Abstract
The Op-iap3 gene from the baculovirus Orgyia pseudotsugata M nucleopolyhedrovirus (OpMNPV) inhibits apoptosis induced by a mutant of Autographa californica MNPV (AcMNPV) that lacks the antiapoptotic gene p35, as well as apoptosis induced by a wide range of other stimuli in both mammalian and insect cells. However, the role of Op-iap3 during OpMNPV infection has not been previously examined. To determine the function of the Op-IAP3 protein during OpMNPV infection, we used RNA interference (RNAi) to silence Op-iap3 expression during OpMNPV infection of Ld652Y cells. Infected cells treated with Op-iap3 double-stranded RNA (dsRNA) did not accumulate detectable Op-iap3 mRNA, confirming that the Op-iap3 gene was effectively silenced. Op-IAP3 protein was found to be a component of the budded virion; however, in OpMNPV-infected cells treated with Op-iap3 dsRNA, the Op-IAP3 protein that was introduced by the inoculum virus decreased to almost undetectable levels by 12 h after dsRNA addition. Apoptosis was observed in infected cells treated with Op-iap3 dsRNA beginning at 12 h, and by 48 h, almost all of the cells had undergone apoptosis. These results show for the first time that Op-IAP3 is necessary to prevent apoptosis during OpMNPV infection. In addition, our results demonstrate that the RNAi technique can be an effective tool for studying baculovirus gene function.
Apoptosis is a cell suicide program that is used by many organisms during normal development and tissue homeostasis and as an antiviral defense mechanism. Infection by many different viruses leads to the initiation of apoptosis in an attempt by the host cell to limit viral replication (34). This results in the activation of a family of cysteine proteases known as caspases, the central executioners of apoptosis (43, 49). To overcome this response, many viruses carry genes whose products inhibit apoptosis. Baculoviruses have two such types of antiapoptotic genes, p35 and inhibitor of apoptosis (iap) (10).
The p35 gene from Autographa californica M nucleopolyhedrovirus (AcMNPV) is required to prevent apoptosis during infection of Sf21 cells, a cell line derived from the lepidopteran insect Spodoptera frugiperda (12). Infection with AcMNPV mutant viruses lacking p35 results in apoptosis and greatly reduced virus yields in Sf21 cells (13, 28). In addition, p35 mutant viruses are significantly attenuated in their ability to infect S. frugiperda larvae (13, 15), and this reduction in infectivity correlates with apoptosis in vivo (9). The P35 protein blocks apoptosis due to its ability to directly inhibit a broad range of caspases (7, 20).
The first iap genes were discovered in Cydia pomonella granulovirus and Orgyia pseudotsugata MNPV (OpMNPV) during a genetic screen for genes that could rescue the AcMNPV p35 mutant phenotype (6, 17). Nearly all baculoviruses that have been sequenced to date contain at least one, and often several, iap-homologous genes, with the sole exception being the highly divergent mosquito baculovirus Culex nigripalpus NPV (1). In addition to baculoviruses, other viruses have also been found to contain genes with homology to baculovirus iap, including African swine fever virus (53), Chilo iridescent virus (32), and Amsacta moorei entomopoxvirus (4). Interestingly, all of the viruses known to date that encode iap genes infect arthropods during at least part of their replication cycle. In addition to viral iap genes, iap-homologous genes have been described for organisms ranging from yeasts to mammals (11). IAP proteins are characterized by having one to three copies of a motif known as the baculovirus IAP repeat (BIR) at their amino termini, which are involved in protein-protein interactions. Certain BIR motifs from several cellular IAP proteins are able to directly interact with and inhibit certain caspases (18). In addition to caspase inhibition, there is evidence for other antiapoptotic functions for IAPs, including interactions between BIRs and proteins in various signal transduction pathways, as well as E3 ubiquitin ligase activity via a carboxy-terminal RING motif found in most, but not all, IAPs (44).
After the discovery of the original Op-iap gene in OpMNPV (6), sequencing of the OpMNPV genome revealed the presence of two additional iap genes (2). The original Op-iap, which was named Op-iap3, is apparently the only OpMNPV gene capable of rescuing AcMNPV mutants lacking p35 (6). It has been shown previously that Op-iap3 can suppress apoptosis in insect and mammalian cells when expressed ectopically (14, 19, 26). Op-IAP3 directly binds to the Drosophila melanogaster apoptosis-inducing proteins Hid, Reaper, and Grim and inhibits the ability of these proteins to induce apoptosis, probably by more than one mechanism (50-52). Although Op-IAP3 has not been shown to directly inhibit caspases, it appears to inhibit an apical caspase activity in Sf21 cells, either directly or indirectly (35, 38, 45).
Besides Op-iap3, the other baculovirus iap genes that have been shown to possess antiapoptotic activity are Cp-iap3 from C. pomonella granulovirus (14) and Eppo-iap1 and Eppo-iap2 from Epiphyas postvittana NPV (37). However, the antiapoptotic activity of each of these viral iap genes has been examined only by expression outside their native context. Thus, to date it has not been shown that any baculovirus iap genes are actually required to prevent apoptosis during infection with their cognate virus. This has been due in large part to difficulties in genetic manipulation that are associated with many baculoviruses.
As an alternative to constructing viral genetic mutants, we have made use of the relatively new technique of gene silencing known as RNA interference (RNAi). RNAi is a sequence-specific gene-silencing mechanism operating in plants, fungi, and multicellular animals (23). Following the introduction of double-stranded RNA (dsRNA) into cells, the degradation of endogenous mRNA with homology to the dsRNA is triggered, effectively silencing expression of the target gene. The mechanisms involved in recognition and cleavage of the target RNAs are only beginning to be elucidated, but it is thought that an endonuclease, termed Dicer (5), processes the trigger dsRNA into ∼22-nucleotide small interfering RNAs. These small interfering RNAs then serve as guide sequences to direct the degradation of homologous RNA molecules by a nuclease activity termed the RNA-induced silencing complex (23). RNAi has been used successfully to suppress the replication of several animal RNA viruses, including human immunodeficiency virus, Rous sarcoma virus, Semliki Forest virus, and dengue virus (8, 16, 30). However, we are not aware of any previous reports of the use of RNAi to silence viral gene expression from DNA viruses.
Here we have used RNAi to silence Op-iap3 and examine its role during OpMNPV infection. Our results demonstrate that, although the Op-IAP3 protein is carried in the OpMNPV budded virion, its continued synthesis after infection is required to suppress apoptosis during OpMNPV replication in Ld652Y cells. This is the first report of a baculovirus iap gene being required to block apoptosis during infection with its cognate virus. Our results also suggest that the RNAi technique may be useful for studying the functions of genes from not only baculoviruses but other viruses as well, especially viruses that are difficult to manipulate by traditional genetic techniques.
MATERIALS AND METHODS
Cells and viruses.
S. frugiperda (Sf21) and Lymantria dispar (Ld652Y) cells were maintained in TC-100 medium (Invitrogen) supplemented with 10% tryptose broth and 10% fetal bovine serum (FBS) (Invitrogen). OpMNPV was obtained from Suzanne Thiem (Michigan State University), and the virus was propagated and its titers were determined by plaque assay in Ld652Y cells, which were obtained from David Theilmann (Pacific Agri-Food Research Center). The recombinant virus vOpIAPR-11 (p35− Op-iap3+) (6) and wild-type AcMNPV (L1 strain) were propagated and their titers were determined in Sf21 cells.
RNAi procedure.
Fragments of the Op-iap3, Sf-actin, and chloramphenicol acetyltransferase (CAT) genes were cloned into the pCRII vector (Invitrogen). The sequences used for Sf-actin and CAT have been described elsewhere (40), while the sequence used for Op-iap3 was the last 511 nucleotides of the Op-iap3 open reading frame (nucleotides 28801 to 29607 on the OpMNPV genome). The plasmids were linearized with PvuII, and the Ampliscribe kit (EpiCentre Technologies) was then used to transcribe cRNA strands in vitro from the T7 and Sp6 promoters according to the protocol supplied with the kit. The resulting single-stranded RNA was purified and resuspended in water as described in the Ampliscribe kit protocol. The concentrations of the RNAs were determined with a spectrophotometer. The appropriate amounts of single-stranded RNAs were then mixed and annealed by being heated to 65οC for 15 min and allowed to cool to room temperature. In addition to the gene-specific sequences, each dsRNA also contained 558 bp derived from the vector. Ld652Y or Sf21 cells were plated in six-well plates at 5 × 105 cells per well in TC-100 medium supplemented with 10% FBS. After 1 h, the cells were infected at a multiplicity of infection (MOI) of 20 PFU per cell. An hour later, the virus inoculum was removed and cells were washed once with 1 ml of TC-100 medium without FBS. Either 80 μg (Fig. 1) or 160 μg (Fig. 2, 3, 5, and 6) of dsRNA was introduced into the cells by lipid-mediated transfection in TC-100 medium without FBS. The lipid used for transfection was a sonicated mixture of 0.6 mg of DOTAP (N-[I-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium chloride salt) (Avanti Polar Lipids, Inc.)/ml and 0.4 mg of DOPE (1,2-di[(cis)-9-octadecanoyl]-sn-glycero-3-phosphoethanolamine) (Sigma)/ml in sterile water. After 4 h, the lipid-RNA mix was replaced with TC-100 medium containing 10% FBS.
FIG. 1.
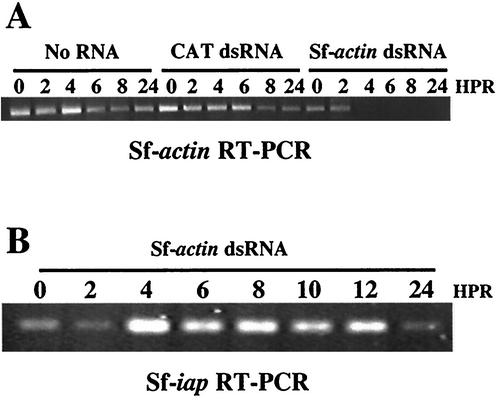
AcMNPV infection does not inhibit RNAi. Sf21 cells were infected with AcMNPV, and 1 h later the cells were mock transfected (No RNA) or transfected with 80 μg of the indicated dsRNAs. Total RNA was isolated at the indicated times, and RT-PCR was performed to detect the presence of Sf-actin (A) or Sf-iap mRNA (B). HPR, hours post-dsRNA addition.
FIG. 2.
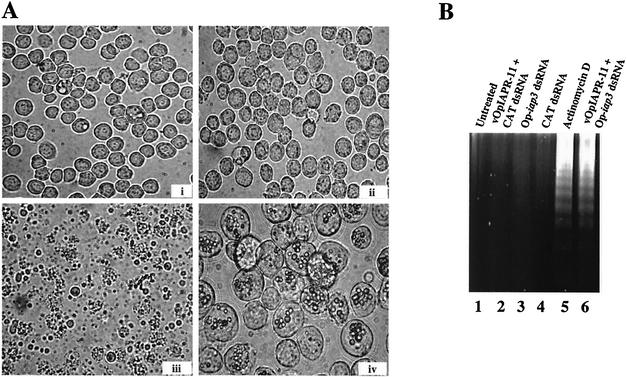
Silencing of Op-iap3 during infection with vOpIAPR-11, an AcMNPV recombinant that expresses Op-iap3 and lacks p35, induces apoptosis in Sf21 cells. (A) Sf21 cells were left untreated (i) or infected with vOpIAPR-11 (ii to iv) and transfected with 160 μg of control CAT dsRNA (ii and iv) or Op-iap3 dsRNA (iii). Photographs were taken at 18 (i to iii) or 74 (iv) h after dsRNA addition. Magnifications, ×400 (i to iii) and ×1,000 (iv). (B) Ethidium bromide-stained agarose gel showing DNA fragmentation in vOpIAPR-11-infected cells transfected with Op-iap3 dsRNA. Lane 1, untreated Sf21 cells; lane 2, vOpIAPR-11-infected Sf21 cells transfected with CAT dsRNA; lane 3, uninfected Sf21 cells transfected with Op-iap3 dsRNA; lane 4, uninfected Sf21 cells transfected with CAT dsRNA; lane 5, Sf21 cells treated with actinomycin D; lane 6, vOpIAPR-11-infected Sf21 cells transfected with Op-iap3 dsRNA.
FIG. 3.
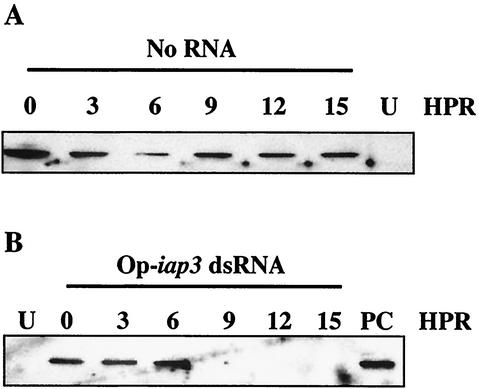
Op-iap3 dsRNA transfection during vOpIAPR-11 infection of Sf21 cells eliminates Op-IAP3 protein expression. The levels of Op-IAP3 protein were determined by immunoblotting with antiserum made against Op-IAP3 at the indicated times following mock transfection of vOpIAPR-11-infected cells (A) or transfection of vOpIAPR-11-infected cells with Op-iap3 dsRNA (B). PC, positive control lysate from Sf21 cells transfected with a vector overexpressing Op-IAP3; U, lysate from untreated Sf21 cells; HPR, hours post-dsRNA addition.
FIG. 5.
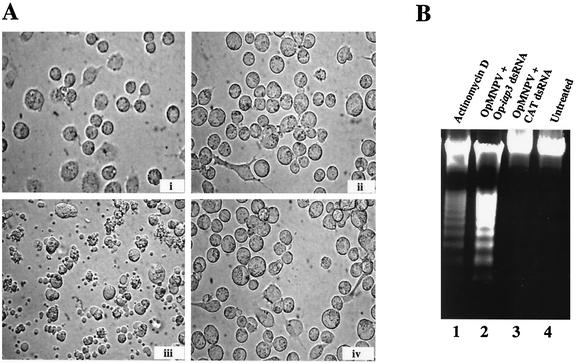
Silencing of Op-iap3 during OpMNPV infection of Ld652Y cells induces apoptosis. (A) Ld652Y cells were either untreated (i), transfected with control CAT dsRNA during OpMNPV infection (ii), transfected with Op-iap3 dsRNA during OpMNPV infection (iii), or transfected with Op-iap3 dsRNA in the absence of virus infection (iv). Photographs were taken at 48 h after the transfection of dsRNA. Magnification, ×400. (B) DNA fragmentation was observed in Ld652Y cells treated with actinomycin D (lane 1) or Op-iap3 dsRNA during OpMNPV infection (lane 2) but not in OpMNPV-infected cells treated with CAT dsRNA (lane 3) or in untreated cells (lane 4).
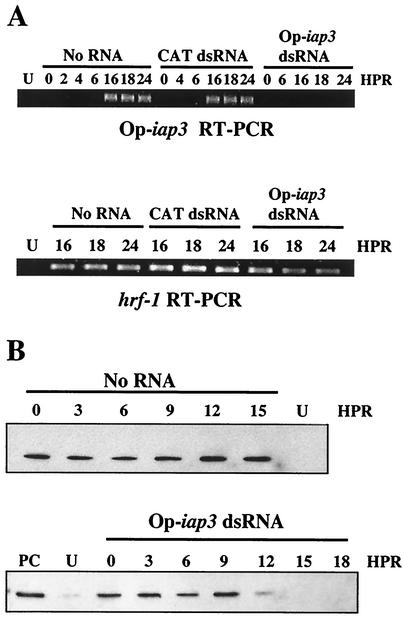
FIG. 6.
Transfection of Op-iap3 dsRNA silences Op-iap3 expression during OpMNPV infection. (A) The presence of Op-iap3 (top panel) and hrf-1 (bottom panel) mRNA was determined by RT-PCR at the indicated times in OpMNPV-infected Ld652Y cells following mock transfection (No RNA) or transfection with CAT or Op-iap3 dsRNA. U, uninfected Ld652Y cells; HPR, hours post-dsRNA addition. (B) The levels of Op-IAP3 protein were determined by immunoblotting in OpMNPV-infected Ld652Y cells at the indicated times following mock transfection (upper panel) or transfection with Op-iap3 dsRNA (lower panel). PC, positive control lysate from Sf21 cells transfected with a vector overexpressing Op-IAP3; U, uninfected Ld652Y cell lysate used as a negative control; HPR, hours post-dsRNA addition.
Purification of BV.
OpMNPV budded virus (BV) was purified essentially as described by Gross et al. (22). In short, a 300-ml culture of Ld652Y cells was infected with OpMNPV BV at an MOI of 10 and BV was isolated at 120 h postinfection. Cells were removed by centrifugation (2,000 × g for 10 min), and the supernatant was then centrifuged for 30 min at 80,000 × g. The resulting pellet was resuspended in TE buffer (10 mM Tris [pH 7.5], 1 mM EDTA) and further purified by centrifugation through a 33 to 55% sucrose gradient at 80,000 × g for 2 h at 4°C. The virion band was then removed, diluted in TE buffer, and sedimented at 80,000 × g for 1 h to remove sucrose. Approximately 24 μg of total protein from either the purified BV sample or the 48-h-infected cell lysate, as determined by the Coomassie Plus protein assay reagent kit (Pierce), was analyzed by immunoblotting.
Immunoblot analysis.
Ld652Y or Sf21 cells were infected with virus and then treated with dsRNA as described above. Samples were collected in sodium dodecyl sulfate-polyacrylamide loading buffer at various times after dsRNA addition. Time zero was defined as the time when dsRNA was added. Cell lysates were analyzed by immunoblotting with polyclonal antisera to Op-IAP3 (raised in rabbits against a peptide corresponding to residues 64 to 76) or gp37 (22) (obtained from George Rohrmann, Oregon State University) or a monoclonal antibody to IE-1 (48) (obtained from David Theilmann) and Supersignal chemiluminescent reagent (Pierce). Before use, the Op-IAP3 antiserum was preabsorbed with Sf21 cell proteins as described elsewhere (24).
Reverse transcription-PCR (RT-PCR).
Ld652Y or Sf21 cells were infected at an MOI of 20 followed by dsRNA addition and harvested at various times after dsRNA treatment. Total RNA was isolated with Trizol reagent (Invitrogen). A gene-specific primer was used in a reverse transcriptase reaction with 3 μg of total RNA. Two microliters of cDNA from a 50-μl reaction mixture was then used as a template in a PCR with nested primers specific for the sequence of interest. In each case, at least one of the primers used for PCR was complementary to a sequence not used to make the dsRNA, such as an untranslated region. The PCR products were separated by agarose gel electrophoresis and stained with ethidium bromide.
DNA laddering.
Cells were treated with dsRNA or 0.2 μg of actinomycin D (Invitrogen)/ml and harvested in 0.4 M Tris (pH 7.5)-0.1% sodium dodecyl sulfate-0.1 M EDTA, followed by phenol-chloroform extraction and ethanol precipitation. DNA was separated by agarose gel electrophoresis and stained with ethidium bromide.
RESULTS
Baculovirus infection does not inhibit RNAi.
Our group has previously used the RNAi technique to silence cellular genes in Sf21 cells (40). However, when we began these studies it was not clear whether RNAi could be used to silence a baculovirus gene, since evidence exists that some viruses are able to inhibit RNAi during infection (3, 36). To determine whether baculovirus infection inhibits RNAi, we first attempted to silence the cellular actin gene in baculovirus-infected cells. Sf21 cells were infected with AcMNPV, and 1 h later, 80 μg of dsRNA corresponding to Sf-actin was transfected into the cells. Total RNA was then extracted at various times after dsRNA addition, and RT-PCR was performed to determine Sf-actin mRNA levels. RT-PCR showed that, within 4 h after dsRNA treatment, Sf-actin transcript levels were undetectable (Fig. 1A). Although silencing was highly effective in infected cells, infection did appear to slightly delay the RNAi effect, since Sf-actin transcript was undetectable at 2 h when uninfected cells were transfected with Sf-actin dsRNA (data not shown). To ensure that silencing was specific, transcript levels for another cellular gene, Sf-iap, were also tested following introduction of Sf-actin dsRNA. RT-PCR showed that the Sf-iap transcript was present throughout the experiment, suggesting that silencing was specific (Fig. 1B); the decrease seen at 24 h was probably due to the ability of AcMNPV to shut off host transcription (41). In addition, Sf-actin mRNA levels were unaffected when dsRNA corresponding to the bacterial CAT gene was transfected as a control (Fig. 1A).
Silencing Op-iap3 in recombinant AcMNPV induces apoptosis.
Infection of Sf21 cells with an AcMNPV mutant that lacks a functional p35 gene induces apoptosis, including the hallmarks of plasma membrane blebbing and DNA fragmentation (12). However, Op-iap3 can functionally substitute for p35 and prevent Sf21 cells from undergoing apoptosis during AcMNPV infection (6). Therefore, we next used RNAi to silence Op-iap3 during infection of Sf21 cells with vOpIAPR-11, a recombinant of AcMNPV that lacks p35 but contains Op-iap3. vOpIAPR-11 was originally isolated as an occlusion-positive plaque following cotransfection of p35 mutant AcMNPV and a plasmid containing Op-iap3 and flanking genomic DNA from OpMNPV (6). Thus, it is assumed that vOpIAPR-11 expresses Op-iap3 from its native OpMNPV promoter. Sf21 cells were infected with vOpIAPR-11, and 1 h after viral infection, dsRNA corresponding to Op-IAP3 was transfected into the cells. Although 80 μg of dsRNA was sufficient to completely silence several different cellular genes in Sf21 cells (Fig. 1) (40), we found that 160 μg of dsRNA was optimal for silencing Op-iap3. Plasma membrane blebbing consistent with apoptosis was observed within 8 h after transfection of 160 μg of Op-iap3 dsRNA (data not shown), and by 18 h 99% of the cells had undergone apoptosis (Fig. 2A). Apoptosis was not observed in infected cells transfected with control CAT dsRNA or in uninfected cells transfected with dsRNA corresponding to Op-iap3, Sf-actin, or CAT (data not shown). The formation of polyhedra was observed in infected cells transfected with CAT dsRNA, indicating that the addition of dsRNA did not inhibit viral replication (Fig. 2A).
DNA laddering was observed in Sf21 cells transfected with Op-iap3 dsRNA during vOpIAPR-11 infection (Fig. 2B), similar to that seen following treatment of uninfected cells with actinomycin D, a known inducer of apoptosis in Sf21 cells (14). This result confirmed that the death observed was due to apoptosis. DNA laddering was not observed for uninfected cells transfected with either Op-iap3 dsRNA or CAT dsRNA or for infected cells transfected with CAT dsRNA (Fig. 2B).
Immunoblot analysis showed that Op-IAP3 protein was present within 1 h after infection with vOpIAPR-11 (at the time of dsRNA addition, shown as time zero in Fig. 3A), and the level of Op-IAP3 appeared to remain relatively constant through the next 15 h (Fig. 3A). However, in vOpIAPR-11-infected cells that received Op-iap3 dsRNA, Op-IAP3 protein disappeared between 6 and 9 h after dsRNA addition (Fig. 3B).
Op-IAP3 is required to prevent apoptosis during OpMNPV infection.
Knowing that we could effectively silence Op-iap3 during vOpIAPR-11 infection in Sf21 cells, we next wanted to determine if Op-iap3 functioned to prevent apoptosis during OpMNPV infection of Ld652Y cells. We first examined the time course of Op-IAP3 expression during OpMNPV infection. Initially, we examined Op-IAP3 expression from 2 to 24 h postinfection (p.i.) and were able to detect Op-IAP3 protein throughout the course of infection (Fig. 4A). Two bands with apparent molecular masses of 35 and 37 kDa were observed; the fainter 37-kDa band is often observed in immunoblots of Op-IAP3, even with the use of different antibodies directed against epitope-tagged versions of the protein, and is presumed to be due to posttranslational processing (52).
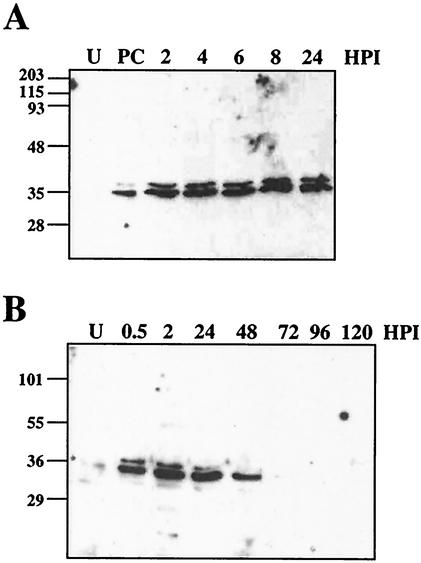
FIG. 4.
Time course of Op-IAP3 expression during OpMNPV infection of Ld652Y cells. Ld652Y cells were infected with OpMNPV and harvested at the indicated hours p.i. (HPI), and the levels of Op-IAP3 protein were determined by immunoblotting with antiserum against Op-IAP3. (A) PC, positive control lysate from Sf21 cells transfected with a plasmid overexpressing Op-IAP3; U, lysate from untransfected Sf21 cells. (B) U, lysate from uninfected Ld652Y cells. Molecular mass markers are indicated to the left of each panel in kilodaltons.
To obtain a more complete profile of Op-IAP3 expression, we also tested earlier and later time points. We were able to detect Op-IAP3 by immunoblotting as early as 30 min p.i. (Fig. 4B). Op-IAP3 levels stayed fairly constant through 24 h p.i., after which they decreased, until by 96 h p.i. Op-IAP3 was undetectable (Fig. 4B; a faint band was visible at 72 h p.i. in the original film). The apparent lack of Op-IAP3 at these late times may be explained by the fact that most of the cells in the culture had lysed by this time.
We then used RNAi to silence Op-iap3 during OpMNPV infection of Ld652Y cells. Ld652Y cells were infected for 1 h with OpMNPV and then transfected with 160 μg of Op-iap3 dsRNA. Plasma membrane blebbing was first observed 12 h after transfection with Op-iap3 dsRNA (data not shown), and by 48 h 98% of the cells had undergone apoptosis (Fig. 5A). Cells transfected in the absence of viral infection with dsRNA corresponding to Op-iap3 or CAT showed no signs of apoptosis, and cells transfected with CAT dsRNA during OpMNPV infection also showed no signs of apoptosis (Fig. 5A and data not shown). DNA laddering was observed in Ld652Y cells treated with Op-iap3 dsRNA during OpMNPV infection, similar to what was observed for actinomycin D-treated cells (Fig. 5B). DNA laddering was not observed in virus-infected cells transfected with CAT dsRNA or uninfected cells transfected with CAT or Op-iap3 dsRNA (Fig. 5B and data not shown).
To confirm that the Op-iap3 dsRNA was indeed silencing Op-IAP3 expression, the levels of Op-iap3 mRNA were examined by RT-PCR (Fig. 6A). In OpMNPV-infected Ld652Y cells receiving no dsRNA or CAT dsRNA, a band corresponding to the Op-iap3 transcript was observed at 16 h p.i. and was present through 24 h p.i. (Fig. 6A, upper panel). In contrast, in infected cells that received Op-iap3 dsRNA, Op-iap3 transcript was not detected through the first 24 h. The presence of a control viral transcript, hrf-1, was unaffected by either CAT or Op-iap3 dsRNA (Fig. 6A, lower panel). Immunoblot analysis showed that Op-IAP3 protein was present in infected cells within 1 h after virus infection, and the levels did not change through the first 16 h of OpMNPV infection (Fig. 6B, upper panel). Transfection of Op-iap3 dsRNA caused Op-IAP3 protein levels to decrease beginning between 9 and 12 h after transfection, and by 15 h Op-IAP3 was undetectable (Fig. 6B, lower panel).
Op-IAP3 copurifies with BV.
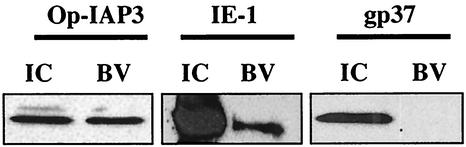
Immunoblot analysis revealed the presence of Op-IAP3 protein in infected cells as early as 30 min p.i. (Fig. 4), but the earliest time at which we were able to detect the presence of Op-iap3 transcript by RT-PCR was 12 h p.i. (data not shown). This led us to hypothesize that Op-IAP3 protein is associated with BV and carried into cells during infection. To test this, OpMNPV budded virions were purified by sucrose gradient centrifugation and analyzed by immunoblotting with antiserum to Op-IAP3. By this technique, Op-IAP3 was readily detectable in the purified BV (Fig. 7). For controls, immunoblot analysis was also carried out on purified BV with antibodies to IE-1 and gp37. Theilmann and Stewart have previously shown that IE-1 copurifies with OpMNPV BV (48), while Gross et al. reported that gp37 is not associated with BV (22). As expected, we were able to detect IE-1 but not gp37 protein in BV by immunoblotting, indicating that our BV purification was successful.
FIG. 7.
Association of Op-IAP3 with purified OpMNPV budded virions. Sucrose gradient-purified BV was analyzed by immunoblotting with polyclonal antisera against Op-IAP3 or OpMNPV gp37 or a monoclonal antibody against OpMNPV IE-1. IC, infected cell lysates harvested at 48 h p.i. An equal amount of total protein was loaded in each lane.
DISCUSSION
In this study we have demonstrated that a lack of expression of the Op-iap3 gene during OpMNPV infection of Ld652Y cells results in extensive apoptosis. To examine the role of Op-IAP3 during OpMNPV infection, we silenced Op-iap3 expression by the relatively new technique of RNAi. To our knowledge this is the first report of using RNAi to silence gene expression from a DNA virus. At the onset of these experiments it was necessary to determine whether baculovirus infection had an inhibitory effect on RNAi. This was a concern since it is thought that RNAi is an ancient antiviral defense mechanism, and some viruses, such as tobacco etch virus and flock house virus, encode genes that when expressed can inhibit RNA silencing (3, 36). Our results demonstrate that, at least in the cases of AcMNPV and OpMNPV, baculovirus infection does not appear to inhibit the RNAi response. Thus, RNAi promises to be an effective tool for silencing either cellular or viral genes during baculovirus infection. In these experiments, we added the dsRNA early in infection, and the viral genes that we attempted to silence are thought to be expressed during the early stage of infection. It thus remains possible that baculoviruses may have the ability to inhibit RNAi late in the infection process, and it is not known at this time whether late viral genes are susceptible to silencing by these techniques.
Although OpMNPV contains three genes with homology to the iap family, it is unlikely that the Op-iap3 dsRNA affected the expression of Op-iap1 or Op-iap2, since significant regions of complete nucleotide identity (greater than 30 to 35 nucleotides) are required for the RNAi effect (46), and Op-iap1 and Op-iap2 do not share any significant regions of identity with Op-iap3. The Op-iap3 open reading frame, of which a portion was used to produce the dsRNA, also does not overlap with any other open reading frames, as is sometimes the case with baculovirus genes. It is possible, however, that transcripts of neighboring genes could overlap with the portion of the Op-iap3 open reading frame used to produce dsRNA and that the expression of neighboring genes could have been affected by the dsRNA used. Transcriptional mapping has not been done in this region of the OpMNPV genome, and most of the genes in this region are of unknown function, although the open reading frame immediately upstream of iap3 has homology to the small subunit of ribonucleotide reductase (2). Given the facts that Op-iap3 is known to have antiapoptotic activity and that silencing of Op-iap3 in the AcMNPV recombinant virus vOpIAPR-11 also resulted in apoptosis, it seems likely that any effects on the expression of neighboring genes (if they occurred) did not contribute to the observed phenotype when Op-iap3 dsRNA was used. However, it is clear that these issues should be carefully considered in future studies using RNAi to silence viral genes.
The mechanism by which Op-IAP3 inhibits apoptosis remains unclear. When AcMNPV infects Sf21 cells, an apoptotic pathway is triggered that leads to the activation of one or more initiator caspases. This then leads to the activation of downstream effector caspases such as Sf-caspase-1, which are then presumably responsible for cleaving various target proteins, leading to apoptosis (47). Previous work has shown that P35 directly binds and inhibits the cleaved, active form of a number of caspases (7). However, p35 expression is unable to prevent the first step in Sf-caspase-1 cleavage (35, 45). On the other hand, expression of Op-iap3 does prevent Sf-caspase-1 cleavage (35, 45), but Op-IAP3 is unable to prevent apoptosis induced by expression of activated Sf-caspase-1 (45). Thus, Op-IAP3 functions upstream of both Sf-caspase-1 activation and the point at which P35 inhibits apoptosis.
Based on the findings described above, it is possible that Op-IAP3 functions by inhibiting an initiator caspase in Sf21 cells. Further support of this hypothesis comes from the report that Cp-IAP can inhibit the mammalian initiator caspase, caspase-9 (31). However, in the same study, Op-IAP3 was not able to inhibit caspase-9. Another possibility is that Op-IAP3 may indirectly inhibit the activation of caspases by interacting with other proapoptotic factors. The baculovirus IAPs Op-IAP3 and Cp-IAP can physically interact with the Drosophila apoptotic inducers Reaper, HID, GRIM, and DOOM (25, 50, 51). Overexpression of these genes in Sf21 cells results in the activation of Sf-caspase-1 and apoptosis, both of which are blocked by coexpression of Op-IAP3 (33). Hence, Op-IAP3 may function by interacting with homologs of these Drosophila apoptotic inducers in Ld652Y cells. However, although the ability to bind to HID appears to be necessary for Op-IAP3 to inhibit HID-induced apoptosis, it is not sufficient, since several mutants have been identified that bind HID normally but are totally inactive in inhibiting HID-induced apoptosis (51, 52). Furthermore, although Op-IAP3 BIR2 is necessary and sufficient for HID binding, BIR2 by itself is only a weak suppressor of HID-induced death compared to the full-length Op-IAP3 protein (51, 52). In addition to BIR2, the presence of the RING motif of Op-IAP3 is required for optimal antiapoptotic activity, and the RING motif has the ability to promote HID ubiquitination both in vitro and in transfected Sf21 cells (M. C. Green, K. Miller, and R. J. Clem, submitted for publication). Thus, Op-IAP3 may function to inhibit apoptosis both by direct caspase inhibition and by ubiquitination of proapoptotic molecules.
Our results also show that Op-IAP3 protein is carried in the budded form of OpMNPV. Op-IAP3 protein was detected in Ld652Y cells within 30 min after addition of virus, but Op-iap3 mRNA was not detected until several hours later. In addition, Op-IAP3 copurified with BV during sucrose gradient centrifugation. We also found that the appearance of Op-IAP3 in infected Ld652Y cells was not affected by the addition of cycloheximide prior to infection (data not shown). Since it has been previously shown that the AcMNPV IE-1 protein induces apoptosis in Sf21 cells under some circumstances (42), and OpMNPV IE-1 is also carried in BV (48), it may be necessary for Op-IAP3 to be carried into cells during infection to immediately inhibit apoptosis stimulated by IE-1. Interestingly, Sf21 cells infected with a recombinant of AcMNPV expressing Op-IAP3 from its native promoter also contained Op-IAP3 protein immediately after infection (Fig. 3), indicating that whatever mechanism results in packaging of Op-IAP3 in OpMNPV is probably conserved in AcMNPV. It has been previously reported that P35 also associates with AcMNPV BV (29). Thus, it appears that the association of an antiapoptotic protein with BV may be common in baculoviruses and is possibly necessary for the prevention of apoptosis early in the infection cycle.
While the finding that Op-IAP3 is necessary to inhibit apoptosis during OpMNPV infection was not unexpected, given its ability to inhibit apoptosis in other contexts, it was nonetheless important to determine whether this was the case. Op-iap3, like several other baculovirus iap genes, has been studied rather extensively outside of the context of infection by its native virus, but it had not been previously shown that any baculovirus iap gene is required to prevent apoptosis during a natural infection. Indeed, mutational analysis of the Ac-iap1 gene indicated that it is not required to prevent AcMNPV-induced apoptosis (21, 39), although Ac-iap1 also lacks the ability to block apoptosis induced by other stimuli (14, 27, 31). Our findings demonstrate that Op-iap3 is important for preventing apoptosis during OpMNPV infection, indicating that other baculovirus iap genes that have the ability to block apoptosis outside of the context of virus infection are probably also required to prevent apoptosis during infection with their native virus. Finally, our results indicate that the RNAi technique shows promise as a useful tool for quickly and easily studying the function of baculovirus genes, and possibly genes from other viruses as well.
Acknowledgments
We are grateful to Thomas Clarke for suggesting the idea of using RNAi to silence baculovirus genes. We also thank Suzanne Thiem, David Theilmann, and George Rohrmann for providing reagents and A. Lorena Passarelli for critical reading of the manuscript.
This work was supported by Public Health Service grants R29 CA78602 from the National Cancer Institute and PO1 RR017686 from the National Center for Research Resources and by the Kansas Agricultural Experiment Station.
Footnotes
Contribution no. 03-165-J from the Kansas Agricultural Experiment Station.
REFERENCES
- 1.Afonso, C. L., E. R. Tulman, Z. Lu, C. A. Balinsky, B. A. Moser, J. J. Becnel, D. L. Rock, and G. F. Kutish. 2001. Genome sequence of a baculovirus pathogenic for Culex nigripalpus. J. Virol. 75:11157-11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahrens, C. H., R. L. Q. Russell, C. J. Funk, J. T. Evans, S. H. Harwood, and G. F. Rohrmann. 1997. The sequence of the Orgyia pseudotsugata multinucleocapsid nuclear polyhedrosis virus genome. Virology 229:381-399. [DOI] [PubMed] [Google Scholar]
- 3.Anandalakshmi, R., G. J. Pruss, X. Ge, R. Marathe, A. C. Mallory, T. H. Smith, and V. B. Vance. 1998. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 95:13079-13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bawden, A. L., K. J. Glassberg, J. Diggans, R. Shaw, W. Farmerie, and R. W. Moyer. 2000. Complete genomic sequence of the Amsacta moorei entomopoxvirus: analysis and comparison with other poxviruses. Virology 274:120-139. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363-366. [DOI] [PubMed] [Google Scholar]
- 6.Birnbaum, M. J., R. J. Clem, and L. K. Miller. 1994. An apoptosis-inhibiting gene from a nuclear polyhedrosis virus encoding a peptide with Cys/His sequence motifs. J. Virol. 68:2521-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bump, N. J., M. Hackett, M. Hugunin, S. Seshagiri, K. Brady, P. Chen, C. Ferenz, S. Franklin, T. Ghayur, P. Li, P. Licari, J. Mankovich, L. Shi, A. H. Greenberg, L. K. Miller, and W. W. Wong. 1995. Inhibition of ICE family proteases by baculovirus antiapoptotic protein p35. Science 269:1885-1888. [DOI] [PubMed] [Google Scholar]
- 8.Caplen, N. J., Z. Zheng, B. Falgout, and R. A. Morgan. 2002. Inhibition of viral gene expression and replication in mosquito cells by dsRNA-triggered RNA interference. Mol. Ther. 6:243-251. [DOI] [PubMed] [Google Scholar]
- 9.Clarke, T. E., and R. J. Clem. 2003. In vivo induction of apoptosis correlating with reduced infectivity during baculovirus infection. J. Virol. 77:2227-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clem, R. J. 2001. Baculoviruses and apoptosis: the good, the bad, and the ugly. Cell Death Differ. 8:137-143. [DOI] [PubMed] [Google Scholar]
- 11.Clem, R. J., and C. S. Duckett. 1997. The iap genes: unique arbitrators of cell death. Trends Cell Biol. 7:337-339. [DOI] [PubMed] [Google Scholar]
- 12.Clem, R. J., M. Fechheimer, and L. K. Miller. 1991. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science 254:1388-1390. [DOI] [PubMed] [Google Scholar]
- 13.Clem, R. J., and L. K. Miller. 1993. Apoptosis reduces both the in vitro replication and the in vivo infectivity of a baculovirus. J. Virol. 67:3730-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clem, R. J., and L. K. Miller. 1994. Control of programmed cell death by the baculovirus genes p35 and iap. Mol. Cell. Biol. 14:5212-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clem, R. J., M. Robson, and L. K. Miller. 1994. Influence of infection route on the infectivity of baculovirus mutants lacking the apoptosis-inhibiting gene p35 and the adjacent gene p94. J. Virol. 68:6759-6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coburn, G. A., and B. R. Cullen. 2002. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J. Virol. 76:9225-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crook, N. E., R. J. Clem, and L. K. Miller. 1993. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J. Virol. 67:2168-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deveraux, Q. L., and J. C. Reed. 1999. IAP family proteins—suppressors of apoptosis. Genes Dev. 13:239-252. [DOI] [PubMed] [Google Scholar]
- 19.Duckett, C. S., V. E. Nava, R. W. Gedrich, R. J. Clem, J. L. Van Dongen, M. C. Gilfillan, H. Shiels, J. M. Hardwick, and C. B. Thompson. 1996. A conserved family of cellular genes related to the baculovirus iap gene and encoding apoptosis inhibitors. EMBO J. 15:2685-2694. [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher, A. J., W. dela Cruz, S. J. Zoog, C. L. Schneider, and P. D. Friesen. 1999. Crystal structure of baculovirus P35: role of a novel reactive site loop in apoptotic caspase inhibition. EMBO J. 18:2031-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths, C. M., A. L. Barnett, M. D. Ayres, J. Windass, L. A. King, and R. D. Possee. 1999. In vitro host range of Autographa californica nucleopolyhedrovirus recombinants lacking functional p35, iap1 or iap2. J. Gen. Virol. 80:1055-1066. [DOI] [PubMed] [Google Scholar]
- 22.Gross, C. H., G. M. Wolgamot, R. L. Q. Russell, M. N. Pearson, and G. F. Rohrmann. 1993. A 37-kilodalton glycoprotein from a baculovirus of Orgyia pseudotsugata is localized to cytoplasmic inclusion bodies. J. Virol. 78:469-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hannon, G. J. 2002. RNA interference. Nature 418:244-251. [DOI] [PubMed] [Google Scholar]
- 24.Harlow, E., and D. Lane. 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Harvey, A., A. Bidwai, and L. Miller. 1997. Doom, a product of the Drosophila mod(mdg4) gene, induces apoptosis and binds to baculovirus inhibitor-of-apoptosis proteins. Mol. Cell. Biol. 17:2835-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hawkins, C., A. Uren, G. Hacker, R. Medcalf, and D. Vaux. 1996. Inhibition of interleukin 1 beta-converting enzyme-mediated apoptosis of mammalian cells by baculovirus IAP. Proc. Natl. Acad. Sci. USA 93:13786-13790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hawkins, C. J., P. G. Ekert, A. G. Uren, S. P. Holmgreen, and D. L. Vaux. 1998. Anti-apoptotic potential of insect cellular and viral IAPs in mammalian cells. Cell Death Differ. 5:569-576. [DOI] [PubMed] [Google Scholar]
- 28.Hershberger, P. A., J. A. Dickson, and P. D. Friesen. 1992. Site-specific mutagenesis of the 35-kilodalton protein gene encoded by Autographa californica nuclear polyhedrosis virus: cell line-specific effects on virus replication. J. Virol. 66:5525-5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hershberger, P. A., D. J. LaCount, and P. D. Friesen. 1994. The apoptotic suppressor P35 is required early during baculovirus replication and is targeted to the cytosol of infected cells. J. Virol. 68:3467-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu, W., C. Myers, J. Kilzer, S. Pfaff, and F. Bushman. 2002. Inhibition of retroviral pathogenesis by RNA interference. Curr. Biol. 12:1301-1311. [DOI] [PubMed] [Google Scholar]
- 31.Huang, Q., Q. L. Deveraux, S. Maeda, G. S. Salvesen, H. R. Stennicke, B. D. Hammock, and J. C. Reed. 2000. Evolutionary conservation of apoptosis mechanisms: Lepidopteran and baculoviral inhibitor of apoptosis proteins are inhibitors of mammalian caspase-9. Proc. Natl. Acad. Sci. USA 97:1427-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jakob, N. J., K. Muller, U. Bahr, and G. Darai. 2001. Analysis of the first complete DNA sequence of an invertebrate iridovirus: coding strategy of the genome of Chilo iridescent virus. Virology 286:182-196. [DOI] [PubMed] [Google Scholar]
- 33.Kaiser, W. J., D. Vucic, and L. K. Miller. 1998. The Drosophila inhibitor of apoptosis D-IAP1 suppresses cell death induced by the caspase drICE. FEBS Lett. 440:243-248. [DOI] [PubMed] [Google Scholar]
- 34.Kannourakis, G., and S. Hay. 2002. A time to kill: viral manipulation of the cell death program. J. Gen. Virol. 83:1547-1564. [DOI] [PubMed] [Google Scholar]
- 35.LaCount, D. J., S. F. Hanson, C. L. Schneider, and P. D. Friesen. 2000. Caspase inhibitor P35 and inhibitor of apoptosis Op-IAP block in vivo proteolytic activation of an effector caspase at different steps. J. Biol. Chem. 275:15657-15664. [DOI] [PubMed] [Google Scholar]
- 36.Li, H., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 37.Maguire, T., P. Harrison, O. Hyink, J. Kalmakoff, and V. K. Ward. 2000. The inhibitors of apoptosis of Epiphyas postvittana nucleopolyhedrovirus. J. Gen. Virol. 81:2803-2811. [DOI] [PubMed] [Google Scholar]
- 38.Manji, G. A., R. R. Hozak, D. J. LaCount, and P. D. Friesen. 1997. Baculovirus inhibitor of apoptosis functions at or upstream of the apoptotic suppressor P35 to prevent programmed cell death. J. Virol. 71:4509-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLachlin, J. R., J. C. Escobar, J. A. Harrelson, R. J. Clem, and L. K. Miller. 2001. Deletions in the Ac-iap1 gene of the baculovirus AcMNPV occur spontaneously during serial passage and confer a cell line-specific replication advantage. Virus Res. 81:77-91. [DOI] [PubMed] [Google Scholar]
- 40.Muro, I., B. A. Hay, and R. J. Clem. 2002. The Drosophila DIAP1 protein is required to prevent accumulation of a continuously generated, processed form of the apical caspase DRONC. J. Biol. Chem. 277:49644-49650. [DOI] [PubMed] [Google Scholar]
- 41.Ooi, B. G., and L. K. Miller. 1988. Regulation of host RNA levels during baculovirus infection. Virology 166:515-523. [DOI] [PubMed] [Google Scholar]
- 42.Prikhodko, E. A., and L. K. Miller. 1996. Induction of apoptosis by baculovirus transactivator IE1. J. Virol. 70:7116-7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roy, N., Q. L. Deveraux, R. Takahashi, G. S. Salvesen, and J. C. Reed. 1997. The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspases. EMBO J. 16:6914-6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salvesen, G. S., and C. S. Duckett. 2002. IAP proteins: blocking the road to death's door. Nat. Rev. Mol. Cell Biol. 3:401-410. [DOI] [PubMed] [Google Scholar]
- 45.Seshagiri, S., and L. K. Miller. 1997. Baculovirus inhibitors of apoptosis (IAPs) block activation of Sf-caspase-1. Proc. Natl. Acad. Sci. USA 94:13606-13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharp, P. A. 2001. RNA interference—2001. Genes Dev. 15:485-490. [DOI] [PubMed] [Google Scholar]
- 47.Stennicke, H. R., and G. S. Salvesen. 2000. Caspases—controlling intracellular signals by protease zymogen activation. Biochim. Biophys. Acta 1477:299-306. [DOI] [PubMed] [Google Scholar]
- 48.Theilmann, D. A., and S. Stewart. 1993. Analysis of the Orgyia pseudotsugata multicapsid nuclear polyhedrosis virus trans-activators IE-1 and IE-2 using monoclonal antibodies. J. Gen. Virol. 74:1819-1826. [DOI] [PubMed] [Google Scholar]
- 49.Thornberry, N. A., and Y. Lazebnik. 1998. Caspases: enemies within. Science 281:1312-1316. [DOI] [PubMed] [Google Scholar]
- 50.Vucic, D., W. J. Kaiser, A. J. Harvey, and L. K. Miller. 1997. Inhibition of Reaper-induced apoptosis by interaction with inhibitor of apoptosis proteins (IAPs). Proc. Natl. Acad. Sci. USA 94:10183-10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vucic, D., W. J. Kaiser, and L. K. Miller. 1998. A mutational analysis of the baculovirus inhibitor of apoptosis Op-IAP. J. Biol. Chem. 273:33915-33921. [DOI] [PubMed] [Google Scholar]
- 52.Wright, C. W., and R. J. Clem. 2002. Sequence requirements for Hid binding and apoptosis regulation in the baculovirus inhibitor of apoptosis Op-IAP: Hid binds Op-IAP in a manner similar to Smac binding of XIAP. J. Biol. Chem. 277:2454-2462. [DOI] [PubMed] [Google Scholar]
- 53.Yanez, R. J., J. M. Rodriguez, M. L. Nogal, L. Yuste, C. Enriquez, J. F. Rodriguez, and E. Vinuela. 1995. Analysis of the complete nucleotide sequence of African swine fever virus. Virology 208:249-278. [DOI] [PubMed] [Google Scholar]