformed BL21 cells; lane 6, solubilized pellet from BL21 cells transformed with the nonrecombinant pGEX plasmid; lane 7, solubilized inclusion bodies from BL21 cells containing the pGEX-lys recombinant plasmid. (B) Recombinant protein was purified by affinity chromatography on glutathione-Sepharose 4B resin and digested with 0.5 U of thrombin per μg of fusion protein. Lane 1 contained molecular weight markers. Lane 2, 20 μg of total proteins from BL21(pGEX-lys) cells sonicated before affinity purification; lane 3, 20 μg of total proteins from sonicated BL21(pGEX-lys) cells, after loading on affinity resin; lane 4, eluate from glutathione-Sepharose 4B resin; lane 5, flowthrough following thrombin digestion of GST-Lys fusion protein bound to glutathione-Sepharose 4B; lane 6, thrombin digest of eluate from glutathione-Sepharose 4B resin. The arrow indicates the position of the 13.5-kDa recombinant Lys protein.
The recombinant protein was purified from bacterial proteins by affinity chromatography by using a gluthatione-Sepharose 4B resin that specifically binds GST. Cleavage of the purified recombinant protein from GST was performed with thrombin. All purification steps were monitored by SDS-PAGE analysis, as shown in Fig. 4. The molecular mass of the thrombin-cleaved recombinant protein was determined to be around 13.5 kDa, as expected by comparison with the theoretical molecular mass of the cloned protein (Fig. 4B, lanes 5 and 6).
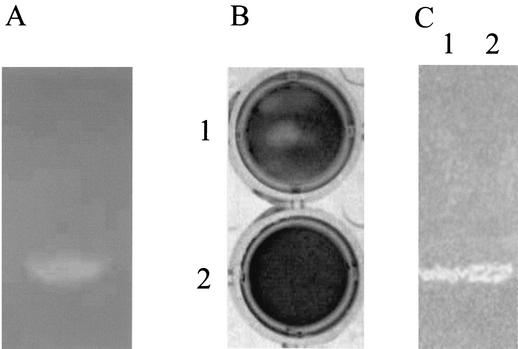
The activity of the recombinant protein was tested by zymogram analysis. The recombinant protein formed a lytic band at 13.5 kDa in gels containing heat-inactivated cells of M. lysodeikticus, indicating that the cloned DNA fragment encodes a protein with cell wall-lytic activity against gram-positive bacteria (Fig. 5A). In contrast, the purified recombinant protein fused with GST did not show any lytic activity in spot and zymogram assays (data not shown). Probably, the fusion protein is expressed in E. coli in an inactive form, and correct refolding of lysozyme protein is prevented by the presence of the GST affinity tail. Moreover, no hydrolytic activity was detected in protein extracts from nontransformed BL21 cells (data not shown).
FIG. 5.
Zymogram analysis of hydrolytic activity against M. lysodeikticus cells (A) and E. coli murein sacculi (C) of recombinant protein Lys and lyso-plate assay with H. pylori sacculi (B). (A) Five micrograms of recombinant Lys protein was loaded on a 12% polyacrylamide gel containing 0.05% (wt/vol) M. lysodeikticus cells, and the gel was stained with methylene blue to visualize lytic bands. (B) Five-micrograms of recombinant Lys protein was spotted on an agar plate containing H. pylori sacculi as the substrate and incubated overnight at 37°C (plate 1). As a negative control, lysate from nontransformed Bl21 cells was spotted on another plate (plate 2). To visualize lytic zones, the gels were stained for 1 h in 0.01% methylene blue and destained with Milli-Q water. (C) Five-micrograms of recombinant Lys protein (lane 1) and 20 μg of total H. pylori proteins (lane 2) were electrophoresed in an SDS-12% polyacrylamide gel containing 0.01% (wt/vol) E. coli murein sacculi. After electrophoresis, the gel was treated to visualize lytic bands as described above.
To test the possible autolytic activity of the recombinant protein on cell walls of H. pylori, a small-scale lyso-plate assay was performed. Purified samples of recombinant enzyme were applied to solidified agar containing peptidoglycan isolated from H. pylori poured onto a 24-well plate. After overnight incubation at 37°C and methylene blue staining, zones of lysis appeared as clear rings around the spots where samples had been applied, indicating the presence of a protein able to digest the peptidoglycan substrate. Extracts from nontransformed BL21 cells were used as negative controls (Fig. 5B).
Attempts to develop a procedure for large-scale cultivation in flasks of our clinical H. pylori strains have been unsuccessful so far. Therefore, it has not been possible to test recombinant enzyme activity on a zymogram gel containing cell walls isolated from H. pylori.
Since gram-negative bacteria are presumed to have structurally homogeneous cell walls (46), murein sacculi from E. coli DH5α cells were extracted and purified. In a zymogram gel analysis performed in the presence of E. coli sacculi as the substrate, both the crude protein extracts from strain HPTS142 (Fig. 5C, lane 2) and the purified recombinant Lys protein (lane 1) yielded a lytic band. The molecular mass of the band is about 13.5 kDa, in perfect agreement with the theoretical calculated molecular mass for this protein (Fig. 5C).
Cloning and expression of the HP0339 gene.
To verify that the absence of the 8-amino-acid insertion in the HP0339 protein could alter the activity, the HP0339 gene was cloned. By using the coding region of the vector construct GHPAE79 obtained from the ATCC, the HP0339 gene was cloned in the pGEX-4T expression vector. The recombinant protein was expressed, purified, and cleaved with thrombin as described above for the Lys protein. As shown in Fig. 6, recombinant HP0339 is an autolytic enzyme that is also capable of degrading M. lysodeikticus cell walls; i.e., it is active on both gram-positive and gram-negative substrates.
FIG. 6.
Zymogram analysis (A) and lyso-plate assay (B) of hydrolytic activity of purified recombinant HP0339 protein. (A) Purified and thrombin-cleaved recombinant HP0339 and Lys proteins were subjected to electrophoresis on a gel containing 0.05% (wt/vol) M. lysodeikticus cells as the substrate. Lytic bands were visualized by renaturing SDS-PAGE as described in Materials and Methods. Lane 1, Lys protein; lane 2, HP0339 protein. MW, molecular weight. (B) Assay of the activity of the HP0339 protein with H. pylori extracted sacculi. The assay plate gel was prepared in 20 mM phosphate buffer (pH 7.0)-10 mM MgCl2 containing H. pylori murein sacculi. Lysates from BL21 nontransformed cells (plate 1, negative control) and purified HP0339 protein (plate 2) were spotted on plates. Each plate was allowed to develop for 16 h before it was stained with methylene blue. Lytic activity is shown by a halo around a well (white spot at the center of a plate).
Molecular modeling of the products of HP0339 and lys.
The structural folding of the product of HP0339 was checked by submitting its amino acid sequence to the 3D-pssm server. The sequence was classified as a member of the phage T4 lysozyme family (Structural Classification Of Proteins family d119l) (37). The expectation value of the match is >95%, suggesting that a good set of templates was found. Two domains were detected, one β domain comprising residues 3 to 27 and one α domain comprising residues 38 to 111. Further modeling was done by using as templates the members of family d119l showing the best alignment with the target amino acid sequence (the atomic coordinates are available in the Research Collaboratory for Structural Bioinformatics protein data bank at PDB # 247L, PDB # 1CU2, PDB # 1CU3, PDB # 236L, PDB # 128L, PDB # 1CU0, PDB # 127L, PDB # 1L59, PDB # 1L25, PDB # 1L32, PDB # 1L27, and PDB # 1L28). The amino acid sequence corresponding to the product of gene HP0339 was threaded on the backbone of the templates and then submitted to Swiss Model. The resulting structure was minimized, and the resulting Ramachandran plot was checked. Some residues (Tyr31, Glu32, Tyr40, Lys108 Ile113, Gly5, Gly16, Gly21, Gly71, and Gly112) were outside the allowed regions. These residues were refined by remodeling the regions to which they belong by making use of the loop library of Swiss-Pdb Viewer. The iterative refinement was continued until only glycine residues were outside the allowed Ramachandran regions. The final structure was obtained by minimizing the refined model.
The tertiary structure of the product of the lys gene was obtained by using the protocol described above. In this case the phage T4 lysozyme family d119l was also identified with an expectation value for the match of >95%; the domains detected were an β domain, formed by residues 3 to 27, and an α domain, formed by residues 56 to 119. A new set of templates (the atomic coordinates are available in the Research Collaboratory for Structural Bioinformatics protein data bank at PDB # 247L, PDB # 1CU2, PDB # 1CU3, PDB # 236L, PDB # 128L, PDB # 1CU0, PDB # 127L, PDB # 1L59, PDB # 1L25, PDB # 1L32, PDB # 1L27, and PDB # 1L28) was used to thread the sequence corresponding to the product of the lys gene. The structure obtained with Swiss Model was minimized and checked for residues in forbidden regions of the Ramachandran plot. After refinement, only residues Ile121, Gly5, Gly16, Gly21, Gly41, Gly79, and Gly120 were found outside the allowed Ramachandran regions.
Superposition of the structures obtained for the HP0339 and Lys proteins onto the templates (Fig. 7) showed that the main features of the typical lysozyme-like fold, such as the distance between the Cα of the catalytic triad, were preserved. In particular, the residues involved in the formation of a complex between T4 lysozyme and its natural substrate, the peptidoglycan, are in general either identical or conserved (27).
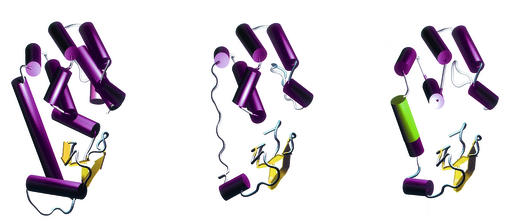
FIG. 7.
Tertiary structures. Left, crystallographic structure of phage T4 lysozyme (PDB ID 2LZM); center, proposed structure of the product of the HP0339 gene; right, proposed structure of the product of the lys gene. The green segment is the 8-amino-acid insert that is characteristic of the Lys protein.
The overall backbone root mean square deviations between the wild-type T4 lysozyme (PDB # 2LZM) and the final models were 3.05 for HP0339 and 2.86 for the Lys protein. However, the root mean square deviations dramatically decreased to 0.62 for HP0339 and to 0.63 for the Lys protein in the regions that could be aligned.
The existence of the 8-amino-acid insertion in the Lys protein allowed us to model a helix in the region connecting the α domain to the β domain, thus increasing the overall structural similarity between the Lys protein and the wild-type T4 lysozyme. The same region was modeled as a loop in HP0339. However, both model structures allow placement of the α and β domains in the same positions relative to each other, thus creating a very reasonable approximation of the active site (Fig. 8).
FIG. 8.
Stereo views of the proposed structure of the product of the lys gene. The crystal structure of phage T4 lysozyme (red) is superimposed over the proposed structure of the product of the lys gene (yellow). The green segment is the 8-amino-acid insert that is characteristic of the Lys protein.
DISCUSSION
Many bacteria have been shown to contain one or more cell wall lytic enzymes. These enzymes, called autolysins, can be as diverse as N-acetylglucosaminidases, N-acetylmuramyl-l-alanine amidases, endopeptidases, transglycosylases, and N-acetylmuramidases (lysozymes) (48).
Despite considerable descriptive information about lytic transglycosylases from numerous bacteria (44), many aspects of the physiological functions of these enzymes remain conjectural; they are described as being involved in cell wall turnover (17), cell separation, competence for genetic transformation, formation of flagella, and sporulation (43).
In this study, we successfully employed a genomics-based approach that allowed us to identify and clone an H. pylori gene that is highly homologous to the T4 phage lysozyme. H. pylori gene HP0339 was described after genomic sequencing of strain ATCC 26695 (53) as a putative gene that codes for a hypothetical lysozyme-like protein. Overall, 27% of its 116 amino acids are identical to those of T4 phage lysozyme. When the fact that 24 of the nonidentical amino acids are conservative substitutions is taken into account, the similarity dramatically increases to 52%. Based on these observations, we hypothesized that the HP0339 gene could code for a protein with lysozyme-like activity.
H. pylori is a gram-negative, spiral, pathogenic bacterium. It specifically colonizes the gastric epithelium of primates and is an etiologic agent of chronic gastritis (10). Bacterial properties and various factors, both factors of the host and factors of the environment, can cause gastritis to progress to more severe diseases over a period of years. These diseases include peptic ulcers, gastric lymphoma, and carcinoma (12, 34). The presence of a lysozyme in H. pylori might result in a number of hypotheses concerning the role of such an enzyme in the life cycle of this extremely important pathogen of the gastric system.
We examined by PCR some unrelated clinical strains of H. pylori, and in only 70% of them was the lysozyme gene detected. This fact was not unexpected, since it is known that at least strain J99, whose complete genome has been sequenced (4), does not have any gene homologous to HP0339. Gene sequencing of six different strains revealed almost complete homology with the HP0339 nucleotide sequence of strain ATCC 26695, except for a conserved insertion consisting of 24 nucleotides present only in the clinical strains. We designated this variant form of the HP0339 gene lys.
These findings confirmed the high level of genetic variation described for H. pylori (23), which is commonly believed to be a very variable species. Moreover, genes that are present in certain isolates of a given bacterial species or strain and that are absent or substantially different in others can be of great biological interest. In some cases, the difference may determine strain-specific features, such as drug resistance (15), bacterial surface structure (49), or restriction modification (26). Strain-specific factors that may influence pathogenicity are especially important (5). Studies of the molecular genetics of H. pylori have revealed that its extensive diversity at the level of individual genes may contribute to the pathogenicity of this bacterium (11, 58). The cag island (1), a 35- to 40-kDa genetic element for which the cagA gene is a marker, is an example of polymorphism that affects clinical outcome. Not all H. pylori strains contain the cag island; its presence is associated with an increase in the production of interleukin 8 and mucosal inflammation and with an increased risk of clinically significant pathology. In addition, there is a strong association between the presence of the cag island and several other genotypes, such as the vacA gene (this is especially true for the s1-m1 type), which codes for a secreted molecule that induces vacuole formation in epithelial cells (13).
In order to validate any statistical correlation between lysozyme expression and clinical outcome, it is necessary to sample a wide variety of strains. In the present study, only CagA+ phenotype strains were available; acquisition of more significant strains is currently being undertaken.
The lys gene, both in the native form and in the recombinant form, codes for an enzyme with lytic activity against cell walls of H. pylori and, in general, gram-positive and gram-negative bacteria.
As indicated above, the HP0339 gene differs from lys in the absence of an insertion consisting of 24 consecutive nucleotides. The absence of an 8-amino-acid insertion in the HP0339 protein does not seem to dramatically alter the activity.
The presence of an autolytic lysozyme in H. pylori is consistent with the hypothesis concerning altruistic behavior (41). According to this hypothesis, H. pylori uses genetically programmed bacterial autolysis of a fraction of the bacterial population to release urease and other antigenic cytoplasmic proteins that become adsorbed onto the outer membrane of the remaining intact bacteria (18). Autolysis of H. pylori could have implications for pathogenesis. It might explain some aspects of the behavior of H. pylori, including how this noninvasive bacterium can present virulence factors and immunogens to its host, inducing a significant inflammatory response (14). Moreover, a massive release of cytoplasmic proteins may overwhelm the immune system, favoring immune evasion by the bacterium (33).
Although we describe only a single lytic activity, at the moment we cannot exclude the possibility that the activity of the Lys lysozyme could be modulated by interactions with a larger complex of other enzymes.
In addition, structural modifications of the cell wall should play a role in H. pylori pathogenesis if cell wall fragments are released. For instance, liberation of peptidoglycan fragments as a consequence of a hydrolytic activity may trigger inflammatory and arthropathic processes (31, 51, 57). Murein fragments from different bacteria are capable of highly specific interactions with particular cell types of a host, as exemplified by Bordetella pertussis tracheal cell cytotoxin [N-acetylglucosaminyl-(1-6)-anhydro-N-acetylmuramyl-l-Ala-d-Glu-meso-diaminopimelyl-d-Ala], which leads to destruction of infected ciliated tracheal cells (32), or by an identical toxin of Neisseria gonorrhoeae that promotes ciliated-cell-specific damage in the fallopian tube mucosa (35). The structural and morphological changes of the cell wall involved in the transition of H. pylori cells from the spiral form to the coccoid form have not been studied in detail. Still, it is worth remarking that the H. pylori cell wall has a unique muropeptide composition and that it undergoes substantial structural modifications. These modifications require activation of specific enzymes when cells stop growing actively and become committed to morphological transition (8, 36).
From an evolutionary point of view, the discovery of prophages that carry drug resistance and virulence genes in different bacterial species (38, 56) and the high sequence homology between T4 lysozyme and the lys (HP0339) gene could lead to the hypothesis that the Helicobacter lysozyme gene had a prophage origin. Prophage genes, when they encode functional proteins, may have an effect on bacterial fitness and consequently may be important as virulence factors or may be useful as markers for epidemiology and evolutionary studies.
From the structural point of view, the models of the proteins encoded by HP0339 and lys have the features typical of the bacterial lysozyme family. Both proteins have α and β domains, forming the catalytic cleft. One interesting characteristic of the protein encoded by lys is the 8-amino-acid insertion, which allows it to form a helix in the region connecting the two domains, whereas the protein encoded by HP0339 seems to form a loop in the same region. One possible effect due to the insertion could be modulation of the domain motions (16) and, consequently, of the shape of the active cleft that might be reflected in variation in the specificity or kinetics of the enzyme. Furthermore, the insertion is an interesting feature whose effect on the structure and function of the enzyme is now being investigated.
Further functional characterization, such as in vivo localization and crystallographic structure analyses, will be performed as soon as a sufficient quantity of protein is available.
Acknowledgments
We thank G. Manzini, G. Tell, F. Fogolari, G. del Sal, and I. Donati for stimulating discussions.
The financial contributions of the Università degli Studi di Trieste (ex 60%) and of F.B.C. s.r.l., Trieste, Italy, are gratefully acknowledged.
REFERENCES
- 1.Akopyants, N. S., S. W. Clifton, D. Kersulyte, J. E. Crabtree, B. E. Youree, C. A. Reece, N. O. Bukanov, E. S. Drazek, B. A. Roe, and D. E. Berg. 1998. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol. Microbiol 28:37-53. [DOI] [PubMed] [Google Scholar]
- 2.Akopyanz, N., N. O. Bukanov, T. U. Westblom, and D. E. Berg. 1992. PCR-based RFLP analysis of DNA sequence diversity in the gastric pathogen Helicobacter pylori. Nucleic Acids Res. 20:6221-6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albertini, A. M., M. Hofer, M. P. Calos, and J. H. Miller. 1982. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell 29:319-328. [DOI] [PubMed] [Google Scholar]
- 4.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 5.Atherton, J. C., R. M. Peek, Jr., K. T. Tham, T. L. Cover, and M. J. Blaser. 1997. Clinical and pathological importance of heterogeneity in vacA, the vacuolating cytotoxin gene of Helicobacter pylori. Gastroenterology 112:92-99. [DOI] [PubMed] [Google Scholar]
- 6.Ausubel, F. M., R. Brent, R. E. Kingston, D. Moore, J. G. Seidmain, J. A. Smith, and S. K. 1993. Current protocols in molecular biology. Greene Publishing & Wiley-Interscience, New York, N.Y.
- 7.Barker, W. C., J. S. Garavelli, Z. Hou, H. Huang, R. S. Ledley, P. B. McGarvey, H. W. Mewes, B. C. Orcutt, F. Pfeiffer, A. Tsugita, C. R. Vinayaka, C. Xiao, L. S. Yeh, and C. Wu. 2001. Protein Information Resource: a community resource for expert annotation of protein data. Nucleic Acids Res. 29:29-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benaissa, M., P. Babin, N. Quellard, L. Pezennec, Y. Cenatiempo, and J. L. Fauchere. 1996. Changes in Helicobacter pylori ultrastructure and antigens during conversion from the bacillary to the coccoid form. Infect. Immun. 64:2331-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bentley, D. R. 2000. The Human Genome Project—an overview. Med. Res. Rev. 20:189-196. [DOI] [PubMed] [Google Scholar]
- 10.Blaser, M. J. 1992. Hypotheses on the pathogenesis and natural history of Helicobacter pylori-induced inflammation. Gastroenterology 102:720-727. [DOI] [PubMed] [Google Scholar]
- 11.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Correa, P. 1995. Helicobacter pylori and gastric carcinogenesis. Am. J. Surg. Pathol. 19:S37-S43. [PubMed] [Google Scholar]
- 13.Cover, T. L. 1996. The vacuolating cytotoxin of Helicobacter pylori. Mol. Microbiol. 20:241-246. [DOI] [PubMed] [Google Scholar]
- 14.Craig, P. M., M. C. Territo, W. E. Karnes, and J. H. Walsh. 1992. Helicobacter pylori secretes a chemotactic factor for monocytes and neutrophils. Gut 33:1020-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies, J. 1994. Inactivation of antibiotics and the dissemination of resistance genes. Science 264:375-382. [DOI] [PubMed] [Google Scholar]
- 16.de Groot, B. L., S. Hayward, D. M. van Aalten, A. Amadei, and H. J. Berendsen. 1998. Domain motions in bacteriophage T4 lysozyme: a comparison between molecular dynamics and crystallographic data. Proteins 31:116-127. [DOI] [PubMed] [Google Scholar]
- 17.Doyle, R. J., J. Chaloupka, and V. Vinter. 1988. Turnover of cell walls in microorganisms. Microbiol. Rev. 52:554-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunn, B. E., N. B. Vakil, B. G. Schneider, M. M. Miller, J. B. Zitzer, T. Peutz, and S. H. Phadnis. 1997. Localization of Helicobacter pylori urease and heat shock protein in human gastric biopsies. Infect. Immun. 65:1181-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 20.Hooft, R. W., G. Vriend, C. Sander, and E. E. Abola. 1996. Errors in protein structures. Nature 381:272.. [DOI] [PubMed] [Google Scholar]
- 21.Hoyle, B. D., and T. J. Beveridge. 1984. Metal binding by the peptidoglycan sacculus of Escherichia coli K-12. Can. J. Microbiol. 30:204-211. [DOI] [PubMed] [Google Scholar]
- 22.Inouye, M., M. Imada, and A. Tsugita. 1970. The amino acid sequence of T4 phage lysozyme. IV. Dilute acid hydrolysis and the order of tryptic peptides. J. Biol. Chem. 245:3479-3484. [PubMed] [Google Scholar]
- 23.Jiang, Q., K. Hiratsuka, and D. E. Taylor. 1996. Variability of gene order in different Helicobacter pylori strains contributes to genome diversity. Mol. Microbiol. 20:833-842. [DOI] [PubMed] [Google Scholar]
- 24.Kelley, L. A., R. M. MacCallum, and M. J. Sternberg. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299:499-520. [DOI] [PubMed] [Google Scholar]
- 25.Kelley, L. A., R. M. MacCallum, and M. J. Sternberg. 1999. Recognition of remote protein homologies using three-dimensional information to generate a position specific scoring matrix in the program 3D-PSSM. The Association for Computing Machinery, New York, N.Y.
- 26.King, G., and N. E. Murray. 1994. Restriction enzymes in cells, not Eppendorfs. Trends Microbiol. 2:465-469. [DOI] [PubMed] [Google Scholar]
- 27.Kuroki, R., L. H. Weaver, and B. W. Matthews. 1993. A covalent enzyme-substrate intermediate with saccharide distortion in a mutant T4 lysozyme. Science 262:2030-2033. [DOI] [PubMed] [Google Scholar]
- 28.Kuroki, R., L. H. Weaver, and B. W. Matthews. 1999. Structural basis of the conversion of T4 lysozyme into a transglycosidase by reengineering the active site. Proc. Natl. Acad. Sci. USA 96:8949-8954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuroki, R., L. H. Weaver, and B. W. Matthews. 1995. Structure-based design of a lysozyme with altered catalytic activity. Nat. Struct. Biol. 2:1007-1011. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 31.Lichtman, S. N., S. Bachmann, S. R. Munoz, J. H. Schwab, D. E. Bender, R. B. Sartor, and J. J. Lemasters. 1993. Bacterial cell wall polymers (peptidoglycan-polysaccharide) cause reactivation of arthritis. Infect. Immun. 61:4645-4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luker, K. E., J. L. Collier, E. W. Kolodziej, G. R. Marshall, and W. E. Goldman. 1993. Bordetella pertussis tracheal cytotoxin and other muramyl peptides: distinct structure-activity relationships for respiratory epithelial cytopathology. Proc. Natl. Acad. Sci. USA 90:2365-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mai, U. E., G. I. Perez-Perez, J. B. Allen, S. M. Wahl, M. J. Blaser, and P. D. Smith. 1992. Surface proteins from Helicobacter pylori exhibit chemotactic activity for human leukocytes and are present in gastric mucosa. J. Exp. Med. 175:517-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McColl, K. E. 1996. Helicobacter pylori infection and its role in human disease—an overview. Pharm. World Sci. 18:49-55. [DOI] [PubMed] [Google Scholar]
- 35.Melly, M. A., Z. A. McGee, and R. S. Rosenthal. 1984. Ability of monomeric peptidoglycan fragments from Neisseria gonorrhoeae to damage human fallopian-tube mucosa. J. Infect. Dis. 149:378-386. [DOI] [PubMed] [Google Scholar]
- 36.Mizoguchi, H., T. Fujioka, K. Kishi, A. Nishizono, R. Kodama, and M. Nasu. 1998. Diversity in protein synthesis and viability of Helicobacter pylori coccoid forms in response to various stimuli. Infect. Immun. 66:5555-5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murzin, A. G., S. E. Brenner, T. Hubbard, and C. Chothia. 1995. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247:536-540. [DOI] [PubMed] [Google Scholar]
- 38.O'Brien, A. D., J. W. Newland, S. F. Miller, R. K. Holmes, H. W. Smith, and S. B. Formal. 1984. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226:694-696. [DOI] [PubMed] [Google Scholar]
- 39.Owen, J. E., D. W. Schultz, A. Taylor, and G. R. Smith. 1983. Nucleotide sequence of the lysozyme gene of bacteriophage T4. Analysis of mutations involving repeated sequences. J. Mol. Biol. 165:229-248. [DOI] [PubMed] [Google Scholar]
- 40.Pearson, W. R., T. Wood, Z. Zhang, and W. Miller. 1997. Comparison of DNA sequences with protein sequences. Genomics 46:24-36. [DOI] [PubMed] [Google Scholar]
- 41.Phadnis, S. H., M. H. Parlow, M. Levy, D. Ilver, C. M. Caulkins, J. B. Connors, and B. E. Dunn. 1996. Surface localization of Helicobacter pylori urease and a heat shock protein homolog requires bacterial autolysis. Infect. Immun. 64:905-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Remington, S. J., W. F. Anderson, J. Owen, L. F. Ten Eyck, C. T. Grainger, and B. W. Matthews. 1978. Structure of the lysozyme from bacteriophage T4: an electron density map at 2.4 A resolution. J. Mol. Biol. 118:81-98. [DOI] [PubMed] [Google Scholar]
- 43.Rogers, H. J. 1979. Microbial polysaccharides and polysaccharidases, p. 237-268. In G. W. Gooday, R. C. W. Berkeley, and D. C. Ellwood (ed.), Microbial polysaccharides and polysaccharidases. Academic Press, London, United Kingdom.
- 44.Rogers, H. J., H. R. Perkins, and J. B. Ward. 1980. Microbial cell walls and membranes, p. 437-460. Chapman & Hall Ltd., London, United Kingdom.
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 46.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwede, T., A. Diemand, N. Guex, and M. C. Peitsch. 2000. Protein structure computing in the genomic era. Res. Microbiol. 151:107-112. [DOI] [PubMed] [Google Scholar]
- 48.Shockman, G. D., and J. V. Holtje. 1994. Microbial peptidoglycan hydrolases, p. 131-166. In J. M. Ghuysen and R. Hakenbeck (ed.), Microbial cell wall. Elsevier, Amsterdam, The Netherlands.
- 49.Stroeher, U. H., and P. A. Manning. 1997. Vibrio cholerae serotype O139: swapping genes for surface polysaccharide biosynthesis. Trends Microbiol. 5:178-180. [DOI] [PubMed] [Google Scholar]
- 50.Sugai, M., T. Akiyama, H. Komatsuzawa, Y. Miyake, and H. Suginaka. 1990. Characterization of sodium dodecyl sulfate-stable Staphylococcus aureus bacteriolytic enzymes by polyacrylamide gel electrophoresis. J. Bacteriol. 172:6494-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takada, H., M. Tsujimoto, K. Kato, S. Kotani, S. Kusumoto, M. Inage, T. Shiba, I. Yano, S. Kawata, and K. Yokogawa. 1979. Macrophage activation by bacterial cell walls and related synthetic compounds. Infect. Immun. 25:48-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, J. C. Venter, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 54.Vetere, A., I. Donati, C. Campa, S. Semeraro, A. Gamini, and S. Paoletti. 2002. Synthesis and characterization of a novel glycopolymer with protective activity toward human anti-α-Gal antibodies. Glycobiology 12:283-290. [DOI] [PubMed] [Google Scholar]
- 55.Vriend, G. 1990. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 8:52-56. [DOI] [PubMed] [Google Scholar]
- 56.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 57.Weidemann, B., J. Schletter, R. Dziarski, S. Kusumoto, F. Stelter, E. T. Rietschel, H. D. Flad, and A. J. Ulmer. 1997. Specific binding of soluble peptidoglycan and muramyldipeptide to CD14 on human monocytes. Infect. Immun. 65:858-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiang, Z., S. Censini, P. F. Bayeli, J. L. Telford, N. Figura, R. Rappuoli, and A. Covacci. 1995. Analysis of expression of CagA and VacA virulence factors in 43 strains of Helicobacter pylori reveals that clinical isolates can be divided into two major types and that CagA is not necessary for expression of the vacuolating cytotoxin. Infect. Immun. 63:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]