Abstract
In vitro experimentation with Plasmodium falciparum has determined that a number of different receptor-ligand interactions are involved in the invasion of erythrocytes. Most culture-adapted parasite isolates use a mechanism of invasion that depends primarily on the erythrocyte sialoglycoprotein glycophorin A (GYPA) and erythrocyte-binding antigen 175 (EBA-175) of the parasite blood-stage merozoite. However, a minority of culture-adapted parasites and a majority of Indian field isolates can apparently invade by other means. Here, erythrocyte invasion phenotypes of P. falciparum field isolates in Africa were studied. For 38 Gambian isolates, invasion of neuraminidase-treated and trypsin-treated erythrocytes was inhibited, on average, by more than 60 and 85%, respectively, indicating a high level of dependence on sialic acid and trypsin-sensitive proteins on the erythrocyte surface. These results support the hypothesis that African P. falciparum parasites use GYPA as a primary receptor for invasion. However, the considerable variation among isolates confirms the idea that alternative receptors are also used by many parasites. Three amino acid polymorphisms in the GYPA-binding region of EBA-175 (region II) were not significantly associated with invasion phenotype. There was variation among isolates in the selectivity index (i.e., a statistical tendency toward aggregation or multiple invasions of host erythrocytes), but this variation did not correlate with enzyme-determined invasion phenotype or with eba-175 alleles. Overall, these invasion phenotypes in Africa support a vaccine strategy of inhibiting EBA-175 binding to GYPA but suggest that parasites with alternative phenotypes would be selected for if this strategy were used alone.
Invasion of the human erythrocyte by the malaria parasite Plasmodium falciparum involves a number of specific receptor-ligand interactions between the blood-stage merozoite and the erythrocyte surface (3). In contrast to erythrocyte invasion by Plasmodium vivax, which is dependent on the interaction between the parasite Duffy binding protein (DBP) (5) and the erythrocyte Duffy antigen (14), P. falciparum is able to use multiple alternative receptor-ligand interactions to achieve successful invasion (7, 12, 19, 28, 30). Investigations of the erythrocyte receptors utilized by the merozoite for invasion have been carried out with erythrocytes deficient in particular surface molecules (8, 12, 25), antibodies that bind to these molecules (13), or enzymes that modify protein and carbohydrate domains on these molecules (4, 7, 8, 19, 26). The principal enzymes used have been neuraminidase (which cleaves sialic acid groups from surface glycoproteins and glycolipids) and trypsin (which cleaves the peptide backbone of a number of surface proteins).
Together, these studies have indicated that at least five separate surface receptors are involved in P. falciparum merozoite invasion of the erythrocyte: glycophorin A (GYPA) (25), which is neuraminidase and trypsin sensitive; glycophorin B (GYPB) (7), which is neuraminidase sensitive but trypsin resistant; a neuraminidase-resistant but trypsin-sensitive receptor, termed X (7); a neuraminidase-sensitive receptor that may involve glycophorin C (GYPC) (18); and a trypsin-resistant receptor that is not GYPB, termed Y (28). Of these surface receptors, GYPA (the most abundant erythrocyte surface sialoglycoprotein [15]) is the best characterized and the only one for which the parasite ligand is known, namely, erythrocyte-binding antigen 175 (EBA-175) (30). The binding of EBA-175 is dependent on the peptide backbone of GYPA, making binding sensitive to trypsin digestion, and on GYPA’s attached sialic acid groups (30), which are removed by neuraminidase. The key binding region of EBA-175, the F2 subdomain (21, 30), is localized in an N-terminal cysteine-rich sequence, termed region II (2). This subdomain is the second in a duplicated DBP-like (DBL) domain with homology to the binding region in the DBPs of P. vivax and the related malaria parasite of macaques, Plasmodium knowlesi (2).
A number of other P. falciparum genes that encode homologues of EBA-175 have been found (1), and their characterization may define other receptor-ligand interactions between the erythrocyte surface and the merozoite. Unrelated genes may also encode proteins involved in the invasion process, even if they do not have DBL domains. One of these proteins without a DBL domain, the normocyte binding protein PfNBP1 (28), interacts with a trypsin-resistant receptor (termed Y) on the erythrocyte surface. Mutations giving rise to premature stop codons in Pfnbp1 in two culture-adapted parasite isolates lead to truncation of the PfNBP1 protein product and are associated with a very poor ability to invade trypsin-treated cells. This is the first reported P. falciparum genotype that apparently correlates with invasion phenotype, albeit in a small number of isolates.
There are major differences in the invasion phenotypes of culture-adapted isolates of P. falciparum. Previous work (4, 7, 19, 26) characterized the sensitivity of diverse cultured isolates to neuraminidase- and trypsin-sensitive receptors on the erythrocyte surface. Invasion by most of these parasites was trypsin sensitive (reducing invasion by >50%), but sensitivity to neuraminidase varied more widely (reducing invasion by 20 to 100%) (4). Although these isolates represented a broad collection of culture-adapted isolates, it is not clear how representative their invasion phenotypes are of P. falciparum in human infections. Recent work on a small number of field isolates from India (23) showed that a majority of the isolates (12 out of 15) were able to invade and grow quite effectively in both neuraminidase- and trypsin-treated erythrocytes, calling into question the primacy of GYPA as a receptor in natural populations.
P. falciparum merozoites are able to invade a large proportion of the erythrocyte population in a human host, enabling infections to reach very high levels of parasitemia; in contrast, P. vivax preferentially invades reticulocytes, the younger erythrocytes (20). However, there is evidence from studies in Thailand that the invasion process may not be random in many P. falciparum infections (6, 31). The degree of aggregation of parasites in erythrocytes with multiple parasite invasions, the selectivity index (SI), has been shown to correlate inversely with the severity of malaria disease (6, 31). The molecular interactions that underlie this SI are unknown, but culture assays suggest that they are to some extent parasite determined (6) and thus are potential virulence determinants. It is likely that they relate to receptor-ligand interactions involved in invasion and therefore may be related to parasite invasion phenotypes assayed by enzyme treatment of erythrocytes.
The parasite ligands necessary for erythrocyte invasion are potential candidates for malaria vaccines (16, 29). As the majority of global P. falciparum malaria cases are in sub-Saharan Africa (11), it is most important to study erythrocyte invasion by African parasites. Here, invasion phenotypes of field isolates from patients in The Gambia were characterized. In vitro assays of the first cycle of invasion of normal and neuraminidase- and trypsin-treated erythrocytes were performed with fresh parasite isolates. The results indicated that the majority of parasites in The Gambia use a neuraminidase- and trypsin-sensitive invasion pathway, although there is also considerable use of alternative invasion pathways. Investigation of the parasite eba-175 alleles and SI in vivo showed no correlation with in vitro invasion phenotypes.
MATERIALS AND METHODS
P. falciparum field isolates.
A 2-ml heparinized venous blood sample was obtained from each of 64 malaria patients presenting at the outpatient clinic of the MRC Hospital in Fajara, The Gambia. All patients or their guardians gave informed consent, and the study was approved by the Joint Gambian Government/MRC Ethics Committee. Samples were collected over a 5-week period from 22 October to 23 November 2001, during the end of the peak of the annual malaria transmission season. Peripheral blood parasitemia was calculated for each patient as an inclusion criterion for allowing informative culture assays to be performed. Samples were obtained from patients only if their parasitemia levels were greater than 100 parasites per high-power (×1,000) microscope field in a thick smear (10), which corresponded to a parasitemia level of about 3% infected erythrocytes in a thin smear. Patients were excluded if they reported having taken an antimalaria treatment during the preceding 2 weeks in an interview. A thin blood smear was prepared to determine the percentage of parasitized erythrocytes and the SI, based on the number of multiple parasite invasions per erythrocyte (see below). Twenty microliters of blood was spotted onto glass fiber filter paper and kept dry at +4°C to be used later for parasite genotyping. The remaining blood sample was washed three times at 500 × g for 5 min in prewarmed RPMI 1640 Invitrogen, Life Technologies, Paisley, United Kingdom) to remove the leukocytes and plasma. The washed, packed, parasitized erythrocytes were resuspended in RPMI 1640 complete medium (containing 25 mM HEPES, 40 μg of gentamicin ml−1, and 10% AB-positive serum) at a 10% hematocrit and added to fluorescein isothiocyanate (FITC)-labeled target erythrocytes for invasion assays (see below).
Enzymatic treatment of erythrocytes.
Blood (O positive, Rhesus factor positive, MN [as determined by agglutination with monoclonal antibodies; see below]) from a single malaria-negative donor who had not taken antimalaria drugs during the preceding 2 months was washed as described above. A 0.1-ml portion of the resulting packed erythrocytes (approximately 109 cells) was treated with 20 mU of neuraminidase (Sigma, Grillingham, United Kingdom) or 1 mg of tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (Sigma) ml−1 in a 1-ml final volume of RPMI 1640 as previously described (4, 23). These mixtures were incubated for 1 h at 37°C with periodic shaking and then were washed three times in RPMI 1640. Soybean trypsin inhibitor (0.5 mg ml−1) (Sigma) was added to the trypsin-treated cells, and the mixture was shaken for 10 min at room temperature to inactivate the enzyme. The cells were washed three times in RPMI 1640. Control (non-enzyme-treated) cells were washed in the same manner as enzyme-treated cells. The efficiency of the enzyme treatments in removing GYPA from the erythrocyte surface was assayed by agglutination tests with monoclonal antibodies directed against the M and N epitopes of GYPA (Biotest, Solihull, United Kingdom).
Fluorescence labeling of target erythrocytes.
In accordance with the method of Lamont et al. (17), 4 μg of filtered FITC (Sigma)/ml in phosphate-buffered saline solution at pH 7.4 (Sigma) was added to washed enzyme-treated and untreated erythrocytes. These mixtures were incubated with shaking for 10 min at room temperature to label target cells, which were then washed three times in RPMI 1640 to remove all unincorporated FITC. In practice, it was not possible to completely dissolve all of the FITC in phosphate-buffered saline; thus, the final concentration in the filtered solution was less than 4 μg/ml. This detail did not affect the ability to obtain fluorescent cells. Labeled erythrocytes were resuspended in RPMI 1640 complete medium at a 10% hematocrit.
In vitro invasion assays.
For invasion assays, 10 μl of target cell suspension (approximately 107 enzyme-treated or untreated control erythrocytes) was added to 10 μl of parasitized donor cell suspension in each of triplicate wells in flat-bottom 96-well microtiter plates (BD Falcon). Prewarmed RPMI 1640 complete medium was added to 200 μl, and the plates were cultured at 37°C in a candle jar with replacement of fresh medium after 24 h. After one invasion cycle, 48 to 54 h in total, a thin smear of a sample from each of the triplicate wells was fixed in methanol and incubated in 4 μg of ethidium bromide (Sigma) ml−1 for 15 min to stain parasite DNA. Slides were washed, air dried, and examined with a vertical fluorescence microscope (Leica Microsystems) at a magnification of ×1,000 under oil immersion. The number of parasites in at least 1,000 FITC-labeled erythrocytes in each of triplicate wells was scored (nonlabeled cells were not counted), and the mean was calculated for each treatment. Percent inhibition by enzyme treatment was determined as [1 − (proportion of enzyme-treated cells invaded/proportion of untreated cells invaded)] × 100 (4).
In vivo parasitemia and SI.
Thin smears of samples from each patient were stained with Giemsa stain and examined at a magnification of ×1,000 to determine parasitemia (the number of parasitized erythrocytes counted among at least 1,000 erythrocytes) and the number of single and multiple infections of erythrocytes (counted among at least 300 infected erythrocytes). To avoid potential biases, all counts were determined for all cells in full microscope fields (thus, the precise numbers of cells counted were greater than or equal to the minimum numbers of 1,000 and 300, respectively). Erythrocytes with multiple infections were defined as those containing two or more individual ring- or trophozoite-stage parasites. The observed number of erythrocytes with multiple infections was compared to the number expected if parasites were randomly distributed. The random probabilities of erythrocytes being infected with single and multiple parasites were calculated according to a Poisson distribution, (e−μμx)/x!, where μ is the mean number of parasites per erythrocyte [−ln(1 − parasitemia)] and x is the number of parasites per erythrocyte; the values obtained were then used to calculate the expected values for each class of parasitized erythrocytes (i.e., with 1, 2, or ≥3 parasites per cell). The difference between the observed and the expected numbers of erythrocytes with single and multiple infections (the pooled number of pooled erythrocytes with two or more parasites) was tested by using the χ2 statistic with the Yates correction calculated for each individual. The degree of deviation of the observed distribution from that randomly expected was calculated as the SI (observed number of cells with multiple infections/number expected under a Poisson distribution) (6, 31). SI values of >1 would indicate aggregation of parasites in erythrocytes with multiple infections, whereas values of <1 would indicate fewer erythrocytes with multiple infections than expected.
Sequencing and genotyping of field isolates.
DNA from isolates that grew successfully in the invasion assays was typed for three nonsynonymous (amino acid-altering) single-nucleotide polymorphisms (SNPs) at nucleotide positions 1731 (codon 577, Lys to Asn), 1750 (codon 584, Lys to Gln or Glu), and 1775 (codon 592, Glu to Ala) of eba-175 (numbering is from the start codon of the reference eba-175 sequence; GenBank accession number X52524). These polymorphic amino acids are located between cysteine residues 4 and 7 of the F2 subdomain, a region homologous to that known to be important for P. vivax and P. knowlesi DBP binding to the erythrocyte surface (27), and are the only polymorphisms occurring in a region that overlaps synthetic peptides of EBA-175 F2 that inhibit human erythrocyte binding and GYPA receptor recognition (21). A short region near the 3′ end of Pfnbp1 in the same isolates was also sequenced. This region contains a reported frameshift stop codon at position 8300 (codon 2767) in the 3D7 clone of P. falciparum that leads to premature truncation of the PfNBP1 ligand and that has been associated with a decreased ability to invade trypsin-treated erythrocytes (28). This position is numbered from the start codon of the predicted Pfnbp1 gene (28), an open reading frame in the P. falciparum chromosome 4 contig MAL4_1. This sequence contig was produced by the P. falciparum Sequencing Group at the Sanger Institute as part of the Malaria Genome Project with support from the Wellcome Trust and can be obtained from the following Internet site: http://www.sanger.ac.uk/Projects/P_falciparum/chr4.
Genomic DNA was used as a template for amplification of an 841-bp fragment of the F2 domain of eba-175 region II with primers designed from the reference sequence (GenBank accession number X52524) and a 540-bp fragment encompassing a region near the 3′ end of Pfnbp1 with primers designed from the publicly available P. falciparum chromosome 4 contig (MAL4_1): eba-175 F2—Fwd (5′-GTTGATACAAACACAAAGGTG-3′) and Rev (5′-CCTTTACTTCTGGACACATCG-3′); and Pfnbp1—Fwd (5′-CAACATCATGCAAAGAATTG-3′) and Rev (5′-GATTATGAACATGATGTGG-3′). The products provided the templates with which a subsequent nested PCR was performed for amplification of a 530-bp fragment of the F2 domain of eba-175 and a 282-bp fragment of Pfnbp1 with the following primers: eba-175 F2-nested—Fwd (5′GATGTATGTGTACCTCCGAG-3′) and Rev (5′CAATTGTCATCTTCACAAGG-3′); and Pfnbp1-nested—Fwd (5′CAATTTGAACACACCTTAG-3′) and Rev (5′GAATTGTTATTTGGCTTGG-3′). Amplification reactions were performed with a total volume of 10 μl containing 0.5 U of BioTaq DNA polymerase (Bioline, London, United Kingdom), reaction buffer with 1.5 mM MgCl2 (Bioline), 200 μM deoxynucleoside triphosphates, 1 μM each oligonucleotide primer, and 10 to 50 ng of extracted DNA. Temperature cycling was as follows: 94°C (2 min); 94°C (30 s), 58°C (45 s), and 72°C (1 min) for 30 cycles; and 5 min at 72°C (final extension). Amplified products from the first round of PCR were diluted 1/10 with water and used as templates for the nested PCR with the same reaction conditions and cycling temperatures.
Purified PCR products (eluted into 30 μl of water by using a spin column [Qiagen, Crawley, United Kingdom]) were sequenced with their respective nested forward primers (eba-175 F2-nested Fwd and Pfnbp1-nested Fwd) by cycle sequencing with a 3′ Big Dye terminator cycle sequencing premixed kit (PE Applied Biosystems, Warrington, United Kingdom). Purified sequencing products were electrophoresed on a Perkin-Elmer ABI Prism 377 DNA sequencer (PE Applied Biosystems), and sequences were checked and assembled by using Sequence Navigator version 1.0.1 (PE Applied Biosystems). All singletons and visibly ambiguous positions were resequenced from new PCR products to confirm their accuracy.
The presence of single or multiple parasite genotypes per isolate was determined by using a convenient method described previously (9, 32); this method involves nested PCR typing of the highly polymorphic loci msp-1 block 2 and msp-2 block 3. The number of clearly visible bands for each polymorphic locus was scored after electrophoresis on a 2% Metaphor agarose gel (FMC Bio-products). Isolates with a single msp-1 block 2 band and a single msp-2 block 3 band were scored as “single-clone” infections (although it is recognized that this typing method cannot completely exclude the possibility that some of these isolates had more than one clone).
Statistical analyses.
Nucleotide polymorphisms in the eba-175 F2 domain and the existence of a potential frameshift nucleotide insertion or deletion in Pfnbp1 were tested for association with the enzyme-determined invasion phenotype by using Mann-Whitney U tests (these treat the values of percent inhibition of invasion in a nonparametric manner). Tests of associations between the multiplicity of parasite genotypes within an isolate (1 or >1 genotype present) and percent inhibition due to enzyme treatment of target erythrocytes were also performed by using Mann-Whitney U tests.
The geometric mean SI for each parasite isolate in vivo was calculated and compared to values reported previously in Thailand (6, 31). Differences in the mean and variance of parasite SI values between isolates with parasitemias of <5%, 5 to 10%, and >10% were tested by using Mann-Whitney U tests and single-factor analysis of variance. The correlation between the SI and the in vitro invasion phenotypes was tested by using the square of Pearson's correlation coefficient (R2). All statistical tests were performed by using Microsoft Excel 97 or SPSS version 10.0.
RESULTS
Invasion phenotypes of Gambian field isolates.
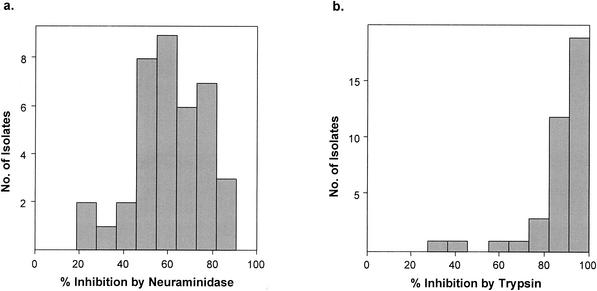
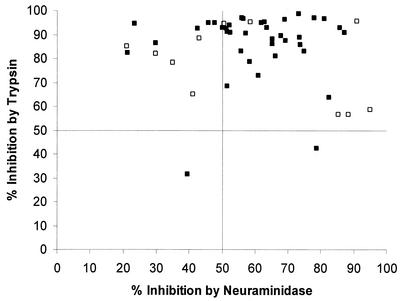
A total of 64 P. falciparum isolates were cultured, and of these, 38 (59%) successfully grew and invaded target erythrocytes in vitro (above 2% parasitemia postinvasion). That a large number of the isolates failed to grow was not unexpected and was probably due to many patients not reporting prior antimalarial treatment. The 64 isolates were from 31 male and 33 female patients (1 to 36 years old; average, 5 years). Inhibition of invasion by treatment of erythrocytes with neuraminidase varied between 21 and 87% for different parasite isolates (Table 1 and Fig. 1a), with a mean percent inhibition of 60%. There was also variation in inhibition by trypsin treatment of erythrocytes, with a range of 32 to 99% (Fig. 1b), but the mean percent inhibition was much higher, at 85%. The percent inhibition of each isolate caused by each enzyme treatment was plotted on two axes (Fig. 2) and compared with data for laboratory-maintained isolates from previous studies (4). This analysis showed that the distribution of sensitivity to both enzymes was similar to the distribution characterized for laboratory-maintained isolates from diverse geographical origins. The majority of parasite isolates were sensitive to both enzyme treatments (>50% inhibition), indicating that they depend largely on the presence of sialic acid groups and trypsin-sensitive proteins on the erythrocyte surface for successful invasion.
TABLE 1.
Invasion by 38 P. falciparum field isolates from The Gambia, West Africa, of untreated and neuraminidase- and trypsin-treated cells
| Sample | Parasitemiaa
|
% Inhibitionb in the following cells:
|
||||
|---|---|---|---|---|---|---|
| In patient | In assays of the following cells:
|
|||||
| Control | Nm treated | Tryp treated | Nm treated | Tryp treated | ||
| JB04 | 11.2 | 5.6 | 2.2 | 1.5 | 61 | 73 |
| JB05 | 11.6 | 4.0 | 2.4 | 2.7 | 39 | 32 |
| JB08 | 9.3 | 4.3 | 0.8 | 1.6 | 83 | 64 |
| JB12 | 4.6 | 3.4 | 1.5 | 0.6 | 56 | 83 |
| JB13 | 7.0 | 2.1 | 1.0 | 0.7 | 52 | 68 |
| JB14 | 12.2 | 4.4 | 1.4 | 0.6 | 69 | 87 |
| JB15 | 9.4 | 2.6 | 0.6 | 1.5 | 79 | 42 |
| JB16 | 3.0 | 2.8 | 0.9 | 0.5 | 66 | 81 |
| JB17 | 9.6 | 3.4 | 0.9 | 0.6 | 75 | 83 |
| JB19 | 9.1 | 5.5 | 2.6 | 0.5 | 52 | 91 |
| JB20 | 10.7 | 3.1 | 2.2 | 0.4 | 30 | 86 |
| JB23 | 4.8 | 1.8 | 0.8 | 0.4 | 58 | 79 |
| JB24 | 6.1 | 1.2 | 0.9 | 0.1 | 23 | 95 |
| JB25 | 7.9 | 6.0 | 2.6 | 0.2 | 56 | 97 |
| JB28 | 3.1 | 3.7 | 1.4 | 0.3 | 64 | 93 |
| JB29 | 10.0 | 5.9 | 1.6 | 0.7 | 74 | 89 |
| JB30 | 14.8 | 3.6 | 0.5 | 0.3 | 87 | 91 |
| JB32 | 5.2 | 4.0 | 2.0 | 0.4 | 52 | 91 |
| JB34 | 15.2 | 6.2 | 2.6 | 0.6 | 57 | 90 |
| JB37 | 6.7 | 4.1 | 2.0 | 0.3 | 52 | 94 |
| JB41 | 10.3 | 4.0 | 1.7 | 0.1 | 56 | 97 |
| JB42 | 2.9 | 2.8 | 0.4 | 0.2 | 86 | 93 |
| JB44 | 8.2 | 3.1 | 0.8 | 0.4 | 74 | 86 |
| JB46 | 3.4 | 2.6 | 1.0 | 0.1 | 62 | 95 |
| JB47 | 7.5 | 8.9 | 3.3 | 0.4 | 63 | 95 |
| JB48 | 11.6 | 2.9 | 1.0 | 0.3 | 65 | 88 |
| JB50 | 7.0 | 4.9 | 1.7 | 0.7 | 65 | 86 |
| JB51 | 1.7 | 3.7 | 2.0 | 0.2 | 48 | 95 |
| JB52 | 5.5 | 2.4 | 1.9 | 0.4 | 21 | 82 |
| JB53 | 5.8 | 2.6 | 1.4 | 0.1 | 46 | 95 |
| JB54 | 8.7 | 3.5 | 1.1 | 0.1 | 69 | 96 |
| JB56 | 3.9 | 1.9 | 0.6 | 0.2 | 68 | 90 |
| JB61 | 8.1 | 3.1 | 1.8 | 0.2 | 43 | 93 |
| JB62 | 10.2 | 3.1 | 0.7 | 0.1 | 78 | 97 |
| JB65 | 6.4 | 3.1 | 1.5 | 0.2 | 51 | 93 |
| JB67 | 3.4 | 2.5 | 0.7 | 0.0 | 73 | 99 |
| JB68 | 3.3 | 2.2 | 1.1 | 0.2 | 50 | 93 |
| JB69 | 7.2 | 4.7 | 0.9 | 0.2 | 81 | 97 |
Calculated as the average number (from triplicate wells) of infected erythrocytes per 1,000 erythrocytes counted. Nm, neuraminidase; Tryp, trypsin.
Calculated as [1 − (parasitemia in enzyme-treated cells/parasitemia in control cells)] × 100.
FIG. 1.
Histogram of percent inhibition of invasion due to neuraminidase (a) and trypsin (b) treatments of target erythrocytes for 38 P. falciparum field isolates from The Gambia.
FIG. 2.
Comparison of in vitro inhibition of invasion due to neuraminidase treatment (x axis) and trypsin treatment (y axis) of target erythrocytes for 38 P. falciparum field isolates from The Gambia (closed squares) and 11 laboratory-maintained isolates from broad geographical origins (open squares) (4). Lines indicate 50% inhibition by enzyme treatment for visual clarity.
Single- and multiple-clone infections.
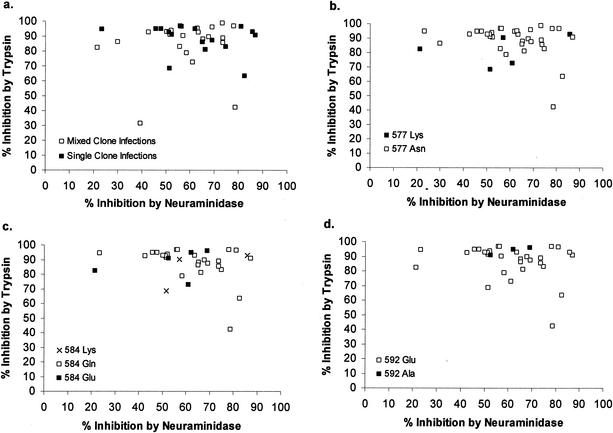
P. falciparum isolates frequently contain multiple distinct genotypes, and so it is possible that invasion phenotypes measured in vitro are often a composite of different parasite types. The presence of single- and multiple-clone infections in each blood sample was determined by PCR amplification of two polymorphic loci, msp-1 block 2 and msp-2 block 3, markers that vary considerably among parasite isolates and that provide a relatively stringent way of detecting multiple genotypes within an infected blood sample (9). Of the 38 blood samples assayed for invasion phenotypes, 17 (45%) were determined to have a single detectable parasite genotype, with the rest having two or more genotypes (some having evidence of at least four genotypes). Figure 3a shows the distribution of invasion phenotypes for multiple-clone (two genotypes or more) and single-clone infections. There was no difference in the mean percent inhibition by neuraminidase treatment in single-clone (mean inhibition, 62%) and multiple-clone (mean inhibition, 58%) infections (P value, 0.56, as determined by a Mann-Whitney U test). Similarly, there was no difference in the mean percent inhibition by trypsin treatment in single-clone (mean inhibition, 89%) and multiple-clone (mean inhibition, 84%) infections (P value, 0.42, as determined by a Mann-Whitney U test).
FIG. 3.
Scatter plots of enzyme-determined invasion phenotypes of all 38 Gambian isolates. Data show multiplicity of P. falciparum infection (a), alleles at codon 577 of eba-175 (b), alleles at codon 584 of eba-175 (c), and alleles at codon 592 of eba-175 (d). Isolates with mixed eba-175 genotypes were omitted.
Tests of association between invasion phenotypes and sequences of eba-175 and Pfnbp1.
Parasite isolates were typed for three nonsynonymous SNPs at codon positions 577, 584, and 592 of eba-175 region II. No association was found between alleles at any of the three SNP loci and invasion phenotypes (Fig. 3b to d) (P value for all comparisons, >0.05, as determined by Mann-Whitney U tests). All 38 parasite isolates had an in-frame sequence of Pfnbp1, i.e., with a C(A10)T sequence and not a C(A11)T sequence at nucleotide position 8300, which was reported to result in a frameshift stop codon in clone 3D7 (28); these data precluded any test of association between Pfnbp1 type and invasion phenotype here.
SI and tests for correlation with invasion phenotype.
Of the thin-film smears of samples from the 64 patients enrolled in the study, 48 (75%) had counts of erythrocytes with single and multiple infections compatible with a random (Poisson) distribution of parasites. In 15 other isolates (23% of the total), significantly more erythrocytes with multiple infections were observed than expected (P value for each isolate, <0.05, as determined by chi-square goodness of fit) (Table 2). A single isolate (JB17) had fewer erythrocytes with multiple infections than expected (Table 2). Overall, the geometric mean SI (which determines the magnitude and direction of the tendency toward erythrocytes with multiple infections) was 1.28 (range, 0.39 to 2.74), indicating a fairly moderate tendency in this direction. The average SI for the 38 isolates that successfully grew in vitro was very similar (1.26). Nonrandomness of invasion did not correlate with parasitemia, as no differences were found among the mean SI values for infections with <5%, 5 to 10%, and >10% parasitemia (P value for all comparisons, >0.05, as determined by a Mann-Whitney U test) or in their variance (P value, 0.790, as determined by a single-factor analysis of variance of the mean SI values for the different levels of parasitemia). The average SI of 1.28 corresponds to the conceptual estimate that about 78% of circulating erythrocytes (the reciprocal of the SI, i.e., 1/1.28) are available to parasites for invasion.
TABLE 2.
SI for Gambian P. falciparum field isolates showing significant nonrandom erythrocyte invasion
| Sample | Parasitemiaa | No. of parasitized erythrocytes containing the indicated no. of parasites:
|
SIc | Pd | |||||
|---|---|---|---|---|---|---|---|---|---|
| Observed
|
Expectedb
|
||||||||
| 1 | 2 | 3+ | 1 | 2 | 3+ | ||||
| JB03 | 8.1 | 290 | 31 | 1 | 309 | 13 | 0 | 2.38 | <0.01 |
| JB04 | 11.2 | 311 | 29 | 0 | 320 | 19 | 1 | 1.47 | 0.04 |
| JB08 | 9.3 | 299 | 25 | 5 | 313 | 15 | 0 | 1.91 | <0.01 |
| JB11 | 3.7 | 328 | 14 | 0 | 336 | 6 | 0 | 2.20 | <0.01 |
| JB17 | 9.6 | 304 | 7 | 0 | 296 | 15 | 1 | 0.45e | 0.02 |
| JB19 | 9.1 | 305 | 23 | 1 | 313 | 15 | 0 | 1.55 | 0.04 |
| JB20 | 0.7 | 306 | 30 | 1 | 318 | 18 | 1 | 1.66 | <0.01 |
| JB32 | 5.2 | 298 | 14 | 2 | 306 | 8 | 0 | 1.93 | 0.01 |
| JB48 | 11.6 | 356 | 34 | 0 | 366 | 23 | 1 | 1.44 | 0.03 |
| JB52 | 5.5 | 303 | 15 | 3 | 312 | 9 | 0 | 2.00 | <0.01 |
| JB56 | 3.9 | 302 | 14 | 0 | 310 | 6 | 0 | 2.26 | <0.01 |
| JB61 | 8.1 | 304 | 20 | 3 | 313 | 13 | 0 | 1.70 | 0.01 |
| JB62 | 10.2 | 329 | 27 | 4 | 341 | 18 | 1 | 1.63 | 0.01 |
| JB66 | 9.9 | 303 | 23 | 3 | 312 | 16 | 1 | 1.54 | 0.03 |
| JB69 | 7.2 | 310 | 31 | 4 | 332 | 12 | 0 | 2.74 | <0.01 |
| JB73 | 17.9 | 322 | 44 | 4 | 335 | 33 | 2 | 1.36 | 0.03 |
Calculated by examining at least 1,000 erythrocytes.
Number of parasitized cells expected in each class if invasion were a purely random process (according to a Poisson distribution).
Calculated as the observed number of cells with multiple invasions divided by the expected number.
As determined by a chi-square goodness-of-fit test of the observed numbers of erythrocytes with single and multiple invasions (pooled number of erythrocytes with two or more parasites) versus the expected numbers, with the Yates correction.
Selectivity was in the opposite direction, showing fewer erythrocytes with multiple invasions than expected.
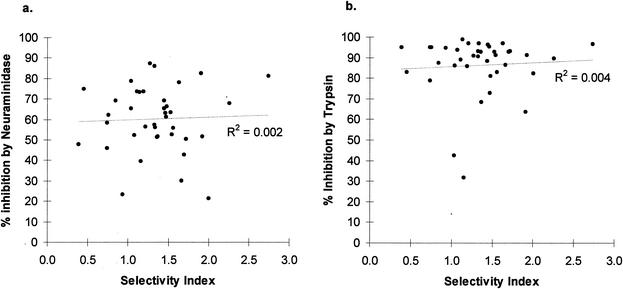
There was no significant association between the SI and percent inhibition of invasion by neuraminidase or trypsin treatment of target erythrocytes for all 38 Gambian isolates that were successfully cultured (R2 = 0.002, P = 0.81, or R2 = 0.004, P = 0.71, respectively) (Fig. 4). Similarly, there was no association between the SI and eba-175 region II SNPs at codons 577, 584, and 592 (P value for all comparisons, >0.05, as determined by a Mann-Whitney U test). Thus, if the SI is due to parasite invasion phenotypes, then the receptors responsible for selectivity on erythrocytes are probably not sensitive to neuraminidase or trypsin treatment, and the parasite determinants do not appear to involve these polymorphisms in eba-175.
FIG. 4.
Test for association between in vivo SI in a peripheral blood smear and percent inhibition by neuraminidase treatment (a) and trypsin treatment (b) of target erythrocytes in cultures.
DISCUSSION
Erythrocyte invasion assays with Gambian P. falciparum field isolates demonstrated that there is considerable variability in the dependence of parasites on the presence of enzyme-sensitive receptors on the erythrocyte surface for invasion. This variability is as extensive as that characterized for laboratory-maintained isolates (4) and for field isolates from India (23). However, the majority of Gambian parasites do depend on neuraminidase- and trypsin-sensitive receptors, as invasion is inhibited, on average, by 60 and 85% in neuraminidase- and trypsin-treated erythrocytes, respectively. These results indicate two features of wild P. falciparum parasites in Africa. First, they are consistent with the hypothesis that the majority of parasites do use the GYPA receptor to invade erythrocytes. These results provide support for the development of a vaccine that elicits antibodies to EBA-175 (the GYPA-binding ligand), which could block erythrocyte invasion via this pathway (16). However, an equally important feature is that none of the parasite isolates are completely dependent on this route of invasion, since none showed 100% inhibition by both enzyme treatments (despite the removal of the GYPA receptor, as determined by antibody agglutination). These results suggest that although a vaccine directed against the GYPA invasion pathway might be partially successful, it could select for parasites using alternative receptor-ligand interactions. These results support the view that an effective vaccine will need to be a multivalent entity to target more than one parasite vulnerability (29).
A previous attempt to correlate parasite genotype with invasion phenotype did not find a correlation between major sequence polymorphisms in P. falciparum merozoite antigens and the ability to invade enzyme-treated erythrocytes (4). Typing of the 38 field isolates here for three amino acid polymorphisms in the putative binding region of the F2 subdomain of region II in EBA-175 also showed no correlation with parasite invasion phenotype. This result suggests that amino acid polymorphisms in EBA-175 region II might not be functionally important with respect to binding to GYPA. However, they could be adaptive if they facilitate evasion of host immune responses; this scenario might explain the recent finding that EBA-175 region II shows significant evidence of being under positive diversifying selection within P. falciparum (3a, 24).
Typing of the 38 isolates for a region of the Pfnbp1 gene reported to have a stop codon in clone 3D7 (potentially conferring trypsin sensitivity of invasion) (28) showed that Gambian parasites all had a read-through allele. The possibility remains that PfNBP1 is truncated in some isolates due to an unidentified stop codon (as in another trypsin-sensitive laboratory isolate, 7G8).
The average SI for all 64 Gambian P. falciparum infections sampled here was 1.28. If we assume that this index reflects the selectivity of erythrocyte invasion in human infections, i.e., that, on average, 78% (∼1/1.28) of circulating erythrocytes are invaded, then most P. falciparum parasites should be able to invade the majority of erythrocytes. This SI is similar to SIs obtained for P. falciparum infections with comparable levels of parasitemia (>2%) in Thailand (6, 31). The Thai studies reported that the SI was inversely associated with the level of parasitemia. However, the majority of this trend was due to lower-parasitemia infections (<2%) that tended to have higher SI values (geometric mean, 2.44) (6, 31). Further studies of lower-density infections are needed to confirm whether this trend is consistent.
The molecular basis of parasite or host determinants of the SI is as yet unknown. The results reported here show an absence of any correlation with the sensitivity of invasion to enzyme treatment of target erythrocytes. These results suggest that the SI is not simply determined by differences in the abilities of parasites to utilize sialic acid or trypsin-sensitive proteins as receptors on the erythrocyte surface. Furthermore, the SI does not appear to be affected by amino acid polymorphisms in the F2 domain of EBA-175 region II. Further investigations of the molecular basis of SI phenotypes could examine variations in other merozoite antigens, in particular, homologues of EBA-175 (1) and PfNBP1 (28).
Gambian field isolates use erythrocyte receptors that are neuraminidase and trypsin sensitive as the major means of erythrocyte invasion. Since GYPA is the dominant sialoglycoprotein on the erythrocyte surface, the data presented here suggest that it is the primary receptor for erythrocyte invasion in West African parasites. Thus, antibodies against the erythrocyte-binding ligand EBA-175 may be effective against these parasites. Antibodies to region II of EBA-175 are common in children in The Gambia, and high levels of these antibodies show a weak association with a lower prospective risk of malaria (22). The role of a homologous antigen, EBA-140, in invasion by wild isolates is also allowed for by these results, since its putative receptor, GYPC (18), is also sensitive to the effects of neuraminidase and trypsin. Despite the variability in the dependence of invasion on neuraminidase- and trypsin-sensitive receptors in Gambian isolates, no parasite isolate is completely dependent on their presence. This finding argues that a vaccine that elicits antibodies against EBA-175 (thereby blocking invasion via GYPA) or EBA-140 (thereby blocking invasion via GYPC) might be initially successful, but protective immune responses to other antigens would also be essential for long-term success.
Acknowledgments
We are grateful to all of the patients who agreed to take part in this study and to Adama Joof-Secka and the staff at the MRC outpatient clinic for helping with patient enrollment. We also thank Demba Jammeh and Kebba Jammeh for diagnostic slide microscopy; Idrissa Sambou, Jamila Ismaili, Simon Correa, and Richard Pearce for help in the laboratory; and Neal Alexander for advice regarding statistical analysis.
This work was supported by the Wellcome Trust (prize studentship for J.B.) and the UK Medical Research Council.
Editor: S. H. E. Kaufmann
REFERENCES
- 1.Adams, J. H., P. L. Blair, O. Kaneko, and D. S. Peterson. 2001. An expanding ebl family of Plasmodium falciparum. Trends Parasitol. 17:297-299. [DOI] [PubMed] [Google Scholar]
- 2.Adams, J. H., B. K. Sim, S. A. Dolan, X. Fang, D. C. Kaslow, and L. H. Miller. 1992. A family of erythrocyte binding proteins of malaria parasites. Proc. Natl. Acad. Sci. USA 89:7085-7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnwell, J. W., and M. R. Galinski. 1998. Merozoite invasion of vertebrate cells: erythrocytes, p. 93-120. In I. W. Sherman (ed.), Malaria: parasite biology, pathogenesis, and protection. ASM Press, Washington, D.C.
- 3a.Baum, J., A. W. Thomas, and D. J. Conway. Evidence for diversifying selection on erythrocyte binding antigens of Plasmodium falciparum and Plasmodium vivax. Genetics, in press. [DOI] [PMC free article] [PubMed]
- 4.Binks, R. H., and D. J. Conway. 1999. The major allelic dimorphisms in four Plasmodium falciparum merozoite proteins are not associated with alternative pathways of erythrocytic invasion. Mol. Biochem. Parasitol. 103:123-127. [DOI] [PubMed] [Google Scholar]
- 5.Chitnis, C. E., and L. H. Miller. 1994. Identification of the erythrocyte binding domains of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion. J. Exp. Med. 180:497-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chotivanich, K., R. Udomsangpetch, J. A. Simpson, P. Newton, S. Pukrittayakamee, S. Looareesuwan, and N. J. White. 2000. Parasite multiplication potential and the severity of falciparum malaria. J. Infect. Dis. 181:1206-1209. [DOI] [PubMed] [Google Scholar]
- 7.Dolan, S. A., J. L. Proctor, D. W. Alling, Y. Okubo, T. E. Wellems, and L. H. Miller. 1994. Glycophorin B as an EBA-175 independent Plasmodium falciparum receptor of human erythrocytes. Mol. Biochem. Parasitol. 64:55-63. [DOI] [PubMed] [Google Scholar]
- 8.Facer, C. A. 1983. Erythrocyte sialoglycoproteins and Plasmodium falciparum invasion. Trans. R. Soc. Trop. Med. Hyg. 77:524-530. [DOI] [PubMed] [Google Scholar]
- 9.Farnert, A., A. P. Arez, H. A. Babiker, H. P. Beck, A. Benito, A. Bjorkman, M. C. Bruce, D. J. Conway, K. P. Day, L. Henning, O. Mercereau-Puijalon, L. C. Ranford-Cartwright, J. M. Rubio, G. Snounou, D. Walliker, J. Zwetyenga, and V. E. do Rosario. 2001. Genotyping of Plasmodium falciparum infections by PCR: a comparative multicentre study. Trans. R. Soc. Trop. Med. Hyg. 95:225-232. [DOI] [PubMed] [Google Scholar]
- 10.Greenwood, B. M., and J. R. Armstrong. 1991. Comparison of two simple methods for determining malaria parasite density. Trans. R. Soc. Trop. Med. Hyg. 85:186-188. [DOI] [PubMed] [Google Scholar]
- 11.Greenwood, B. M., and T. Mutabingwa. 2002. Malaria in 2002. Nature 415:670-672. [DOI] [PubMed] [Google Scholar]
- 12.Hadley, T. J., F. W. Klotz, G. Pasvol, J. D. Haynes, M. H. McGinniss, Y. Okubo, and L. H. Miller. 1987. Falciparum malaria parasites invade erythrocytes that lack glycophorin A and B (MkMk). Strain differences indicate receptor heterogeneity and two pathways for invasion. J. Clin. Investig. 80:1190-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holt, E. H., M. E. Nichols, Z. Etzion, and M. E. Perkins. 1989. Erythrocyte invasion by two Plasmodium falciparum isolates differing in sialic acid dependency in the presence of glycophorin A antibodies. Am. J. Trop. Med. Hyg. 40:245-251. [DOI] [PubMed] [Google Scholar]
- 14.Horuk, R., C. E. Chitnis, W. C. Darbonne, T. J. Colby, A. Rybicki, T. J. Hadley, and L. H. Miller. 1993. A receptor for the malarial parasite Plasmodium vivax: the erythrocyte chemokine receptor. Science 261:1182-1184. [DOI] [PubMed] [Google Scholar]
- 15.Huang, C. H., and O. O. Blumenfeld. 1995. MNSs blood groups and major glycophorins: molecular basis for allelic variation, p. 153-188. In J. P. Cartron and P. Rouger (ed.), Blood cell biochemistry: molecular basis of major human blood group antigens, vol. 6. Plenum Press, New York, N.Y.
- 16.Jones, T. R., D. L. Narum, A. S. Gozalo, J. Aguiar, S. R. Fuhrmann, H. Liang, J. D. Haynes, J. K. Moch, C. Lucas, T. Luu, A. J. Magill, S. L. Hoffman, and B. K. Sim. 2001. Protection of Aotus monkeys by Plasmodium falciparum EBA-175 region II DNA prime-protein boost immunization regimen. J. Infect. Dis. 183:303-312. [DOI] [PubMed] [Google Scholar]
- 17.Lamont, G., A. Saul, and C. Kidson. 1981. Plasmodium falciparum: assay of invasion of erythrocytes. Exp. Parasitol. 51:74-79. [DOI] [PubMed] [Google Scholar]
- 18.Mayer, D. C., O. Kaneko, D. E. Hudson-Taylor, M. E. Reid, and L. H. Miller. 2001. Characterization of a Plasmodium falciparum erythrocyte-binding protein paralogous to EBA-175. Proc. Natl. Acad. Sci. USA 98:5222-5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell, G. H., T. J. Hadley, M. H. McGinniss, F. W. Klotz, and L. H. Miller. 1986. Invasion of erythrocytes by Plasmodium falciparum malaria parasites: evidence for receptor heterogeneity and two receptors. Blood 67:1519-1521. [PubMed] [Google Scholar]
- 20.Mons, B., W. E. Collins, J. C. Skinner, W. van der Star, J. J. Croon, and H. J. van der Kaay. 1988. Plasmodium vivax: in vitro growth and reinvasion in red blood cells of Aotus nancymai. Exp. Parasitol. 66:183-188. [DOI] [PubMed] [Google Scholar]
- 21.Ockenhouse, C. F., A. Barbosa, D. P. Blackall, C. I. Murphy, O. Kashala, S. Dutta, D. E. Lanar, and J. R. Daugherty. 2001. Sialic acid-dependent binding of baculovirus-expressed recombinant antigens from Plasmodium falciparum EBA-175 to glycophorin A. Mol. Biochem. Parasitol. 113:9-21. [DOI] [PubMed] [Google Scholar]
- 22.Okenu, D. M., E. M. Riley, Q. D. Bickle, P. U. Agomo, A. Barbosa, J. R. Daugherty, D. E. Lanar, and D. J. Conway. 2000. Analysis of human antibodies to erythrocyte binding antigen 175 of Plasmodium falciparum. Infect. Immun. 68:5559-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okoyeh, J. N., C. R. Pillai, and C. E. Chitnis. 1999. Plasmodium falciparum field isolates commonly use erythrocyte invasion pathways that are independent of sialic acid residues of glycophorin. A. Infect. Immun. 67:5784-5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozwara, H., C. H. Kocken, D. J. Conway, J. M. Mwenda, and A. W. Thomas. 2001. Comparative analysis of Plasmodium reichenowi and P. falciparum erythrocyte-binding proteins reveals selection to maintain polymorphism in the erythrocyte-binding region of EBA-175. Mol. Biochem. Parasitol. 116:81-84. [DOI] [PubMed] [Google Scholar]
- 25.Pasvol, G., J. S. Wainscoat, and D. J. Weatherall. 1982. Erythrocytes deficiency in glycophorin resist invasion by the malarial parasite Plasmodium falciparum. Nature 297:64-66. [DOI] [PubMed] [Google Scholar]
- 26.Perkins, M. E., and E. H. Holt. 1988. Erythrocyte receptor recognition varies in Plasmodium falciparum isolates. Mol. Biochem. Parasitol. 27:23-34. [DOI] [PubMed] [Google Scholar]
- 27.Ranjan, A., and C. E. Chitnis. 1999. Mapping regions containing binding residues within functional domains of Plasmodium vivax and Plasmodium knowlesi erythrocyte-binding proteins. Proc. Natl. Acad. Sci. USA 96:14067-14072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rayner, J. C., E. Vargas-Serrato, C. S. Huber, M. R. Galinski, and J. W. Barnwell. 2001. A Plasmodium falciparum homologue of Plasmodium vivax reticulocyte binding protein (PvRBP1) defines a trypsin-resistant erythrocyte invasion pathway. J. Exp. Med. 194:1571-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richie, T. L., and A. Saul. 2002. Progress and challenges for malaria vaccines. Nature 415:694-701. [DOI] [PubMed] [Google Scholar]
- 30.Sim, B. K. L., C. E. Chitnis, K. Wasniowska, T. J. Hadley, and L. H. Miller. 1994. Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science 264:1941-1944. [DOI] [PubMed] [Google Scholar]
- 31.Simpson, J. A., K. Silamut, K. Chotivanich, S. Pukrittayakamee, and N. J. White. 1999. Red cell selectivity in malaria: a study of multiple-infected erythrocytes. Trans. R. Soc. Trop. Med. Hyg. 93:165-168. [DOI] [PubMed] [Google Scholar]
- 32.Viriyakosol, S., N. Siripoon, C. Petcharapirat, P. Petcharapirat, W. Jarra, S. Thaithong, K. N. Brown, and G. Snounou. 1995. Genotyping of Plasmodium falciparum isolates by the polymerase chain reaction and potential uses in epidemiological studies. Bull. W. H. O. 73:85-95. [PMC free article] [PubMed] [Google Scholar]