Abstract
The Ca2+ signal is essential for the activation of plant defense responses, but downstream components of the signaling pathway are still poorly defined. Here we demonstrate that specific calmodulin (CaM) isoforms are activated by infection or pathogen-derived elicitors and participate in Ca2+-mediated induction of plant disease resistance responses. Soybean CaM (SCaM)-4 and SCaM-5 genes, which encode for divergent CaM isoforms, were induced within 30 min by a fungal elicitor or pathogen, whereas other SCaM genes encoding highly conserved CaM isoforms did not show such response. This pathogen-triggered induction of these genes specifically depended on the increase of intracellular Ca2+ level. Constitutive expression of SCaM-4 and SCaM-5 in transgenic tobacco plants triggered spontaneous induction of lesions and induces an array of systemic acquired resistance (SAR)-associated genes. Surprisingly, these transgenic plants have normal levels of endogenous salicylic acid (SA). Furthermore, coexpression of nahG gene did not block the induction of SAR-associated genes in these transgenic plants, indicating that SA is not involved in the SAR gene induction mediated by SCaM-4 or SCaM-5. The transgenic plants exhibit enhanced resistance to a wide spectrum of virulent and avirulent pathogens, including bacteria, fungi, and virus. These results suggest that specific CaM isoforms are components of a SA-independent signal transduction chain leading to disease resistance.
The resistance of plants to invading pathogens is accompanied by the deployment of a complex array of defense responses (1). These responses include rapid death of challenged cells leading to the formation of local lesions (termed the hypersensitive response, HR) (2) and nonspecific immunity to subsequent infection by a variety of pathogens known as systemic acquired resistance (SAR) (3). Accumulating evidence implicates the involvement of a Ca2+ signal in certain plant defense responses. A Ca2+ ion influx is one of the earliest events in challenged cells (4–9) and has been shown to be essential for the activation of defense responses such as phytoalexin biosynthesis, induction of defense-related genes, and hypersensitive cell death (10–13). However, the molecular target(s) of this Ca2+ signal and how it regulates downstream events in the defense signaling pathway is not well understood. In particular, little is known about the Ca2+ signal receptors and the mechanisms by which these Ca2+-modulated proteins transduce the Ca2+ signal into defense responses. Based on pharmacological studies with calmodulin (CaM) antagonists (10–12), it has been proposed that CaM, a major Ca2+ signal transducer in both animals and plants (14), is involved. However, CaM antagonists also can influence other cellular processes not related to the Ca2+/CaM signaling (15). Thus, whether CaM is an actual component of plant defense signaling and, if so, what is the identity of the CaM-modulated enzymes and/or proteins involved remain to be determined.
Previously, we cloned five CaM isoform genes from soybean (SCaM-1 through SCaM-5) (16). Although some of these isoforms (SCaM-1, -2, and -3) are >96% identical with mammalian CaM, two isoforms (SCaM-4 and SCaM-5) exhibit only ≈78% identity and are thus the most divergent isoforms reported thus far in the plant and animal kingdom. The ability of these two divergent isoforms to activate two CaM-dependent enzymes in vitro is very different from that of the highly conserved isoforms, SCaM-1, -2, and -3 (16). This difference in their ability to activate a variety of target enzymes is presumably because of their divergent primary structure (17). These data suggest that SCaM-4 and SCaM-5 may play different roles in the plant cell.
In this paper, we describe a role of the divergent CaM isoforms in plant disease resistance responses, as evidenced by pathogen-induced rapid increase in gene expression level and activation of plant defense responses in the absence of pathogens. We provide in vivo evidence not only for functional differences among CaM isoforms but also for a potential role of SCaM-4 and SCaM-5 in plant defense signaling.
MATERIALS AND METHODS
Plant Materials and Treatments.
Soybean suspension cell culture (SB-P) was maintained in KN-1 medium as described (18). A 10-day-old subculture that had been dark-adapted for 3 days was used for the treatments. Fungal elicitor was prepared from Fusarium solani or Phytophthora parasitica var. nicotianae as described (19) and added to the cell culture at a final total reducing sugar concentration of 50 μg/ml.
RNA Blot Analysis.
Isolation of total RNA and Northern hybridization were performed as described (16). The tobacco pathogenesis-related (PR)-protein probes were kindly provided by J. Ryals (CIBA–Geigy), and the phenylalanine ammonia-lyase gene cDNA probe was generously donated by C. Lamb (Salk Institute, San Diego, CA).
Construction of Transgenic Plants.
For construction of the transgenic tobacco plants (Nicotiana tabacum cv. Xanthi-nc), the SCaM-4 or -5 cDNA was ligated into a plant binary vector, pGA643 (20), in which the SCaM cDNAs were placed under the control of the cauliflower mosaic virus (CaMV) 35S promoter in a sense orientation. The recombinant plasmids were introduced into Agrobacterium tumefacience EHA101, and transformed tobacco plants were obtained by a standard leaf disc transformation method (21). R1 progeny of transgenic plants expressing high levels of SCaM-4 or SCaM-5 were used for the experiments and maintained at 25°C day and 20°C night temperature, a 16-h photoperiod, and 65% relative humidity. To generate transgenic tobacco plants coexpressing SCaM-4, SCaM-5, and nahG genes, the NahG-10 tobacco plants (gift from J. Ryals) were transformed with the CAMBIA vector (pCAMBIA1300) harboring hpt as a selection marker, in which SCaM inserts were placed under the control of the CaMV 35S promoter. Plant transformation was performed as described above, and transformed plants were selected by kanamycin and hygromycin resistance.
Analysis of Transgenic Plants.
Bacterial pathogens were grown in King’s B medium supplemented with appropriate antibiotics (22) and pressure-inoculated into leaves by using a 1-ml syringe. Infection of tobacco mosaic virus (TMV) and Phytophthora parasitica pv. nicotianae were performed as described (23). Salicylic acid (SA) levels were determined from the fourth or fifth leaves of mature tobacco plants as described by Meuwly and Métraux (24).
RESULTS
Rapid Induction of Specific CaM Isoforms By Pathogen.
To investigate whether CaM is involved in plant defense signaling, we first examined expression of the five SCaM genes encoding different CaM isoforms (SCaM-1, -2, -3, -4, and -5) (16) after treatment with a fungal elicitor (19). Treatment of soybean cell suspension cultures with a nonspecific fungal elicitor prepared from Fusarium solani resulted in a dramatic rise (>10 fold) in mRNA encoded by SCaM-4 and SCaM-5 (Fig. 1A), the two SCaM genes whose sequences are most diverged from other CaM genes and subsequently will be referred to as the divergent CaM. Their mRNA levels peaked at 1 h and then slowly declined to basal levels by 12 h. The basal expression levels of these two SCaM genes in untreated cells were very low in comparison to those of the highly conserved SCaM genes, SCaM-1, -2, and -3. In contrast to SCaM-4 and SCaM-5, expression of the three conserved SCaM genes was not activated by the elicitor treatment (Fig. 1A). Consistent with the changes in SCaM mRNA levels, the protein levels of SCaM-4 and -5 but not SCaM-1, -2, and -3 also increased within 30 min of treatment (Fig. 1B). SCaM-4 and -5 protein levels reached their maximum of approximately 0.5 μg/mg of soluble protein after 2 h and then slowly declined after 12 h. In contrast, SCaM-1, -2, and -3 protein levels were not changed significantly by the fungal elicitor treatment. Interestingly, the induction of the SCaM-4 and SCaM-5 genes expression preceded that of the phenylalanine ammonia-lyase gene whose mRNA level began to increase 3 h after treatment (see Fig. 1A). An elicitor prepared from Phytophthora parasitica var. nicotianae had a similar effect on the expression of these CaM genes (see Fig. 1C).
Figure 1.
Rapid induction of SCaM-4 and SCaM-5 during plant defense response. (A) Effect of a fungal elicitor on the expression of SCaM genes. Soybean cell suspension culture (SB-P) was treated with fungal elicitor prepared from Fusarium solani, and total RNA was isolated at the indicated times and analyzed for SCaM-1, SCaM-4, SCaM-5, phenylalanine ammonia-lyase (PAL), and β-tubulin mRNA. (B) Changes in SCaM protein levels upon fungal elicitor treatment. SB-P cells were treated as described in A and relative protein levels of SCaM-4/SCaM-5 or SCaM-1/SCaM-2/SCaM-3 were examined by immunoblot analysis using either anti-SCaM-4 or anti-SCaM-1 antibody, respectively (16). Relative protein levels of SCaM isoforms were calculated by comparing band intensities and areas in autoradiograms with those of known quantities of standard SCaM-1 or SCaM-4 proteins by using scanning densitometry. (C) Effect of various defense signaling molecules on the expression of SCaM genes. SB-P cells were treated for 1 h as indicated above the lanes, and the mRNA levels of the SCaM genes were examined by Northern blot analysis. dH2O, water control; Psg, P. syringae pv. glycinea carrying avrC; FE, fungal elicitor prepared from P. parasitica var. nicotianae; FE+CHX, fungal elicitor plus 1 μg/ml of cycloheximide; FE+BAPTA, fungal elicitor plus 5 mM 1,2-bis-(o-aminophenoxy)-ethane-N,N,N′,N′-tetraacetic acid; Ca2++A23187, 25 μM Ca2+ ionophore A23187 plus 5 mM CaCl2; H2O2, 2 mM hydrogen peroxide; G-GO, glucose and glucose oxidase system; X-XO, xanthine and xanthine oxidase system; SA, 2 mM salicylic acid; JA, 100 μM jasmonic acid; ABA, 100 μM abscisic acid.
Ca2+-Dependent Induction of SCaM-4 and SCaM-5.
We next examined the effect of several potential inducers of plant defense-related genes to gain further insight into the molecular signals involved in the induction of SCaM-4 and SCaM-5 gene expression. Both P. parasitica elicitor and a bacterial pathogen, Pseudomonas syringae pv. glycinea (Psg) carrying avrC, effectively induced SCaM-4 and SCaM-5 gene expression (Fig. 1C). Cycloheximide, a protein synthesis inhibitor, did not block SCaM-4 and SCaM-5 induction by the P. parasitica elicitor. In contrast, addition of a Ca2+-chelator, 1,2-bis-(o-aminophenoxy)-ethane-N,N,N′,N′-tetraacetic acid, abolished the induction of SCaM-4 and SCaM-5, whereas the Ca2+-ionophore A23187 alone was sufficient to induce SCaM-4 and SCaM-5 expression as effectively as the fungal elicitor. However, application of exogenous SA (1–5 mM) or hydrogen peroxide (H2O2; 1–10 mM) did not induce the expression of SCaM-4 and SCaM-5 genes during a 24-h time course (data not shown). A H2O2-generating system (glucose and glucose oxidase; ref. 22) and a superoxide-generating system (xanthine and xanthine oxidase; ref. 25) also failed to induce SCaM-4 and SCaM-5 gene expression. Two unrelated signal molecules, jasmonic acid (JA) (26) and abscisic acid (ABA) (27), also were unable to induce SCaM-4 and SCaM-5. These results indicate that transcriptional activation of SCaM-4 and SCaM-5 genes does not require protein synthesis and either precedes synthesis and accumulation of reactive oxygen species (ROS) and SA or is in another disease-resistance signaling pathway. Furthermore, their activation was specific; signaling compounds associated with other stress responses such as wounding (JA) and drought (ABA) did not induce SCaM-4 and SCaM-5. However, their induction appears to be mediated by an increase of intracellular Ca2+ concentration.
Spontaneous Lesion Formation in Transgenic Tobacco Plants Constitutively Expressing SCaM-4 or SCaM-5.
To examine the biological functions of the divergent SCaM isoforms in plant defense responses, we constructed transgenic tobacco plants that constitutively expressed SCaM-4 or SCaM-5 under the control of the constitutive cauliflower mosaic virus 35S promoter. Transgenic plant lines expressing SCaM-4 or SCaM-5 first were selected by Northern analysis and verified by immunoblot analysis (16). Interestingly, the transgenic SCaM-4 and SCaM-5 plants often spontaneously formed disease-like necrotic spots on their leaves (Fig. 2 A and B). The lesions appeared first in the oldest mature leaves, whereas the top 3–4 young leaves never developed lesions. Lesions may form on older leaves as they senesce. Untransformed wild-type and empty vector-transformed control transgenic plants grown under identical conditions did not show these symptoms. To determine whether these lesions were HR-like lesions, they were examined with a fluorescence microscope. HR-like lesions accumulate fluorescent material in the cell walls that is readily visible under UV illumination (28, 29), whereas necrotic regions generated by mechanical wounding or freezing and thawing have no such fluorescence (23). Leaves of the transgenic plants exhibited accumulation of bright UV-excitable fluorescent material in their lesions, suggesting that these necrotic lesions resemble HR-like lesions (Fig. 2 C and D). Similar spontaneous lesions have been found in a variety of lesion-mimic mutants of Arabidopsis and maize (30).
Figure 2.
Phenotypes of transgenic tobacco plants constitutively expressing SCaM-4 or SCaM-5. (A) Formation of spontaneous disease lesion-like necrotic regions in the transgenic plants. The whole plant morphology of an 8-week-old representative transgenic SCaM-4 plant (Right) shows that lesions appeared only in the older leaves. A normal wild-type plant of similar age is shown (Left). (B) A closer look at the spontaneous lesions formed on the leaf of a 10-week-old SCaM-5 transgenic plant (Right). A normal leaf of a similar age is shown (Left) for comparison. (C and D) Accumulation of UV-excitable fluorescent material in the cell walls of spontaneously formed lesions. Microscopic examination of a spontaneous lesion under differential interference contrast (C) and epifluorecence (D) optics after nuclear staining of leaf tissues with 4′, 6-diamidino-2-phenylindole. Magnifications: ×50.
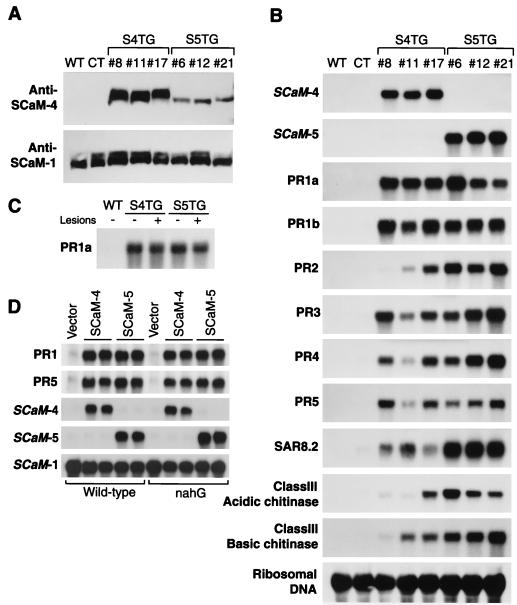
The constitutive expression of these divergent CaM isoforms, SCaM-4 and SCaM-5, did not affect the level of the highly conserved endogenous tobacco CaM recognized by the anti-SCaM-1 antibody (Fig. 3A). The anti-SCaM-1 antibody recognized equally the highly conserved CaM isoforms SCaM-1, SCaM-2, and SCaM-3 protein but not the divergent SCaM-4 and SCaM-5. The anti-SCaM-4 antibody cross-reacted with SCaM-5 but not with the highly conserved CaM isoforms (ref. 16, data not shown). The amounts of SCaM-4 and SCaM-5 protein in these transgenic plants were estimated to be 0.3–0.5 μg/mg total soluble leaf protein, which is similar to the maximal level of the SCaM-4/SCaM-5 protein induced by a fungal elicitor in soybean cells (see Fig. 1B). Furthermore, the levels of SCaM-4 and SCaM-5 in transgenic plants did not exceed the protein levels of the highly conserved CaM isoforms (SCaM-1, SCaM-2, and SCaM-3). Thus, it is likely that the formation of spontaneous lesions in the transgenic plants is not the result of high-level overexpression of SCaM-4 or SCaM-5 per se, which could lead to abnormal cellular responses (30, 31), but rather is caused by levels of these CaM isoforms that are similar to those after activation of these genes by pathogen attack.
Figure 3.
Constitutive expression of PR protein genes in transgenic SCaM-4 (S4TG) and SCaM-5 (S5TG) tobacco plants. (A) Immunoblots showing elevated SCaM-4 and SCaM-5 protein levels in transgenic plants. Total soluble protein (50 μg) from three independent transgenic plant lines was analyzed for CaM protein levels by using either the anti-SCaM-4 or anti-SCaM-1 antibody. (B) Constitutive expression of PR protein genes in the transgenic plants. Total RNA was isolated from a wild type (WT), a control transgenic plant harboring an empty vector (CT), and three representative independent transgenic plants expressing SCaM-4 (S4TG) or SCaM-5 (S5TG) and examined for the mRNA levels of tobacco SAR genes (31). # indicates designation of transgenic plant line. (C) Comparison of PR-1a gene expression in lower old leaves that formed lesions (+) and upper young leaves that did not form lesions (−) from SCaM-4 and SCaM-5 transgenic plants. The lesion (+) and lesion (−) leaf samples were taken from the same transgenic plants. (D) SA-independent PR protein gene expression. The effect of constitutive expressing SCaM-4 or SCaM-5 in wild-type plants or NahG transgenic plants (33) on PR protein gene expression was examined by RNA gel blot analysis using PR1a and PR5 probes. Data shown are representative results obtained from at least 10 respective independent transgenic plant lines.
Constitutive Expression of SAR Genes in Transgenic SCaM-4 and SCaM-5 Plants.
Plants that produce spontaneous lesions often show elevated expression of genes encoding PR proteins and increased resistance to pathogens (30, 31). Interestingly, in the absence of pathogens, the transgenic SCaM-4 and SCaM-5 plants expressed at high levels all nine of the SAR marker genes in tobacco that normally are activated during the development of SAR (3, 32) (Fig. 3B). Wild-type plants and control empty vector transgenic plants grown under identical conditions did not express these genes. The SAR-associated genes examined were constitutively expressed throughout the growth and development of the SCaM-4 and SCaM-5 transgenic plants and were apparent even in the leaves of axenically grown young plants that are free of any visible lesions when examined under the microscope (Fig. 3C). Thus, expression of PR genes in the transgenic SCaM-4 and SCaM-5 plants was observed to be independent of lesion formation, suggesting that PR gene expression in the transgenic plants may not be a consequence of cell death.
SA-Independent SAR Gene Induction in Transgenic SCaM-4 and SCaM-5 Plants.
SA is known to be a natural signaling molecule for the activation of certain plant defense responses (3, 31, 33). The application of exogenous SA to tobacco leaves mimics the pathogen-induced SAR responses such as PR protein synthesis. In addition, endogenous SA levels in several lesion-mimic mutants are substantially higher than those of wild-type plants in the absence of pathogen challenge (3, 31). Furthermore, transgenic plants that constitutively expressed the bacterial nahG gene, which encodes a SA-degrading enzyme, are defective in their ability to induce SAR (34, 35). We therefore examined whether the elevated levels of PR gene expression in SCaM-4 or SCaM-5 transgenic plants might be the result of elevation of endogenous SA levels. The levels of SA were assayed in the TMV-infected leaves, and the leaves were harvested 10 days after TMV infection. Surprisingly, SA levels in SCaM-4 and SCaM-5 transgenic plants were similar to those of wild type. Moreover, infection with TMV resulted in similar increases in SA levels in the transgenic plants and wild-type plants, demonstrating that the expression of SCaM-4 and SCaM-5 in the transgenic plants did not affect endogenous SA accumulation (Table 1). Consistent with these observations, the expression of nahG gene did not suppress the constitutive expression of PR genes in SCaM-4 and SCaM-5 transgenic plants (Fig. 3D). Furthermore, the infection of TMV on SCaM-4 or SCaM-5 transgenic plants did not show significant additive effect on the levels of PR gene expression (data not shown). These results strongly argue that SA is not involved in the induction of PR gene expression mediated by SCaM-4 or SCaM-5.
Table 1.
Endogenous SA levels in transgenic plants
| Plants | Uninfected
|
TMV-infected
|
|
|---|---|---|---|
| Free SA | Free SA | Bound SA | |
| Wild type | 0.042 ± 0.005 | 2.845 ± 0.5 | 2.254 ± 0.574 |
| SCaM-4 | 0.031 ± 0.010 | 4.121 ± 1.0 | 2.465 ± 0.832 |
| SCaM-5 | 0.035 ± 0.005 | 2.523 ± 0.5 | 1.621 ± 0.564 |
| NahG | 0.029 ± 0.025 | 0.067 ± 0.5 | 0.085 ± 0.014 |
| SCaM-4 + NahG | 0.038 ± 0.072 | 0.083 ± 0.7 | 0.079 ± 0.021 |
| SCaM-5 + NahG | 0.036 ± 0.056 | 0.072 ± 0.4 | 0.065 ± 0.035 |
The levels of SA (μg/g fresh weight) were determined from the top forth or fifth middle leaves of mature plants as described (41). SCaM-4, SCaM-5, and NahG indicate transgenic SCaM-4, SCaM-5, and NahG plants, respectively. SCaM-4 + NahG and SCaM-5 + NahG indicate transgenic plants that coexpress SCaM-4 and nahG gene, and SCaM-5 and nahG gene. TMV was infected in the leaves of 6-week-old transgenic plants that did not form lesions and leaf samples for SA assay were taken from TMV-infected leaves 10 days after inoculation, which appeared as HR lesions on leaves. Data are the means of two determinations from five independent transgenic plant lines. Bound SA levels were below detection limit in uninfected plants.
Enhanced Disease Resistance of Transgenic SCaM-4 and SCaM-5 Plants to a Wide Spectrum of Pathogens.
Finally, we assessed whether transgenic SCaM-4 and SCaM-5 plants had altered resistance to pathogens. We used an oomycete fungal pathogen, Phytophthora parasitica var. nicotianae (the causal agent of black shank disease) to inoculate the transgenic and wild-type plants. At 5 days after P. parasitica inoculation, disease symptoms started to appear on the wild-type plants but not on the transgenic plants. By 7 days after inoculation, the wild-type plants had severe disease symptoms, such as leaf wilting and stem rot, and eventually died by 8 days after inoculation. However, the transgenic plants remained healthy without appreciable disease symptoms. The inoculated SCaM-4 or SCaM-5 transgenic plants did not show necrotic lesions in the 4- to 5-week-old plants. However, the lesions started to develop in the bottom leaves of 7- to 8-week-old transgenic plants similarly to the uninoculated transgenic plants (Fig. 4A). The leaves of the fungus-infected wild-type plants showed uninterrupted spreading of fungal hyphae with little callose deposition (Fig. 4B). In contrast, leaves of the inoculated transgenic plants had no such growth of fungal hyphae but instead exhibited bright fluorescence related to callose deposition and accumulation of autofluorescent material in the cells surrounding the fungal penetration sites (Fig. 4C), which is characteristic of an HR. Interestingly, P. parasitica var. nicotianae is a virulent pathogen on the parental, nontransformed Xanthi-nc tobacco cultivar and does not normally trigger an HR. The transgenic plants also showed enhanced resistance to a virulent bacterial pathogen, Pseudomonas syringae pv. tabaci (Pst). They successfully blocked development of disease symptoms, and the in planta growth of Pst was retarded greater than 10-fold compared with their growth in wild-type plants (Fig. 4D). In addition, transgenic SCaM-4 and SCaM-5 plants exhibited increased resistance to an avirulent viral pathogen, TMV. The transgenic plants developed TMV-induced HR lesions approximately 12 h earlier than wild-type plants. Furthermore, the transgenic plants had fewer (≈5-fold) and smaller TMV-induced lesions (Fig. 4E), which are two characteristics associated with enhanced SAR to TMV.
Figure 4.
Enhanced disease resistance of transgenic tobacco plants constitutively expressing SCaM-4 or SCaM-5. (A) Disease responses to the virulent fungal pathogen, P. parasitica var. nicotianae. At 7 days after inoculation, plants were examined for disease symptoms. Representative results of wild type (WT), transgenic plants expressing SCaM-4 (S4TG) or SCaM-5 (S5TG) are shown. (B and C) Fluorescence micrographs of leaves infected with the virulent P. parasitica var. nicotianae. Infected leaves were cleared, stained with aniline blue, and examined under an UV-light epifluorescence microscope. In leaves of the wild-type plants (B) the spreading of fungal hyphae is evident, but leaves of the transgenic plants (C) show the accumulation of UV-excitable fluorescent material in the cells surrounding the fungal penetration sites without appreciable growth of fungal hyphae. (Scale bars represent 100 μm.) (D) In planta bacterial growth. Pst was inoculated into leaves of mature wild-type (WT) and transgenic SCaM-4 (S4TG) and SCaM-5 (S5TG) plants at 105 cfu/ml, and in planta bacterial growth was monitored over 5 days. Data points represent means of two determinations from five independent lines. (E) Elevated resistance of SCaM-4 and SCaM-5 transgenic plants to the avirulent pathogen TMV. The second or third fully expanded young leaves from the uppermost part of plants were inoculated with TMV by gently rubbing leaves with carborundum and 2 μg/leaf TMV, and the development of HR lesions were monitored over 5 days. Data shown are the numbers of HR lesions formed in these plants. In all of these pathogen tests, control transgenic plants transformed with an empty vector showed results essentially similar to those of wild-type plants (data not shown).
DISCUSSION
In this report, we have described a potential role for the major Ca2+ signal transducer, CaM, in plant defense signaling. Our results argue that the divergent CaM isoforms act as both signal receptor and transmitter of the pathogen-induced Ca2+ signal. SCaM-4 and SCaM-5 resemble immediate early gene products such as fos and jun in animal systems in that certain external stimuli immediately activate their expression, which then leads to cellular responses (36, 37). Thus, SCaM-4 and SCaM-5 are two of the inducible components in plant defense responses.
Transgenic plants that constitutively expressed SCaM-4 and SCaM-5 had phenotypes similar to those of spontaneous lesion-mimic (e.g., lsd and acd) mutants (28, 29); however, there are several notable differences. The first concerns the causal relationship between cell death and PR gene expression. Although PR gene expression is tightly linked to cell death in lsd and acd mutants, it is observed to be independent of microscopic and macroscopic lesion formation in the SCaM-4 and SCaM-5 transgenic plants. The second major difference is SA dependence. SA levels in most lesion-mimic mutants are substantially higher than those in normal plants (3, 30, 31). In contrast, in the SCaM-4 and SCaM-5 transgenic plants disease resistance responses were activated without concurrent elevation of endogenous SA level. In addition, removal of SA in these transgenic plants by coexpression of the nahG gene failed to block the constitutive expression of PR genes (Fig. 3D). Furthermore, activation of SCaM-4 and -5 gene expression was independent of SA accumulation; exogenous supply of SA to the SB-P cells failed to induce these genes (Fig. 1C), and the induction was evident in NahG transgenic plants (data not shown). The enhanced disease resistance by constitutive expression of SCaM-4 or SCaM-5 is also evident in the transgenic plants coexpressing nahG gene and SCaM-4 or nahG gene and SCaM-5 (unpublished data). These observations strongly suggest that SCaM-4 and SCaM-5 activate plant disease resistance responses via a SA-independent pathway(s).
Our results provide in vivo evidence for functional differences among plant CaM isoforms. Only SCaM-4 and SCaM-5 isoforms are induced by pathogens and enhanced defense responses in transgenic plants, whereas the other highly conserved CaM isoforms such as SCaM-1 and SCaM-2 did not have these properties (data not shown). During preparation of this paper, a role for CaM in the ROS production during the plant defense response was reported (38). Those authors propose that Ca2+ activation of CaM stimulates NAD kinase and the resulting increase of cellular NADPH levels then activates NADPH oxidase, which produces ROS. This Ca2+/CaM pathway in ROS production is thought to be mediated by the highly conserved CaM isoforms because the divergent CaM isoforms are unable to activate NAD kinase (16, 17). These observations support a model for concerted roles of CaM isoforms in plant defense response against pathogens, in which the highly conserved CaM isoforms mediate ROS increases, whereas the divergent CaM isoforms activate programmed cell death and defense gene expression.
Transgenic plants constitutively expressing several other transgenes also have been shown to have lesion-mimic phenotype and altered disease resistance. These genes include Halobacterium opsin, mutant ubiquitin ubR48, cholera toxin subunit A1, and yeast invertase (23, 39–41). These transgenic plants have elevated levels of endogenous SA, which argues that their altered disease-resistance responses result from SA accumulation that may be ascribed to the metabolic stress induced by these transgenes (30, 31). Indeed, it is unlikely that these genes are bona fide components of the plant defense response pathway(s). In contrast, the divergent CaM isoforms represent one of the “natural” pathogen-inducible components in plant defense signaling whose constitutive expression leads to enhanced disease resistance. However, at the present time, it is not clear what the exact role of SCaM-4 and SCaM-5 is in various plant defense responses against pathogens because our knowledge relies solely on the gain-of-function experiment. Further loss-of-function studies such as antisense transgenic soybean plants or knockout mutations will help to establish further the physiological role of SCaM-4 and SCaM-5. Nevertheless, the results presented here not only enhance our understanding of the pathway(s) leading to plant disease resistance but also may provide opportunities to genetically engineer plants with resistant to a wide spectrum of pathogens.
Acknowledgments
We thank Drs. C. Lamb and J. Ryals for generously providing us with PR protein cDNAs, Dr. J. Ryals for NahG plant seeds, and Dr. J. Widholm for SB-P suspension cells. We also thank Dr. W.M. Park for providing TMV, C. Lamb for Psg and Pst, and S.J. Kim for technical assistance in testing viral resistance. We are grateful to Drs. J. Schell, I. Somssich, C. Lamb, and D. Klessig for their helpful comments on the manuscript. This work was supported by a grant from the Korea Science and Engineering Foundation to the Plant Molecular Biology and Biotechnology Research Center, a nondirected research fund from the Korea Research Foundation (1996), and a G7 grant (08-04-A27) to M.J.C.
ABBREVIATIONS
- CaM
calmodulin
- SCaM
soybean CaM
- HR
hypersensitive response
- JA
jasmonic acid
- PR
pathogenesis-related
- Psg
Pseudomonas syringae pv. glycinea
- Pst
P. syringae pv. tabaci
- ROS
reactive oxygen species
- SA
salicylic acid
- SAR
systemic acquired resistance
- TMV
tobacco mosaic virus
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Jackson A O, Tayler C B. Plant Cell. 1996;8:1651–1668. doi: 10.1105/tpc.8.10.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodman R N, Novacky A J. The Hypersensitive Reaction in Plants to Pathogens: A Resistance Phenomenon. St. Paul, MN: APS Press; 1994. [Google Scholar]
- 3.Ryals J A, Neuenschwander U H, Willitis M G, Molina A, Steiner H-Y, Hunt M D. Plant Cell. 1996;8:1809–1819. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dixon R A, Harrison M J, Lamb C J. Annu Rev Phytopathol. 1994;32:479–501. [Google Scholar]
- 5.Knight M R, Campbell A K, Smith S M, Trewavas A J. Nature (London) 1991;352:524–526. doi: 10.1038/352524a0. [DOI] [PubMed] [Google Scholar]
- 6.Nürnberger T, Nennstiel D, Jabs T, Sacks W R, Hahlbrock K, Scheel D. Cell. 1994;78:449–460. doi: 10.1016/0092-8674(94)90423-5. [DOI] [PubMed] [Google Scholar]
- 7.Atkinson M M, Midland S L, Sims J J, Keen N T. Plant Physiol. 1996;112:297–302. doi: 10.1104/pp.112.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gelli A, Higgins V J, Blumwald E. Plant Physiol. 1997;113:269–279. doi: 10.1104/pp.113.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zimmermann S, Nürnberger T, Frachisse J-M, Wirts W, Guern J, Hedrich R, Scheel D. Proc Natl Acad Sci USA. 1997;94:2751–2755. doi: 10.1073/pnas.94.6.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sacks W R, Ferreira P, Hahlbrock K, Jabs T, Nürnberger T, Renelt A, Scheel D. In: Advances in Molecular Genetics of Plant-Microbe Interactions. Nester E W, Verma D P S, editors. Vol. 2. Dordrecht, The Netherlands: Kluwer; 1993. pp. 485–495. [Google Scholar]
- 11.Vogeli U, Vogeli-Lanfe R, Chappel J. Plant Physiol. 1992;100:1369–1376. doi: 10.1104/pp.100.3.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tavernier E, Wendehenne D, Blein J-P, Pugin A. Plant Physiol. 1995;109:1025–1031. doi: 10.1104/pp.109.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levine A, Pennell R I, Alvarez M E, Palmer R, Lamb C. Curr Biol. 1996;6:427–437. doi: 10.1016/s0960-9822(02)00510-9. [DOI] [PubMed] [Google Scholar]
- 14.Roberts D M, Harmon A C. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:375–414. [Google Scholar]
- 15.Roberts D M, Lukas T J, Watterson D M. Crit Rev Plant Sci. 1986;4:311–339. [Google Scholar]
- 16.Lee S H, Kim J C, Lee M S, Heo W D, Seo H Y, Yoon H W, Hong J C, Lee S Y, Bahk J D, Hwang I, Cho M J. J Biol Chem. 1995;270:21806–21812. doi: 10.1074/jbc.270.37.21806. [DOI] [PubMed] [Google Scholar]
- 17.Lee S H, Seo H Y, Kim J C, Heo W D, Chung W S, Lee K J, Kim M C, Cheong Y H, Choi J Y, Lim C O, Cho M J. J Biol Chem. 1997;272:9252–9259. doi: 10.1074/jbc.272.14.9252. [DOI] [PubMed] [Google Scholar]
- 18.Horn M E, Widholm J M, Ogren W L. Plant Physiol. 1989;72:426–429. doi: 10.1104/pp.72.2.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simmons C R, Litts J C, Huang N, Rodriguez R L. Plant Mol Biol. 1992;18:33–45. doi: 10.1007/BF00018454. [DOI] [PubMed] [Google Scholar]
- 20.An G, Ebert P R, Mitra A, Ha S B. In: Plant Molecular Biology Manual. Gelvin S B, Schilperoort R A, editors. Dordrecht, The Netherlands: Kluwer; 1988. p. A3. [Google Scholar]
- 21.Horsch R B, Fry J, Hoffmann N, Neidermeyer J, Rogers S G, Fraley R T. In: Plant Molecular Biology Manual. Gelvin S B, Schilperoort R A, editors. Dordrecht, The Netherlands: Kluwer; 1988. p. A5. [Google Scholar]
- 22.Levine A, Tenhaken R, Dixon R, Lamb C. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- 23.Mittler R, Shulaev V, Lam E. Plant Cell. 1995;7:29–42. doi: 10.1105/tpc.7.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meuwly P, Métraux J–P. Anal Biochem. 1993;214:500–505. doi: 10.1006/abio.1993.1529. [DOI] [PubMed] [Google Scholar]
- 25.Jabs T, Dietrich R A, Dangl J L. Science. 1996;273:1853–1856. doi: 10.1126/science.273.5283.1853. [DOI] [PubMed] [Google Scholar]
- 26.Bergey D R, Howe G A, Ryan C A. Proc Natl Acad Sci USA. 1996;93:12053–12508. doi: 10.1073/pnas.93.22.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giraudat J, Parcy F, Bertauche N, Gosti F, Leung J, Morris P-C, Bouvier-Durand M, Vartanian N. Plant Mol Biol. 1994;26:1557–1578. doi: 10.1007/BF00016490. [DOI] [PubMed] [Google Scholar]
- 28.Dietrich R A, Delaney T P, Uknes S J, Ward E R, Ryals J A, Dangl J L. Cell. 1994;77:565–577. doi: 10.1016/0092-8674(94)90218-6. [DOI] [PubMed] [Google Scholar]
- 29.Greenberg J T, Guo A, Klessig D F, Ausubel F M. Cell. 1994;77:551–564. doi: 10.1016/0092-8674(94)90217-8. [DOI] [PubMed] [Google Scholar]
- 30.Dangl J L, Dietrich R A, Richberg M H. Plant Cell. 1996;8:1793–1807. doi: 10.1105/tpc.8.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durner J, Shah J, Klessig D F. Trends Plant Sci. 1997;2:266–274. [Google Scholar]
- 32.Ward E R, Uknes S J, Williams S C, Dincher S S, Wiederhold D L, Alexander D C, Ahl-Goy P, Métraux J-P, Ryals J A. Plant Cell. 1991;3:1085–1094. doi: 10.1105/tpc.3.10.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raskin I. Plant Physiol. 1992;99:799–803. doi: 10.1104/pp.99.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaffney T, Friedrich L, Vernooji B, Negrotto D, Nye G, Uknes S, Ward E, Ryals J. Science. 1993;261:754–756. doi: 10.1126/science.261.5122.754. [DOI] [PubMed] [Google Scholar]
- 35.Delaney T P, Uknes S, Vernooji B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessman H, Ward E, Ryals J. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- 36.Greenberg M E, Thompson M A, Sheng M. J Physiol (Paris) 1992;86:99–108. doi: 10.1016/s0928-4257(05)80013-0. [DOI] [PubMed] [Google Scholar]
- 37.Morgan J I, Curran T. Trends Neurosci. 1989;12:459–462. doi: 10.1016/0166-2236(89)90096-9. [DOI] [PubMed] [Google Scholar]
- 38.Harding S A, Oh S–H, Roberts D M. EMBO J. 1997;16:1137–1144. doi: 10.1093/emboj/16.6.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Becker F, Buschfeld E, Schell J, Bachmair A. Plant J. 1993;3:875–881. [Google Scholar]
- 40.Beffa R, Szell M, Meuwly P, Pay A, Vogeli-Lange R, Métraux J-P, Neuhaus G, Meisn F, Nagy F. EMBO J. 1995;14:5753–5761. doi: 10.1002/j.1460-2075.1995.tb00264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herbers K, Meuwly P, Frommer W B, Métraux J-P, Sonnewald U. Plant Cell. 1996;8:793–803. doi: 10.1105/tpc.8.5.793. [DOI] [PMC free article] [PubMed] [Google Scholar]