Abstract
The known envelope membrane proteins of the chloroplastic protein import apparatus lack sequence similarity to proteins of other eukaryotic or prokaryotic protein transport systems. However, we detected a putative homolog of the gene encoding Toc75, the protein-translocating channel from the outer envelope membrane of pea chloroplasts, in the genome of the cyanobacterium Synechocystis sp. PCC 6803. We investigated whether the low sequence identity of 21% reflects a structural and functional relationship between the two proteins. We provide evidence that the cyanobacterial protein is also localized in the outer membrane. From this information and the similarity of the predicted secondary structures, we conclude that Toc75 and the cyanobacterial protein, referred to as SynToc75, are structural homologs. synToc75 is essential, as homozygous null mutants were not recovered after directed mutagenesis. Sequence analysis indicates that SynToc75 belongs to a family of outer membrane proteins from Gram-negative bacteria whose function is not yet known. However, we demonstrate that these proteins are related to a specific group of prokaryotic secretion channels that transfer virulence factors, such as hemolysins and adhesins, across the outer membrane.
Keywords: chloroplasts, Synechocystis, protein import
About 100 years ago, Schimper (1) and Mereschowsky (2) proposed that plant cells developed from colorless hosts that engulfed cyanobacteria, which subsequently evolved to plastids. The endosymbiotic origin of plastids is now widely accepted (3), although some details remain unresolved. Subsequent to endosymbiosis, many cyanobacterial genes were transferred to the host nucleus (4), requiring an envelope machinery for transporting the gene products back into the endosymbiotic organelle. Details of how this protein transport machinery evolved are unclear. Most chloroplastic proteins of higher plants are targeted to chloroplasts by a cleavable N-terminal transit sequence (5) and are imported posttranslationally into the organelle. The chloroplastic protein import apparatus consists of at least three outer membrane proteins (OMPs), Toc86, Toc75, and Toc34 (6–10), four inner membrane proteins, Tic110, Tic55, Tic22, and Tic20 (11–15), and one stromal molecular chaperone, ClpC (16, 17). Toc75 is an integral membrane protein that has been postulated to form the protein-translocating channel of the outer membrane translocon (10). This hypothesis is supported by electrophysiological experiments demonstrating that Toc75 forms a relatively small, ion-permeable channel (18).
Neither Toc75 nor other membrane proteins of the two chloroplastic translocons show sequence similarity to known components of functionally similar protein transport systems from mitochondria, the endoplasmic reticulum, peroxisomes, or Gram-negative bacteria, including cyanobacteria. However, genome sequencing of the cyanobacterium Synechocystis sp. PCC 6803 (19) allowed the identification of putative cyanobacterial homologs of Toc75 [Slr1227, E value = 2⋅10−34 (E value reports the number of hits expected to be found by chance)]. Tic55 (Slr1747, 7⋅10−34), Tic20 (Sll1737, 4⋅10−7), Tic22 (Slr0924, 6⋅10−5), and several possible candidates for Toc34 (E values ≥ 1⋅10−3), all of which are open reading frames (ORFs) of unknown function. In this report, we provide evidence that ORF slr1227 encodes a structural and potential functional homolog of Toc75.
MATERIALS AND METHODS
Chemicals.
DNA primers were synthesized at the Michigan State University Macromolecular Facility. The polyclonal antibodies against PsaD, SomA, and NrtA were gifts from L. McIntosh (Michigan State University), T. Mizuno (Nagoya University), and L. Sherman (Purdue University), respectively. RSF1010-based plasmid pRL1342 and Escherichia coli strain J53 (RP4) were kindly provided by C. P. Wolk (Michigan State University).
Cloning of synToc75.
A 4501-bp product containing the synToc75 (slr1227) ORF as well as 950 bp of the upstream and 965 bp of the downstream flanking sequences was amplified from genomic DNA of Synechocystis sp. PCC 6803 by PCR, while creating two unique terminal XhoI restriction sites. The PCR product was inserted into the XhoI site of plasmid pBSII SK (+) (Stratagene), yielding pBSsynToc75. The plasmid was cut with BamHI (bp 653 of the 4501-bp fragment) and XhoI, and the 3848-bp fragment was inserted into pRL1342, yielding pRL1342synToc75.
Generation of Antibodies Recognizing SynToc75.
The 1038-bp ScaI/SnaBI fragment encoding nucleotides 742-1786 of the synToc75 ORF was inserted into the SmaI site of the glutathione S-transferase gene fusion vector pGEX2X (Pharmacia). Expression and purification of the resulting fusion protein and immunization of rabbits were performed as described previously (17).
Creation of Plasmids for Directed Disruption of synToc75.
Insertional mutagenesis vector pBSsynToc75-kanr was created by inserting, in either direction, the HincII fragment (1252 bp) of the kanamycin-resistance gene (kanr), including its promoter region from plasmid pUC4K (Pharmacia) into the SnaBI site 1783 bp downstream from the start of the synToc75 ORF of pBSsynToc75. ORF replacement vector pBSΔsynToc75-kanrORF was created by replacing the synToc75 ORF with the kanr ORF. The kanr ORF from pUK4K was amplified by PCR, while changing the three bases immediately upstream of the ATG start codon to CAT to create an NdeI site and adding the 39-bp region downstream from the synToc75 ORF stop codon after the stop codon of the kanr ORF. This PCR product was ligated into the EcoRV site of pBSII SK (+). The resulting plasmid was cut with HpaI and EcoRI and ligated with a HpaI/EcoRI fragment from pBSsynToc75, containing the 3′ synToc75 flanking sequence (926 bp). The resulting plasmid was cut with XhoI and NdeI and then ligated with a PCR product containing the 5′ flanking sequence of synToc75. This PCR product was generated from pBSsynToc75, while creating an NdeI site at the ORF start.
Separation of Cyanobacterial Membranes by Sucrose Density Gradient Fractionation.
Cyanobacterial membranes were separated according to Omata and Murata (20) with the modification that all fractions subsequent to cell lysis contained 0.7 mM phenylmethanesulfonyl fluoride (PMSF) and 1 μg⋅ml−1 each of aprotinin, pepstatin, leupeptin, and ɛ-aminocaproic acid. Synechocystis sp. PCC 6803 cells grown mixotrophically (21) were lysed in a French press and separated from unbroken cells by low-speed centrifugation, and the supernatant was loaded on a discontinuous sucrose gradient (20). The fractions of subcellular membranes were collected at the interphases of the sucrose fractions, diluted, collected by ultracentrifugation (1 hr at 140,000 × g, Beckman SW-28), and resuspended in SDS/PAGE loading buffer (22). Equal-sized portions of each fraction were analyzed. To investigate whether SynToc75 is a membrane protein, a portion of the supernatant from the first centrifugation (80 μg of protein) was diluted (≥1:5) in 20 mM Tes buffer, pH 7.0, and centrifuged (30 min at 200,000 × g, Sorvall AT-2). The supernatant and pellet fractions were loaded onto the gel by equal portions. For NaCl and Na2CO3 treatment, outer membranes (20 μg of protein) were incubated in 1.0 M NaCl (in Tes buffer) or 100 mM Na2CO3, pH 11.5 (30 min on ice). Soluble and membrane-associated proteins were separated by ultracentrifugation (1 hr at 200,000 × g, Sorvall AT-2), and the fractions were analyzed in equal-sized portions.
The amount of protein was determined according to Lowry et al. (23). Proteins of the soluble fraction were precipitated with chloroform/methanol (24). The proteins were separated on an SDS/7.5–15% polyacrylamide gradient gel (22) and transferred to poly(vinylidene difluoride) (PVDF) membrane (Immobilon-P, Millipore), which was subsequently incubated with polyclonal antibodies raised against SynToc75, SomA (25), NrtA [= CbpA (26)], or PsaD (27). Primary antibodies were detected with horseradish peroxidase-conjugated goat anti-rabbit or anti-mouse sera and enhanced chemiluminescence (Pierce).
Transformation of Synechocystis sp. PCC 6803 and Serial Passaging.
Plasmid DNA (1 μg) was added to 500 μl of photoautotrophically grown Synechocystis sp. PCC 6803 at early logarithmic phase. After 48 hr, an aliquot (100 μl) was streaked on BG-11 plates containing 5 μg⋅ml−1 kanamycin (21). For mixotrophic growth, 5 mM glucose was added. A colony was transferred to a new plate after 10–14 days. For isolation of genomic DNA, cells were suspended in 50 ml of BG-11 medium containing 5 μg⋅ml−1 kanamycin and grown to late logarithmic phase. Bacteria that were transformed with a plasmid containing psaC-kanr were grown under photoautotrophic or light-activated heterotrophic growth conditions (27).
Plasmid pRL1342synToc75 (12.2 kbp) was introduced into Synechocystis sp. PCC 6803 that had a partially segregated synToc75-genotype by triparental mating using the broad-host-range conjugal plasmid RP4. After being washed, a few microliters of a suspension culture of E. coli strain DH5α with pRL1342-synToc75, E. coli strain J53 (RP4), and the Synechocystis transformants were mixed and spotted on a filter placed on top of plates of BG-11 agar that lacked antibiotics and had been overlaid with 5% Luria–Bertani (LB) agar. After 24 hr the filters were transferred to BG-11 plates supplemented with 5 μg⋅ml−1 each of kanamycin and erythromycin. Single colonies were picked after 13 days and grown in suspension cultures in the presence of 10 μg⋅ml−1 kanamycin and 5 μg⋅ml−1 erythromycin.
DNA Gel Blot Analysis.
Genomic DNA was isolated by the method of Chrisholm (28). Equal amounts of genomic DNA (about 1 μg) were digested by EcoRI, and the DNA fragments were fractionated on a 0.8% (wt/vol) agarose gel and transferred to a nitrocellulose membrane (Hybond N+, Amersham) by capillary blotting. The membrane was processed according to the manufacturer’s protocol and hybridized under stringent conditions (65°C, 5× SSC) with a digoxigenin-labeled probe (Boehringer Mannheim) corresponding to the 3′ and 5′ flanking regions of synToc75.
Sequence Analysis.
The nonredundant database at the National Center for Biotechnology Information was searched for similar proteins by using blast 2.0 (Gapped Blast) or psi-blast (blosum62, gap penalties: existence, 11; extension, 1) (http://www.ncbi.nlm.nih.gov/BLAST/) (29). In the blast search with SynToc75 as a query, the E values and GenBank accession numbers of the D15-related proteins are as follows: OMP1 of Brucella abortus (2⋅10−22; U51683), OMP85 of Neisseria meningitis (5⋅10−13; AF021245), OMP of Neisseria gonorrhoeae (1⋅10−12; U81959), OMP of Aquifex aeolicus (3⋅10−11; AE000733), unknown protein of E. coli (6⋅10−9; Swiss-Prot P39170), putative OMP of Treponema pallidum (8⋅10−9; AF042789), D15_1 of Haemophilus influenzae (4⋅10−6; Swiss-Prot P46024), OMP of Helicobacter pylori (8⋅10−5; AE000579), Oma87 of Pasteurella multocida (1⋅10−3; U60439), OMP of Borrelia burgdorferi (3⋅10−3; AE001178), and OMP85 analog of Chlamydia trachomatis (6⋅10−3; AE001297). In the psi-blast search with SynToc75 as a query, five iterations with an E value threshold of 1⋅10−3 were performed, after which the detected proteins converged. In the psi-blast search with the virulence factors, ShlA, EthA, HpmB, HhdA, FHA, and HecA were included. Overall sequence identity and similarity were determined with gap from the Genetics Computer Group (Wisconsin package version 9.0). The complete sequence alignment is published as supplemental data on the PNAS web site (www.pnas.org). Binary sequence alignment of pea Toc75 and SynToc75 (blosum62; gap penalties: existence, 11; extension, 1) was performed by using sim at Expasy (http://expasy.hcuge.ch/sprot/sim-prot.html) (30). Multiple sequence alignment (blosum62, gap penalties: existence, 12; extension, 2) was performed by using multalin at the Institut National de la Recherche Agronomique (http://www.toulouse.inra.fr/multalin.html) (31). Motif specificity was determined by using PatternFind at the Institut Suisse de Recherche Experimentale sur le Cancer (http://www.isrec.isb-sib.ch/software/PATFND_form.html) (32). Protein targeting was analyzed by using psort (http://www.imcb.osaka-u.ac.jp/nakai/psort.html) (33) and SignalP (http://www.cbs.dtu.dk/services/SignalP/) (34). Secondary structure was analyzed with TopPred II (35) and PhD (http://www.embl-heidelberg.de/predictprotein/predictprotein.html) (36).
RESULTS AND DISCUSSION
Toc75 and Slr1227 Are Structurally Homologous.
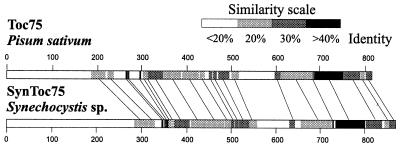
Pea Toc75 and the cyanobacterial protein encoded by slr1227 share 21% overall sequence identity (24% identity and 41% similarity between amino acids 187–809 of Toc75, 14% gap frequency). They can be aligned over the entire sequence of the two proteins, which are similar in size (Toc75 precursor contains 809 amino acids; Slr1227, 861). The regions of highest sequence similarity are located in the middle and the C-terminal regions (Fig. 1). Toc75 is targeted to the chloroplastic outer envelope by a bipartite cleavable transit peptide (37); Slr1227 is predicted to contain an N-terminal cleavable signal sequence of 30–32 amino acids. As with Toc75, secondary structure prediction for Slr1227 indicates a high number of amphiphilic β-strands, especially in the C-terminal part, suggesting a transport function similar to that of Toc75. However, polyclonal antibodies against Slr1227 crossreact only weakly with pea Toc75 (data not shown).
Figure 1.
Schematic representation of the sequence alignment between pea Toc75 and SynToc75 (Slr1227) from Synechocystis sp. PCC 6803. The degree of sequence similarity between certain segments of the proteins is indicated by gray shading.
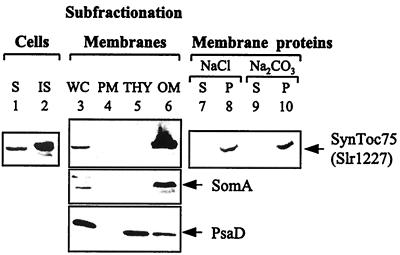
The chloroplast outer membrane is thought to have evolved, at least in part, from the outer membrane of the bacterial symbiont (38–40). Thus, a cyanobacterial homolog of Toc75 is expected to be located in the outer membrane of the Gram-negative bacterium. We determined experimentally the subcellular location of Slr1227 by fractionation of membranes from Synechocystis sp. PCC 6803, by using sucrose density gradients (20), followed by immunolocalization with polyclonal antibodies raised against a heterologously expressed fusion protein of Slr1227. After cell lysis, a single crossreacting protein of a molecular mass of approximately 93 kDa was detected in the fraction containing total cell membranes (Fig. 2, lane 2), indicating that Slr1227 is membrane associated. The small amount of crossreacting protein in the soluble fraction (Fig. 2, lane 1) probably represents small membrane vesicles that were not sedimented. The apparent molecular mass of Slr1227 of 93 kDa is close to that calculated for the predicted mature protein (89 kDa).
Figure 2.
Localization of SynToc75 (Slr1227) in the outer membrane of Synechocystis sp. PCC 6803. After cell disruption, the cell components were fractionated either by ultracentrifugation into soluble (S) and insoluble (IS) fractions or by sucrose density gradients (20) into remaining whole cells (WC), plasma membranes (PM), thylakoids (THY), and outer membranes (OM). The outer membrane fraction was incubated in 1.0 M NaCl or 0.1 M Na2CO3, pH 11.5, and soluble (S) and pellet fractions (P) were separated by ultracentrifugation. Proteins in each fraction were analyzed by SDS/PAGE and immunoblotting. SomA and PsaD were used as marker proteins for outer and thylakoid membranes, respectively.
Fractionated cyanobacterial membranes were analyzed with polyclonal antibodies against marker proteins, namely the 57-kDa OMP of Synechococcus sp. PCC 7942, SomA (25), the 15.5-kDa thylakoid membrane protein of Nostoc sp. PCC 8009, PsaD (27) (Fig. 2, lanes 3–6), and the 42-kDa plasma membrane protein of Synechococcus sp. PCC 7942, NrtA (26) (data not shown). Analysis of the markers showed that OMPs were highly enriched in the fraction designated as outer membranes and absent from the fractions containing plasma and thylakoid membranes. Thylakoid membrane proteins were concentrated in the fraction designated as thylakoid membrane with some contamination of the outer membrane fraction. Plasma membrane proteins were about equally distributed in all fractions (data not shown), as reported earlier (41); however, they were the only membrane proteins found in the fraction designated as plasma membrane. The distribution of Slr1227 paralleled that of the outer membrane marker SomA (Fig. 2, lanes 3–6). Furthermore, Slr1227 was not extracted from the membrane by NaCl or Na2CO3 (Fig. 2, lanes 8 and 10). From these results, we conclude that Slr1227 is an integral protein of the outer membrane of Synechocystis sp. PCC 6803. Soll and colleagues have independently reached a similar conclusion (42). On the basis of similar primary structure, similar actual and predicted protein properties, and analogous localization of Toc75 and Slr1227, we conclude that these two proteins are structurally homologous and refer to the cyanobacterial homolog as SynToc75.
Mutagenesis of SynToc75.
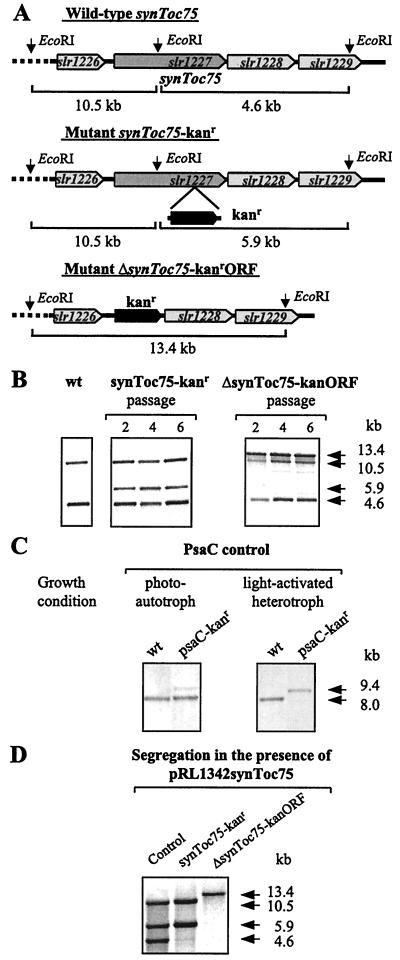
We attempted to assess the function of SynToc75 genetically by the generation of null mutants based on gene replacement through homologous recombination. A plasmid copy of wild-type synToc75 was either disrupted by insertion of the kanamycin-resistance gene or replaced with the ORF of the resistance gene and used to transform Synechocystis sp. PCC 6803 (Fig. 3A). Transcription of synToc75 as a monocistronic message was verified by RNA gel blot analysis (data not shown). Because Synechocystis sp. PCC 6803 maintains about 12 genome copies per cell (43), segregation of mutant and wild-type genomes was determined by DNA gel blot analysis after several passages. In transformed cells grown photoautotrophically, neither type of gene disruption caused disappearance of the wild-type gene even after six passages (Fig. 3B). Nonessential wild-type genes segregate after a few passages (21, 44), as confirmed for a disruption of psaC of transformants grown under light-activated heterotrophic growth conditions (Fig. 3C; ref. 27). Mixotrophically grown synToc75 mutants also retained a similar high content of wild-type synToc75 (data not shown), excluding the possibility of a specific function of SynToc75 in photoautotrophic growth. An increase of the selection pressure by raising the kanamycin concentration (50 μg⋅ml−1) did not reduce the proportion of wild-type synToc75 (data not shown).
Figure 3.
Generation of synToc75 (slr1227) mutants. (A) The wild-type gene synToc75 (slr1227) of Synechocystis sp. PCC 6803 was disrupted either by an insertion of the kanamycin-resistance gene, kanr, into synToc75 (synToc75-kanr) or by a replacement of the synToc75 ORF by the kanr ORF (ΔsynToc75-kanrORF). Only the data obtained for the sense orientation of the kanr insertion are shown (synToc75-kanr). (B) Segregation of wild-type synToc75 was analyzed by DNA gel blotting. Transformed cells were grown under photoautotrophic conditions and transferred to new plates for the number of passages indicated. (C) As a control for genome segregation, wild-type cells were transformed with a plasmid copy of psaC, containing an insertion of the kanr gene, increasing the size of the EcoRI fragment from 8.0 kb (wild type, wt) to 9.4 kb. Transformants were grown under photoautotrophic or light-activated heterotrophic conditions; data of the sixth and the third passage, respectively, are presented. (D) In the presence of pRL1342-encoded SynToc75 and selection with erythro- mycin and kanamycin, the chromosomal wild-type copies of synToc75 disappeared, as indicated by the absence of the 4.6-kb EcoRI fragment (synToc75-kanr) and also the 10.5-kb fragment (ΔsynToc75-kanrORF). The control (synToc75-kanr) shows the presence of chromosomal wild-type and mutant synToc75. Only data for photoautotrophically grown Synechocystis transformants are presented.
To investigate a possible position effect of the mutagenesis on the expression of essential genes located up- or downstream of synToc75, wild-type synToc75 was cloned into the self-replicating plasmid pRL1342 and transferred into Synechocystis transformants by conjugation. In the presence of plasmid-localized synToc75 and selection for the marker interrupting the chromosomal copy, the chromosomal wild-type copies of synToc75 disappeared, as indicated by the absence of the 4.6-kb EcoRI fragment in insertion mutants (synToc75-kanr) and the absence of the 4.6- and 10.5-kb fragments in ORF replacement mutants (ΔsynToc75-kanrORF, Fig. 3D). Complementation of the synToc75 mutants by the plasmid copy of the wild-type gene confirmed that synToc75 is essential in Synechocystis sp. PCC 6803.
Most OMPs only provide growth advantages for the prokaryote rather than being essential for growth per se. Thus, the most likely explanations for the essentiality of SynToc75 are that it mediates the secretion of either a required surface protein or a protein whose cytoplasmic accumulation is lethal. The latter effect has been observed with PilE in PilC-insertional mutants deficient in pili assembly (45) and is even more plausible with channel proteins secreting virulence factors (see below).
SynToc75 Belongs to a Family of Prokaryotic OMPs.
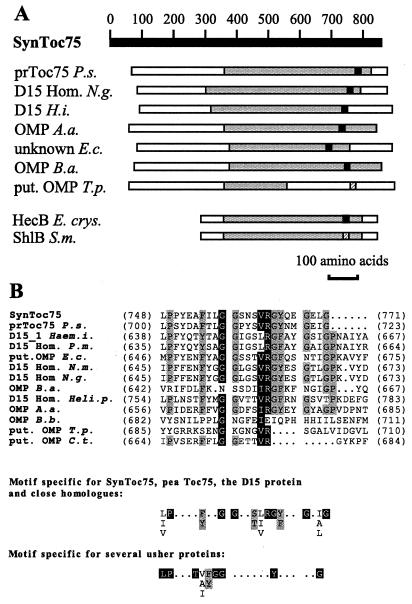
To gain information about the function of the ancestral protein of Toc75, SynToc75 was used to search databases by using the blast algorithm (Blast 2.0). The protein with greatest similarity to SynToc75, pea Toc75 (Blast score = 123, E value = 5⋅10−27), is followed by several OMPs from different Gram-negative bacteria, with Blast scores ranging from 108 (E value = 2⋅10−22, OMP of Brucella abortus) to 44 (E value = 6⋅10−3, OMP85 analog of Chlamydia trachomatis; a complete list of all homologs is presented in Materials and Methods). Despite the low primary sequence similarity, several properties support a relationship among these proteins. These include (i) their similar size (780–920 amino acids); (ii) their proven or predicted outer membrane localization; (iii) the predicted presence of a cleavable N-terminal signal sequence, indicating a Sec-dependent biogenesis pathway; (iv) the secondary structure prediction of a large number of amphiphilic β-strands; and (v) a parallel alignment to SynToc75 with similar middle and C-terminal regions of sequence identity (Fig. 4). A C-terminal motif was deduced that identifies the D15 protein and close homologs as well as pea and Synechocystis Toc75 with high specificity (Fig. 4B). Interestingly, this motif is closely related to a motif that we identified for several usher proteins (Fig. 4B), such as AFAC, FIMD, SFMD, and FOCD, which mediate the export of fimbriae subunits across the outer membrane. Finally, all these OMPs represent the proteins of highest sequence similarity to one another, confirming that they form a distinct protein family.
Figure 4.
Multiple sequence alignments of SynToc75 with related proteins. (A) Summary of sequence alignments of pea Toc75, the D15 homologs, and D15-related proteins with SynToc75. The gray shaded areas represent the regions of sequence similarity with SynToc75, as indicated by blast 2.0 (pea Toc75, D15, and D15-related proteins) or psi-blast (HecB of Erwinia chrysanthemi, ShlB of Serratia marcescens). The black boxes indicate the location of the C-terminal motif. The C-terminal motifs of the putative (put.) OMP of Treponema pallidum and ShlB are less conserved, as indicated by a different pattern. (B) Multiple sequence alignment of the C-terminal regions of SynToc75 and the D15-related proteins. The black boxes indicate high sequence similarity (>90%), the gray shading indicates low sequence similarity (>40%) (31).
In addition to the parallel alignment (Fig. 4A), the middle region of SynToc75 (amino acids 300–500) shows a lower but detectable sequence similarity to the N-terminal region of pea Toc75 (amino acids 50–250) and proteins that are closely related to SynToc75, such as those of Neisseria gonorrhoeae, N. meningitis, and E. coli. This two-fold sequence similarity suggests that these proteins of about 800 amino acids evolved from a smaller ancient protein by duplication of the N-terminal region. This region of highest sequence variability (Fig. 4A) may be adapted to the different transport functions of the various organisms.
The SynToc75-related proteins include the D15 protective surface antigen of Haemophilus influenzae (46, 47) and its homologs identified by annotation, Oma87 of Pasteurella multocida (48) and the OMPs of Helicobacter pylori, N. gonorrhoeae, and N. meningitis. However, the functions of these proteins, referred to as the D15-related proteins, are currently unknown. Interestingly, a member of this protein family is present in all eight Gram-negative bacteria whose genomes have been completely sequenced. Because these bacteria cover a wide range of families, the presence of a similar protein suggests a basic rather than a physiologically specialized function of these OMPs. Taken together, the structural evidence and predictions suggest that SynToc75 belongs to a family of OMPs common to Gram-negative bacteria.
SynToc75 and the D15 Homologs Are Related to Specific Secretion Channels.
To detect more distantly related proteins, possibly including some of known function, a database search was performed by using the psi-blast program (Position-specific iteration blast) (29). Pea and Synechocystis Toc75 and the D15-related proteins were found to be related to a group of specific prokaryotic secretion channels, most of which transfer virulence factors, such as hemolysins and adhesins, across the outer membrane (Table 1). The specificity of the search process is supported by the high stringency of the search (E value threshold 1⋅10−3), the convergence of proteins, and the fact that no proteins known to have a different function were identified as being related. Lower sequence similarity is detected to several usher proteins (700–800 amino acids in size), such as FimC, FASD, AFAC, and CSSD.
Table 1.
psi-blast search with SynToc75
| Protein | Organism | Accession no. | Size, aa | Localization | Function |
|---|---|---|---|---|---|
| HecB | Erwinia chrysanthemi | gi|1772622 | 558 | OM | Probably HecA secretion |
| HpmB | Proteus mirabilis | sp|P16465 | 561 | OM | Activation and secretion of the hemolysin HpmA |
| ShlB | Serratia marcescens | sp|P15321 | 557 | OM | Activation and secretion of the hemolysin ShlA |
| EthB | Edwardsiella tarda | gi|2244626 | 559 | OM | Activation and secretion of the hemolysin EthA |
| HxuB1/2 | Haemophilus influenzae | sp|P45356/sp|P44601 | 565 | OM | Secretion of heme–hemopexin-binding protein HxuA1/2 |
| HhdB | Haemophilus ducreyi | gi|1151071 | 532 | OM | Activation and secretion of the hemolysin HhdA |
| FhaC | Bordetella pertussis | sp|P35077/pir∥S41526 | 584 | OM | Secretion of filamentous hemagglutinin (FHA) |
| HmwB1/2 | Haemophilus influenzae | gi|475772/gi|482842 | 545 | OM | Probably secretion of the adhesin HmwA1/2 |
The nonredundant database was searched with SynToc75 for related proteins by using psi-blast (29). The search converged after five iterations. Pea Toc75, the D15-related proteins, and seven proteins of unknown function are not listed. Databases are gi, GenBank; sp, Swiss-Prot; and pir, Protein Identification Resource. OM, outer membrane.
All these accessory proteins secreting virulence factors also consist of amphiphilic β-strands and have a similar but smaller size (530–580 amino acids, Table 1), compared with the D15-related proteins. The region of sequence similarity with SynToc75 is the stretch of about 450 amino acids that is conserved between Synechocystis and pea Toc75 and the D15 homologs (Fig. 4A). This small family of channel proteins (49, 50) has not yet been assigned to any of the four known types of prokaryotic secretion systems. Compared with the type II and type III secretion apparatus, these channel proteins are characterized by (i) a remarkable simplicity (1–3 components instead of 12–14); (ii) a high substrate specificity, restricted to a single protein; and (iii) a high transport efficiency, especially when considering the high molecular mass of many of their substrates (100–370 kDa).
In addition to the sequence similarity found among the channel proteins, a remarkable sequence similarity is also exhibited by the proteins secreted via these channels—i.e., filamentous hemagglutinin and the hemolysins, particularly in the N-terminal domain (49–51), which interacts with the respective accessory protein during secretion (49, 52). Surprisingly, the plant protein with highest sequence similarity (psi-blast, E value = 2⋅10−4) to this family of virulence factors is the Arabidopsis homolog of Toc86 (GenBank accession no. AC002330). The idea that the receptor of the chloroplastic protein import apparatus could be related to the substrate of the Toc75 ancestor is intriguing, but it cannot be substantiated by further computer analysis at this point because of the lack of a cyanobacterial homolog of Toc86.
In summary, we have provided evidence that a cyanobacterium contains a structural homolog of Toc75 in its outer membrane. We propose that the current prokaryotic channels that secrete hemolysins and adhesins, the D15-related proteins, SynToc75, and Toc75 of higher plants are derived from a common ancient channel protein. We posit that partial gene duplication of the N-terminal region gave rise to the larger proteins. The functions of the D15-related proteins remain to be elucidated. Electrophysiological studies, which indicate that SynToc75 forms a voltage-gated pore (42), support a transport function for SynToc75.
Because Toc75 itself is encoded in the nucleus, we speculate that transfer of its ancestral gene was an event early in endosymbiosis, and the starting point for development of the chloroplastic translocon. A rather simple but efficient secretion system could easily have adapted to the new function of importing nucleus-encoded proteins into the evolving organelle. Because the topology of membrane proteins is usually determined by their direction of insertion into a given membrane (53), expression of the Toc75 ancestor in the eukaryotic cytosol could have led to an inverted orientation of the channel in the outer membrane of the endosymbiont. Exposure of a previously periplasmic binding site for precursor proteins to the eukaryotic cytosol would have facilitated precursor import into primitive chloroplasts.
Although some data support the idea that chloroplastic transit peptides evolved by mutation, other lines of evidence suggest that an ancient targeting signal was added to the N-terminal end of the transferred symbiont genes by exon shuffling (54, 55). If the latter hypothesis holds, we postulate that the transit peptides of chloroplastic proteins are derived from the substrate secreted by the ancestor of SynToc75. Therefore, the identification of the postulated peptide secreted by SynToc75 might give interesting insights into the evolution of chloroplastic transit peptides. The availability of Synechocystis mutants lacking chromosomal synToc75 but harboring a plasmid whose synToc75 expression is regulated by an inducible promoter might allow the identification of the postulated substrate of SynToc75 by second-site mutagenesis.
Supplementary Material
Acknowledgments
We thank Drs. C. P. Wolk and L. McIntosh for stimulating discussions regarding cyanobacterial biology and genetics, Dr. J. Yu and R. Nichols for technical assistance in cyanobacterial culture, and Drs. C. Wilkerson and C. Delwiche for assistance in computer analysis. Drs. T. Mizuno and L. Sherman kindly provided the antisera against SomA and NrtA, respectively. Finally, we thank Dr. S. He, K. Bird, and members of the Keegstra laboratory for helpful comments. This work was supported by grants from the National Science Foundation and the Division of Energy Biosciences of the U.S. Department of Energy (to K.K.) and a fellowship from the Deutsche Forschungsgemeinschaft (to S.R.).
ABBREVIATIONS
- OMP
outer membrane protein
- SynToc75
Synechocystis homolog of pea Toc75
References
- 1.Schimper A F W. Bot Z. 1883;41:105–114. [Google Scholar]
- 2.Mereschowsky C. Biol Zentralbl. 1905;25:593–604. , 689–691. [Google Scholar]
- 3.Gray M W. Int Rev Cytol. 1992;141:233–357. doi: 10.1016/s0074-7696(08)62068-9. [DOI] [PubMed] [Google Scholar]
- 4.Martin W, Stoebe B, Goremykin V, Hansmann S, Hasegawa M, Kowallik K V. Nature (London) 1998;393:162–165. doi: 10.1038/30234. [DOI] [PubMed] [Google Scholar]
- 5.Keegstra K. Cell. 1989;56:247–253. doi: 10.1016/0092-8674(89)90898-2. [DOI] [PubMed] [Google Scholar]
- 6.Hirsch S, Muckel E, Heemeyer F, von Heijne G, Soll J. Science. 1994;266:1989–1992. doi: 10.1126/science.7801125. [DOI] [PubMed] [Google Scholar]
- 7.Kessler F, Blobel G, Patel H A, Schnell D J. Science. 1994;266:1035–1039. doi: 10.1126/science.7973656. [DOI] [PubMed] [Google Scholar]
- 8.Perry S E, Keegstra K. Plant Cell. 1994;6:93–105. doi: 10.1105/tpc.6.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schnell D J, Kessler F, Blobel G. Science. 1994;266:1007–1012. doi: 10.1126/science.7973649. [DOI] [PubMed] [Google Scholar]
- 10.Tranel P J, Froehlich J, Goyal A, Keegstra K. EMBO J. 1995;14:2436–2446. doi: 10.1002/j.1460-2075.1995.tb07241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kessler F, Blobel G. Proc Natl Acad Sci USA. 1996;93:7684–7689. doi: 10.1073/pnas.93.15.7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lübeck J, Soll J, Akita M, Nielsen E, Keegstra K. EMBO J. 1996;15:4230–4238. [PMC free article] [PubMed] [Google Scholar]
- 13.Caliebe A, Grimm R, Kaiser G, Lübeck J, Soll J, Heins L. EMBO J. 1997;16:7342–7350. doi: 10.1093/emboj/16.24.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kouranov A, Schnell D J. J Cell Biol. 1997;139:1677–1685. doi: 10.1083/jcb.139.7.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kouranov A, Chen X, Fuks B, Schnell D J. J Cell Biol. 1998;143:991–1002. doi: 10.1083/jcb.143.4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akita M, Nielsen E, Keegstra K. J Cell Biol. 1997;136:983–994. doi: 10.1083/jcb.136.5.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nielsen E, Akita M, Davila-Aponte J, Keegstra K. EMBO J. 1997;16:935–946. doi: 10.1093/emboj/16.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinnah S C, Hill K, Wagner R, Schlicher T, Soll J. EMBO J. 1997;16:7351–7360. doi: 10.1093/emboj/16.24.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, et al. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 20.Omata T, Murata N. Arch Microbiol. 1984;139:113–116. [Google Scholar]
- 21.Williams J G K. Methods Enzymol. 1988;167:766–778. [Google Scholar]
- 22.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Lowry O H, Rosebrough N J, Farr A L, Randall R J. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 24.Wessel D, Flügge U I. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 25.Umeda H, Aiba H, Mizuno T. Microbiology. 1996;142:2121–2128. doi: 10.1099/13500872-142-8-2121. [DOI] [PubMed] [Google Scholar]
- 26.Reddy K J, Masamoto K, Sherman D M, Sherman L A. J Bacteriol. 1989;171:3486–3493. doi: 10.1128/jb.171.6.3486-3493.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu J, Smart L B, Jung Y-S, Golbeck J, McIntosh L. Plant Mol Biol. 1995;29:331–342. doi: 10.1007/BF00043656. [DOI] [PubMed] [Google Scholar]
- 28.Chrisholm D. Cyanonews. 1990;6:7. [Google Scholar]
- 29.Altschul S F, Madden T L, Schaeffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duret L, Gasteiger E, Perriere G. Comput Appl Biosci. 1996;12:507–510. doi: 10.1093/bioinformatics/12.6.507. [DOI] [PubMed] [Google Scholar]
- 31.Corpet F. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bairoch A, Bucher P, Hofmann K. Nucleic Acids Res. 1997;24:217–221. doi: 10.1093/nar/25.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakai K, Kanehisa M. Proteins. 1991;11:95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 35.Claros M G, von Heijne G. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 36.Rost B, Schneider R, Sander C. Comput Appl Biosci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- 37.Tranel P J, Keegstra K. Plant Cell. 1996;8:2093–2104. doi: 10.1105/tpc.8.11.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cavalier-Smith T. Ann N Y Acad Sci. 1987;503:55–71. doi: 10.1111/j.1749-6632.1987.tb40597.x. [DOI] [PubMed] [Google Scholar]
- 39.Keegstra K, Werner-Washburne M, Cline K, Andrews J. J Cell Biochem. 1984;24:55–68. doi: 10.1002/jcb.240240105. [DOI] [PubMed] [Google Scholar]
- 40.Joyard J, Block M A, Douce R. Eur J Biochem. 1991;199:489–509. doi: 10.1111/j.1432-1033.1991.tb16148.x. [DOI] [PubMed] [Google Scholar]
- 41.Sonoda M, Kitano K, Katoh A, Katoh H, Ohkawa H, Ogawa T. J Bacteriol. 1997;179:3845–3850. doi: 10.1128/jb.179.12.3845-3850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bölter B, Soll J, Schulz A, Hinnah S, Wagner R. Proc Natl Acad Sci USA. 1998;95:15831–15836. doi: 10.1073/pnas.95.26.15831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Labarre J, Chauvat F, Thuriaux P. J Bacteriol. 1989;171:3449–3457. doi: 10.1128/jb.171.6.3449-3457.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smart L B, Anderson S L, McIntosh L. EMBO J. 1991;10:3289–3296. doi: 10.1002/j.1460-2075.1991.tb04893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jonsson A B, Pfeifer J, Normark S. Proc Natl Acad Sci USA. 1992;89:3204–3208. doi: 10.1073/pnas.89.8.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flack F S, Loosmore S, Chong P, Thomas W R. Gene. 1995;156:97–99. doi: 10.1016/0378-1119(95)00049-c. [DOI] [PubMed] [Google Scholar]
- 47.Loosmore S M, Yang Y P, Coleman D C, Shortreed J M, England D M, Klein M H. Infect Immun. 1997;65:3701–3707. doi: 10.1128/iai.65.9.3701-3707.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruffolo G C, Adler B. Infect Immun. 1996;64:3161–3167. doi: 10.1128/iai.64.8.3161-3167.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Willems R J L, Geuijen C, van der Heide H G J, Renauld G, Bertin P, van den Akker W M R, Locht C, Mooi F R. Mol Microbiol. 1994;11:337–347. doi: 10.1111/j.1365-2958.1994.tb00314.x. [DOI] [PubMed] [Google Scholar]
- 50.Jacob-Dubuisson F, Buisine C, Willery E, Renauld-Mongenie G, Locht C. J Bacteriol. 1997;179:775–783. doi: 10.1128/jb.179.3.775-783.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delisse-Gathoye A M, Locht C, Jacob F, Raaschou-Nielsen M, Heron I, Ruelle J L, de Wilde M, Cabezon T. Infect Immun. 1990;58:2895–2905. doi: 10.1128/iai.58.9.2895-2905.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ondraczek R, Hobbie S, Braun V. J Bacteriol. 1992;174:5086–5094. doi: 10.1128/jb.174.15.5086-5094.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.von Heijne G. BioEssays. 1995;17:25–30. doi: 10.1002/bies.950170107. [DOI] [PubMed] [Google Scholar]
- 54.Wolter F P, Fritz C C, Willmitzer L, Schell J, Schreier P H. Proc Natl Acad Sci USA. 1988;85:846–850. doi: 10.1073/pnas.85.3.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gantt J S, Baldauf S L, Calie P J, Weeden N F, Palmer J D. EMBO J. 1991;10:3073–3078. doi: 10.1002/j.1460-2075.1991.tb07859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.