Abstract
Feline immunodeficiency virus (FIV), like other members of the lentivirus subfamily, such as human immunodeficiency virus type 1 (HIV-1), can infect nondividing and terminally differentiated cells. The transport of the preintegration complex into the nucleus is cell cycle-independent, but the mechanism is not well understood. Integrase is a key component of the complex and has been suggested to play a role in nuclear import during HIV-1 replication. To determine its karyophilic property, FIV integrase fused with glutathione S-transferase and enhanced green fluorescent protein was expressed in various feline and human cells and the subcellular localization was visualized by fluorescence microscopy. Wild-type FIV integrase was karyophilic in all cell lines tested and capable of targeting the fusion protein to the nuclei of transfected cells. Analysis of deletion and point mutation variants of FIV integrase failed to reveal any canonical nuclear localization signal, and the karyophilic determinant was mapped to the highly conserved N-terminal zinc-binding HHCC motif. A region near the C-terminal domain enriched with basic amino acid residues also affected the nuclear import of integrase. However, the role of this region is only modulatory in comparison to that of the zinc-binding domain. The N-terminal zinc-binding domain does not bind DNA and instead is essential in integrase multimerization. We therefore postulate that the karyophilic property of FIV integrase requires subunit multimerization promoted by the HHCC motif. Alternatively, the HHCC motif may directly promote interaction between FIV integrase and cellular proteins involved in nuclear import.
Retroviruses are a family of viruses that store genetic information as RNA. To establish a productive infection, a retrovirus must first reverse transcribe its RNA genome to form a double-stranded DNA copy that must then be integrated into a host cell chromosome (reviewed in reference 3). Since the integration reaction occurs in the nucleus, an important step prior to integration is the transport from the cytoplasm to the nucleus of the reverse-transcribed viral cDNA in the form of a large nucleoprotein complex termed the preintegration complex (PIC). The assembly of the PIC allows for a stable association between the viral cDNA and the integration machinery during the translocation of the cDNA within the host cell (42).
Lentiviruses and oncoretroviruses are two major subfamilies of retroviruses. Lentiviruses, including human immunodeficiency virus type 1 (HIV-1) and feline immunodeficiency virus (FIV), are distinguished from oncoretroviruses by their ability to establish productive infections in nondividing cells, such as macrophages, mucosal dendritic cells, and quiescent T lymphocytes (16, 25, 43). The ability of HIV-1 to infect nondividing cells is largely attributed to the karyophilic property of the virus's PIC that allows it to traverse the nuclear envelope in a cell cycle-independent fashion (4, 26). In contrast, infection by oncoretroviruses is restricted to proliferating cells. Presumably, the PICs of oncoretroviruses lack nuclear localization signals (NLS) and the accessibility of the host chromosomes to these PICs relies on the transient dissolution of the nuclear envelope during mitosis (26, 35).
HIV-1 appears to employ multiple signals to facilitate the nuclear import of its PIC in nondividing cells. Matrix, Vpr, and integrase are known constituents of the HIV-1 PIC, and each has been shown to display karyophilic properties (reviewed in reference 13). Besides the viral proteins, a cis-acting factor named central DNA flap, a partial three-stranded intermediate formed during reverse transcription of the plus strand, also contributes to the nuclear import of the HIV-1 PIC (45). Such redundancy may function to ensure successful infection of various nondividing cells irrespective of their cellular environments, a step that is likely critical for viral transmission and reservoir establishment. However, the presence of multiple factors makes it difficult to dissect their respective contributions in the nuclear import of the PIC. The ability of matrix to localize in the nucleus remains controversial (7, 14, 15, 18, 33), and Vpr may play only a modulatory role in the nuclear entry process. Indeed, viruses with mutations affecting both the matrix NLS and Vpr demonstrated only a partial or no reduction in viral replication in nondividing cells (16, 20), suggesting that integrase has a primary role in nuclear import (2).
HIV-1 integrase is karyophilic (7, 16, 31), though conflicting results exist on the identity of its karyophilic determinant and the import mechanism. Gallay et al. (16) showed that the NLS of integrase belongs to the basic bipartite type and comprises residues 186 to 188 and 211 to 219. The bipartite NLS is recognized by karyopherin α, a member of a family of related proteins involved in nucleocytoplasmic transport (reviewed in reference 28). However, others could not confirm the role of this bipartite NLS in the nuclear import of integrase (40) or the PIC (30), and another study reported that the nuclear localization of integrase does not involve karyopherins α, β1, or β2 and that it is independent of GTP hydrolysis and Ran (6). A separate study found that the karyophilic determinant of HIV-1 integrase is a short, noncanonical NLS located in residues 161 to 173 within the catalytic core domain (2). Mutation of this core NLS results in diminished nuclear import of the PIC and suppresses viral replication in both nondividing and dividing cells.
Like HIV-1, FIV can infect nondividing cells, such as neurons and macrophages (9, 12). However, the factor responsible for the nuclear entry of FIV is not known. Whether or not FIV integrase is karyophilic has not been determined. Compared with that of HIV-1, the FIV genome lacks a vpr equivalent, and the presence of a functional NLS in the matrix has not been reported (39). Due to an apparent lack of redundant karyophilic factors, we hypothesized that FIV may be a more amenable system for investigating the role of integrase in the nuclear import of the lentiviral PIC. In addition, the availability of a small natural host for FIV will provide an in vivo system to address the importance of nuclear import in nondividing cells in viral transmission and pathogenesis (10).
To characterize the karyophilic property of FIV integrase, we have expressed integrase as a fusion protein with enhanced green fluorescence protein (EGFP) in human and feline cells. The subcellular localization of the expressed fusion protein was determined by fluorescence microscopy. Our results showed that FIV integrase was able to direct the fusion protein to the nuclei of transfected cells, thereby indicating that FIV integrase is karyophilic. Subsequent mutational analyses of the protein found that the karyophilic property of FIV integrase depends on the integrity of the conserved N-terminal zinc-binding domain. We postulate that the zinc-binding motif promotes protein-protein interaction between the cellular nuclear import machinery and FIV integrase, resulting in the nuclear entry of FIV integrase.
MATERIALS AND METHODS
Cloning of fusion proteins consisting of FIV integrase and EGFP.
Wild-type or mutant FIV integrase was fused to the N terminus of EGFP, a redshifted variant of green fluorescence protein optimized for expression in eukaryotic cells (Clontech, Palo Alto, Calif.). For the fusion protein containing the full-length wild-type integrase, the DNA encoding FIV integrase was amplified by PCR by using the FIV molecular clone 34TF10 (37) as the template. The 5′ and 3′ primers used to amplify the FIV integrase gene were F-IN5′EcoR and 3′FIV-INL-AgeI, respectively. All oligonucleotides used as PCR primers were purchased from Operon (Alameda, Calif.), and their sequences are shown in Table 1. The amplification generated a fragment encoding all 281 amino acid residues of FIV integrase plus an additional eight alternating Gly and Ala residues at the 3′ end to function as a flexible linker between FIV integrase and EGFP. The PCR product was cloned into the TOPO TA vector (Invitrogen Corp., Carlsbad, Calif.) and sequenced by the UCLA Sequencing Core Facility with the ABI 3700 DNA Analyzer (PE Applied Biosystems, Foster City, Calif.). After sequence confirmation, the integrase fragment was obtained by digestion with EcoRI and AgeI and purified with a gel extraction kit (Qiagen, Chatsworth, Calif.). The purified EcoRI-AgeI fragment was then ligated with the pEGFP-NI vector (Clontech) previously digested with EcoRI and AgeI to form pF-IN/EGFP.
TABLE 1.
DNA sequences of PCR primers used to generate fusion proteins consisting of wild-type or mutant FIV integrase and EGFP with or without GST
| Primer name | Primer sequencea | Restriction enzyme |
|---|---|---|
| F-IN5′EcoR | 5′-CTCGAATTCGCCACCATGGGATCCTCTTGGGTTGACAGAATT-3′ | EcoRI |
| 3′FIV-INL-AgeI | 5′-CTCACCGGTGCGCCAGCACCAGCACCAGCGCC-3′ | AgeI |
| FINΔC3′Kpn | 5′-TGGTCCGGTACCTTTCTTATCTTTTTGATCTTTATAAT-3′ | KpnI |
| F-INΔN5′Eco | 5′-CTCGAATTCGCCACCATGGGAGGACAATTGAAAATAGG-3′ | EcoRI |
| FIV-IN3′Kpn | 5′-CAGTCAGGTACCCTCATCCCCTTCAGG-3′ | KpnI |
| GST5′Kpnl | 5′-TCGGGTACCGGCGCGGGTGCTGGAGCAGGAGCAATGTCCCCTATACTAG-3′ | KpnI |
| GST3′KpnI | 5′-TCGGGTACCTTTTGGAGGATGGTCGCCACCA-3′ | KpnI |
| FINΔC3′Kpn6 | 5′-TGGTCCGGTACCTACCACATCTTTTTGATCTTTATAAT-3′ | KpnI |
| FINΔC3′Kpn7 | 5′-TGGTCCGGTACCTACCACATCTGCTTGATCTGCATAATAAATCCACTG-3′ | KpnI |
| FINΔC3′Kpn3 | 5′-CCTCCGGTACCTTCTCCTCTGATTCTGCATAC-3′ | KpnI |
| FINΔC3′Kpn5 | 5′-ATTGCGGTACCATAATCTTGTATTCTTAAGG-3′ | KpnI |
| F-C42/45Has | 5′-CCTCTGATTCTGTGTACTGGGTGTTTTCGTCTTATC-3′ | |
| F-C42/45Hss | 5′-AAACACCCAGTACACAGAATCAGAGGAGAA-3′ | |
| 3ΔN213LNK | 5′-TGCAGAAAAATATGCGCCAGCACCAGCACCAGCGCC-3′ | |
| 5LNKΔN213 | 5′-GCTGGTGCTGGCGCATATTTTTCTGCAATACCACAAAAATTG-3′ |
Restriction sites are underlined, and nucleotide substitutions for the desired codon changes are identified in bold type.
To generate the fusion protein consisting of EGFP and the C-terminus-truncated FIV integrase 1-235, the FIV integrase gene was amplified with F-IN5′EcoR as the 5′ primer and FINΔC3′Kpn as the 3′ primer, which anneals to the codon for amino acid residue 235. PCR products generated were cloned into the TOPO TA vector, sequenced, and digested with EcoRI and KpnI. The EcoRI-KpnI-digested fragment was purified with the Qiagen gel extraction kit and ligated with pF-IN/EGFP previously digested with EcoRI and KpnI to form p1-235/EGFP. This digestion strategy takes advantage of a KpnI site at the 5′ end of the linker sequence between the DNA encoding FIV integrase and that encoding EGFP and allows the in-frame cloning of mutant FIV integrase sequences upstream of the 5′ end of the DNA encoding EGFP.
An N-terminus-truncated mutant of FIV integrase, residues 53 to 281, and an N- and C-terminus-truncated mutant, residues 53 to 235, were constructed with F-INΔN5′Eco as the 5′ primer for the amplification of the integrase gene at the position corresponding to residue 53 and FIV-IN3′Kpn and FINΔC3′Kpn as 3′ primers that anneal to the codons for amino acid residues 281 and 235, respectively. The PCR-amplified fragments were cloned into pF-IN/EGFP as described earlier for p1-235/EGFP.
Cloning of fusion proteins consisting of FIV integrase, GST, and EGFP.
To ensure that the size of each fusion protein was above the estimated limit for passive diffusion across the nuclear envelope, we added the gene encoding glutathione S-transferase (GST; 25 kDa) derived from Schistosoma japonicum to the fusion constructs. The GST gene was first amplified by PCR by using pGEX (Amersham Biosciences, Piscataway, N.J.) as the template and GST5′KpnI and GST3′KpnI as the 5′ and 3′ primers, respectively. The resulting PCR product encoded residues 1 to 218 of GST with the addition of eight alternating Gly and Ala residues at the 5′ end to serve as a linker between FIV integrase and GST. The PCR-amplified products were cloned into the TOPO TA vector, sequenced as previously described, and then digested with KpnI and purified. The GST fragment was cloned into each of the previously described pF-IN/EGFP constructs prepared by digestion with KpnI. This resulted in the in-frame insertion of GST between integrase and EGFP. Proper orientation of the GST fragment was confirmed by restriction analysis.
In our hands, the expression of the fusion proteins containing GST was improved when the pcDNA4 vector (Invitrogen Corp.) was used instead of the pEGFP-NI vector. Therefore, all mutant FIV integrases fused to GST and EGFP were cloned into pcDNA4 by digesting each of the fusion constructs cloned in pEGFP-NI with EcoRI and NotI. This digestion yielded an approximately 2-kbp DNA fragment encoding the FIV integrase variant fused to GST and EGFP. These fragments were purified and ligated with the pcDNA4 vector previously digested with EcoRI and NotI. Constructs containing FIV integrase fused to GST and EGFP are identified by the addition of g before the construct name.
The fusion constructs 1-235KV and 1-235KAV containing the mutant FIV integrases fused to GST and EGFP were made in a manner similar to that described for the C-terminus-truncated mutant 1-235, except a 3′ mutagenic primer that anneals to the codon for residue 235 of FIV integrase was used. The mutagenic primer FINΔC3′Kpn6 introduced nucleotide substitutions that changed Lys residues at positions 234 and 235 to Val (g1-235KV), whereas primer FINΔC3′Kpn7 introduced substitutions that changed Lys at positions 229 and 232 to Ala and Lys at positions 234 and 235 to Val (g1-235KAV). The PCR fragments were cloned and sequenced to confirm mutations and then digested with EcoRI and KpnI. The purified EcoRI-KpnI fragment was then cloned into the 5′ end of the GST-EGFP fusion construct in pcDNA4 prepared by digestion with EcoRI and KpnI.
To generate the C-terminus-truncated FIV integrase fusion constructs g1-50 and g1-213, the FIV integrase gene was amplified with F-IN5′EcoR as the 5′ primer and FINΔC3′Kpn3 (anneals to the codon for amino acid residue 50) and FINΔC3′Kpn5 (anneals to the codon for amino acid residue 213) as the 3′ primers, respectively. The PCR products generated were sequenced and cloned as described above for g1-235KV and g1-235KAV.
Mutations changing the Cys residues at positions 42 and 45 to His were also introduced into the full-length FIV integrase (g1-281CH) as well as the C-terminus-truncated mutant (g1-235CH), and the mutant integrases were fused to GST and EGFP. Both constructs were prepared by using an overlapping PCR (1) that generated 5′ and 3′ fragments encoding integrase, each encoding the Cys-to-His changes in the shared region of homology. In the first set of PCRs, the 5′ fragment was generated by using pF-IN/EGFP as the template and F-IN5′EcoR and F-C42/45Has as the 5′ and 3′ primers, respectively. The 3′ fragment was amplified by using F-C42/45Hss as the 5′ primer and FIV-IN3′Kpn and FINΔC3′Kpn as the 3′ primers for g1-281CH and g1-235CH, respectively. The two PCR products containing a common overlapping region were annealed together and extended in the presence of 1 mM deoxynucleoside triphosphates (United States Biochemicals) and 2.5 U of PFU polymerase (PFU Turbo; Stratagene). The extension reaction was carried out in the thermocycler (MJ Research, Inc.) programmed for 30 cycles, with each cycle consisting of 1 min of denaturation at 95°C, 2 min of annealing at 62°C, and 4 min of extension at 72°C. The extended products were subjected to another round of PCR with F-IN5′EcoR as the 5′ primer and FIV-IN3′Kpn and FINΔC3′Kpn as the 3′ primers for g1-281CH and g1-235CH, respectively. The PCR products were cloned in the TOPO TA vector and sequenced as described previously and then cloned as an EcoRI-KpnI fragment into the 5′ end of the GST-EGFP fusion construct in pcDNA4.
The mutant construct containing integrase residues 1 to 50 and 213 to 235 fused to GST and EGFP was made by using the aforementioned overlapping PCR method. In the first round of PCR, the construct g1-50 was used as the template and F-IN5′EcoR and 3ΔN213LNK were used as the 5′ and 3′ primers, respectively. The reaction generated a 5′ fragment that encodes residues 1 to 50 of integrase, followed by a flexible linker sequence and residues 213 to 216 of integrase at the 3′ end. Amplification of g1-235 with 5LNKΔN213 and GST3′KpnI as the 5′ and 3′ primers, respectively, generated a 3′ fragment. This fragment encodes part of the flexible linker at the 5′ end, followed by integrase residues 213 to 235 fused to GST with a flexible linker between the protein domains. The 5′ and 3′ fragments, which share homology across the linker sequences and the regions corresponding to residues 213 to 216 of integrase, were annealed and extended as described above for g1-281CH and g1-235CH. The extension product was then amplified with F-IN5′EcoR and GST3′KpnI as the 5′ and 3′ primers, respectively. The resulting PCR product was cloned into the TOPO TA vector as described previously and sequenced. The construct was subsequently inserted as an EcoRI-SwaI fragment into the gEGFP fusion construct previously prepared by digestion with EcoRI and SwaI.
Cell transfection.
HeLa-Tat, 293T, and Crendall feline kidney (CrFK) cells were grown as a monolayer on glass coverslips (22 mm2; Corning, Big Flats, N.Y.) in Dulbecco's modified Eagle medium (Mediatech, Inc., Herndon, Va.) containing 10% fetal bovine serum (HyClone, Logan, Utah), 100 U of penicillin/ml, and 0.1 mg of streptomycin (Mediatech)/ml. Typically, 6 × 105 cells were transferred onto a coverslip in 6-well plates. After 24 h or when the culture reached approximately 70% confluency, cells were transfected by using Polyfect (Qiagen) according to the manufacturer's protocol. Twenty-four hours after transfection, the medium was aspirated and coverslips were washed once with phosphate-buffered saline (PBS) and treated with a mild fixative solution (2% [vol/vol] formaldehyde and 0.05% [vol/vol] glutaraldehyde in PBS). To minimize photobleaching, coverslips were prepared for fluorescence microscopy by using the SlowFade Antifade kit (Molecular Probes, Eugene, Oreg.) and then mounted on a drop of 50% glycerol in PBS. Coverslip edges were sealed with nail polish.
Western blotting.
293T cells were grown in 6-well plates and transfected as described above. Twenty-four hours after transfection, the medium was aspirated from the wells and cells were washed once with PBS. Cells were dislodged from the well bottoms by a rinsing with 1.5 ml of fresh PBS, and cell suspensions were transferred to 1.5-ml centrifuge tubes and pelleted by spinning at 3,000 × g for 5 min. The supernatant fractions were aspirated, and pellets were transferred to dry ice for 10 min. Cell pellets were either stored at −80°C or lysed immediately by adding 100 μl of loading dye containing 10% sodium dodecyl sulfate (SDS), 0.5% bromophenol blue, and 5% β-mercaptoethanol and boiling for 10 min. Typically, 3 ×106 cells were collected per transfected sample. The lysate from the equivalent of approximately 1.5 × 105 cells was loaded onto a SDS-13% polyacrylamide gel for separation and then transferred to nitrocellulose membrane (pore size, 0.45 μm; Osmonics, Minnetonka, Minn.). Expressed fusion proteins were immunoblotted with a 1:1,000 dilution of mouse monoclonal antibody against EGFP (JL-8; Clontech) and a secondary goat anti-mouse horseradish peroxidase-conjugated antibody diluted at 1:50,000 (Zymed Laboratories). Detection was carried out with a horseradish peroxidase detection kit (Super Signal; Pierce, Rockford, Ill.) according to the manufacturer's instructions.
Fluorescence microscopy.
The subcellular localization of EGFP-containing fusion proteins in transfected cells was determined by fluorescence microscopy. Samples were evaluated for green fluorescence protein fluorescence on an Olympus IX-70 microscope stand, equipped for epifluorescence illumination from a 100-W mercury arc lamp, by using a standard fluorescein isothiocyanate filter set (528 ± 19-nm-wavelength emission filters and a polychroic mirror; excitation at 490 ± 10 nm) and a 60× PlanAPO oil immersion objective. Images were recorded on a liquid-cooled −40°C charge-coupled device camera (CH350L; Photometrics, Tucson, Ariz.) and analyzed with SoftWoRx software (Applied Precision Inc., Issaquah, Wash.).
RESULTS
FIV integrase is karyophilic.
To determine the karyophilic property of FIV integrase, we generated a plasmid DNA construct expressing full-length FIV integrase fused to the N terminus of EGFP, with an 8-amino-acid flexible linker separating the two protein domains (Fig. 1A, construct b). The DNA construct encoding either EGFP alone or FIV integrase fused to EGFP was transfected into HeLa-Tat cells. Twenty-four hours posttransfection, cells were analyzed by fluorescence microscopy for the subcellular localization of EGFP or the expressed integrase-EGFP fusion protein.
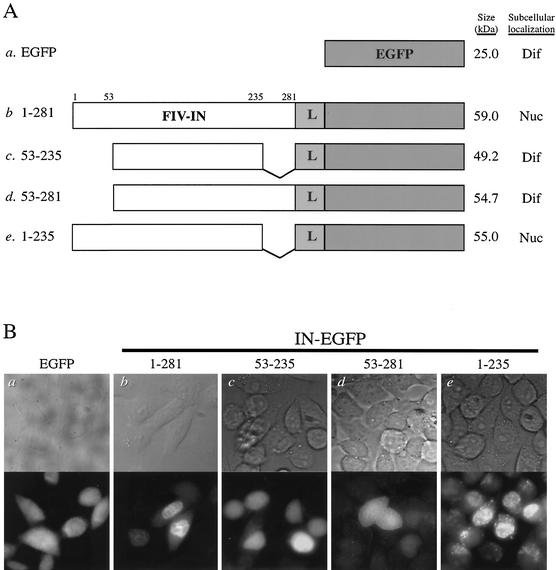
FIG. 1.
FIV integrase is karyophilic and targets EGFP fusion proteins to the nuclei of transfected cells. (A) DNA constructs of fusion proteins. Full-length wild-type (construct b) or truncated (constructs c to e) FIV integrases (FIV-IN; white boxes) were cloned into the pEGFP-NI plasmid and expressed as N-terminal fusion proteins with EGFP (gray boxes). An 8-amino-acid flexible linker with alternating Gly and Ala residues (L) was inserted between the two proteins. The N- and C-terminus-truncated mutant 53-235 (construct c), N-terminus-truncated mutant 53-281 (construct d), and C-terminus-truncated mutant 1-235 (construct e) were constructed based on biochemical and functional studies of FIV integrase (37). The amino acid positions for FIV integrase are given at the top. The expected molecular mass (in kilodaltons) and subcellular localization of each fusion protein are indicated to the right of the corresponding construct. Nuc indicates that fluorescence was localized predominantly to the nuclei of transfected cells, and Dif indicates that fluorescence was distributed evenly throughout transfected cells. A V-shaped line represents the region deleted from the sequence of FIV integrase. (B) Subcellular localization of fusion proteins consisting of wild-type or mutant derivatives of FIV integrase and EGFP. Briefly, 1.5 μg of the wild-type or mutant DNA constructs was individually transfected into approximately 4 × 105 HeLa-Tat cells by using the transfection reagent Polyfect (Qiagen). Twenty-four hours posttransfection, cells were fixed and the localization of the expressed fusion protein was visualized by fluorescence microscopy with a 60× oil immersion objective (images are shown at a magnification of ∼30×). Top panels are phase contrast images and bottom panels are fluorescence images showing the same fields selected for the indicated constructs to demonstrate representative fluorescent patterns. IN-EGFP, FIV integrase-EGFP fusion proteins.
Previous studies have shown that for proteins of <50 kDa, passive diffusion into the nucleus can occur via the aqueous channel of the nuclear pore complex (reviewed in reference 17). Consistent with this characteristic of the nuclear pore complex and previous work done with EGFP (for example, see reference 41), HeLa-Tat cells transfected with the plasmid expressing EGFP only (Fig. 1B, panel a) demonstrated a diffuse fluorescent pattern, indicating that EGFP, a 25-kDa protein, was able to diffuse freely between the nucleus and cytoplasm. In contrast, HeLa-Tat cells transfected with the plasmid expressing the fusion protein comprising full-length FIV integrase fused to EGFP demonstrated a nuclear fluorescent pattern (Fig. 1B, panel b). The result indicated that FIV integrase can target the fusion protein to the nuclei of transfected cells and showed that FIV integrase is karyophilic.
The karyophilic determinant of FIV integrase is located in the N-terminal domain.
Next, we sought to map the karyophilic determinant of FIV integrase by analyzing the subcellular localization of truncated variants of FIV integrase fused to EGFP. Biochemical and functional studies (36, 37) identified three discrete domains of FIV integrase: the N-terminal domain, comprising residues 1 to 52; the nonspecific DNA-binding domain, residues 236 to 281, in the C terminus; and the catalytic domain, located in the central region of the protein. We constructed mutant FIV integrases that represented N- or C-terminal deletion variants and fused them to the N terminus of EGFP (Fig. 1A, constructs c to e).
Previous studies examining the karyophilic property of HIV-1 integrase have implicated amino acid residues located in the core domain as important karyophilic determinants (2, 40). When the corresponding domain of FIV integrase (residues 53 to 235) was expressed as a fusion protein with EGFP (Fig. 1A, construct c), a diffuse fluorescent pattern was observed (Fig. 1B, panel c). The diffuse fluorescent signal was presumably a result of passive diffusion due to the size of the truncated fusion protein (49 kDa). However, the lack of nuclear localization of the fluorescent signal suggested that, unlike HIV-1 integrase, the karyophilic determinant for FIV integrase does not reside in this region. Another study based on HIV-1 integrase has suggested that stretches of basic amino acids, located in both the core and C-terminal domains, function as a bipartite NLS in HIV-1 integrase (16). For FIV integrase, however, the expression of residues 53 to 281 fused to EGFP (Fig. 1A, construct d) resulted in a diffuse fluorescent pattern (Fig. 1B, panel d), suggesting that the expression of the central catalytic domain together with the C terminus does not reconstitute the NLS of FIV integrase. In contrast, when the C-terminus-truncated FIV integrase (residues 1 to 235) was fused with EGFP (Fig. 1A, construct e), the expressed fusion protein demonstrated a nuclear fluorescent pattern similar to that of the fusion construct containing wild-type integrase (Fig. 1B, panels b and e). The results suggested that the karyophilic determinant of FIV integrase resides in the N-terminal domain.
Subcellular localization of mutant FIV integrases fused to GST and EGFP.
The diffuse fluorescent signals of the fusion constructs 53-235 and 53-281 suggested that the N-terminal deletion disrupted the karyophilic property of FIV integrase. However, the conclusion was based on the assumption that the sizes of the fusion proteins containing the truncated integrases were below the cutoff for passive diffusion across the nuclear envelope, thereby resulting in a diffuse fluorescent pattern. To confirm the mapping results and to eliminate the possible artifacts resulting from passive diffusion, we increased the size of the fusion proteins by cloning the gene encoding the 25-kDa GST from S. japonicum into the integrase-EGFP fusion constructs. The GST was inserted between FIV integrase and EGFP, with a flexible linker between each of the protein domains (Fig. 2A, constructs c to f). When fused to GST and EGFP, deletion mutants of FIV integrase incapable of nuclear import should be excluded from the nucleus based on size and demonstrate a cytoplasmic fluorescent pattern. The construct containing EGFP and GST only (Fig. 2A, construct a) served as a control for the nuclear exclusion of GST-containing fusion proteins that were not capable of nuclear import and showed the expected cytoplasmic fluorescent pattern (Fig. 2B, panel a). The GST-EGFP fusion construct containing the NLS sequence of the simian virus 40 (SV40) large T antigen (Fig. 2A, construct b) represented a positive control for the nuclear import of fusion proteins containing GST (Fig. 2B, panel b). The fluorescent signal of full-length wild-type FIV integrase expressed as a fusion protein with GST and EGFP (Fig. 2A, construct c) was localized predominantly in the nucleus (Fig. 2B, panel c), indicating that FIV integrase is capable of targeting this large heterologous protein to the nuclei of transfected cells and further supporting the conclusion that FIV integrase is karyophilic.
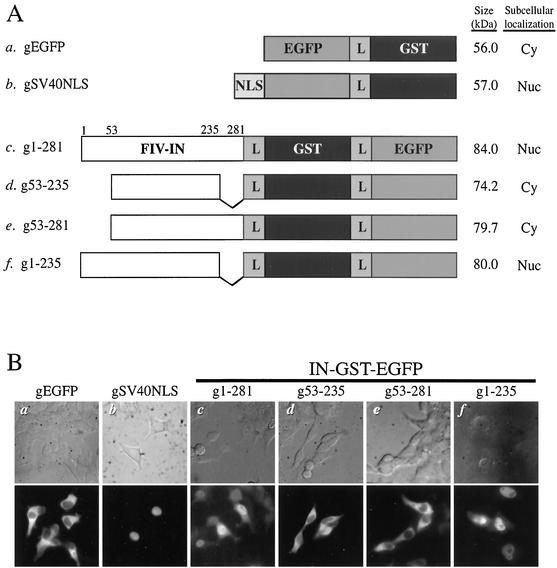
FIG. 2.
Subcellular localization of wild-type or truncated FIV integrase (FIV-IN) fused with GST and EGFP. (A) DNA constructs of fusion proteins. GST was added to the full-length wild-type integrase (construct c) or to deletion mutants of pF-IN/EGFP constructs (constructs d to f), with the 8-amino-acid flexible linker (L) inserted between the adjoining protein domains (for details, see Materials and Methods). The domains of integrase fused to GST and EGFP were as described in the legend for Fig. 1. The black and gray boxes indicate GST and the NLS of the SV40 large T antigen, respectively. The peptide containing the SV40 large T antigen NLS (MPKKKRKVEDPGT) was inserted upstream of EGFP at the EcoRI-KpnI restriction sites of pEGFP-N1. EGFP-GST fusion protein alone (construct a) and EGFP-GST fused with the SV40 large T antigen NLS (construct b) served as negative and positive controls, respectively, for subcellular localization. The subcellular localization of transfected fusion proteins is described as Nuc and Cy, indicating that fluorescence was localized primarily to the nuclei and cytoplasm of transfected cells, respectively. Numbers have the same connotation as those in Fig. 1. (B) Subcellular localization. Plasmid DNA (2 μg) encoding the various fusion constructs was individually transfected into approximately 6 × 105 293T cells by using the transfection reagent Polyfect (Qiagen). The remaining procedure was as described in the legend for Fig. 1. Top and bottom panels depict the phase contrast and fluorescence images, respectively, of the same fields. Images are shown at a magnification of ∼340×. IN-GST-EGFP, integrase-GST-EGFP fusion proteins.
When expressed as fusion proteins with GST and EGFP, the FIV integrase truncation variants 53-235 and 53-281 (Fig. 2A, constructs d and e, respectively) demonstrated fluorescent patterns restricted largely to the cytoplasm instead of the previously observed diffuse fluorescent patterns (compare Fig. 1B, panels c and d, with Fig. 2B, panels d and e). These results agree with our earlier conclusion that residues 53 to 281 are insufficient to target the fusion protein to the nucleus. Consistent with the results observed for the 1-235 fusion construct (Fig. 1B, panel e), the fluorescent signal of the g1-235 fusion protein (Fig. 2A, construct f) localized primarily to the nuclei of transfected cells (Fig. 2B, panel f). Together, these results further indicated that the karyophilic determinant for FIV integrase is not located in the central catalytic or C-terminal domain but is contained in the N-terminal domain.
Transfection assays were performed with both HeLa-Tat and 293T cell lines. For each construct, and under the described experimental conditions, the expression levels of the fusion proteins were higher in the 293T cell line. However, the subcellular localization patterns of the expressed fusion proteins were identical in both of these human cell lines.
Observed fluorescent signals correlate with expression of full-length fusion proteins.
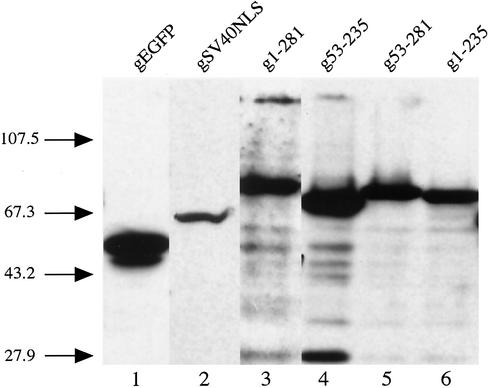
In addition to size, another possible explanation for the diffuse fluorescent signal, as seen in Fig. 1B, is protein instability. Degradation might destroy the NLS or decrease the size of the fusion protein, leading to a fluorescent signal that did not represent the direct effect of the mutation on the karyophilic property of FIV integrase. To ensure that degradation of the fusion proteins did not compromise our microscopy data, Western blot analysis was used to confirm that the observed fluorescent patterns correlated with the expression of the full-length fusion proteins (Fig. 3). Western blot analysis with a monoclonal antibody against EGFP detected full-length fusion proteins in lysates derived from cells transfected with either wild-type integrase (Fig. 3, lane 3) or one of the mutant integrases (Fig. 3, lanes 4 to 6). The majority of the expressed fusion proteins were intact, and the amount of degradation was minimal. The result verified that the observed fluorescent pattern indeed reflects the subcellular localization of the fusion protein and is not an artifact due to degradation.
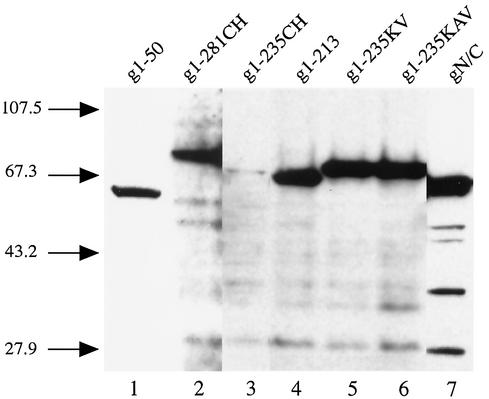
FIG. 3.
Western blot analysis of 293T cells expressing the various integrase-GST-EGFP fusion proteins. The lysate from approximately 1.5 × 105 293T cells transfected with 2 μg of DNA encoding the indicated fusion proteins was resolved by electrophoresis through a SDS-13% polyacrylamide gel at 150 V for approximately 2 h. After separation, the proteins were transferred to a nitrocellulose membrane and analyzed by immunoblotting with a monoclonal antibody against EGFP (Clontech). The numbers on the left indicate the molecular masses, in kilodaltons, of size markers (BRL).
The karyophilic determinant of FIV integrase maps to the N-terminal zinc-binding motif.
Because the results of our initial experiments using truncation mutants suggested that the karyophilic determinant of FIV integrase resides in the N-terminal domain, we sought to further characterize the role of the N terminus in nuclear import by preparing additional mutant FIV integrases as fusion proteins with GST and EGFP (Fig. 4A). To investigate whether the N-terminal domain alone could target the fusion protein to the nucleus, we made a construct comprising the N-terminal domain (residues 1 to 50) fused to GST and EGFP (Fig. 4A, construct a). The fluorescence pattern of this fusion protein was predominantly cytoplasmic (Fig. 4B, panel a), indicating that the N-terminal domain, although necessary, was by itself insufficient to direct the fusion protein to the nucleus. This result also suggested that the karyophilic determinant for FIV integrase is unlikely to be a simple signal sequence harbored in the N-terminal domain.
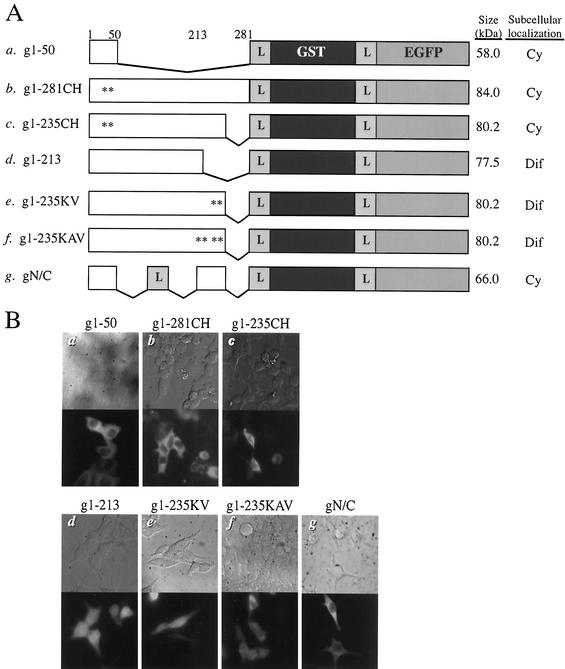
FIG. 4.
Role of the N-terminal zinc-binding domain and Lys residues within the region from 214 to 235 of FIV integrase in nuclear import. (A) DNA constructs. Mutant FIV integrases containing amino acid substitutions or deletions were fused with GST and EGFP and cloned into pcDNA4 (see Materials and Methods). The mutant derivatives of FIV integrase tested were as follows: g1-50, N-terminal domain (construct a); g1-281CH, full-length integrase containing Cys-to-His changes at positions 40 and 43 (construct b); g1-235CH, C-terminus-truncated mutant with the Cys-to-His changes (construct c); g1-213, C-terminus-truncated mutant (construct d); g1-235KV, C-terminus-truncated mutant with Lys-to-Val changes at positions 234 and 235 (construct e); g1-235KAV, C-terminus-truncated mutant with Lys-to-Val changes at positions 234 and 235 and Lys-to-Ala changes at positions 229 and 232 (construct f); and gN/C, bidomain mutant containing the N-terminal domain and residues 213 to 235 separated by the flexible linker (construct g). An asterisk indicates a single amino acid substitution, and other symbols and numbers have the same connotation as those in Fig. 1. (B) Subcellular localization. 293T cells were transfected with the indicated DNA constructs and analyzed as described in the legend for Fig. 2B.
The N terminus of integrase contains the zinc-binding HHCC motif that is highly conserved among retroviral integrases (34). For HIV-1 and murine leukemia virus integrases, the N-terminal HHCC motif promotes the multimerization of integrase upon coordination of zinc by the conserved His and Cys residues (24, 44, 46). The replacement of the Cys of the HHCC motif of FIV integrase (residues 42 and 45) with His abolishes the enzymatic activity of the protein (R. S. Appa and S. A. Chow, unpublished results). To examine the possible role of the zinc-binding motif in the nuclear import of FIV integrase, the Cys residues of the HHCC motif were changed to His in the context of full-length FIV integrase, g1-281CH, and its C-terminus-truncated mutant, g1-235CH (Fig. 4A, constructs b and c, respectively). In contrast to that of their respective wild-type counterparts, the expression of each of these fusion proteins resulted in a cytoplasmic fluorescent pattern, suggesting that the nuclear import of FIV integrase requires an intact zinc-binding domain (Fig. 4B, panels b and c).
FIV integrase contains an additional karyophilic determinant near the C terminus.
Based on the results observed for g1-281CH and g1-235CH, we concluded that the N-terminal zinc-binding domain plays a primary role in the nuclear import of the FIV integrase fusion proteins. However, the cytoplasmic fluorescent signals observed for the g1-50 fusion protein indicated that the zinc-binding domain alone does not constitute the karyophilic determinant for FIV integrase and that an additional determinant may exist elsewhere.
To map the additional determinant, we made the fusion construct g1-213 in which residues 214 to 281 of FIV integrase were deleted (Fig. 4A, construct d). We chose residues 214 to 235 because they are enriched with basic amino acids, a feature characteristic of canonical NLS (17). This region also corresponds to the distal sequence of the purported bipartite NLS identified for HIV-1 integrase (16). The presence of fluorescent signals in the nucleus indicated that the g1-213 variant is still karyophilic. However, a significant level of fluorescence was also observed in the cytoplasm, suggesting that the fusion protein was targeted to the nucleus in a less efficient manner than wild-type FIV integrase or the g1-235 mutant. Western blot analysis showed that the majority of the g1-213 was full-length and eliminated the possibility that the diffuse fluorescent pattern was a result of degradation (Fig. 5, lane 4).
FIG. 5.
Analysis of fusion proteins containing mutant FIV integrases and GST-EGFP by Western blotting. Separation of the cell lysate and immunoblotting with anti-EGFP monoclonal antibody were performed as described in the legend for Fig. 3. The identity of the fusion proteins is as labeled on top, and the numbers on the left are the sizes in kilodaltons of the molecular mass markers.
To further characterize the role of residues 214 to 235 in nuclear import, selected basic residues within this region of FIV integrase were mutated. Within the 214-235 region of FIV integrase, we targeted the 229KDQKDKK235 sequence for mutational analysis to determine if these basic residues might constitute an NLS sequence. The replacement of Lys residues 234 and 235 with Val in construct g1-235KV (Fig. 4A, construct e) and the replacement of Lys residues 229 and 232 with Ala, in addition to Val-for-Lys substitutions at positions 234 and 235, in construct g1-235KAV (Fig. 4A, construct f) had effects similar to that of the deletion mutant g1-213 and resulted in diffuse fluorescent signals (Fig. 4B, panels e and f). In the case of both g1-235KV and 1-235KAV, Western blot analysis ruled out the possibility that the observed fluorescent patterns were a result of degradation (Fig. 5, lanes 5 and 6, respectively). Although the presence of the Lys residues in the sequence 229KDQKDKK235 did not mediate the nuclear import of FIV integrase in the context of the zinc-binding domain mutants, g1-281CH and g1-235CH, the increased nuclear fluorescence for constructs g1-235KV and g1-235KAV suggested that these basic amino acid residues may function as an accessory karyophilic determinant and play a modulatory role in the nuclear import of FIV integrase.
Although the region containing residues 214 to 235 is not karyophilic by itself, it may function in conjunction with the N-terminal domain as a nonconventional, bipartite NLS in FIV integrase. To test this possibility, we generated a DNA construct encoding residues 1 to 50 and residues 214 to 235 separated by a flexible linker (Fig. 4A, construct g). When expressed as a fusion protein with GST and EGFP, this bidomain construct was unable to direct the fusion protein to the nuclei of transfected 293T cells (Fig. 4B, panel g). We therefore conclude that the zinc-binding domain and 214-235 region of FIV integrase do not function together as a bipartite NLS to mediate the nuclear import of FIV integrase.
Subcellular localization of fusion constructs expressed in feline cells.
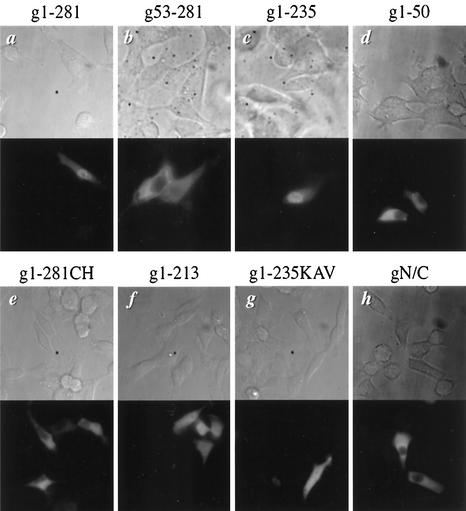
Both 293T and HeLa-Tat cells used in the previous experiments were derived from humans. To ensure that the results observed for the FIV integrase fusion proteins were not artifacts of the human cell system, we repeated the transfection experiments in a feline kidney cell line, CrFK. Consistent with the results obtained with human cells, full-length wild-type FIV integrase expressed as a fusion protein with GST and EGFP localized primarily to the nuclei of transfected CrFK cells (Fig. 6a), thereby offering further evidence that FIV integrase is karyophilic. Analysis of the fluorescent patterns demonstrated by the fusion constructs g53-281 and g1-235 expressed in CrFK cells corroborated our previous conclusion that the N terminus harbors the karyophilic determinant for FIV integrase. Truncation of the C-terminal domain (g1-235) did not affect the nuclear localization of the fusion protein (Fig. 6c), whereas the deletion of the N terminus prohibited the nuclear import of g53-281 in feline cells (Fig. 6b). Similar to that in human cells, the expression of g1-281CH in feline cells resulted in a cytoplasmic fluorescent pattern (Fig. 6e). These results underscore the importance of the N-terminal zinc-binding domain to the nuclear import of FIV integrase and suggest that similar mechanisms for the import of the protein are used in both feline and human cells.
FIG. 6.
Karyophilic property of FIV integrase and its mutant derivatives in feline cells. CrFK cells were transfected with the indicated DNA constructs and analyzed for distribution of fluorescent signals as described in the legend for Fig. 1. Images are shown at a magnification of ∼450×.
As in human cells, the N-terminal domain of FIV integrase by itself was insufficient to target the fusion protein to the nuclei in feline cells (Fig. 6d). Residues 214 to 235, near the C terminus, also played a role in the nuclear import of FIV integrase in CrFK cells. The expression of g1-213 and g1-235KAV in CrFK cells resulted in diffuse fluorescent patterns similar to those observed with human cells (compare Fig. 4B, panels d and f, with Fig. 6f and g). These results further suggested that these basic residues have a modulatory role in the nuclear import of FIV integrase. Also consistent with our observations in human cells was the result for the bidomain mutant, gN/C (Fig. 4A, construct g). When expressed in feline cells, gN/C failed to localize to the nuclei of transfected cells (Fig. 6h), indicating that the zinc-binding domain and the basic residues near the C terminus do not function as a bipartite NLS in feline cells.
DISCUSSION
We have demonstrated that FIV integrase is karyophilic based on its ability to localize a fusion protein to the nuclei of transfected human and feline cells. The karyophilic property of FIV integrase is consistent with the karyophilic property described for other retroviral integrases. The integrases of both HIV-1 and simian immunodeficiency virus can target heterologous fusion proteins to the nuclei of transfected cells (2, 7, 16, 30, 31, 40). Using a similar approach, avian sarcoma virus integrase fused with β-galactosidase also localizes in the nucleus (21, 22). Although the nuclear localization of several retroviral integrases has been described in the context of transfection assays, the identity of the NLS is not well defined and the mechanism of import remains poorly understood (2, 19, 30). In the case of HIV-1 integrase, a putative bipartite NLS comprising residues 186 to 188 and 211 to 219 has been identified and shown to facilitate interaction between integrase and karyopherin α (16). However, the function of these residues as an NLS and the involvement of karyopherins in the nuclear import of HIV-1 integrase have been refuted (6). In a separate study, V165 and R166 within a 13-residue peptide located in the core domain of HIV-1 integrase, but not bipartite, have been found to be critical for nuclear import (2). For avian sarcoma virus integrase, the karyophilic determinant has been proposed to be a noncanonical NLS that maps to basic amino acids located within residues 207 to 235 of the protein (22).
In this study, based on the subcellular localization of truncated and mutant variants of FIV integrase fused to GST and EGFP, we mapped the karyophilic determinant of FIV integrase to the highly conserved N-terminal zinc-binding motif. For full-length FIV integrase and its C-terminus-truncated variant, the replacement of two highly conserved Cys residues in the N-terminal zinc-binding domain (g1-281CH and g1-235CH, respectively) results in the nuclear exclusion of the fusion protein and indicates that the zinc-binding motif plays an essential role in the nuclear import of FIV integrase. However, the N terminus alone (g1-50) was insufficient to direct the fusion protein to the nucleus, suggesting that the karyophilic determinant is not a simple signal sequence harbored in the N-terminal domain.
In addition to the N terminus, we also analyzed the central catalytic and C-terminal domains of FIV integrase for their potential contribution to nuclear import. As discussed earlier, a putative bipartite NLS and a 13-residue peptide in the core domain of HIV-1 integrase have been proposed to function in nuclear import (2, 16). However, the central catalytic domain of FIV integrase is not karyophilic as determined by our fusion protein assay (Fig. 1 and 2). Interestingly, the noncanonical 13-residue NLS identified in the core domain of HIV-1 integrase is conserved among HIV-1, HIV-2, and simian immunodeficiency virus isolates (2) but not in FIV integrase. Like those of other retroviral integrases, the C terminus of FIV integrase is capable of binding DNA in a sequence-independent manner (37). Given its DNA-binding property, an alternative explanation for the nuclear localization of FIV integrase is the trapping of the protein in the nucleus as a result of DNA binding by the C-terminal domain. We think that this scenario is unlikely because of the nuclear fluorescent pattern demonstrated by the truncation variants 1-235 and g1-235, both of which lack the C-terminal DNA-binding domain. Although the C-terminus-truncated FIV integrase can still bind DNA, the binding affinity is greater than 20-fold less than that of the full-length protein (37). Furthermore, the DNA-binding property of the HIV-1 core domain is limited to the branched “Y-mer” disintegration substrate (11). Therefore, our results indicate that the nuclear import of FIV integrase does not depend on the C terminus and that trapping is not a likely mechanism by which the protein accumulates in the nucleus.
Because the N terminus of FIV integrase is not karyophilic per se, we postulated that the nuclear import of FIV integrase might require both the N-terminal zinc-binding domain and an additional region elsewhere in the protein. Since the fusion proteins without the last 50 amino acids of FIV integrase are karyophilic, we sought to identify the additional region by further truncating the C-terminal end of FIV integrase. The diffuse fluorescence observed for the mutant g1-213, compared to the nuclear fluorescence observed for mutants 1-235 and g1-235, suggests a role for residues 214 to 235 in the nuclear import of integrase. Further supporting the role of this region in nuclear import are the results obtained for mutants g1-235KV and g1-235KAV. Because basic amino acid residues such as Lys and Arg are known to constitute canonical NLS (17), we targeted the Lys residues in the 214-235 region for mutational analysis. For both mutant fusion proteins, the replacement of these Lys residues with Ala and Val led to the detection of fluorescence in both the nucleus and cytoplasm. The immunoblot analysis showed that the majority of the expressed fusion proteins were full-length, ruling out the possibility that the diffuse fluorescent patterns were a result of protein degradation. Since the sizes of g1-235KV and 1-235KAV (both 80 kDa) far exceed the cutoff limit for passive diffusion across the nuclear envelope, the presence of the mutant fusion proteins in the nucleus indicates that they are still capable of nuclear import. However, the increased fluorescent signals in the cytoplasm, in comparison to that of the wild type, suggests that the efficiency of the nuclear import of the fusion proteins containing the Lys mutations is reduced. Though these results underscore the importance of these basic residues in the nuclear import of FIV integrase, the lack of fluorescent signals in the nucleus for g1-281CH and g1-235CH indicates that in the absence of the zinc-binding motif, the Lys residues near the C terminus are insufficient to direct the nuclear import of the fusion protein. Therefore, we conclude that the role of the Lys residues in the nuclear import of FIV integrase is secondary to that of the zinc-binding motif.
We also tested the possibility that the N-terminal zinc-binding domain and the region consisting of residues 214 to 235 near the C terminus, each by itself incapable of mediating nuclear import, may together constitute a functional NLS. This is analogous to the bipartite NLS of nucleoplasmin, which consists of two noncontiguous clusters of basic amino acids (8). The cytoplasmic fluorescence demonstrated by the bidomain construct gN/C indicates that these two regions do not form a bipartite NLS to mediate the nuclear import of FIV integrase.
Our work identifies FIV integrase as an example of a protein whose karyophilic property is attributed to a zinc-binding motif. The NLS function depends on the integrity of the zinc-binding motif, as substitutions that alter the coordination of zinc disrupt the nuclear localization of the protein. For many zinc-binding proteins, the metal-coordination domain adopts a fingerlike structure and is involved in DNA binding (for a review, see reference 23). However, for HIV-1 integrase, the zinc-binding motif has a helix-turn-helix structure (5) that does not bind DNA but functions to promote multimerization of integrase monomers (24, 46). The amino acid sequences of the N-terminal domains of FIV and HIV-1 integrases share 40% identity (36). The N-terminal domains of the two integrases possess the same biochemical properties (37) and presumably have similar structures. Given the role of the zinc-binding domain in multimer formation, one possible explanation for the requirement of the HHCC motif in nuclear import is that FIV integrase is preferentially transported into the nucleus as a multimer. The assembly of higher-order multimers of integrase may be required for interaction with host proteins involved in nuclear import or for the stabilization of structural features of integrase critical for its karyophilic property. Alternatively, the nuclear import of FIV integrase is accomplished by a direct interaction between the HHCC motif and the cellular nuclear trafficking machinery.
According to the above model, it is possible that the basic residues located within the region from 214 to 235 may contribute to the stability of a structural feature involved in nuclear import. Such a role would explain why these residues alone do not constitute an NLS but their deletion or replacement interferes with the efficient nuclear import of the fusion protein and leads to a diffuse fluorescent pattern in our assay. Alternatively, the increased number of these sequences in the context of an integrase multimer may provide a stronger signal for a more efficient transport into the nucleus.
We also investigated the role of the N-terminal zinc-binding domain in the nuclear import of HIV-1 integrase. In contrast to FIV, HIV-1 integrase did not require an intact zinc-binding domain for nuclear import. When the conserved Cys residues in the N-terminal zinc-binding domain of HIV-1 integrase were replaced with His, the mutant was still able to target an EGFP fusion protein to the nuclei of transfected cells in a manner indistinguishable from that of the wild type (data not shown). Furthermore, the deletion of the entire N terminus (residues 1 to 50) of HIV-1 integrase still resulted in the nuclear localization of the fusion protein (data not shown). These results indicate that, for HIV-1 integrase, the N-terminal zinc-binding motif is not involved in mediating the nuclear import of the integrase. The result suggests that the mechanism for nuclear entry of HIV-1 integrase is different from that of FIV integrase. Alternatively, according to the model proposed for FIV integrase, the two integrases share the same mechanism but the zinc-binding motif of HIV-1 integrase either does not directly interact with cellular nuclear import factors or is not required to stabilize structural features of the integrase that are involved in nuclear import.
The model we have presented here for the nuclear import of FIV integrase requires a specific interaction between integrase and a host factor. It is therefore necessary to confirm the validity of our observations for feline cells. The facts that FIV integrase is imported into the nuclei in both feline and human cells and that in both cell types the import depends on the zinc-binding domain suggest that the mechanism of nuclear entry does not rely on import factors present only in feline cells. We hypothesize that FIV integrase interacts with yet-to-be-identified cellular factors that are conserved in both feline and human cells. Potential host candidates include members of the karyopherin or importin families of nuclear import factors, as well as components of the nucleoporin complex (for reviews, see references 17 and 28).
An increasing number of karyophilic proteins with their karyophilic determinants mapped to a zinc-binding domain have been characterized. In some cases, the structure of the zinc-binding domain has been separated from its NLS function. For example, two members of the lipid-dependent serine/threonine protein kinase C family that shuttle between the nucleus and cytoplasm, ξPKC and λPKC, require the N-terminal part of their zinc-finger domains for nuclear import but the NLS function of the zinc-binding domain can be separated from the integrity of the zinc-finger structure (29). Similarly, the karyophilic property of TIS11, a member of the CCCH zinc-finger family that functions as a positive transcriptional regulator, is maintained even when the Cys residues required for zinc binding are mutated to Ala (27). In contrast, some proteins seem to depend on the integrity of the zinc-finger structure for nuclear import. For the Krüppel-like transcription factor family, whose members include erythroid Krüppel-like factor (EKLF/KLF1) and RANTES factor of late activated T lymphocytes-1 (RFLAT-1/KLF13), the highly conserved zinc fingers common to this protein family act as strong NLS (32, 38). No canonical NLS sequence is present within the three zinc fingers of RFLAT-1, but the deletion of the last zinc finger significantly reduces the nuclear import of the protein. In the case of EKLF, all three zinc fingers are required for them to function as an NLS. In addition to the zinc-finger NLS, a canonical basic NLS is present in both RFLAT-1 and EKLF. However, in both instances the zinc-finger NLS is able to direct nuclear import in the absence of the canonical basic NLS. Our characterization of the nuclear import of FIV integrase adds another member to this growing family of proteins whose karyophilic property is closely tied to a zinc-binding domain.
One critical and salient feature of lentiviruses is their ability to infect nondividing and terminally differentiated cells. Some of the key questions include the mechanism of nuclear import and the role of integrase in enabling lentiviral particles to infect susceptible cells independent of the cell cycle phase. Further studies on the trafficking of FIV integrase and the subviral particle will shed light on the nuclear entry process of lentiviruses and identify host factors involved at this important step of viral replication.
Acknowledgments
We thank Xin Liu and Hong Wu for the use of a fluorescence microscope and Stephanie Betancourt and Hoorig Nassanian for technical assistance.
This work was supported by NIH grants CA68859 (S.A.C.) and GM053933 (W.J.D.), the Margaret E. Early Medical Research Trust (H.H.), and NIGMS predoctoral training grant GM08652 (C.L.W.).
REFERENCES
- 1.Ausubel, F. A., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1999. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Bouyac-Bertoia, M., J. D. Dvorin, R. A. Fouchier, Y. Jenkins, B. E. Meyer, L. I. Wu, M. Emerman, and M. H. Malim. 2001. HIV-1 infection requires a functional integrase NLS. Mol. Cell 7:1025-1035. [DOI] [PubMed] [Google Scholar]
- 3.Brown, P. O. 1997. Integration, p. 161-204. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 4.Bukrinsky, M. I., N. Sharova, M. P. Dempsey, T. L. Stanwick, A. G. Bukrinskaya, S. Haggerty, and M. Stevenson. 1992. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc. Natl. Acad. Sci. USA 89:6580-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai, M., R. Zheng, M. Caffrey, R. Craigie, G. M. Clore, and A. M. Gronenborn. 1997. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat. Struct. Biol. 4:567-577. [DOI] [PubMed] [Google Scholar]
- 6.Depienne, C., A. Mousnier, H. Leh, E. Le Rouzic, D. Dormont, S. Benichou, and C. Dargemont. 2001. Characterization of the nuclear import pathway for HIV-1 integrase. J. Biol. Chem. 276:18102-18107. [DOI] [PubMed] [Google Scholar]
- 7.Depienne, C., P. Roques, C. Creminon, L. Fritsch, R. Casseron, D. Dormont, C. Dargemont, and S. Benichou. 2000. Cellular distribution and karyophilic properties of matrix, integrase, and Vpr proteins from the human and simian immunodeficiency viruses. Exp. Cell Res. 260:387-395. [DOI] [PubMed] [Google Scholar]
- 8.Dingwall, C., and R. A. Laskey. 1998. Nuclear import: a tale of two sites. Curr. Biol. 8:R922-R924. [DOI] [PubMed]
- 9.Dow, S. W., M. J. Dreitz, and E. A. Hoover. 1992. Feline immunodeficiency virus neurotropism: evidence that astrocytes and microglia are the primary target cells. Vet. Immunol. Immunopathol. 35:23-35. [DOI] [PubMed] [Google Scholar]
- 10.Elder, J. H., G. A. Dean, E. A. Hoover, J. A. Hoxie, M. H. Malim, L. Mathes, J. C. Neil, T. W. North, E. Sparger, M. B. Tompkins, W. A. Tompkins, J. Yamamoto, N. Yuhki, N. C. Pedersen, and R. H. Miller. 1998. Lessons from the cat: feline immunodeficiency virus as a tool to develop intervention strategies against human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 14:797-801. [DOI] [PubMed] [Google Scholar]
- 11.Engelman, A., A. B. Hickman, and R. Craigie. 1994. The core and carboxy-terminal domains of the integrase protein of human immunodeficiency virus type 1 each contribute to nonspecific DNA binding. J. Virol. 68:5911-5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.English, R. V., C. M. Johnson, D. H. Gebhard, and M. B. Tompkins. 1993. In vivo lymphocyte tropism of feline immunodeficiency virus. J. Virol. 67:5175-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouchier, R. A., and M. H. Malim. 1999. Nuclear import of human immunodeficiency virus type-1 preintegration complexes. Adv. Virus Res. 52:275-299. [DOI] [PubMed] [Google Scholar]
- 14.Fouchier, R. A., B. E. Meyer, J. H. Simon, U. Fischer, and M. H. Malim. 1997. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 16:4531-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freed, E. O., G. Englund, F. Maldarelli, and M. A. Martin. 1997. Phosphorylation of residue 131 of HIV-1 matrix is not required for macrophage infection. Cell 88:171-174. [DOI] [PubMed] [Google Scholar]
- 16.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 18.Haffar, O. K., S. Popov, L. Dubrovsky, I. Agostini, H. Tang, T. Pushkarsky, S. G. Nadler, and M. Bukrinsky. 2000. Two nuclear localization signals in the HIV-1 matrix protein regulate nuclear import of the HIV-1 pre-integration complex. J. Mol. Biol. 299:359-368. [DOI] [PubMed] [Google Scholar]
- 19.Katz, R. A., J. G. Greger, K. Darby, P. Boimel, G. F. Rall, and A. M. Skalka. 2002. Transduction of interphase cells by avian sarcoma virus. J. Virol. 76:5422-5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kootstra, N. A., and H. Schuitemaker. 1999. Phenotype of HIV-1 lacking a functional nuclear localization signal in matrix protein of gag and Vpr is comparable to wild-type HIV-1 in primary macrophages. Virology 253:170-180. [DOI] [PubMed] [Google Scholar]
- 21.Kukolj, G., K. S. Jones, and A. M. Skalka. 1997. Subcellular localization of avian sarcoma virus and human immunodeficiency virus type 1 integrases. J. Virol. 71:843-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kukolj, G., R. A. Katz, and A. M. Skalka. 1998. Characterization of the nuclear localization signal in the avian sarcoma virus integrase. Gene 223:157-163. [DOI] [PubMed] [Google Scholar]
- 23.Laity, J. H., B. M. Lee, and P. E. Wright. 2001. Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11:39-46. [DOI] [PubMed] [Google Scholar]
- 24.Lee, S. P., J. Xiao, J. R. Knutson, M. S. Lewis, and M. K. Han. 1997. Zn2+ promotes the self-association of human immunodeficiency virus type-1 integrase in vitro. Biochemistry 36:173-180. [DOI] [PubMed] [Google Scholar]
- 25.Lewis, P., M. Hensel, and M. Emerman. 1992. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 11:3053-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis, P. F., and M. Emerman. 1994. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J. Virol. 68:510-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murata, T., Y. Yoshino, N. Morita, and N. Kaneda. 2002. Identification of nuclear import and export signals within the structure of the zinc finger protein TIS11. Biochem. Biophys. Res. Commun. 293:1242-1247. [DOI] [PubMed] [Google Scholar]
- 28.Nakielny, S., and G. Dreyfuss. 1999. Transport of proteins and RNAs in and out of the nucleus. Cell 99:677-690. [DOI] [PubMed] [Google Scholar]
- 29.Perander, M., G. Bjorkoy, and T. Johansen. 2001. Nuclear import and export signals enable rapid nucleocytoplasmic shuttling of the atypical protein kinase C lambda. J. Biol. Chem. 276:13015-13024. [DOI] [PubMed] [Google Scholar]
- 30.Petit, C., O. Schwartz, and F. Mammano. 2000. The karyophilic properties of human immunodeficiency virus type 1 integrase are not required for nuclear import of proviral DNA. J. Virol. 74:7119-7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pluymers, W., P. Cherepanov, D. Schols, E. De Clercq, and Z. Debyser. 1999. Nuclear localization of human immunodeficiency virus type 1 integrase expressed as a fusion protein with green fluorescent protein. Virology 258:327-332. [DOI] [PubMed] [Google Scholar]
- 32.Quadrini, K. J., and J. J. Bieker. 2002. Kruppel-like zinc fingers bind to nuclear import proteins and are required for efficient nuclear localization of erythroid Kruppel-like factor. J. Biol. Chem. 277:32243-32252. [DOI] [PubMed] [Google Scholar]
- 33.Reil, H., A. A. Bukovsky, H. R. Gelderblom, and H. G. Gottlinger. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 17:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rice, P., R. Craigie, and D. R. Davies. 1996. Retroviral integrases and their cousins. Curr. Opin. Struct. Biol. 6:76-83. [DOI] [PubMed] [Google Scholar]
- 35.Roe, T., T. C. Reynolds, G. Yu, and P. O. Brown. 1993. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 12:2099-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shibagaki, Y., and S. A. Chow. 1997. Central core domain of retroviral integrase is responsible for target site selection. J. Biol. Chem. 272:8361-8369. [DOI] [PubMed] [Google Scholar]
- 37.Shibagaki, Y., M. L. Holmes, R. S. Appa, and S. A. Chow. 1997. Characterization of feline immunodeficiency virus integrase and analysis of functional domains. Virology 230:1-10. [DOI] [PubMed] [Google Scholar]
- 38.Song, A., A. Patel, K. Thamatrakoln, C. Liu, D. Feng, C. Clayberger, and A. M. Krensky. 2002. Functional domains and DNA-binding sequences of RFLAT-1/KLF13, a Kruppel-like transcription factor of activated T lymphocytes. J. Biol. Chem. 277:30055-30065. [DOI] [PubMed] [Google Scholar]
- 39.Talbott, R. L., E. E. Sparger, K. M. Lovelace, W. M. Fitch, N. C. Pedersen, P. A. Luciw, and J. H. Elder. 1989. Nucleotide sequence and genomic organization of feline immunodeficiency virus. Proc. Natl. Acad. Sci. USA 86:5743-5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsurutani, N., M. Kubo, Y. Maeda, T. Ohashi, N. Yamamoto, M. Kannagi, and T. Masuda. 2000. Identification of critical amino acid residues in human immunodeficiency virus type 1 IN required for efficient proviral DNA formation at steps prior to integration in dividing and nondividing cells. J. Virol. 74:4795-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wachsmuth, M., W. Waldeck, and J. Langowski. 2000. Anomalous diffusion of fluorescent probes inside living cell nuclei investigated by spatially-resolved fluorescence correlation spectroscopy. J. Mol. Biol. 298:677-689. [DOI] [PubMed] [Google Scholar]
- 42.Wei, S. Q., K. Mizuuchi, and R. Craigie. 1997. A large nucleoprotein assembly at the ends of the viral DNA mediates retroviral DNA integration. EMBO J. 16:7511-7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinberg, J. B., T. J. Matthews, B. R. Cullen, and M. H. Malim. 1991. Productive human immunodeficiency virus type 1 (HIV-1) infection of nonproliferating human monocytes. J. Exp. Med. 174:1477-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang, F., O. Leon, N. J. Greenfield, and M. J. Roth. 1999. Functional interactions of the HHCC domain of Moloney murine leukemia virus integrase revealed by nonoverlapping complementation and zinc-dependent dimerization. J. Virol. 73:1809-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zennou, V., C. Petit, D. Guetard, U. Nerhbass, L. Montagnier, and P. Charneau. 2000. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101:173-185. [DOI] [PubMed] [Google Scholar]
- 46.Zheng, R., T. M. Jenkins, and R. Craigie. 1996. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc. Natl. Acad. Sci. USA 93:13659-13664. [DOI] [PMC free article] [PubMed] [Google Scholar]