Abstract
A gene encoding a 23.5-kDa ehrlichial morula membrane protein designated MmpA was cloned by screening an Ehrlichia canis expression library with convalescent dog sera, which resulted in three positive clones. Sequence analysis of the insert DNAs from all three clones indicated an open reading frame with a size of 666 bp that encodes MmpA. The structural analysis of MmpA indicated that it is a transmembrane protein with extreme hydrophobicity. Southern blot analysis of the HindIII-digested chromosomal DNA demonstrated the presence of a single copy of the mmpA gene in E. canis and Ehrlichia chaffeensis but not in the human granulocytic ehrlichiosis agent. The mmpA gene was amplified, cloned, and expressed as a fusion protein. Polyclonal antibodies to the recombinant protein (rMmpA) were raised in rabbits. Western blot analysis of E. canis and E. chaffeensis lysates with the anti-rMmpA serum resulted in the presence of an MmpA band only in E. canis, not in E. chaffeenesis. Sera from dogs which were either naturally or experimentally infected with E. canis recognized the recombinant protein. Double immunofluorescence confocal microscopy studies demonstrated that MmpA was localized mainly on the morula membrane of E. canis. Since the morula membrane is the interface between the ehrlichial growing environment and the host cytoplasm, MmpA may play a role in bacterium-host cell interactions.
Canine monocytic ehrlichiosis is a potentially fatal tick-borne disease caused by the rickettsia Ehrlichia canis (33). E. canis is an obligate intracytoplasmic rickettsia invading reticuloendothelial cells of the liver, spleen, and lymph nodes (33). The organism replicates primarily in monocytes and lymphocytes (33), and it is transmitted by the brown dog tick, Rhipicephalus sanguineus.
Donatien and Lestoquard first recognized canine monocytic ehrlichiosis in Algeria (17). Since then, it has been recognized worldwide as an important disease causing extensive morbidity and mortality among domestic dogs and other canids (24, 28, 33). In humans, the etiological agent of monocytic ehrlichiosis is Ehrlichia chaffeensis (31), whereas the causative organism of human granulocytic ehrlichiosis (HGE) was temporarily named the HGE agent (18, 23, 31) and in 2001 was named Anaplasma phagocytophila (18a). The phylogenetic analysis of 16S rRNA indicates that E. canis and E. chaffeensis have 98.2% homology (3). Western blot analysis of E. canis and E. chaffeensis lysates with antisera to E. canis and E. chaffeensis also revealed close antigenic similarity (14).
Similarly to all intracellular bacterial pathogens that form membrane-bound vesicles in the host cells, E. canis organisms form microcolonies inside cellular vacuoles (morulae) that harbor many individual ehrlichiae. Several survival strategies have been identified in various intracellular bacterial pathogens, such as escaping from vacuoles, avoidance of lysosomal fusion, and tolerance of the lysosomal environment (19). Ehrlichia risticii-containing morulae evade lysosomal fusion (44). The morula membrane provides a permissive environment not only for ehrlichial survival but also for replication. To date, several inclusion membrane proteins from Chlamydia spp. have been identified, but none in E. canis (5-7, 35, 37).
In order to identify ehrlichial antigens, an E. canis genomic library was constructed and screened with convalescent-phase dog sera. The screening resulted in the isolation of a gene encoding a protein that is localized on the morula membranes of E. canis-infected cells. The gene encoding this protein is named mmpA (for morula membrane protein A).
MATERIALS AND METHODS
Bacterial strains, plasmids, culture conditions, and an anti-E. canis monoclonal antibody (MAb).
The E. canis Oklahoma strain and E. chaffeensis (ATCC CRL-10679) were cultured in the DH82 dog macrophage cell line (ATCC CCL-10389), and the HGE agent strain WI-1 was cultured in the HL-60 cell line (ATCC CCL-240) as previously described (15, 16, 23). Bacterial infection rates were determined by LeukoStat staining (Fisher Scientific, Pittsburgh, Pa.), and bacteria were counted under a microscope. Escherichia coli TB1 hsdR (27) and E. coli DH5α [F− Φ80d lacZ ΔM15(lacZYA-argF) U169 endA1 recA1 hsdR17(rk−mk+) deoR thi-1 phoA supE44λ-gyrA96 relA1)] were used for plasmids pHG165 (38) and pRSETB (Invitrogen, Carlsbad, Calif.) manipulations. E. coli strain BL21(DE3)(pLysS) [F− ompT hsdSB (rB−mB−) dcm gal λ(DE3) pLysS Cmr] (Invitrogen) served as a host for the expression of the mmpA gene. E. canis was purified by Renografin gradient centrifugation as described elsewhere (42, 43).
An anti-E. canis MAb (anti-E. canis; 62.7-kDa protein mass [data not shown]) was a kind gift of Fort Dodge Laboratories, Fort Dodge, Iowa.
DNA manipulations.
All standard DNA manipulations and analyses, except where mentioned, were performed according to the procedures described by Sambrook et al. (36).
Library construction and immunoscreening.
E. canis DNA was extracted from purified organisms as previously described (11, 13). The purified DNA was subjected to Sau3A digestion, and the 3- to 8-kb fragments were isolated and ligated to plasmid pHG165 (38) and then transformed into E. coli TB1 (13). The recombinants were screened (colony blotting) with dog anti-E. canis antisera as previously described (13). Dog anti-E. canis antisera were prepared in beagles infected with live E. canis by intravenous injection as previously described (8, 41). These beagles were checked by detection of morulae in the monocytes. The antiserum was preabsorbed with E. coli TB1 lysates before use.
Southern blot analysis.
E. canis, E. chaffeensis, and the HGE agent genomic DNAs were prepared as previously described (11, 13), digested with HindIII, electrophoresed through a 0.7% agarose gel, transferred to nitrocellulose membranes, and probed with a 427-bp DNA fragment containing the mmpA gene amplified by PCR with a primer pair (1RACE1, 5′-GCTGCATTCTTGTTTGCTGC-3′, and 4F, 5′-ACGTGAGTTTGTTTATCTGGAC-3′) (see Fig. 2). The DNA fragment was labeled with a nonradioactive labeling kit (ECL direct nucleic acid labeling and detection systems; Amersham, Little Chalfont, Buckinghamshire, England) (12). Southern blot hybridization and detection were performed as described by the manufacturer.
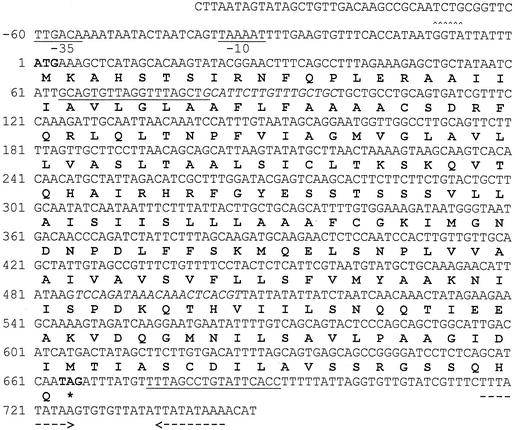
FIG. 2.
Nucleotide sequences of the mmpA gene and its coded protein. Nucleotide numbers are indicated on the left. The start and stop codons are indicated in boldface type. A promoter-like region proximal to mmpA is underlined. The potential ribosome-binding site preceding mmpA is indicated by carets. A potential transcription terminator of mmpA is indicated by dashed arrows. The underlined nucleotides indicate the primer annealing sites for generation of an mmpA gene fragment for insertion into pRESTB. The italicized nucleotides indicate the primer annealing sites for the generation of the DNA probes used for Fig. 4. The stop codon (amino acid) is indicated by an asterisk.
PCR procedures and subcloning of mmpA gene.
We designed a primer pair to amplify the whole mmpA gene with the exception of the first 63 bp. The primer pair consisted of a sense primer, EC2F3 (5′-CAGAATTCGCAGTGTTAGGTTTAGCT-3′) and an antisense primer, EC2R2 (5′-GCAAGCTTAGGTGAATACAGGCTAAA −3′) (see Fig. 2). PCR was carried out in a Perkin-Elmer Gene Amp PCR system 9600 thermal cycler. The amplification reaction was performed in a final volume of 50 μl containing 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 0.4 mM (each) deoxynucleoside triphosphate (Pharmacia, Piscataway, N.J.), primers (0.2 μM), 1.25 U of Taq polymerase (Gibco BRL, Grand Island, N.Y.), 1 μl of template, and 33 μl of distilled water. The template (pCH2) was denatured at 94°C for 30 s, and 30 amplification cycles were performed as follows: 30 s of denaturation at 94°C, 45 s of annealing at 56°C, and 30 s of primer extension at 72°C, followed at 72°C for 15 min and held at 4°C. The PCR product containing the mmpA gene was cut with EcoRI and HindIII and ligated to pRSETB cut with the same restriction enzymes (Invitrogen) and was designated pTEC2. pTEC2 was transferred into E. coli strain BL21(DE3)(pLysS).
Purification of MmpA protein and antiserum production in rabbits.
E. coli BL21(DE3)(pLysS) harboring pTEC2 was grown in Luria-Bertani medium to an optical density at 600 nm of ∼0.5. IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 1 mM, and the culture was grown for 3 h. The cells were harvested by centrifugation and resuspended in phosphate-buffered saline (PBS) containing 0.5% Triton X-100 and 10 mM EDTA. The cells were ruptured by a French press at 8,000 lb/in2. The total lysate was then spun at 3,000 × g for 5 min to separate cell debris (10) and then centrifuged at 12,000 × g for 5 min at 4°C. The pellets were washed with 9 volumes of PBS containing 0.5% Triton X-100 and 10 mM EDTA, incubated at room temperature for 5 min, and then centrifuged at 12,000 × g for 5 min at 4°C. The washing step was repeated once. The protein samples (∼500 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were visualized after being stained with Coomassie blue. The recombinant MmpA (rMmpA) band was excised, mixed with an equal volume of PBS, and ground to homogeneity for injection into rabbits. Polyclonal antiserum to rMmpA was raised in New Zealand White rabbits by three subcutaneous injections at 3-week intervals with ∼100 μg of rMmpA in Freund's incomplete adjuvant as previously described (13).
SDS-PAGE and Western and dot blotting.
The procedures for the SDS-PAGE and Western blot analyses were as previously described (9, 11). For dot blotting, E. canis- or E. chaffeensis-infected DH82 cells (4 to 5 days old with an ∼50 to 60% infection rate) were gently scraped from the bottom of a 25-cm2 flask (6 ml) with a rubber policeman. The cells were pelleted (6,000 × g; 10 min), washed twice with PBS (pH 7.2), resuspended with 1 ml of PBS, and subjected to sonication at setting 4 for 2 min on ice (model W-185 Sonifier; Branson Ultrasonics Corp., Danbury, Conn.). Unbroken cells were removed by centrifugation at 1,000 × g for 10 min at 4°C, and the lysate was subjected to dot blot analysis. For identification of rMmpA expression, MAbs against the Xpress epitope (Invitrogen) were used as primary antibodies. The secondary antibody, alkaline phosphatase-conjugated affinity-purified anti-mouse immunoglobulin G (IgG) (KPL, Gaithersburg, Md.) was used at a dilution of 1:5,000. For testing the expression of MmpA in E. canis and of possible MmpA homologues in E. chaffeensis and HGE, the rabbit anti-rMmpA antiserum served as the primary antibody (1:500). Goat alkaline phosphatase-conjugated anti-rabbit IgG (KPL) was used as a secondary antibody (1:5,000). To determine whether the naturally and experimentally infected dog sera contained anti-MmpA antibodies, rMmpA protein (∼1 μg/lane) was used as an antigen and subjected to SDS-PAGE and Western blot analysis. Test sera (1:500) from experimentally or naturally infected animals were used as the first antibody, followed by goat anti-dog IgG conjugated to alkaline phosphatase (1:5,000) (KPL) as the second antibody.
Location of the MmpA protein in E. canis.
To localize the MmpA protein, E. canis was cultured on DH82 cells and subjected to double immunofluorescence labeling and confocal microscopy (MRC 600; Bio-Rad, Hercules, Calif.) examination. Briefly, E. canis-infected DH82 cells (∼60 to 70% infected) 4 to 5 days after infection were gently scraped from the bottom of a 25-cm2 flask (6 ml) with a rubber policeman. The cells were harvested by centrifugation at 6,000 × g for 10 min and resuspended with 2 ml of fresh culture medium. The cell suspensions (5 μl) were loaded into individual wells of 12-well Teflon-coated slides (Erie Scientific, Portsmouth, N.H.), left to air dry, and fixed in acetone for 15 min. The slides were maintained at −20°C until they were used.
For the first immunofluorescence labeling, the frozen slides were air dried again and then fixed in methanol for 3 min. The fixed cells were incubated with 20 μl of rabbit antiserum to MmpA in each well at a 1:100 dilution (PBS with 0.05% SDS and 5% fetal calf serum) for 30 min at 37°C. After incubation, the slides were washed by dipping them twice into PBS followed by immersion in fresh PBS for 5 min and then were washed twice with distilled water and air dried. Fluorescein isothiocyanate-conjugated anti-rabbit IgG (Sigma, St. Louis, Mo.), a second antibody, was prepared at a 1:30 dilution with the same buffer used for the primary antibody. The labeling process for the second antibody was similar to that for the primary antibody.
For the second immunofluorescence labeling, after the steps above, the cells were sequentially incubated with the anti-E. canis MAb and then with lissamine-rhodamine-conjugated anti-mouse IgG (Jackson) in PBS with 0.05% Tween 20 and 5% fetal calf serum at a 1:40 dilution. All other conditions were identical to the procedures described above for the first immunofluorescence labeling. After all the procedures, the slides were air dried and mounted with mounting fluid (2.5 μl/well; Becton Dickinson, Sparks, Md.). The tests were repeated five times. Each time, 100 morulae were counted.
5′-RACE.
To determine the transcription site, 5′ rapid amplification of cDNA ends (5′-RACE) was followed as previously described (21) with the following modifications. Briefly, RNA was extracted by using RNAzol (Tel-test, Friendswood, Tex.), and the first-strand cDNA (antisense) was synthesized by using an mmpA-specific primer (2RACE1, 5′-CTTTCCACAAAATGCTGCAG-3′). The RNA template was then degraded with RNase H (BRL, Life Technologies, Rockville, Md.), and single-stranded cDNA was purified with the PRC purification kit (Gibco BRL, Life Technologies, Rockville, Md.). An oligo(dC) anchor sequence was added to the 3′ end of the cDNA using terminal deoxynucleotidyl- transferase (TdT) and dCTP. PCR amplification was accomplished with two primers: a deoxynosine-containing anchor primer (Qc, 5′-CCAGTGAGCAGAGTGACGAGGACTCGAGCTCAAGCTTGGGIIGGGIIGGGIIG-3′) that annealed to the poly(C) tail of the cDNA and a nested mmpA-specific primer (2RACE2, 5′-TTGCAAGCAGTACAGAAGAAG-3′). Following that, a second-round nested PCR was performed with primer Qo (5′-CCAGTGAGCAGAGTGACG-3′), which was complementary to the 5′ end of Qc, and the other mmpA-specific primer (2RACE3, 5′-GTGCTTGACTCGTATCCAAAG-3′). The resulting PCR product was cloned in a TA cloning vector (Invitrogen) and subjected to DNA sequencing. We repeated 5′-RACE three times, and one clone from each repetition was subjected to DNA sequencing.
DNA sequencing and analysis.
Automated DNA sequencing was performed on an Applied Biosystems model 373A DNA system by using the lac universal primer and primers complementary to regions already sequenced in the plasmid. The thermal cycling of the sequencing reactions utilized the Taq DyeDeoxy Terminator cycle-sequencing kit. Both strands of the cloned DNA were completely sequenced. The nucleotide sequence was analyzed using the BLAST programs of the National Center for Biotechnology Information (NCBI) (1, 2).
Nucleotide sequence accession number.
The GenBank accession number of the nucleotide sequence of pCH4 determined in this study is AF219120.
RESULTS
Immunoscreening of an E. canis genomic library.
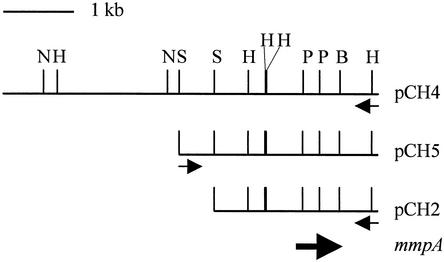
A plasmid library of E. canis genomic DNA was screened using pooled convalescent dog sera, and three clones that reacted strongly to the antisera were identified. These clones contained the plasmids pCH2, pCH4, and pCH5 with DNA insertions of 2.4, 2.8, and 5.3 kbp, respectively (Fig. 1).
FIG. 1.
Ehrlichial DNA inserts in pCH4, pCH5, and pCH2 with their restriction map. The location and direction of the mmpA gene is indicated by the large arrow. The direction of the lac promoter from plasmid pHG165 is indicated by small arrows. B, BamHI; H, HindIII; N, NdeI; P, PstI; and S, Sau3A.
Sequence analysis of mmpA genomic clones.
The E. canis DNAs in pCH2, -4, and -5 were sequenced, and the results revealed the presence of an overlapping open reading frame (ORF) in all three clones. This ORF, with a size of 666 bp, was designated mmpA (Fig. 2).
The DNA sequence of mmpA was searched for E. coli promoter consensus sequences using the homology score method (32). Putative −10 (TAAAAT) and −35 (TTGACA) promoter regions were identified in an upstream region of the initiation codon (Fig. 2). A ribosome-binding site (Shine-Dalgarno sequence) was also evident upstream of the initiation codon, and an inverted repeat sequence that is probably capable of forming a stem-loop (ΔG = −7.4 kCal/mol) and thus terminating transcription was also found downstream of the stop codon of mmpA (Fig. 2).
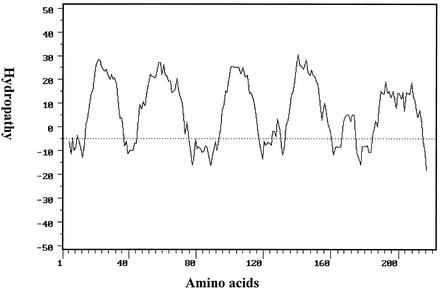
The deduced amino acid sequence was also used to search for protein homologues in the NCBI database, but there was no significant homology to any other proteins in the database. The hydrophobicities of the deduced amino acids of MmpA and its potential transmembrane regions were analyzed using previously described methods (30), and the analysis predicted MmpA to be extremely hydrophobic, with five main potential transmembrane regions (Fig. 3). Based on the method developed by Hopp and Woods, three potential antigenic determinants were identified (25). Three potential protein kinase C phosphorylation sites located in positions 7 (Ser), 37 (Ser), and 213 (Ser) and one possible casein kinase II phosphorylation site in position 177 (Thr) were found by using the program PROSITE (PCgene), as previously described (20).
FIG. 3.
Hydropathy plot of the predicted amino acids of MmpA. The vertical axis represents the scale of the hydrophobic (positive) and hydrophilic (negative) values established for each window of nine amino acids as previously described (30).
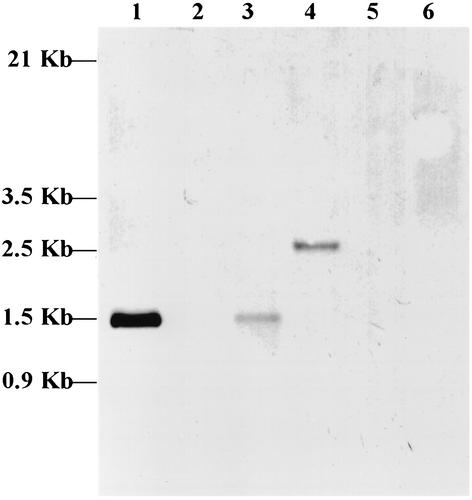
Southern blot analysis.
To address the frequency and distribution of the mmpA gene in E. canis, E. chaffeensis, and the HGE agent, Southern blot analyses of HindIII-digested genomic DNAs of E. canis-infected DH82 cells, E. chaffeensis-infected DH82 cells, and HGE agent-infected HL60 cells were performed. The digested genomic DNAs from DH82 and HL60 cells were used as negative controls, and pCH4 was used as a positive control. A single band with a size of 1.5 kb was detected from E. canis-infected DH82 cells (Fig. 4, lane 3) and pCH4 (Fig. 4, lane 1), whereas a 2.5-kb band was detected from E. chaffeensis-infected DH82 cells (Fig. 4, lane 4). The HGE agent-infected HL60 cells (Fig. 4, lane 6) and controls, such as DH82 and HL60 cells (Fig. 4, lanes 2 and 5), did not show any bands. The results suggest that the mmpA gene is present as a single copy in E. canis and E. chaffeensis but is not present in the HGE agent.
FIG. 4.
Southern blot hybridization with the PCR products (427 bp) derived from mmpA against total genomic DNA digests (∼20 μg of chromosomal DNA was digested with HindIII) from DH-82 cells (lane 2), E. canis-infected cells (lane 3), E. chaffeensis-infected DH-82 cells (lane 4), HL-60 cells (lane 5), and HGE agent-infected HL-60 cells (lane 6). pCH4 digested with HindIII, served as a positive control (lane 1).
5′-RACE for determination of the transcriptional start site.
To determine the transcriptional start site of mmpA, 5′-RACE was performed. The cDNA product was cloned in a TA cloning vector and subjected to DNA sequence analysis. We repeated RACE three times, and one clone from each repetition was subjected to DNA sequencing. Based on the sequencing results, the transcriptional start site was located at either a G or a T 7 or 8 nucleotides downstream from the promoter −10 region, respectively.
Expression of MmpA and Western blotting analysis.
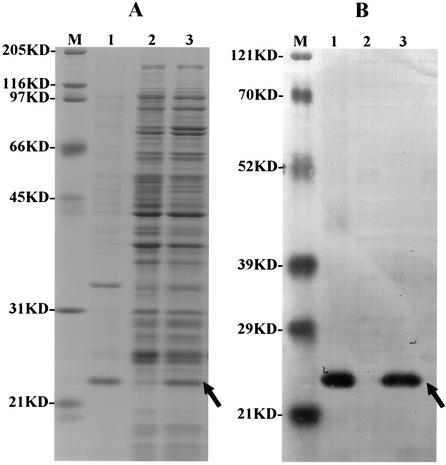
A 0.6-kb mmpA gene fragment was amplified and cloned into pRSETB as described in Materials and Methods. This construct (pTEC2) was transferred into E. coli and expressed as a recombinant protein (rMmpA). rMmpA contains a six-His Tag and an Xpress epitope on its N terminus. A partial purification of the recombinant protein from the E. coli lysate was accomplished by washing it with Triton X-100 buffer, since the molecule formed insoluble inclusion bodies. The molecular mass of the partially purified recombinant protein was found by SDS-PAGE to be ∼26 kDa (Fig. 5A). Western blot analysis with anti-Xpress antibodies also confirmed the in-frame fusion of the His tag and rMmpA (Fig. 5, B).
FIG. 5.
Identification of rMmpA by SDS-PAGE (A) and Western immunoblot assay with MAb against the Xpress epitope (B). Lanes M, mass marker protein; lanes 1, partially purified rMmpA (inclusion bodies); lanes 2, E. coli lysate (negative control); lanes 3, IPTG-induced pTEC2-transformed E. coli lysate. The expressed protein band is indicated by an arrow.
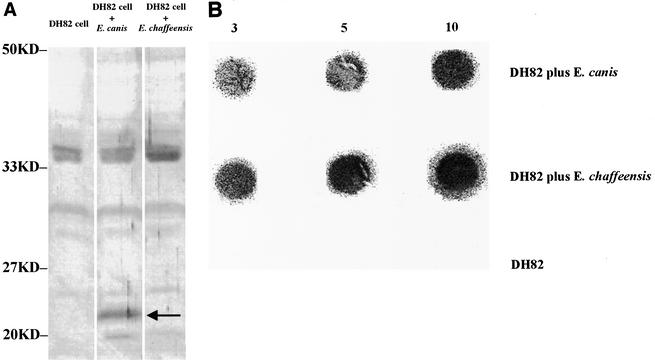
To determine whether E. chaffeensis expressed MmpA, lysates from E. canis-infected DH82 cells, E. chaffeensis-infected DH82 cells, and uninfected DH82 cells (negative control) were probed with anti-rMmpA antibodies in a Western blot analysis. The results showed only a protein band of ∼23.5 kDa closely correlated with the deduced molecular mass of MmpA in E. canis-infected cells. No protein band was detected from either E. chaffeensis-infected DH82 cells or uninfected DH82 cells by anti-MmpA antibodies (Fig. 6A). However, dot blot analysis indicated the presence of an MmpA homologue in E. chaffeensis-infected DH82 cells (Fig. 6B).
FIG. 6.
Western (A) and dot (B) immunoblot assays of DH82 cells, E. canis-infected DH82 cells, and E. chaffeensis-infected DH82 cells with rabbit anti-rMmpA serum. The molecular masses of markers (Bio-Rad) are indicated on the left. For dot blot analysis (Bio-Dot; Bio-Rad), 3, 5, and 10 μl of lysate (see Materials and Methods) were adjusted to 10 μl using PBS and subjected to analysis following the manufacturer's instructions.
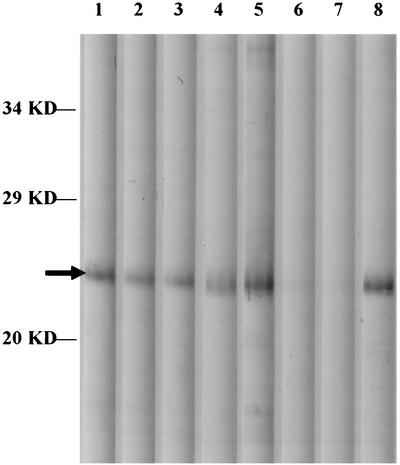
To evaluate whether naturally and experimentally infected dog sera contained anti-MmpA antibodies, four naturally and one experimentally infected dog serum plus one uninfected and two specific-pathogen-free (SPF) dog sera were used for Western blot analysis. The results showed that partially purified rMmpA was recognized by all the naturally and experimentally E. canis-infected dog antisera, but not by the normal serum or the SPF dog sera (Fig. 7). These data suggest in vivo expression of MmpA and its involvement in canine ehrlichiosis.
FIG. 7.
Western blot of sera obtained from dogs experimentally or naturally infected with E. canis. The lanes show reactivity with rMmpA. Serum from a dog experimentally infected with E. canis (lane 1), sera from four different naturally E. canis-infected dogs (lanes 2 to 5), sera from two SPF dogs (lanes 6 and 7), and pooled convalescent-phase sera (lane 8) were used to screen the library.
Subcellular localization of MmpA.
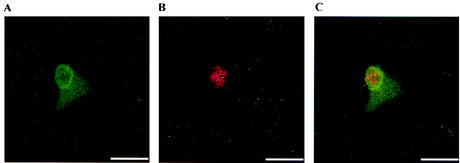
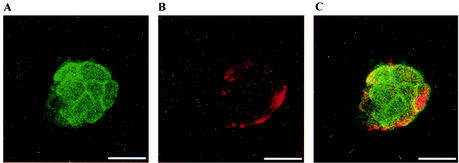
The intracellular location of MmpA was determined by using anti-rMmpA antibodies and anti-E. canis MAbs for double immunofluorescence labeling of E. canis-infected DH82 cells under confocal microscopy. The results revealed that the MmpA antigens were detectable mostly on the morula membrane, displaying a nonuniform fluorescent-staining pattern in ∼80% of the morulae examined (Fig. 8). Repeated attempts with increasing or decreasing concentrations of primary or secondary antibodies or affinity-purified primary antibodies could not eliminate the background staining, but the prominent staining of the morula membrane suggests that the MmpA antigen is mainly located on the morula membrane (Fig. 9). In contrast, the MAb stained only the E. canis organisms within the morulae. Normal rabbit serum (preimmune rabbit serum) did not stain either infected or uninfected DH82 cells (data not shown).
FIG. 8.
(A and B) Confocal micrographs of a single plane (0.5 μm thick) showing the localization of MmpA antigen on the morula membrane by probing with anti-rMmpA serum (A) and a MAb recognized by E. canis antigen in the organisms (B). (C) Merged images from panels A and B. Antibody labels: MmpA (green), anti-E. canis antigen (red). Bars = 18.5 μm.
FIG. 9.
(A and B) Confocal micrographs of a single plane (0.5 μm thick) showing the localization of MmpA antigen on the morula membrane by probing with anti-rMmpA serum (A) and a MAb recognized by E. canis antigen in the organisms (B). (C) Merged images from panels A and B. Antibody labels: MmpA (green), anti-E. canis antigen (red). Bars = 18.5 μm.
DISCUSSION
Ehrlichia spp. reside mainly in monocytes, macrophages, and neutrophils, which are considered primary effector cells of antimicrobial defense. Within the host cell, the bacteria reside within inclusion bodies (morulae), which provide a hospitable environment for their survival. The intracellular events leading to the establishment and maintenance of ehrlichia morulae in the host cells, especially for E. canis, is poorly understood. In this study, we have identified and partially characterized an ehrlichial morula membrane protein, MmpA, which was obtained by screening an E. canis genomic library with convalescent-phase dog sera. Only three positive clones were identified, and all of them contained an ORF that encodes MmpA. Neither the mmpA gene nor the MmpA protein shows any significant homology with other genes or proteins in the NCBI databases. The structural analysis of MmpA predicts that it is extremely hydrophobic, with five transmembrane segments (Fig. 3) and three potential antigenic determinants. We found that in vitro-grown (DH82 cells) E. canis expressed MmpA. In addition, the sera obtained from dogs naturally and experimentally infected with E. canis recognized MmpA. Taken together, these results confirm not only the antigenicity but also the in vivo and in vitro expression of MmpA.
Repeated attempts with 5′-RACE indicated that the mmpA transcription start site was either a G or T located downstream of the −10 promoter. Since the G is located at the corresponding G of the template, and also adjacent to the appended poly(G) tail, it is difficult to say whether the G is derived from poly(C) addition during 5′-RACE or from the cDNA. However, this G or T is in good agreement with the consensus spacing of 6 to 8 bp between the promoter and the transcription start site.
We found the mmpA gene in E. canis and E. chaffeensis but not in the HGE agent. This is not surprising, since there is a higher homology between E. canis and E. chaffeensis than between E. canis and the HGE agent, according to either 16S rRNA or groEL heat shock gene analysis (39, 40). Since this gene is not present in the HGE agent, it may serve as a genetic marker to differentiate the HGE agent from the above-mentioned monocytic ehrlichiae. However, this conclusion must be viewed cautiously, since we have examined only one strain of each organism. Western blot analysis of lysates from E. canis and E. chaffeensis with anti-MmpA sera demonstrated the presence of MmpA in E. canis at the predicted molecular mass of 23.5 kDa but not in E. chaffeensis (Fig. 6A). However, the dot blot analysis revealed the presence of an MmpA-like protein in E. chaffeensis culture (Fig. 6B), and it indicates that SDS may denature the MmpA homologue during Western blot analysis. Further cloning and sequencing of the E. chaffeensis homologous gene is needed to answer this question.
Confocal fluorescence microscopy of E. canis-infected cells revealed that MmpA was localized primarily in the morula membrane. Several attempts to determine the subcellular location of the MmpA protein by immunogold labeling were unsuccessful (unpublished data). Since the MmpA appears to be localized on the morula membrane, it is likely to require transport across the ehrlichial membranes. The secretory pathways for some of the intracellular pathogens have been elucidated, but none has been reported in Ehrlichia spp. (4, 10, 22, 26). Based on the predicted amino acid sequence, MmpA may not use a classic type II secretion system, because it lacks the typical type II signal sequence at its amino terminus. Several other secretion systems, such as type III, which do not require N-terminal signal sequences may be involved in MmpA secretion. The secretory pathways used by the ehrlichiae await elucidation.
To our knowledge, MmpA is the first E. canis protein to be found on the morula membrane. A 44-kDa protein in the HGE agent, a member of the genus Ehrlichia, is the only other protein that has been reported to partially localize on the morula membrane (29). Several other proteins that are also localized in inclusion membranes have been identified in other intracellular bacteria. For example, in Chlamydia psittaci and Chlamydia trachomatis, three products of three genes, incA, -B, and -C, have been shown to localize to the inclusion membrane. The deduced amino acid sequences of these genes show less similarity to each other, but all of them have a common bilobed hydrophobic motif (6, 37). Based on the hydrophobic motif of Inc proteins, Bannantine et al. recently predicted that there are probably 46 inclusion membrane proteins in C. trachomatis (5). In contrast to chlamydia proteins, the MmpA protein of E. canis contains five major hydrophobic domains, and this motif may serve as a model to find other morula membrane proteins. Furthermore, we identified a putative operon with two genes (probably encoding endopeptidases) upstream of mmpA, while downstream of this gene is a probable lipoprotein signal peptidase gene (this study). Likewise, in chlamydia, the regions upstream of the incA, -B, and -C genes have two ORFs encoding 40- and 44-kDa proteins that are similar to amino acid transporters and Na+-dependent transporters, respectively (6). The function of MmpA, together with those of most of the identified chlamydial inclusion membrane proteins, remains to be characterized. Phosphorylation of one of the chlamydial inclusion membrane proteins, IncA, by the host cells indicates communication between the bacteria and the host cells (34). MmpA also possesses three potential phosphorylation sites, indicating the possibility of a similar communication strategy between E. canis and host cells.
In conclusion, we have identified and expressed a novel ehrlichial gene (mmpA) coding for a protein (MmpA) that is localized on the morula membrane of E. canis. This gene is present in E. canis and E. chaffeensis but not in the agent of HGE. However, the encoded protein is detected only in E. canis. Since it is an immunodominant protein, it may also be a candidate for recombinant or DNA vaccine development and/or a serologic reagent. Further characterization of MmpA function may offer significant insights into the molecular mechanisms of ehrlichial pathogenesis.
Acknowledgments
This work was supported by grants from Fort Dodge Animal Health, a Division of American Home Products Corporation, and the New York State Science and Technology Foundation.
Editor: J. T. Barbieri
REFERENCES
- 1.Altschul, S. F., and E. V. Koonin. 1998. Iterated profile searches with PSI-BLAST—a tool for discovery in protein databases. Trends Biochem. Sci. 23:444-447. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, B. E., J. E. Dawson, D. C. Jones, and K. H. Wilson. 1991. Ehrlichia chaffeensis, a new species associated with human ehrlichiosis. J. Clin. Microbiol. 29:2838-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balakrishnan, L., C. Hughes, and V. Koronakis. 2001. Substrate-triggered recruitment of the TolC channel-tunnel during type I export of hemolysin by Escherichia coli. J. Mol. Biol. 313:501-510. [DOI] [PubMed] [Google Scholar]
- 5.Bannantine, J. P., R. S. Griffiths, W. Viratyosin, W. J. Brown, and D. D. Rockey. 2000. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell Microbiol. 2:35-47. [DOI] [PubMed] [Google Scholar]
- 6.Bannantine, J. P., D. D. Rockey, and T. Hackstadt. 1998. Tandem genes of Chlamydia psittaci that encode proteins localized to the inclusion membrane. Mol. Microbiol. 28:1017-1026. [DOI] [PubMed] [Google Scholar]
- 7.Bannantine, J. P., W. E. Stamm, R. J. Suchland, and D. D. Rockey. 1998. Chlamydia trachomatis IncA is localized to the inclusion membrane and is recognized by antisera from infected humans and primates. Infect. Immun. 66:6017-6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buhles, W. C., D. L. Huxsoll, and M. Ristic. 1974. Tropical canine pancytopenia: Clinical, hematologic, and serologic response of dogs to Ehrlichia canis infection, tetracycline therapy, and challenge inoculation. J. Infect. Dis. 130:357-367. [DOI] [PubMed] [Google Scholar]
- 9.Chang, Y. F., M. J. Appel, R. H. Jacobson, S. J. Shin, P. Harpending, R. Straubinger, L. A. Patrican, H. Mohammed, and B. A. Summers. 1995. Recombinant OspA protects dogs against infection and disease caused by Borrelia burgdorferi. Infect. Immun. 63:3543-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, Y.-F., T.-L. Lauderdale, W. Y. Lee, S. J. Shin, R. H. Jacobson, M. J. Appel, and D. H. Lein. 1992. Expression and secretion of outer surface protein (Osp-A) of Borrelia burgdorferi from Escherichia coli. FEMS Microbiol. Lett. 109:297-302. [DOI] [PubMed] [Google Scholar]
- 11.Chang, Y. F., J. Shi, D. P. Ma, S. J. Shin, and D. H. Lein. 1993. Molecular analysis of the Actinobacillus pleuropneumoniae RTX toxin-III gene cluster. DNA Cell Biol. 12:351-362. [DOI] [PubMed] [Google Scholar]
- 12.Chang, Y.-F., R. K. Straubinger, R. H. Jacobson, J. B. Kim, T. J. Kim, D. Kim, S. J. Shin, and M. J. G. Appel. 1996. Dissemination of Borrelia burgdorferi after experimental infection in dogs. J. Spiroch. Tick-borne Dis. 3:80-86. [Google Scholar]
- 13.Chang, Y. F., R. Young, D. Post, and D. K. Struck. 1987. Identification and characterization of the Pasteurella haemolytica leukotoxin. Infect. Immun. 55:2348-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, S. M., J. S. Dumler, H. M. Feng, and D. H. Walker. 1994. Identification of the antigenic constituents of Ehrlichia chaffeensis. Am. J. Trop. Med. Hyg. 50:52-58. [PubMed] [Google Scholar]
- 15.Dawson, J. E., B. E. Anderson, D. B. Fishbein, J. L. Sanchez, C. S. Goldsmith, K. H. Wilson, and C. W. Duntley. 1991. Isolation and characterization of an Ehrlichia sp. from a patient diagnosed with human ehrlichiosis. J. Clin. Microbiol. 29:2741-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dawson, J. E., F. J. Candal, V. G. George, and E. W. Ades. 1993. Human endothelial cells as an alternative to DH82 cells for isolation of Ehrlichia chaffeensis. E. canis, and Rickettsia rickettsii. Pathobiology 61:293-296. [DOI] [PubMed] [Google Scholar]
- 17.Donatien, A., and F. Lestoquard. 1935. Existence en Algerie d'une Rickettsia du chien. Bull. Soc. Pathol. Exot. 28:418-419. [Google Scholar]
- 18.Dumler, J. S., and J. S. Bakken. 1996. Human granulocytic ehrlichiosis in Wisconsin and Minnesota: a frequent infection with the potential for persistence. J. Infect. Dis. 173:1027-1030. [DOI] [PubMed] [Google Scholar]
- 18a.Dumler, J. S., A. F. Barbet, C. P. Bekker, G. A. Dasch, G. H. Palmer, S. C. Ray, Y. Rikihisa, and F. R. Rurangirwa. 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 51:2145-2165. [DOI] [PubMed]
- 19.Ernst, R. K., T. Guina, and S. I. Miller. 1999. How intracellular bacteria survive: surface modifications that promote resistance to host immune responses. J. Infect. Dis. 179:S326-S330. [DOI] [PubMed] [Google Scholar]
- 20.Falquet, L., M. Pagni, P. Bucher, N. Hulo, C. J. Sigrist, K. Hofmann, and A. Bairoch. 2002. The PROSITE database, its status in 2002. Nucleic Acids Res. 30:235-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frohman, M. 1993. Rapid amplification of complementary DNA ends for generation of full-length complementary DNAs: thermal RACE. Methods Enzymol. 218:340-356. [DOI] [PubMed] [Google Scholar]
- 22.Galan, J. E., and A. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322-1328. [DOI] [PubMed] [Google Scholar]
- 23.Goodman, J. L., C. Nelson, B. Vitale, J. E. Madigan, J. S. Dumler, T. J. Kurtti, and U. G. Munderloh. 1996. Direct cultivation of the causative agent of human granulocytic ehrlichiosis. N. Engl. J. Med. 334:209-215. [DOI] [PubMed] [Google Scholar]
- 24.Groves, M. G., G. L. Dennis, H. L. Amyx, and D. L. Huxsoll. 1975. Transmission of Ehrlichia canis to dogs by ticks (Rhipicephalus sanguineus). Am. J. Vet. Res. 36:937-940. [PubMed] [Google Scholar]
- 25.Hopp, T. P., and K. R. Woods. 1981. Prediction of protein antigenic determinants from amino acid sequences. Proc. Natl. Acad. Sci. USA 78:3824-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, T. C., R. B. Thompson, and T. O. Baldwin. 1986. Nucleotide sequence of the luxB gene of Vibrio harveyi and the complete amino acid sequence of the β subunit of bacterial luciferase. J. Biol. Chem. 261:4805-4811. [PubMed] [Google Scholar]
- 28.Keefe, T. J., C. J. Holland, P. E. Salyer, and M. Ristic. 1982. Distribution of Ehrlichia canis among military working dogs in the world and selected civilian dogs in the United States. J. Am. Vet. Med. Assoc. 181:236-238. [PubMed] [Google Scholar]
- 29.Kim, H. Y., and Y. Rikihisa. 1998. Characterization of monoclonal antibodies to the 44-kilodalton major outer membrane protein of the human granulocytic ehrlichiosis agent. J. Clin. Microbiol. 36:3278-3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 31.Maeda, K., N. Markowitz, R. C. Hawley, M. Ristic, D. Cox, and J. E. McDade. 1987. Human infection with Ehrlichia canis, a leukocytic rickettsia. N. Engl. J. Med. 316:853-856. [DOI] [PubMed] [Google Scholar]
- 32.Mulligan, M. E., D. K. Hawley, R. Entriken, and W. R. McClume. 1984. Analysis of the occurrence of promoter-sites in DNA. Nucleic Acids Res. 12:789-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ristic, M., and C. J. Holland. 1993. Canine ehrlichiosis. Pergamon Press, Oxford, United Kingdom.
- 34.Rockey, D. D., D. Grosenbach, D. E. Hruby, M. G. Peacock, R. A. Heinzen, and T. Hackstadt. 1997. Chlamydia psittaci IncA is phosphorylated by the host cell and is exposed on the cytoplasmic face of the developing inclusion. Mol. Microbiol. 24:217-228. [DOI] [PubMed] [Google Scholar]
- 35.Rockey, D. D., R. A. Heinzen, and T. Hackstadt. 1995. Cloning and characterization of a Chlamydia psittaci gene coding for a protein localized in the inclusion membrane of infected cells. Mol. Microbiol. 15:617-626. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Scidmore-Carlson, M. A., E. I. Shaw, C. A. Dooley, E. R. Fischer, and T. Hackstadt. 1999. Identification and characterization of a Chlamydia trachomatis early operon encoding four novel inclusion membrane proteins. Mol. Microbiol. 33:753-765. [DOI] [PubMed] [Google Scholar]
- 38.Stewart, G. S., S. Lubinsky-Mink, C. G. Jackson, A. Cassel, and J. Kuhn. 1986. pHG165: a pBR322 copy number derivative of pUC8 for cloning and expression. Plasmid 15:172-181. [DOI] [PubMed] [Google Scholar]
- 39.Sumner, J. W., W. L. Nicholson, and R. F. Massung. 1997. PCR amplification and comparison of nucleotide sequences from the groESL heat shock operon of Ehrlichia species. J. Clin. Microbiol. 35:2087-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walker, D. H., and J. S. Dumler. 1996. Emergence of the ehrlichioses as human health problems. Emerg. Infect. Dis. 2:18-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weisiger, R. M., M. Ristic, and D. L. Huxsoll. 1975. Kinetics of antibody response to Ehrlichia canis assayed by the indirect fluorescent antibody method. Am. J. Vet. Res. 36:689-694. [PubMed] [Google Scholar]
- 42.Weiss, E., G. A. Dasch, Y. H. Kang, and H. N. Westfall. 1988. Substrate utilization by Ehrlichia sennetsu and Ehrlichia risticii separated from host constituents by renografin gradient centrifugation. J. Bacteriol. 170:5012-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiss, E., J. C. Williams, G. A. Dasch, and Y. H. Kang. 1989. Energy metabolism of monocytic Ehrlichia. Proc. Natl. Acad. Sci. USA 86:1674-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wells, M. Y., and Y. Rikihisa. 1988. Lack of lysosomal fusion with phagosomes containing Ehrlichia risticii in P388D1 cells: abrogation of inhibition with oxytetracycline. Infect. Immun. 56:3209-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]