Abstract
Retroviral integration in vivo is mediated by preintegration complexes (PICs) derived from infectious virions. In addition to the integrase enzyme and cDNA substrate, PICs contain a variety of viral and host cell proteins. Whereas two different cell proteins, high-mobility group protein A1 (HMGA1) and the barrier-to-autointegration factor (BAF), were identified as integration cofactors based on activities in in vitro PIC assays, only HMGA1 was previously identified as a PIC component. By using antibodies against known viral and cellular PIC components, we demonstrate here functional coimmunoprecipitation of endogenous BAF protein with human immunodeficiency virus type 1 (HIV-1) PICs. Since integrase protein and integration activity were also coimmunoprecipitated by anti-BAF antibodies, we conclude that BAF is a component of HIV-1 PICs. These data are consistent with the model that BAF functions as an integration cofactor in vivo.
Retroviral replication requires the integration of the viral cDNA made by reverse transcription into a chromosome of an infected cell. Although the viral integrase protein is necessary and sufficient to catalyze the DNA cleavage and joining steps of retroviral integration in in vitro DNA recombination assays (10, 22), numerous studies indicate that provirus formation in vivo likely requires the action of host cell proteins. In vivo, integration is mediated by large subviral preintegration complexes (PICs) that can integrate their endogenous cDNA into an added target DNA in vitro (1, 2, 11, 13, 25). In addition to the integrase enzyme, retroviral PICs contain a variety of viral and host cell proteins (1, 3, 14, 20, 27-29, 35).
Human immunodeficiency virus type 1 (HIV-1) and Moloney murine leukemia virus (MoMLV) PICs treated with relatively high concentrations of salt no longer integrated into exogenous target DNA, and extracts of uninfected host cells restored intermolecular DNA recombination to salt-stripped PICs (7, 12, 23, 26). Since extracts of MoMLV (23) and HIV-1 (12) virions failed to reconstitute integration activity under these assay conditions, it was concluded that host cell proteins play essential roles in retroviral PIC function. Two different proteins, high-mobility group protein A1 (HMGA1) (4, 12) and barrier-to-autointegration factor (BAF) (24), were purified from crude cell extracts by using functional PIC reconstitution assays.
A variety of results address whether HMGA1 and/or BAF might participate in virus integration in vivo. These include the specific activities of the proteins in in vitro integration assays and their identification as a component of PICs isolated from infected cells. Studies that analyzed the in vitro activity of purified integrase protein have yielded varied results. Whereas one report indicated that recombinant HMGA1 stimulated the activity of purified HIV-1 integrase perhaps 10-fold (21), a separate study failed to detect stimulation by either HMGA1 or BAF (6). In contrast to these results, BAF was substantially (about 500-fold) more active than HMGA1 in HIV-1 (7, 12) and MoMLV (26, 36) PIC reconstitution assays. However, only HMGA1 has thus far been identified as a component of HIV-1 and MoMLV PICs (12, 28). We report here that human BAF protein is a component of PICs isolated from HIV-1-infected cells.
Experimental strategy.
In order to analyze endogenous BAF protein in cell lysates, rabbits were inoculated with purified recombinant human BAF protein (19), and the resulting serum was affinity purified by using Sepharose beads conjugated to a 13-residue amino-terminal human BAF peptide (5). Although the purified serum readily detected recombinant BAF by Western blotting, it failed to immunoprecipitate the protein (data not shown). Immunodepletion of functional reconstitution activity from high-salt extracts of HIV-1 PICs with anti-HMGA1 antibodies (12) as well as direct immunoprecipitation of functional MoMLV PICs (28) previously demonstrated that HMGA1 is a component of HIV-1 and MoMLV PICs. However, the inability to immunoprecipitate BAF protein precluded the use of our serum in similar analyses. Although we eventually obtained an anti-BAF serum with immunoprecipitation activity (see below), we initially devised a novel functional coimmunoprecipitation strategy to investigate the association of human BAF with HIV-1 PICs (Fig. 1).
FIG. 1.
Functional coimmunoprecipitation strategy. Cytoplasmic extracts (fraction I) of acutely infected T cells were subjected to a variety of purification steps to investigate the association of human BAF with HIV-1 PICs. In arm A of the scheme, fraction I was directly immunoprecipitated. Extracts purified by either ultrafiltration or gel filtration chromatography (fraction II) were immunoprecipitated in arm B of the scheme. Whereas integration activity was quantified following Southern blotting as previously described (7, 8), levels of integrase and BAF proteins in fraction PB were detected by Western blotting. PA and SA, pellet and supernatant fractions following immunoprecipitation in arm A of the scheme, respectively; PB and SB, pellet and supernatant fractions after immunoprecipitation of fraction II, respectively.
The coimmunoprecipitation strategy took advantage of the different viral and host cell proteins known to comprise HIV-1 PICs (3, 12, 14, 20, 27, 29). Monoclonal antibodies (MAb) and polyclonal antibodies against known PIC components were tested for their ability to immunoprecipitate viral cDNA and integration activity by Southern blotting, and integrase and BAF protein were tested by Western blotting (Fig. 1). PICs were isolated from cytoplasmic extracts of C8166 T cells 7 h after infection with cell-free HIV-1NL4-3 as previously described (8, 9), with the modification that the cell lysis buffer (buffer K; 20 mM HEPES [pH 7.6], 150 mM KCl, 5 mM MgCl2, 0.5% [vol/vol] Triton X-100, 1 mM dithiothreitol, 20 μg of aprotinin/ml) contained Triton X-100 in place of digitonin. Integration into linearized φX174 target DNA was assayed as described previously (7-9). Activity was quantified by using a PhosphorImager with ImageQuant version 1.11 (Molecular Dynamics, Sunnyvale, Calif.) as the percentage of total HIV-1 cDNA converted into the 15.1-kb integration product.
Coimmunoprecipitation of BAF protein in active PIC samples with an anti-matrix antibody.
HIV-1 matrix is a PIC component (3, 20, 29), and we began by analyzing the activity of anti-matrix MAb 3H7 (30) in PIC immunoprecipitation assays. Supernatant of 3H7 hybridoma cells (a generous gift of Wayne Marasco, Dana-Farber Cancer Institute) was purified by using protein-A Sepharose (Amersham Pharmacia Biotech, Piscataway, N.J.) essentially as previously described (18). The concentration of purified 3H7 was estimated by spectrophotometry at 0.6 mg/ml.
Antibody (10 μl) was prebound to Dynabeads protein A (15 μl; Dynal Biotech Inc., Lake Success, N.Y.) in 100 μl of buffer K for 1 h at room temperature. Beads washed with 1 ml of buffer K were added to 0.5 ml of fraction I, the mixture was rocked for 1 h at room temperature, and immune complexes were recovered with a magnet. The SA supernatant fraction was saved, the beads were washed (1 ml of buffer K) three times, and DNA in unbound SA and pelleted PA fractions was recovered by digestion with proteinase K, extraction with phenol-chloroform, and precipitation with ethanol essentially as previously described (8).
MAb 3H7 immunoprecipitated 6 to 16% of input HIV-1 cDNA under these conditions; the average value of four measurements was 10.0% ± 3.9% (Fig. 2A, lanes 4 and 5; data not shown). Importantly, 3H7-precipitated PICs (lane 4) catalyzed integration at levels similar to those of fraction I (lane 2) and the unbound SA fraction (lane 5), demonstrating that PICs bound to 3H7 protein A beads were competent for intermolecular DNA recombination. Since immunoglobulin G3(κ) ([IgG3(κ)]) isotype control antibodies (Sigma-Aldrich, St. Louis, Mo.) failed to recover detectable cDNA levels (lane 3), we concluded that the specific interaction between MAb 3H7 and residues 113 to 122 of the HIV-1 matrix (30) mediated PIC immunoprecipitation.
FIG. 2.
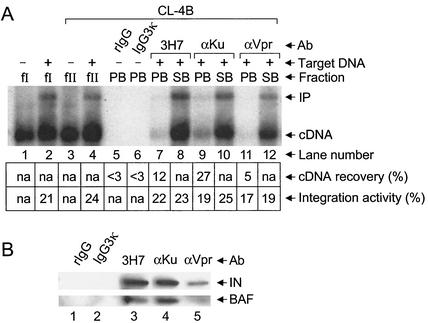
Analysis of PIC activity and integrase and BAF proteins following immunoprecipitation with MAb 3H7. (A) Levels of HIV-1 cDNA synthesis in fraction I (lane 1) and integration activity of various fractions analyzed by Southern blotting. Target DNA was added to the in vitro integration assays shown in lanes 2 and 4 to 7 as indicated. The migration positions of the 9.7-kb HIV-1 substrate and the 15.1-kb integration product are marked cDNA and IP, respectively. Percentage of total cDNA recovered in pellet fractions following immunoprecipitation is indicated beneath lanes 3, 4, and 6. Levels of fraction I (lane 2), PA (lane 4), SA (lane 5), PB (lane 6), and SB (lane 7) integration activities are also indicated beneath the blot. (B) Western blot analysis of integrase and BAF proteins. Whereas integrase was detected by using MAb 8E5 (31) as previously described (8), BAF was detected by using affinity-purified rabbit antiserum at 1:1,000 dilution. The identities of the integrase and BAF proteins in the lane marked 3H7 were confirmed by comigration with purified recombinant proteins. fI, fraction I; Ab, antibody; IN, integrase; n.a., not applicable. Other labeling is the same as that defined in the legend to Fig. 1.
HIV-1 virions contain about 2,000 copies of the matrix protein (37), but only a small fraction of this population, perhaps 1%, remains PIC associated after virus uncoating and reverse transcription (16). We therefore tested whether separating PICs from the bulk of cell and viral proteins in fraction I would increase the level of cDNA recovery by MAb 3H7. Fraction I mixed with an equal volume of buffer K was centrifuged at 1,000 × g for 1 h at 4°C in a Centricon 100 concentrator (Millipore Corp., Bedford, Mass.). Whereas proteins with molecular masses of less than 100 kDa pass through this ultrafilter, large proteins and PICs remain in the retentate (12). After centrifugation the sample volume was readjusted to 0.5 ml with buffer K, and immunoprecipitation was performed as described above. Since six independent measurements yielded an average value of 22.2% ± 7.7% cDNA recovery (Fig. 2A, lanes 6 and 7; data not shown), we concluded that ultrafiltration yielded an approximate twofold increase in the efficiency of PIC immunoprecipitation. As was observed with fractions PA and SA (lanes 4 and 5), fraction PB PICs (lane 6) catalyzed integration at a level similar to that of the unbound SB fraction (lane 7).
Fraction PB samples were next analyzed for integrase and BAF protein content by Western blotting. As expected from the results of Southern blotting, MAb 3H7 coimmunoprecipitated integrase protein under conditions where IgG3(κ) failed to recover appreciable levels of integrase (Fig. 2B). Since BAF was also detected in 3H7 immunoprecipitates under conditions where the isotype control failed to recover detectable BAF levels, anti-matrix MAb 3H7 specifically immunoprecipitated integration-competent PICs (Fig. 2A), HIV-1 integrase, and endogenous human BAF protein (Fig. 2B).
Coimmunoprecipitation of integrase, BAF, and PIC activity by using antibodies against Ku-80 and Vpr.
We next investigated the coimmunoprecipitation properties of antibodies against two other known PIC components, HIV-1 Vpr (20) and cellular Ku-80, the 86-kDa subunit of human DNA-dependent protein kinase (27). Whereas the anti-Ku serum was purchased from a previously described (27) source (Serotec Inc., Raleigh, N.C.), the anti-Vpr serum was a generous gift from Ulrich Schubert (National Institute of Allergy and Infectious Diseases).
For this experiment, fraction I was purified by spin column chromatography. We previously determined that CL-4B Sepharose (Amersham-Pharmacia) spin columns removed 80 to 90% of total protein from fraction I without affecting PIC recovery or integration activity (8). Whereas small proteins like free BAF (10.1 kDa) are retained in these columns, large complexes like PICs pass through. Preliminary experiments (n = 5) with MAb 3H7 yielded an average value of 18.2% ± 6.8% cDNA recovery in fraction PB (Fig. 3A, lanes 7 and 8; data not shown).
FIG. 3.
Coimmunoprecipitation of integration activity and integrase and BAF proteins by using anti-Ku and anti-Vpr antibodies. (A) Southern blot analysis of HIV-1 cDNA synthesis (lane 1) and PIC activity. Target DNA was included in the in vitro integration reactions shown in lanes 2, 4, and 7 to 12. Whereas fraction I was analyzed in lanes 1 and 2, the fraction II samples shown in lanes 3 to 12 were purified by spin column chromatography. We note that the faint integration product in lane 11 was not revealed by this level of exposure. (B) Western blot analysis of integrase and BAF proteins. fII, fraction II; rIgG, normal rabbit IgG. Other labeling is the same as that described in the legend to Fig. 2.
Each rabbit serum (10 μl) was prebound to Dynabeads protein A (15 μl) as described above. Whereas the anti-Ku serum recovered about twice as many PICs as MAb 3H7 in two different experiments, the anti-Vpr serum recovered less PICs than did 3H7 (Fig. 3A, lanes 7 to 12; data not shown). As was observed with 3H7-precipitated fractions (Fig. 2A, lanes 6 and 7, and Fig. 3A, lanes 7 and 8), anti-Ku (Fig. 3A, lane 9) and anti-Vpr (lane 11) PB fractions catalyzed integration at levels similar to those of fraction I (lane 2), fraction II (lane 4), and unbound SB fractions (lanes 10 and 12). Since normal rabbit IgG (Sigma-Aldrich) failed to recover detectable cDNA levels under these conditions (lane 5), we concluded that cDNA recovery was mediated by specific interactions between anti-Ku and anti-Vpr antibodies and the Ku-80 (27) and Vpr (20) components of HIV-1 PICs, respectively.
Samples of these immunoprecipitates were next analyzed for integrase and BAF protein by Western blotting. As observed in Fig. 2B, MAb 3H7 coimmunoprecipitated integrase and BAF protein under conditions where IgG3(κ) failed to recover appreciable levels of either protein (Fig. 3B, lanes 2 and 3). Similarly, anti-Ku and anti-Vpr each coimmunoprecipitated integrase and BAF under conditions where normal rabbit IgG failed to recover detectable levels of either protein (compare lanes 4 and 5 to lane 1). We note that the levels of BAF recovered by MAb 3H7 and the anti-Ku and anti-Vpr sera roughly paralleled the levels of active PICs immunoprecipitated by these antibodies (compare lanes 3 to 5 in the lower panel of Fig. 3B to lanes 7, 9, and 11 in panel A). These results are consistent with the interpretation that BAF is a component of functional HIV-1 PICs.
Coimmunoprecipitation of active PICs and integrase protein by anti-BAF antibodies.
Antibodies capable of immunoprecipitating BAF protein were unavailable at the start of this project. However, a rabbit serum raised against a peptide comprised of residues 4 to 20 (15, 17) was subsequently shown to display immunoprecipitation activity (Y. Suzuki and R. Craigie, personal communication). A sample of this serum, referred to here as anti-BAF4-20 to distinguish it from our original serum, was a generous gift from Robert Craigie (National Institute of Diabetes, Digestive, and Kidney Diseases).
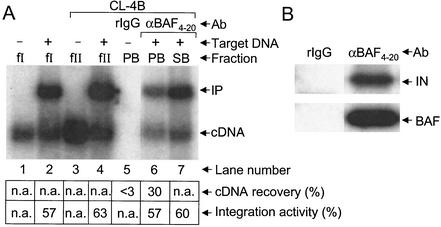
Anti-BAF4-20 (20 μl) prebound to Dynabeads protein A (15 μl) recovered about 30% of fraction II PICs under conditions where normal rabbit IgG failed to recover detectable cDNA levels (Fig. 4A, lanes 5 to 7). In addition, akin to the results presented by using antibodies against previously known PIC components (Fig. 2A and 3A), anti-BAF4-20-immunoprecipitated PICs (Fig. 4A, lane 6) catalyzed integration at a level similar to that of the PICs in fraction I (lanes 1 and 2), fraction II (lanes 3 and 4), and unbound fraction SB (lane 7). As expected from these results, anti-BAF4-20 coimmunoprecipitated integrase under conditions where rabbit IgG failed to recover detectable levels of integrase or BAF (Fig. 4B).
FIG. 4.
Coimmunoprecipitation of integration activity and integrase protein by anti-BAF4-20 antibodies. (A) Southern blot analysis. Target DNA was included in the integration reactions depicted in lanes 2, 4, 6, and 7. Whereas fraction I was analyzed in lanes 1 and 2, the fraction II samples in lanes 3 to 7 were purified by spin column chromatography. (B) Western blot analysis. Labeling is the same as that described in the legends to Fig. 2 and 3.
We next tested the effects of adding purified recombinant BAF protein as a competitor during the immunoprecipitation reaction. Fraction II samples recovered from CL-4B spin columns were mixed with a fixed level of anti-BAF4-20-protein A beads and various levels of recombinant BAF protein; in this case, PIC recovery was monitored by Western blotting for integrase protein. Whereas the addition of 10 nM recombinant BAF did not affect the level of integrase recovery, 100 nM BAF resulted in an approximate twofold reduction in integrase coimmunoprecipitation (Fig. 5A, compare the integrase signal in lanes 2 and 3 to the signal in lane 1). Although the excess BAF protein (7, 19) in lane 3 was expected to effectively inhibit immunoprecipitation, we speculate that cDNA binding by recombinant BAF and/or recombinant BAF-anti-BAF4-20 complexes yielded residual PIC recovery (see below). We note that Li et al. (28) observed residual MoMLV PIC recovery by using excess recombinant HMGA1 as a competitor in anti-HMGA1-based immunoprecipitation assays.
FIG. 5.
Competition of anti-BAF4-20-mediated PIC immunoprecipitation by recombinant BAF proteins. (A) Western blot analysis of integrase coimmunoprecipitation in the absence of added BAF (lane 1) or in the presence of 10 nM (lane 2) and 100 nM (lane 3) recombinant human BAF. The excess recombinant BAF in the reaction shown in lane 3 contributed to the increased BAF signal. (B) Differential effects of BAF DNA binding mutants on PIC immunoprecipitation. For lane 1, normal rabbit IgG was used in the immunoprecipitation reaction; lanes 2 to 5, anti-BAF4-20 was used in the immunoprecipitation reaction. Whereas recombinant BAF protein was omitted from the reactions shown in lanes 1 and 2; lanes 3 to 5 contained 100 nM wild-type, K6A, and K18A BAF, respectively. The longer exposure of the anti-BAF Western shown in the lower boxed panel highlights the remaining endogenous BAF protein in lane 5. WT, wild-type; rBAF, recombinant BAF. Other labeling is the same as that described in the legends to Fig. 2 and 3.
To circumvent the likely binding of recombinant BAF to HIV-1 cDNA during immunoprecipitation, we next analyzed BAF mutant proteins defective for DNA binding activity. We previously characterized a number of BAF missense mutants as defective for DNA binding (19). A subset of these, however, was primarily defective for BAF protein folding, and because of this we avoided using these mutants in the PIC immunoprecipitation assay. In contrast, two of our previously described mutant proteins, K6A and K18A, were properly folded and thus specifically defective for DNA binding. Whereas the K6A mutant failed to detectably bind a 30-bp oligonucleotide by electrophoretic mobility shift, K18A BAF displayed weak but detectable activity in this assay (19). However, we note that Segura-Totten et al. (32) recently ascribed wild-type levels of DNA binding to K18A BAF by using substantially longer (600 to 2,000 bp) substrates under conditions where K6E BAF was severely defective for DNA binding. We further noted that anti-BAF4-20 failed to immunoprecipitate either K6A or K18A BAF under conditions where wild-type recombinant BAF was efficiently immunoprecipitated (data not shown). Based on this we speculate that Lys-6 and Lys-18 each contribute significantly to the epitope recognized by anti-BAF4-20 antibodies. Thus, despite our goal to utilize DNA binding-defective BAF mutants as competitors in the immunoprecipitation assay, we highlight that neither K6A nor K18A BAF directly competed with PIC-associated endogenous BAF for antibody binding, because anti-BAF4-20 antibodies failed to recognize either of these recombinant proteins.
The addition of K6A BAF (100 nM) to the immunoprecipitation reaction mixture had no effect on integrase recovery (Fig. 5B, compare lane 4 to lane 2), as predicted for a mutant defective for both DNA binding and antibody recognition. In contrast, K18A BAF reduced the level of integrase recovery to a level similar to that observed with the wild-type competitor (compare lane 5 to lane 3). Since less endogenous BAF protein was recovered under these conditions (lane 5), we conclude that excess K18A BAF displaced the majority of PIC-associated wild-type BAF from HIV-1 cDNA, and this reduced level of endogenous BAF protein (Fig. 5B, lane 5 of the lower boxed panel) resulted in less integrase coimmunoprecipitation. This interpretation is consistent with the results that (i) K18A BAF displayed the wild-type level of binding to relatively long DNA fragments (32) and (ii) anti-BAF4-20 failed to immunoprecipitate recombinant K18A BAF protein.
BAF is a component of HIV-1 PICs.
Retroviral PICs treated with relatively high concentrations of salt and purified by size separation to remove potentially dislodged proteins lost their ability to integrate into exogenous target DNA in vitro (7, 12, 23, 26). Since extracts of uninfected host cells restored intermolecular DNA recombination under conditions where virion extracts failed to function (12, 23), these studies indicated that host cell proteins play essential roles in retroviral PIC function. HMGA1 and BAF were each previously purified from cell extracts by using functional PIC reconstitution assays (12, 24).
A variety of studies have addressed whether BAF and/or HMGA1 might function as retroviral integration cofactor(s) in vivo. One obvious issue is whether the protein(s) is associated with PICs during integration. Since anti-HMGA1 antibodies depleted the functional reconstitution activity from high-salt extracts of HIV-1 PICs (12) and directly immunoprecipitated active MoMLV complexes (28), it was concluded that HMGA1 was a component of HIV-1 and MoMLV PICs. Here we used similar immunoprecipitation strategies to investigate the association of endogenous human BAF protein with HIV-1 PICs.
Since anti-matrix MAb 3H7 specifically coimmunoprecipitated integration activity, viral integrase, and endogenous BAF protein (Fig. 2), we inferred that BAF was a component of functional HIV-1 PICs. Finding that the levels of BAF in 3H7-, anti-Vpr-, and anti-Ku-80-precipitated PB fractions mirrored the levels of active PICs recovered by these antibodies (Fig. 3) bolstered this contention. Additionally, anti-BAF4-20 antibodies directly immunoprecipitated fraction II PICs under conditions where normal rabbit serum failed to recover detectable cDNA levels (Fig. 4A). Although previous studies concluded that capsid (1), integrase (3, 14, 28), matrix (3, 20), Vpr (20), HMGA1 (28), and Ku-80 (27) proteins were components of retroviral PICs by using direct immunoprecipitation strategies, our study revealed human BAF protein as a component of active HIV-1 PICs by using a novel coimmunoprecipitation strategy in addition to direct immunoprecipitation by anti-BAF antibodies.
Although only about 30% of our fraction II PICs were immunoprecipitated by anti-BAF4-20 (Fig. 4A), we note that the anti-Ku serum (Fig. 3A and reference 27) and 3H7 MAb (results reported here) yielded similar levels of PIC recovery. It is unclear why the anti-Vpr serum yielded only about 5% of input PIC recovery (Fig. 3A). Since this is the first study to quantify the level of PIC recovery by anti-Vpr antibodies, it is possible that Vpr is less available than either matrix or cellular Ku-80 for antibody binding under the immunoprecipitation conditions used here. Alternatively, it is possible that the anti-Vpr serum tested here was a suboptimal immunoprecipitation reagent.
Two additional findings here parallel results from the previous analysis of HMGA1's association with MoMLV (28). First, excess recombinant host factor reduced the level of PIC immunoprecipitation by anti-host factor antibodies (Fig. 5A). Since excess K18A BAF displaced the majority of endogenous BAF protein from HIV-1 cDNA (Fig. 5B, lane 5), we speculate that efficient cDNA binding by recombinant wild-type BAF contributed to the residual level of integrase coimmunoprecipitation observed here (Fig. 5A, lane 3). The second similar finding was the inability to immunoprecipitate salt-stripped PICs, as anti-BAF4-20 failed to immunoprecipitate detectable levels of HIV-1 cDNA after salt stripping (data not shown). On the basis of this finding we conclude that the association of human BAF with HIV-1 PICs is sensitive to high concentrations of salt.
Potential mechanism of BAF action.
BAF is apparently excluded from MoMLV (23, 24) and HIV-1 (12) particles. Since recombinant human BAF failed to gel shift single-stranded RNA, single-stranded DNA, or an RNA/DNA hybrid under conditions where binding to double-stranded DNA was readily detected (19), it was previously proposed that endogenous BAF was recruited by infectious retroviruses relatively late in the process of reverse transcription, perhaps coincident with PIC formation (19, 38). Since Suzuki and Craigie (33) recently identified murine BAF protein as a component of MoMLV PICs, we speculate that BAF is likely to mediate the early replication events of a variety of retroviruses.
Since endogenous rat BAF was previously reported as solely nuclear (15), it seems curious that we identified human BAF as a component of PICs isolated from cytoplasmic extracts of infected cells. Consistent with our finding, results of Western blotting revealed approximately one-half of cellular BAF in cytoplasmic extracts of human 293T cells (M. Owens and A. Engelman, unpublished results). We therefore continue to favor the model whereby BAF is incorporated into PICs during or shortly after their formation in the cytoplasm of newly infected cells. Whether the primary role of PIC-associated human BAF is to protect HIV-1 cDNA from suicidal autointegration as proposed for MoMLV (24), to promote efficient intermolecular DNA recombination once a suitable chromosomal target site is located, or both (33) awaits further experimentation. Present attempts to interfere with the normal expression pattern of human BAF via small-interfering RNA technology (reviewed in reference 34) in both transformed T-cell lines and primary cells may help elucidate the precise role(s) of the BAF protein in the HIV-1 life cycle.
Acknowledgments
We thank W. A. Marasco for 3H7 hybridoma cells, U. Schubert for anti-Vpr antiserum, R. Craigie for anti-BAF4-20, and Y. Suzuki and R. Craigie for sharing results prior to publication. We also thank E. Devroe, A. Limón, and R. Lu for their comments on the manuscript.
This work was supported by NIH grants AI39394, AI53812, and AI28691 (Dana-Farber Cancer Institute Center for AIDS Research).
REFERENCES
- 1.Bowerman, B., P. O. Brown, J. M. Bishop, and H. E. Varmus. 1989. A large nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 3:469-478. [DOI] [PubMed] [Google Scholar]
- 2.Brown, P. O., B. Bowerman, H. E. Varmus, and J. M. Bishop. 1987. Correct integration of retroviral DNA in vitro. Cell 49:347-356. [DOI] [PubMed] [Google Scholar]
- 3.Bukrinsky, M. I., N. Sharova, T. L. McDonald, T. Pushkarskaya, W. G. Tarpley, and M. Stevenson. 1993. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. USA 90:6125-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bustin, M. 2001. Revised nomenclature for high mobility group (HMG) chromosomal proteins. Trends Biochem. Sci. 26:152-153. [DOI] [PubMed] [Google Scholar]
- 5.Cai, M., Y. Huang, R. Zheng, S.-Q. Wei, R. Ghirlando, M. S. Lee, R. Craigie, A. M. Gronenborn, and G. M. Clore. 1998. Solution structure of the cellular factor BAF responsible for protecting retroviral DNA from autointegration. Nat. Struct. Biol. 5:903-909. [DOI] [PubMed] [Google Scholar]
- 6.Carteau, S., R. J. Gorelick, and F. D. Bushman. 1999. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J. Virol. 73:6670-6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, H., and A. Engelman. 1998. The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc. Natl. Acad. Sci. USA 95:15270-15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, H., S.-Q. Wei, and A. Engelman. 1999. Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type 1 intasome. J. Biol. Chem. 274:17358-17364. [DOI] [PubMed] [Google Scholar]
- 9.Chen, H., and A. Engelman. 2001. Asymmetric processing of human immunodeficiency virus type 1 cDNA in vivo: implications for functional end coupling during the chemical steps of DNA transposition. Mol. Cell. Biol. 21:6758-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craigie, R., T. Fujiwara, and F. Bushman. 1990. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell 62:829-837. [DOI] [PubMed] [Google Scholar]
- 11.Ellison, V., H. Abrams, T. Roe, J. Lifson, and P. O. Brown. 1990. Human immunodeficiency virus integration in a cell-free system. J. Virol. 64:2711-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farnet, C. M., and F. D. Bushman. 1997. HIV-1 cDNA integration: requirement for HMG I(Y) protein for function of preintegration complexes in vitro. Cell 88:483-492. [DOI] [PubMed] [Google Scholar]
- 13.Farnet, C. M., and W. A. Haseltine. 1990. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc. Natl. Acad. Sci. USA 87:4164-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farnet, C. M., and W. A. Haseltine. 1991. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J. Virol. 65:1910-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furukawa, K. 1999. LAP2 binding protein 1 (L2BP1/BAF) is a candidate mediator of LAP2-chromatin interaction. J. Cell Sci. 112:2485-2492. [DOI] [PubMed] [Google Scholar]
- 16.Gallay, P., S. Swingler, C. Aiken, and D. Trono. 1995. HIV-1 infection of nondividing cells: C-terminal tyrosine phosphorylation of the viral matrix protein is a key regulator. Cell 80:379-388. [DOI] [PubMed] [Google Scholar]
- 17.Haraguchi, T., T. Koujin, M. Segura-Totten, K. K. Lee, Y. Matsuoka, Y. Yoneda, K. L. Wilson, and Y. Hiraoka. 2001. BAF is required for emerin assembly into the reforming nuclear envelope. J. Cell Sci. 114:4575-4585. [DOI] [PubMed] [Google Scholar]
- 18.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Harris, D., and A. Engelman. 2000. Both the structure and DNA binding function of the barrier-to-autointegration factor contribute to reconstitution of HIV type 1 integration in vitro. J. Biol. Chem. 275:39671-39677. [DOI] [PubMed] [Google Scholar]
- 20.Heinzinger, N. K., M. I. Bukrinsky, S. A. Haggerty, A. M. Ragland, V. Kewalramani, M. Lee, H. E. Gendelman, L. Ratner, M. Stevenson, and M. Emerman. 1994. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc. Natl. Acad. Sci. USA 91:7311-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hindmarsh, P., T. Ridky, R. Reeves, M. Andrake, A. M. Skalka, and J. Leis. 1999. HMG protein family members stimulate human immunodeficiency virus type 1 and avian sarcoma virus concerted DNA integration in vitro. J. Virol. 73:2994-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katz, R. A., G. Merkel, J. Kulkosky, J. Leis, and A. M. Skalka. 1990. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell 63:87-95. [DOI] [PubMed] [Google Scholar]
- 23.Lee, M. S., and R. Craigie. 1994. Protection of retroviral DNA from autointegration: involvement of a cellular factor. Proc. Natl. Acad. Sci. USA 91:9823-9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, M. S., and R. Craigie. 1998. A previously unidentified host protein protects retroviral DNA from autointegration. Proc. Natl. Acad. Sci. USA 95:1528-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, Y. M. H., and J. M. Coffin. 1991. Relationship of avian retrovirus DNA synthesis to integration in vitro. Mol. Cell. Biol. 11:1419-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, L., C. M. Farnet, W. F. Anderson, and F. D. Bushman. 1998. Modulation of activity of Moloney murine leukemia virus preintegration complexes by host factors in vitro. J. Virol. 72:2125-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, L., J. M. Olvera, K. E. Yoder, R. S. Mitchell, S. L. Butler, M. Lieber, S. L. Martin, and F. D. Bushman. 2001. Role of the non-homologous end joining pathway in the early steps of retroviral infection. EMBO J. 20:3272-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, L., K. Yoder, M. S. T. Hansen, J. Olvera, M. D. Miller, and F. D. Bushman. 2000. Retroviral cDNA integration: stimulation by HMG I family proteins. J. Virol. 74:10965-10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency type 1 preintegration complexes: studies of organization and function. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niedrig, N., J. Hinkula, W. Weigelt, J. L'Age-Stehr, G. Pauli, J. Rosen, and B. Wahren. 1989. Epitope mapping of monoclonal antibodies against human immunodeficiency virus type 1 structural proteins using peptides. J. Virol. 63:3525-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nilsen, B. M., I. R. Haugan, K. Berg, L. Olsen, P. O. Brown, and D. E. Helland. 1996. Monoclonal antibodies against human immunodeficiency virus type 1 integrase: epitope mapping and differential effects on integrase activities in vitro. J. Virol. 70:1580-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segura-Totten, M., A. K. Kowalski, R. Craigie, and K. L. Wilson. 2002. Barrier-to-autointegration factor: major roles in chromatin decondensation and nuclear assembly. J. Cell Biol. 158:475-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki, Y., and R. Craigie. 2002. Regulatory mechanisms by which barrier-to-autointegration factor blocks autointegration and stimulates intermolecular integration of Moloney murine leukemia virus preintegration complexes. J. Virol. 76:12376-12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tuschl, T. 2002. Expanding small RNA interference. Nat. Biotechnol. 20:446-448. [DOI] [PubMed] [Google Scholar]
- 35.Wei, S.-Q., K. Mizuuchi, and R. Craigie. 1997. A large nucleoprotein assembly at the ends of the viral DNA mediates retroviral DNA integration. EMBO J. 16:7511-7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei, S.-Q., K. Mizuuchi, and R. Craigie. 1998. Footprints on the viral DNA ends in Moloney murine leukemia virus preintegration complexes reflect a specific association with integrase. Proc. Natl. Acad. Sci. USA 95:10535-10540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilk, T., and S. D. Fuller. 1999. Towards the structure of the human immunodeficiency virus: divide and conquer. Curr. Opin. Struct. Biol. 9:231-243. [DOI] [PubMed] [Google Scholar]
- 38.Zheng, R., R. Ghirlando, M. S. Lee, K. Mizuuchi, M. Krause, and R. Craigie. 2000. Barrier-to-autointegration factor (BAF) bridges DNA in a discrete, higher-order nucleoprotein complex. Proc. Natl. Acad. Sci. USA 97:8997-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]