Abstract
DNA looping occurs in many important protein–DNA interactions, including those regulating replication, transcription, and recombination. Recent theoretical studies predict that tension of only a few piconewtons acting on DNA would almost completely inhibit DNA looping. Here, we study restriction endonucleases that require interaction at two separated sites for efficient cleavage. Using optical tweezers we measured the dependence of cleavage activity on DNA tension with 15 known or suspected two-site enzymes (BfiI, BpmI, BsgI, BspMI, Cfr9I, Cfr10I, Eco57I, EcoRII, FokI, HpaII, MboII, NarI, SacII, Sau3AI, and SgrAI) and six one-site enzymes (BamHI, EcoRI, EcoRV, HaeIII, HindIII, and DNaseI). All of the one-site enzymes were virtually unaffected by 5 pN of tension, whereas all of the two-site enzymes were completely inhibited. These enzymes thus constitute a remarkable example of a tension sensing “molecular switch.” A detailed study of one enzyme, Sau3AI, indicated that the activity decreased exponentially with tension and the decrease was ≈10-fold at 0.7 pN. At higher forces (≈20–40 pN) cleavage by the one-site enzymes EcoRV and HaeIII was partly inhibited and cleavage by HindIII was enhanced, whereas BamHI, EcoRI, and DNaseI were largely unaffected. These findings correlate with structural data showing that EcoRV bends DNA sharply, whereas BamHI, EcoRI, and DNaseI do not. Thus, DNA-directed enzyme activity involving either DNA looping or bending can be modulated by tension, a mechanism that could facilitate mechanosensory transduction in vivo.
Keywords: DNA looping, protein–DNA interactions, single-molecule manipulation
Restriction endonucleases (REases) are prokaryotic enzymes that act to “restrict” invasion of foreign DNA by cleaving phosphodiester bonds (1). These enzymes also serve as indispensable tools in molecular biology research and are used in procedures such as DNA cloning, fingerprinting, mapping, and sequencing (2). From the perspective of molecular biophysics, these enzymes are excellent model systems for studying basic principles of protein–DNA interactions (3, 4).
The most commonly studied REases are of the type II variety, which cleave within or near specific recognition sites, usually require Mg2+ ions as a cofactor, and do not hydrolyze ATP. More than 3,500 different type II REases having >200 different binding specificities have been identified (5). Of particular interest in our present study are the many unorthodox type II REases that do not cleave DNA efficiently if the template contains only one recognition site (6, 7). Efficient cleavage is only observed with templates containing two or more sites, suggesting that the active complex binds at two sites and the intervening DNA is looped out (7). This phenomenon of DNA looping is of broad importance in molecular biology and plays a role in many key processes including DNA transcription, replication, recombination, and repair (8–13). Looping of DNA by the Lac and Gal repressor proteins in Escherichia coli, for example, is well demonstrated and has recently been studied at the single DNA level (14–16). Looping also occurs with many eukaryotic transcription factors (8, 17). It has been proposed that the requirement for two recognition sites by some REases may have evolved as an additional control mechanism to protect against the possibility that a single unmethylated site in the host genome could result in undesired cleavage (18).
It has recently been proposed that internal forces associated with chromatin expansions and contractions during the eukaryotic cell cycle may play an important role in governing chromosome function, and that such forces could transmit signals to “molecular stress sensors” (19). In addition, certain types of cells, such as vascular endothelial cells, can sense extracellular stresses (20). Here, we investigate the possibility that DNA looping interactions could provide a mechanism for a molecular stress transducer.
Several recent theoretical studies have considered the effect of tension on DNA looping (21–23). As discussed by Marko and coworkers (22), the probability of forming a loop of size ΔL via thermal fluctuations against an applied tension F would be expected to be suppressed by a factor proportional to exp(−FΔL/kT). Detailed calculations based on a worm-like chain (WLC) model, which accounts for DNA bending rigidity, predict that a few piconewtons of tension would decrease the probability of looping by at least several orders of magnitude over the tension-free probability (21, 22). An independent theoretical study by Blumberg et al. (23) reached essentially the same conclusion. Free energy differences between looped and unlooped DNA were calculated in a simple two-state WLC model. For loop sizes >100 bp a few piconewtons of DNA tension is predicted to increase the time required for loop closure by at least two orders of magnitude and the degree of inhibition is predicted to increase rapidly with loop size.
Here, we study the effect of tension on DNA cleavage by one-site and two-site REases through manipulation of single DNA molecules with force-measuring optical tweezers. Partial inhibition of one-site cleavage by EcoRV at very high DNA tension was recently reported in an independent study (24). We confirm and extend those findings on one-site enzymes and report a much stronger and universal inhibitory effect of tension on activity with two-site enzymes.
Results and Discussion
Tension-Dependent Inhibition of Two-Site Enzymes.
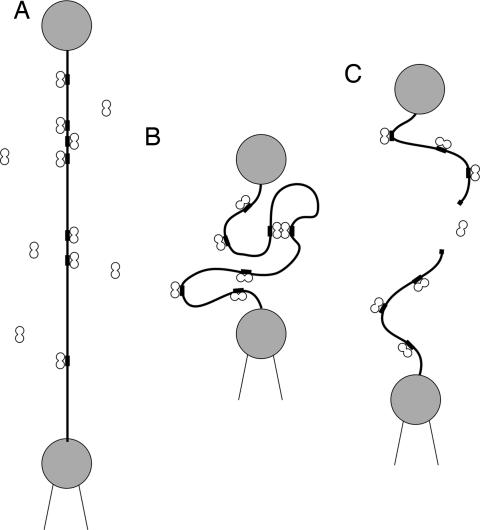
Our technique for measuring DNA cleavage activity is shown schematically in Fig. 1. We held a single DNA molecule taut at an end-to-end extension of ≈95% (corresponding to a tension of 5 pN) while the enzyme solution was flowed into the sample chamber. Information on the enzymes and DNA templates is given in Materials and Methods. The flow was continued for 30 s to ensure that the entire chamber was uniformly filled with enzyme solution, and then stopped. Measurements were made with the DNA either held stretched at 5 pN or relaxed to an end-to-end extension of 35%, corresponding to a tension of ≈0.06 pN. When the DNA was held taut, cleavage events were detected as a sudden drop in the measured tension, rendering the event observable in real time. With the DNA relaxed, cleavage was monitored by testing for the presence of the DNA tether every 30 s by quickly separating the microspheres. If the molecule was cleaved, the measured force remained zero as the microspheres were separated. If the molecule remained intact, however, a tension of up to a few piconewtons was sensed and the molecule was then quickly relaxed for another incubation period. This procedure was continued until either the molecule was cleaved or 5 min had elapsed. This process was repeated many times for both one-site and two-site endonucleases (see Materials and Methods).
Fig. 1.
Schematic of the single DNA cleavage measurements. DNA is tethered between two microspheres, one manipulated by optical tweezers and the other manipulated by a micropipette positioned by a piezoelectric transducer. (A) The DNA is held taut at an end-to-end extension of 95% (≈5 pN) while a solution containing the enzyme is introduced. (B) The molecule is then relaxed to 35% extension (≈0.06 pN) so that the active enzyme complex may form via DNA looping (in the case of two-site enzymes). (C) Upon separating the microspheres we detect whether or not cleavage has occurred.
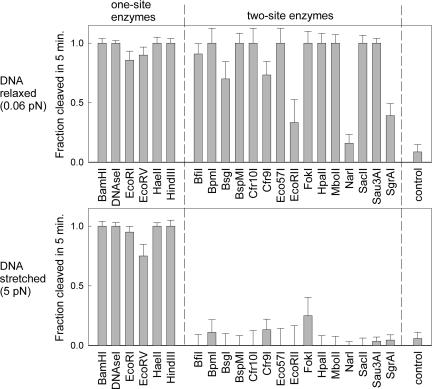
The cleavage statistics are shown in Fig. 2. One-site enzymes were largely unaffected by an applied tension of 5 pN, whereas all of the known or suspected two-site enzymes were strongly inhibited. The two-site enzymes differ from the one-site enzymes in that they must loop the DNA and therefore this inhibition is consistent with the theoretical predictions for tension-dependent looping (21–23).
Fig. 2.
Dependence of cleavage activity on DNA tension. Fraction of molecules that were cleaved after a 5-min incubation with each enzyme is graphed for one-site enzymes (left side of each plot) and known or suspected two-site enzymes (right side of each plot). (Upper) The results with the relaxed DNA. (Lower) Results with the DNA held stretched at an end-to-end extension of 95%, corresponding to a tension of 5 pN. The error bars were calculated as the standard deviation of the binomial distribution (P(1 − P)/N)1/2, where P is the probability of a molecule being cut and N is the number of trials. In the special cases where P = 0 or 1 the error was estimated as 1/N. The control bar indicates measurements with no enzyme, showing that a small fraction of molecules spontaneously detach from the microspheres even at zero tension.
Three of the 15 enzymes studied (EcoRII, NarI, and SgrAI) did not show complete cleavage even with the DNA relaxed, but there was still a clear drop in activity between the relaxed and stretched trials. In the cases of SgrAI and NarI the relative lack of activity may be attributable to the DNA template having relatively few recognition sites [only four and five sites, respectively, with minimum separations of 1,321 and 428 bp; looping of sites separated by >1,000 bp is generally very slow (25)]. In the case of NarI this result is also consistent with the recent finding that, unlike most other type II REases, this enzyme only cleaves one strand of DNA per binding event and therefore has to bind twice to cleave dsDNA (26). Incomplete cleavage was also observed with EcoRII, for which the DNA template contained a similar number of sites and a similar distribution of separations as those of the enzymes for which complete cleavage was observed. Interestingly, this result might be explained by a recent report (27) that EcoRII appears to require interaction at three sites for full activity.
We note that a small fraction of tethers appeared to be cleaved with two-site REases even when the DNA was held under tension. This finding, however, can be completely attributed to spontaneous unbinding of the link holding the DNA to the anti-digoxygenin (DIG) microsphere rather than actual cleavage of the DNA by the enzyme. As shown in Fig. 2, approximately the same background level of breakage was observed in control experiments in which the reaction buffer without enzyme was flowed into the sample chamber. Thus our results are consistent with complete inhibition of cleavage by two-site REases at 5 pN of tension.
Control experiments with the two-site enzymes were done by using templates with zero or one site. Cfr9I, NarI, SacII, SfiI, and SgrAI were tested on a template with zero sites and showed no activity. BfiI was tested on a template with only one site and also showed no activity. SacII cleaved a template with one copy of its recognition sequence very infrequently. Cleavage by two-site enzymes on templates containing only one site has been reported in bulk studies with Acc36I, BpmI, BsgI, BspMI, and FokI. In accord with our finding with SacII, it was found that these enzymes cleave more than an order of magnitude more slowly than they do on a template containing two sites (28).
Several thousand type II REases have been discovered, and further discoveries are occurring at a rapid pace. Although only a small number of these enzymes are currently known or suspected to operate by a two-site mechanism, it is likely that many more do as well (5). We note that our assay provides a way to rapidly screen enzymes for two-site behavior without needing engineered DNA templates. Our measurements provide a unique form of evidence for two-site behavior with these enzymes.
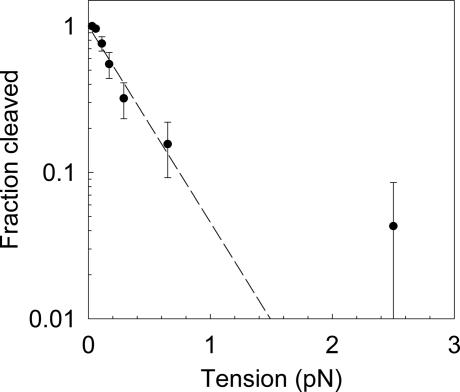
For one enzyme, Sau3AI, we studied the dependence of cleavage activity on DNA tension in more detail. To quantify the activity we reduced the incubation time to 30 s so that the fraction cleaved was <100% over the entire range of applied tension. As the tension was increased from 0.06 to ≈0.7 pN we found that the activity decreased exponentially and the decrease was ≈10-fold at 0.7 pN (Fig. 3). We note that the measurements done at 2.5 and 5 pN indicated only one and zero cleavage events, respectively, in N ≈ 30 trials. These data points are thus also consistent to within their error bars with the plotted exponential decrease. The experimental trend is thus in accord with the dependence predicted by theory, although a 10-fold inhibition was predicted to occur at somewhat smaller force, of order 0.1 pN, for an optimum-sized teardrop-shaped loop (≈500 bp) (21). In our experiment a quasi-continuum of loop sizes is possible, including many in the range predicted to be favorable (≈200–1,000 bp). Notably, theory predicts that the degree of inhibition by tension would decrease with decreasing loop size, suggesting that we are inhibiting loops that are predominantly smaller than an optimum-sized teardrop loop. Although loops shorter than the persistence length of DNA (≈150 bp) are predicted to be unfavorable by classical models because of the bending rigidity of DNA, cleavage of templates with site separations ranging from 10 to 200 bp has been reported to occur quite readily with the two-site REase EcoRII (25). The lower than predicted degree of inhibition we observed with Sau3AI may therefore be indicative of effects that facilitate formation of smaller loops, such as protein-induced or spontaneous DNA bending, nonclassical loop geometry, finite protein span, and/or protein flexibility (21, 22, 29–31).
Fig. 3.
Detailed study of cleavage activity versus DNA tension for the two-site enzyme Sau3AI. Activity is reported as fraction of DNA molecules cleaved in 30 s. The error bars are calculated as the standard deviation of the binomial distribution (P(1 − P)/N)½, where P is the probability of a molecule being cut and N is the number of trials (N ≈ 30 for each point). The dashed line is an exponential decay fit to the data, indicating a 1/e point of ≈0.3 pN. Measurements done at 2.5 and 5 pN indicated only one and zero cleavage events, respectively, in n = 30 trials. These data points are thus also consistent to within their error bars with the plotted exponential decrease.
Effect of Tension on One-Site Enzymes.
Our primary purpose for studying one-site REases was to contrast their behavior with that of two-site enzymes. However, as some REases are known to induce sharp bending of DNA (32), we hypothesized that cleavage activity might also be inhibited in such cases, although to a lesser extent than with two-site enzymes. Tension would be expected to interfere with the binding of enzymes that induce DNA bending because it would increase the free energy change required for binding.
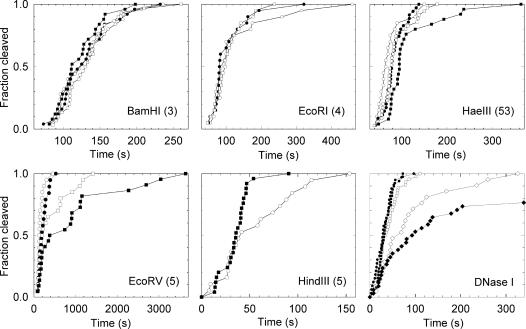
Unlike the two-site REases all of the one-site enzymes were largely unaffected by 5 pN of tension, although some were affected by higher tensions (Fig. 4). Clear inhibition of EcoRV was observed at 20 and 40 pN, whereas very little inhibition was observed with BamHI, EcoRI, or DNaseI. These findings support the hypothesis that this inhibition is related to DNA bending because crystal structure data show that EcoRV bends DNA sharply through an angle of ≈50°, whereas neither EcoRI, BamHI, or DNaseI bends DNA significantly (32–34). Our findings on BamHI and EcoRV are also in agreement with findings on these two enzymes recently reported in an independent study (24). Here, we provide additional results on four other one-site enzymes. We observed an inhibitory effect of tension with HaeIII, but only at 40 pN. No structure for a HaeIII–DNA complex has been reported to our knowledge, but on the basis of our measurements, we predict that it bends DNA, although to a lesser extent than EcoRV.
Fig. 4.
Tension dependence of DNA cleavage with one-site endonucleases. Measurements with relaxed DNA (fractional extension 35%, 0.06 pN, ○), and tensions of 5 pN (●), 20 pN (□), 40 pN (■). The pBAC-A construct was used in all of these measurements and the number of recognition sites is listed in parentheses after the enzyme name. Enzyme concentrations were 200 units/ml for BamHI, EcoRI, EcoRV, and HindIII, 500 units/ml for HaeIII, and 60 units/ml for DNaseI. Note that HaeIII and EcoRV show tension-induced partial inhibition, whereas HindIII and, to a lesser extent, EcoRI shown tension-induced enhancement. DNaseI (Lower Right) showed a slight enhancement at 5 and 40 pN, and partial inhibition at very high tension of 75 pN (◇), where the DNA is overstretched. To quantify spontaneous unbinding from the microspheres at high force, negative controls without enzyme were also carried out at 75 pN (♦).
With HindIII we observed a novel effect of tension: enhancement rather than inhibition. A tension of 40 pN dramatically increased the rate of DNA cleavage, and control experiments without enzyme added confirmed that this effect was not merely caused by increased unlinking of the DNA tether from the microspheres. We interpret this result as indicating that the applied stress is directly transmitted to the phosphodiester backbone of the DNA. Such stress would accelerate cleavage as it acts in a direction favoring the formation of the final products of the reaction. Furthermore, upon close inspection of the data with BamHI, EcoRI, and DNaseI one sees a small degree of tension-induced enhancement, although it is not as dramatic as that observed with HindIII. No crystal structures for HindIII–DNA complexes have been reported, but we predict that they do not involve significant DNA bending. Our findings with DNaseI are in accord with crystal structure data, which show that this enzyme binds in the minor groove of DNA and causes only minor distortions (35).
As DNaseI cleaved very rapidly even at high tension, and as it is a nonspecific endonuclease, we wondered whether extreme distortion of the double helix would have an effect on its activity. When held at a tension greater than ≈65 pN, dsDNA undergoes a structural transition in which it lengthens by 70% (36). Interestingly, when the DNA was held in this overstretched configuration at 75 pN, we observed strong, but surprisingly only partial, inhibition of DNaseI. Thus, DNaseI appears to be very permissive in its interaction with DNA.
Conclusions
Cleavage of DNA by endonucleases was studied via optical tweezers manipulation of single DNA molecules by using a large number of different one-site and two-site enzymes. The specific activity of two-site enzymes was universally “switched off” by application of 5 pN of tension to the DNA, whereas that tension had virtually no effect on one-site enzymes. This finding is in accord with several recent theories that predict a strong tension dependence of DNA looping. Inhibition was also observed with certain one-site enzymes, but only at much higher tensions, and this inhibition was correlated with protein-induced DNA bending. Our results indicate that DNA looping provides a mechanism for a tension-sensing switch that is sensitive to quite low tensions. It is conceivable that this mechanism could act in vivo to facilitate intracellular and/or extracellular mechanosensory transduction (19, 20, 23).
Materials and Methods
DNA Constructs.
LBAC-A was prepared by ligating a 4,282-bp DIG-labeled PCR fragment to a 10,845-bp biotin-end-labeled restriction fragment of pBACe3.6. The PCR fragment was generated by amplification of a sequence from pFastBac HT-b (Invitrogen) by using the primers 5′-GTGGTATGGCTGATTATGATC-3′ and 5′-GCAGCCTGAATGGCGAATGG-3′ and was labeled by incorporation of 20 μM of dUTP-11-DIG (Roche Applied Science) and 200 μM each of dATP, dCTP, dGTP, and dTTP in the PCR. The multiple DIG labeling was used to provide a stronger attachment in some experiments. The 10,845-bp fragment was produced by digesting pBACe3.6 (Children’s Hospital Oakland Research Institute, Oakland, CA) with BsrGI (NEB, Beverly, MA) and end-labeling by using the Klenow fragment of E. coli DNA polymerase I, exo− (NEB) to incorporate dATP-14-biotin (Invitrogen). Both fragments were purified by using the Qiagen (Valencia, CA) PCR purification kit and digested with XhoI (NEB). To isolate the desired product the samples were run on a 1% agarose gel in 1× TAE buffer (40 mM Tris·acetate/1 mM EDTA, pH 8) and purified by using the Qiagen gel extraction kit. The two fragments were then ligated with a T4 DNA ligase (NEB).
LBAC-B was prepared by labeling the aforementioned biotin-labeled, 10,845-bp XhoI fragment of pBACe3.6 with the Klenow fragment of DNA polymerase I, exo− to incorporate dUTP-11-DIG.
½-λ-L was prepared by using the Klenow fragment of DNA polymerase I, exo− to fill in the ends of methyladenine-free DNA (NEB) with biotin-dATP and dCTP (Invitrogen). The DNA was then digested by XbaI and purified with the Promega Wizard DNA clean-up kit. A second fill-in was then done with DIG-labeled dUTP (Roche Applied Science). The fragments were then digested with XhoI to select the left end (24,508 bp).
The 15-kbp human DNA sequence, 14-kbp Drosophila sequence, and 10-kbp bacteriophage λ sequence were generated by PCR amplification with genomic DNA using biotin and DIG-labeled primers as described (37).
In each case the DNA template contained multiple recognition sites for the enzymes being tested with separations ranging from ≈200 to 1,000 bp, predicted by theory to be favorable for looping (21, 23). Information on the recognition sites is given in Table 1. The total lengths of the molecules were much greater (by ≈20- to 60-fold) than the length of loops favored to occur. In this limit theory predicts that looping is not strongly influenced by the length of the DNA (21). In all cases having nonpalandromic recognition sequences the templates contained both repeating and inverted sites. We note that the enzyme we studied in detail, Sau3AI, has a palindromic recognition sequence, such that the sites do not have alternative orientations.
Table 1.
Properties of the two-site endonucleases studied in the cleavage experiments, as reported in REBASE (5), the DNA templates used in cleavage experiments, number of recognition sites, and concentration of enzyme used
| Enzyme | Type | Molecular mass, kD | Form in solution | Active complex | Recognition sequence | DNA (# sites) | C, units/ml |
|---|---|---|---|---|---|---|---|
| BfiI | IIS | 40 | Dimer | Dimer | ACTGGG (5/4) | Human 15 kb (5) | 0.63 |
| BpmI | IIE, G, S | 117 | * | * | CTGGAG(N)16↓ | ½-λ-L (17) | 20 |
| BsgI | IIE, G, S | 121 | * | * | GTGCAG(N)16↓ | ½-λ-L (31) | 30 |
| BspMI | IIE | 222 | Tetramer | Tetramer | ACCTGC(N)4↓ | ½-λ-L (24) | 20 |
| Cfr9I | IIE | 37 | * | * | C↓CCGGG | Human 15 kb (7) | 200 |
| Cfr10I | IIF, P | 320 | Tetramer | Tetramer | R↓CCGGY | ½-λ-L (56) | 100 |
| Eco57I | IIE, G | 117 | Monomer | * | CTGAAG(N)16↓ | ½-λ-L (25) | 50 |
| EcoRII | IIE, P | 92 | Dimer | Dimer | ↓CCWGG | ½-λ-L (36) | 50 |
| FokI | IIS | 66 | Monomer | Dimer | GGATG(N)9↓ | LBAC-B (26) | 40 |
| HpaII | IIE | 41 | * | * | C↓CGG | LBAC-B (49) | 100 |
| MboII | IIS | 49 | Monomer | Dimer | GAAGA (8/7) | LBAC-B (27) | 50 |
| NarI | IIE | * | * | * | GG↓CGCC | Drosophila 14 kb (5) | 80 |
| SacII | IIE | * | * | * | CCGC↓GG | Drosophila 14 kb (5) | 100 |
| Sau3AI | IIE | 56 | Monomer | Dimer | ↓GATC | LBAC-A (55) | 40 |
| SgrAI | IIP | 38 | Dimer | Tetramer | C(A/G)↓CCGG(C/T)G | λ 10 kb (4) | 200 |
Type IIE REases bind at two sites, but only one is cleaved, whereas type IIF cleave coordinately at both binding sites. Type IIG enzymes have restriction and modification activities in the same subunit, and type IIS enzymes recognize asymmetric sequences and cleave at least one strand outside of the recognition sequence. Entries marked with an asterisk are those for which no information was available.
Enzymes.
We sought to examine as many two-site REases as possible given the available recognition sites on our DNA templates (Table 1). Evidence in the literature for two-site behavior of REases comes from a variety of studies. In several cases looped complexes have been directly imaged by electron microscopy (NaeI, Cfr10I, EcoRII, and Sau3AI) (38–41). DNA looping by SfiI and Cfr10I has been inferred in DNA recombination and gel mobility-shift measurements (42–44), by NgoMIV in FRET measurements (45), by BspMI in magnetic tweezers experiments (46), and by NarI in tethered particle experiments (47). Evidence in other cases comes from the comparison of DNA cleavage rates on templates containing two sites versus only one site, which has been reported for BfiI, BsgI, BpmI, FokI, MboII, NarI, and SgrAI (26, 28, 48, 49). Finally, stimulation of activity upon addition of short oligonucleotide duplexes containing the recognition sequence has been reported for Eco57I, HpaII, Cfr9I, and SacII. Such stimulation provides evidence that an enzyme complex can bind in trans (i.e., across two sites on different molecules) (50–53).
BamHI, BpmI, BsgI, BspMI, EcoRI, EcoRV, FokI, HaeIII, HindIII, HpaII, MboII, NarI, SacII, Sau3AI, SfiI, and SgrAI were obtained from NEB. BfiI, Cfr9I, Cfr10I, and Eco57I were obtained from Fermentas (Hanover, MD). EcoRII was obtained from Roche Applied Science. Bovine pancreatic DNaseI was obtained from Calbiochem. Six of these (BamHI, EcoRI, EcoRV, HaeIII, HindIII, and DNaseI) are common one-site endonucleases, which were chosen for comparison with the two-site enzymes. With each enzyme we used the reaction buffer recommended by the manufacturer. The DNaseI buffer contained 50 mM Tris·HCl (pH 7.8), 50 μg/ml BSA, and 10 mM MgCl2, which facilitates nicking of DNA. All measurements in the optical tweezers were made at room temperature (≈20°C). Relevant data on the enzymes are given in Table 1. With each enzyme we aimed to increase the concentration to the point where we observed complete cleavage when the DNA was held slack and then repeated the measurements using the same concentration but with the DNA stretched. In a few cases we did not achieve complete cleavage in 5 min with the DNA slack, although a decrease consistent with total inhibition at 5-pN tension was still observed.
DNA Tethering.
Streptavidin-coated microspheres [200 μl of 0.5% (wt/vol), 2.2-μm diameter, Spherotech, Libertyville, IL] were washed by twice centrifuging at 10,000 × g and resuspended in 200 μl of PBS, pH 7.4 (Fisher Scientific) and 0.1 mg/ml BSA (NEB). Five microliters of diluted DNA (≈10–100 ng/μl) was mixed with 5 μl of microspheres and incubated for ≈45 min at room temperature on a slowly rotating mixer. Five to 10 μl of these microspheres was diluted in 0.5 ml of PBS and loaded into a 1-ml tuberculin syringe for injection into the sample chamber.
Protein G-coated microspheres [200 μl of 0.5% (wt/vol), 2.8-μm diameter, Spherotech] were washed in the same manner and resuspended in 20 μl of PBS. Then, 5 μl of 200 μg/ml of anti-DIG (Roche Applied Science) was added. The microspheres were incubated on the mixer for ≈45 min and then washed three more times and resuspended in 20 μl of PBS. Five microliters of the microspheres was loaded into a syringe for injection into the sample chamber.
Our optical tweezers instrument has been described (54). The anti-DIG-coated microsphere was held by a micropipette while the microsphere carrying the DNA was trapped with the optical tweezers. The two microspheres were brought into proximity such that the DIG-labeled end of one DNA molecule bound to the anti-DIG-coated bead, forming a DNA tether between them.
Acknowledgments
We thank D. Fuller, E. P. Geiduschek, K. Haushalter, J. Kadonaga, P. Kaiser, J. P. Rickgauer, and R. Sim for assistance and discussions. This research was supported by a Burroughs Wellcome Fund Career Award, a Searle Scholars Award from the Kinship Foundation, and a Young Investigator Award from the Arnold and Mabel Beckman Foundation.
Abbreviation
- DIG
digoxygenin.
Note Added in Proof.
We (55) have recently found that substitution of Ca2+ for Mg2+ permits measurement of stable loops with many of these enzymes. This technique has allowed us to measure loop size distributions and confirm the inference that loop sizes are generally smaller than predicted by the simple worm-like chain models.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Halford S. E. Biochem. Soc. Trans. 2001;29:363–374. doi: 10.1042/bst0290363. [DOI] [PubMed] [Google Scholar]
- 2.Roberts R. J. Proc. Natl. Acad. Sci. USA. 2005;102:5905–5908. doi: 10.1073/pnas.0500923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pingoud A., Jeltsch A. Nucleic Acids Res. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pingoud A., Fuxreiter M., Pingoud V., Wende W. Cell Mol. Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PubMed] [Google Scholar]
- 5.Roberts R. J., Vincze T., Posfai J., Macelis D. Nucleic Acids Res. 2005;33:D230–D232. doi: 10.1093/nar/gki029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kruger D. H., Barcak G. J., Reuter M., Smith H. O. Nucleic Acids Res. 1988;16:3997–4008. doi: 10.1093/nar/16.9.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halford S. E., Welsh A. J., Szczelkun M. D. Annu. Rev. Biophys. Biomol. Struct. 2004;33:1–24. doi: 10.1146/annurev.biophys.33.110502.132711. [DOI] [PubMed] [Google Scholar]
- 8.Schleif R. Annu. Rev. Biochem. 1992;61:199–223. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]
- 9.Mukherjee S., Erickson H., Bastia D. Cell. 1988;52:375–383. doi: 10.1016/s0092-8674(88)80030-8. [DOI] [PubMed] [Google Scholar]
- 10.Su W., Middleton T., Sugden B., Echols H. Proc. Natl. Acad. Sci. USA. 1991;88:10870–10874. doi: 10.1073/pnas.88.23.10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oehler S., Amouyal M., Kolkhof P., von Wilcken-Bergmann B., Muller-Hill B. EMBO J. 1994;13:3348–3355. doi: 10.1002/j.1460-2075.1994.tb06637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiner B. M., Kleckner N. Cell. 1994;77:977–991. doi: 10.1016/0092-8674(94)90438-3. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y., Rice P. A. Annu. Rev. Biophys. Biomol. Struct. 2003;32:135–159. doi: 10.1146/annurev.biophys.32.110601.141732. [DOI] [PubMed] [Google Scholar]
- 14.Finzi L., Gelles J. Science. 1995;267:378–380. doi: 10.1126/science.7824935. [DOI] [PubMed] [Google Scholar]
- 15.Wong O. K., Guthold M., Erie D., Gelles J. FASEB J. 2000:A1445–A1445. [Google Scholar]
- 16.Lia G., Bensimon D., Croquette V., Allemand J. F., Dunlap D., Lewis D. E., Adhya S., Finzi L. Proc. Natl. Acad. Sci. USA. 2003;100:11373–11377. doi: 10.1073/pnas.2034851100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blackwood E. M., Kadonaga J. T. Science. 1998;281:60–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- 18.Mucke M., Kruger D. H., Reuter M. Nucleic Acids Res. 2003;31:6079–6084. doi: 10.1093/nar/gkg836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleckner N., Zickler D., Jones G. H., Dekker J., Padmore R., Henle J., Hutchinson J. Proc. Natl. Acad. Sci. USA. 2004;101:12592–12597. doi: 10.1073/pnas.0402724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y. S., Haga J. H., Chien S. J. Biomech. 2005;38:1949–1971. doi: 10.1016/j.jbiomech.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 21.Sankararaman S., Marko J. F. Phys. Rev. E. 2005;71:021911. doi: 10.1103/PhysRevE.71.021911. [DOI] [PubMed] [Google Scholar]
- 22.Yan J., Kawamura R., Marko J. F. Phys. Rev. E. 2005;71:061905. doi: 10.1103/PhysRevE.71.061905. [DOI] [PubMed] [Google Scholar]
- 23.Blumberg S., Tkachenko A. V., Meiners J. C. Biophys. J. 2005;88:1692–1701. doi: 10.1529/biophysj.104.054486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van den Broek B., Noom M. C., Wuite G. J. Nucleic Acids Res. 2005;33:2676–2684. doi: 10.1093/nar/gki565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reuter M., Kupper D., Meisel A., Schroeder C., Kruger D. H. J. Biol. Chem. 1998;273:8294–8300. doi: 10.1074/jbc.273.14.8294. [DOI] [PubMed] [Google Scholar]
- 26.Gowers D. M., Bellamy S. R., Halford S. E. Nucleic Acids Res. 2004;32:3469–3479. doi: 10.1093/nar/gkh685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamulaitis G., Sasnauskas G., Mucke M., Siksnys V. J. Mol. Biol. 2006;358:406–419. doi: 10.1016/j.jmb.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 28.Bath A. J., Milsom S. E., Gormley N. A., Halford S. E. J. Biol. Chem. 2002;277:4024–4033. doi: 10.1074/jbc.M108441200. [DOI] [PubMed] [Google Scholar]
- 29.Merlitz H., Rippe K., Klenin K. V., Langowski J. Biophys. J. 1998;74:773–779. doi: 10.1016/S0006-3495(98)74002-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rippe K. Trends Biochem. Sci. 2001;26:733–740. doi: 10.1016/s0968-0004(01)01978-8. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y. L., McEwen A., Crothers D. M., Levene S. Biophys. J. 2006;90:1903–1912. doi: 10.1529/biophysj.105.070490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winkler F. K., Banner D. W., Oefner C., Tsernoglou D., Brown R. S., Heathman S. P., Bryan R. K., Martin P. D., Petratos K., Wilson K. S. EMBO J. 1993;12:1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim Y. C., Grable J. C., Love R., Greene P. J., Rosenberg J. M. Science. 1990;249:1307–1309. doi: 10.1126/science.2399465. [DOI] [PubMed] [Google Scholar]
- 34.Newman M., Strzelecka T., Dorner L. F., Schildkraut I., Aggarwal A. K. Science. 1995;269:656–663. doi: 10.1126/science.7624794. [DOI] [PubMed] [Google Scholar]
- 35.Suck D. J. Mol. Recognit. 1994;7:65–70. doi: 10.1002/jmr.300070203. [DOI] [PubMed] [Google Scholar]
- 36.Smith S. B., Cui Y., Bustamante C. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 37.Fuller D. N., Gemmen G. J., Rickgauer J. P., Dupont A., Millin R., Recouvreux P., Smith D. E. Nucleic Acids Res. 2006;34:e15. doi: 10.1093/nar/gnj016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Topal M. D., Thresher R. J., Conrad M., Griffith J. Biochemistry. 1991;30:2006–2010. doi: 10.1021/bi00221a038. [DOI] [PubMed] [Google Scholar]
- 39.Siksnys V., Skirgaila R., Sasnauskas G., Urbanke C., Cherny D., Grazulis S., Huber R. J. Mol. Biol. 1999;291:1105–1118. doi: 10.1006/jmbi.1999.2977. [DOI] [PubMed] [Google Scholar]
- 40.Mucke M., Lurz R., Mackeldanz P., Behlke J., Kruger D. H., Reuter M. J. Biol. Chem. 2000;275:30631–30637. doi: 10.1074/jbc.M003904200. [DOI] [PubMed] [Google Scholar]
- 41.Friedhoff P., Lurz R., Luder G., Pingoud A. J. Biol. Chem. 2001;276:23581–23588. doi: 10.1074/jbc.M101694200. [DOI] [PubMed] [Google Scholar]
- 42.Szczelkun M. D., Halford S. E. EMBO J. 1996;15:1460–1469. [PMC free article] [PubMed] [Google Scholar]
- 43.Milsom S. E., Halford S. E., Embleton M. L., Szczelkun M. D. J. Mol. Biol. 2001;311:515–527. doi: 10.1006/jmbi.2001.4893. [DOI] [PubMed] [Google Scholar]
- 44.Watson M. A., Gowers D. M., Halford S. E. J. Mol. Biol. 2000;298:461–475. doi: 10.1006/jmbi.2000.3676. [DOI] [PubMed] [Google Scholar]
- 45.Katiliene Z., Katilius E., Woodbury N. W. Biophys. J. 2003;84:4053–4061. doi: 10.1016/S0006-3495(03)75131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan J., Skoko D., Marko J. F. Phys. Rev. E. 2004;70:011905. doi: 10.1103/PhysRevE.70.011905. [DOI] [PubMed] [Google Scholar]
- 47.van den Broek B., Vanzi F., Normanno D., Pavone F. S., Wuite G. J. Nucleic Acids Res. 2006;34:167–174. doi: 10.1093/nar/gkj432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bilcock D. T., Daniels L. E., Bath A. J., Halford S. E. J. Biol. Chem. 1999;274:36379–36386. doi: 10.1074/jbc.274.51.36379. [DOI] [PubMed] [Google Scholar]
- 49.Soundararajan M., Chang Z., Morgan R. D., Heslop P., Connolly B. A. J. Biol. Chem. 2002;277:887–895. doi: 10.1074/jbc.M109100200. [DOI] [PubMed] [Google Scholar]
- 50.Reuter M., Kupper D., Pein C. D., Petrusyte M., Siksnys V., Frey B., Kruger D. H. Anal. Biochem. 1993;209:232–237. doi: 10.1006/abio.1993.1113. [DOI] [PubMed] [Google Scholar]
- 51.Kupper D., Moncke-Buchner E., Reuter M., Kruger D. H. Anal. Biochem. 1999;272:275–277. doi: 10.1006/abio.1999.4170. [DOI] [PubMed] [Google Scholar]
- 52.Vanamee E. S., Santagata S., Aggarwal A. K. J. Mol. Biol. 2001;309:69–78. doi: 10.1006/jmbi.2001.4635. [DOI] [PubMed] [Google Scholar]
- 53.Roberts R. J., Belfort M., Bestor T., Bhagwat A. S., Bickle T. A., Bitinaite J., Blumenthal R. M., Degtyarev S., Dryden D. T., Dybvig K., et al. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gemmen G. J., Sim R., Haushalter K. A., Ke P. C., Kadonaga J. T., Smith D. E. J. Mol. Biol. 2005;351:89–99. doi: 10.1016/j.jmb.2005.05.058. [DOI] [PubMed] [Google Scholar]
- 55.Gemmen G. J., Millin R., Smith D. E. Nucleic Acids Res. 2006;34:2864–2877. doi: 10.1093/nar/gkl382. [DOI] [PMC free article] [PubMed] [Google Scholar]