Abstract
Assembly of 73,000 expressed sequence tags (ESTs) representing multiple organs and developmental stages of maize (Zea mays) identified approximately 22,000 tentative unique genes (TUGs) at the criterion of 95% identity. Based on sequence similarity, overlap between any two of nine libraries with more than 3,000 ESTs ranged from 4% to 20% of the constituent TUGs. The most abundant ESTs were recovered from only one or a minority of the libraries, and only 26 EST contigs had members from all nine EST sets (presumably representing ubiquitously expressed genes). For several examples, ESTs for different members of gene families were detected in distinct organs. To study this further, two types of micro-array slides were fabricated, one containing 5,534 ESTs from 10- to 14-d-old endosperm, and the other 4,844 ESTs from immature ear, estimated to represent about 2,800 and 2,500 unique genes, respectively. Each array type was hybridized with fluorescent cDNA targets prepared from endosperm and immature ear poly(A+) RNA. Although the 10- to 14-d-old postpollination endosperm TUGs showed only 12% overlap with immature ear TUGs, endosperm target hybridized with 94% of the ear TUGs, and ear target hybridized with 57% of the endosperm TUGs. Incomplete EST sampling of low-abundance transcripts contributes to an underestimate of shared gene expression profiles. Reassembly of ESTs at the criterion of 90% identity suggests how cross hybridization among gene family members can overestimate the overlap in genes expressed in micro-array hybridization experiments.
A central goal of genome analysis is to identify and classify all the genes of a particular species. Functional genomics seeks to understand the precise roles of these genes, including unique and redundant functions. Apart from Arabidopsis, for which the complete genome is already available, gene discovery in most plants is primarily based on sample sequencing of expressed sequence tags (ESTs) prepared as cDNA to polyadenylated mRNA (Lim et al., 1996; Delseny et al., 1997; Covitz et al., 1998; Sterky et al., 1998; Ewing et al., 1999). The frequency of EST recovery for individual genes in diverse cDNA libraries can be used to estimate the expression patterns of individual genes. This “electronic RNA” analysis is limited in scope by the diversity of biological samples used to generate the cDNA libraries (e.g. developmental stage, tissue type, and growth conditions). A second limitation lies in the difficulty of sampling a particular cDNA library to sufficient depth to identify low-abundance transcript types. The primary value of accurate EST sequencing is that the expression of closely related genes can be distinguished based on limited nucleotide polymorphisms. In principle, EST sampling should be more precise in pinpointing both qualitative and quantitative differences in the expression of individual loci within a gene family compared with standard RNA hybridization methods when each tissue is sampled deeply.
Alternative and potentially more powerful methods for profiling gene expression require prior knowledge of the gene sequences garnered from an EST or genome sequencing project, but measure RNA expression more directly. One such method relies on PCR amplification of mRNA and restriction digestion patterns of the resulting cDNAs to enumerate expressed genes identified by the lengths of fragments generated (Bruce et al., 2000). A more widely adopted approach employs micro-arrays of EST elements deposited onto a glass slide followed by scoring hybridization signals with RNA from diverse tissues (Schena et al., 1995). Both of these experimental methods are limited to identification of transcripts already defined by cDNA or predicted from genomic sequences. On the other hand, RNA samples can be prepared from tissues and treatments that were not subjected to EST sequence analysis for global assessment of the overlap in gene expression between defined elements and any RNA sample. Variations of these methods, such as serial analysis of gene expression and total gene expression analysis, have been used for profiling expression of virtually all genes, including those without previously discovered mRNA (Matsumura et al., 1999; Sutcliffe et al., 2000).
Complete interpretation of gene profiling results depends on knowledge of the underlying genome structure. Ideally, the complete genome sequence would be available, with accurate prediction of all the genes and their alternative transcripts. For Arabidopsis, a near complete genome sequence is available, but current annotation is incomplete (for review, see Cho and Walbot, 2001). One of the major surprises evident in this genome is the extent of gene duplication. Local duplications exist for nearly 20% of the genes (Arabidopsis Genome Initiative, 2000) and several ancient polyploidization events are represented today by interstitial chromosome duplications (Vision et al., 2000).
Global analysis of maize (Zea mays) genome structure indicates that a relatively recent allotetraploidization event occurred approximately 11.5 million years ago (MYA) between grass species that had diverged approximately 20 MYA (Gaut and Doebley, 1997). Even earlier in the grass lineage, there were several genome-wide duplications (Wilson et al., 1999). As a consequence, small gene families are expected to encode similar products for most maize functions. Historically, however, maize genetic analysis identified many single gene mutations conferring an obvious phenotype (see http://www.agron.missouri.edu/locus.html). With the advent of molecular cloning, loci defined by mutation led to analysis of other gene family members. To be amenable to single-gene genetic analysis, such duplicated loci must be expressed in different parts or stages of the plant or have acquired distinct biochemical functions. For example, R and B-I are interchangeable helix-loop-helix transcription factors regulating the anthocyanin biosynthetic genes in the aleurone (R) or in leaves (B-I; Chandler et al., 1989). In other cases, gene families exhibit functional redundancy such that elimination of two or more members are required to see a phenotype in a particular organ; for example, chalcone synthase function in anthers specified by both C2 and Whp (Coe et al., 1981; Franken et al., 1991).
Given the complication of recent duplications within the modern maize genome, we were interested in comparing gene expression profiling results and conclusions based on EST sampling with results and conclusions derived from micro-array hybridization. Here, we report gene discovery results, list widely expressed genes, and determine the extent of overlap of EST sequence representation in maize based on about 73,000 ESTs drawn mainly from nine developmental stages. Micro-arrays fabricated with ESTs from either developing endosperm or immature ear were hybridized with the source and heterologous RNA samples. These micro-arrays were analyzed for reproducibility of hybridization results, quantification of transcript levels compared with EST recovery, and the extent of overlap in RNA expression profile between endosperm and ear. Both the EST and initial micro-array analyses demonstrate quantitative differences in expression profiles, but micro-array analyses detected a much higher qualitative overlap in gene expression between the tissues, relative to that observed by EST sequencing.
RESULTS
Assignment of EST as Singlets or Members of Contigs
The publicly available maize ESTs are periodically assembled into unique contigs, annotated, and made available via the ZmDB database (http://www.zmdb. iastate.edu; Gai et al., 2000). Based on a set of about 73,000 ESTs, including approximately 68,000 ESTs from the Maize Gene Discovery Project, 22,532 tentative unique genes (TUGs) were defined in the September 2000 assembly at the criteria of >95% identity in a 40-bp overlap (details in “Materials and Methods”). We found these criteria reliable in separating members of many gene families. The TUGs consist of tentative unique singlets (TUSs; ESTs lacking significant similarity to other maize EST sequences) and tentative unique contigs (TUCs; groups of ESTs sharing significant sequence similarity). Singlets comprised about one-half (51.4%) of all TUGs. In addition, 27.0% of the contigs contained only two or three ESTs. About one-half of the sequences recovered in the final stages of most of our EST sequencing projects were unique within that project, indicating that sampling was far from complete.
In the following sections, we report more detailed analyses of nine cDNA projects with at least 3,000 EST entries each; the projects are listed in Table I. They collectively define 17,096 TUGs composed of 9,597 singlets and 7,499 contigs. Contig assembly depends on both EST length, which averaged from 380 to 520 nucleotides in the Maize Gene Discovery projects considered here, and sequence quality, which is very high (Table I). Plasmid templates were sequenced from only one end in most cases; bidirectional sequencing was used throughout projects 707 and 946 and on a limited basis in other projects as an aid in contig assembly. Of the 12,208 pairs of 5′ and 3′ sequences available, 8,882 were grouped into single contigs.
Table I.
Characteristics of ESTs analyzed
| Project | Source | No. of ESTs | Average Length | Average Phred Quality Score | Source-Specific ESTs |
|---|---|---|---|---|---|
| nt | % | ||||
| 486 | Leaf primordia | 5,867 | 386 | N.D. | 35 |
| 605 | 10- to 14-d-old endosperm | 6,566 | 510 | 30.9 | 25 |
| 606 | 1- to 2-cm immature ear | 5,518 | 526 | 32.9 | 24 |
| 614 | 4-d-old root | 10,635 | 460 | 43.3 | 34 |
| 618 | <2-cm tassel | 3,407 | 378 | 45.3 | 27 |
| 660 | Mixed tassel stages | 6,444 | 444 | 45.1 | 31 |
| 687 | Mixed embryo stages | 4,765 | 458 | 45.2 | 33 |
| 707 | Mixed adult organs | 8,688 | 439 | 41.4 | 43 |
| 946 | 1-mm tassel | 10,646 | 412 | 38.9 | 26 |
All EST sequences were submitted to GenBank and are also available with annotation at ZmDB (http://zmdb.iastate.edu). Minimal requirements for submission were a sequence length of at least 100 consecutive nucleotides (nt) with a minimum Phred quality score of 15 in each position for sequences determined with capillary electrophoresis technology. N.D., Not determined because 486 samples were sequenced using both gel and capillary electrophoresis technologies. The percentage of source-specific ESTs was calculated as the fraction of ESTs within each project that formed singlets or contigs with ESTs exclusively from the same project after assembly of all ESTs from all projects.
Comparing the ESTs recovered from different cDNA libraries, 24% to 43% of the ESTs from a given library were apparently unique to their specific source (Table I). Moreover, about two-thirds (11,280) of the TUGs are comprised of sequences from a single library (Table II). Within each library, the majority (54%–78%) of TUGs were accounted for by singlets and by contigs with two or three ESTs only from that library. These ESTs should represent high to moderately expressed genes in that tissue source because ESTs of rarely expressed genes are unlikely to be sampled.
Table II.
Diversity of EST sources in the TUGs
| TUGs | No. of Libraries Represented |
|---|---|
| 11,280 | 1 |
| 3,133 | 2 |
| 1,353 | 3 |
| 628 | 4 |
| 323 | 5 |
| 191 | 6 |
| 111 | 7 |
| 51 | 8 |
| 26 | 9 |
| 17,096 | Total |
The no. of libraries contributing ESTs to a TUG. For example, there are 11,280 TUGs found in only one of the nine cDNA libraries. Only 26 TUGs have ESTs from all nine libraries.
Pair-Wise Comparison of EST Representation
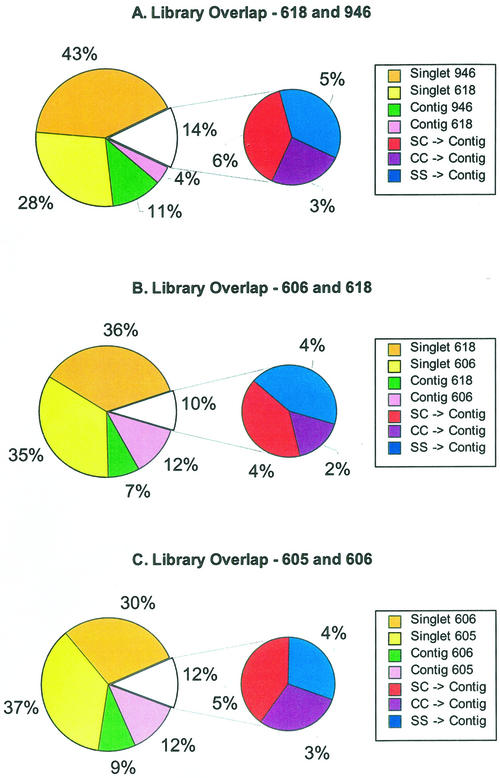
More detailed pair-wise comparisons between individual projects are presented in Figure 1. The shared TUGs are shown between two stages of tassel development (618 and 946) in Figure 1A, between tassel (618) and ear (606) inflorescences at the stage of spikelet formation in Figure 1B, and between immature ear (606) and 10- to 14-d-old endosperm (605) in Figure 1C. The smaller pie chart in each Figure 1, A through C, represents the fraction of TUGs containing at least one EST from each of the two libraries being compared as a measure of the overlap between the two EST projects. The “C + C” slice (purple) comprises contigs (C) with at least two ESTs from each of the two libraries. The “S + S” and “S + C” slices in blue and red, respectively, indicate contigs comprised of either two singlets (S) or a singlet from one of the libraries and a contig from the other library. The primary pie chart has four colored slices. Using Figure 1A as an example, the “contig 618” (pink) and “contig 946” (green) slices indicate contigs comprised of ESTs from just that library. The largest slices in the pair-wise analysis are the singlets in each library: “singlet 618” in yellow and “singlet 946” in orange.
Figure 1.
Comparison of TUG overlap between three pairs of cDNA libraries. The first section (starting at the top and moving counterclockwise) of the larger pie chart in each panel represents the singlets unique to the second library. The second section represents the singlets unique to the first library. The third and fourth sections represent the contigs unique to the second and first libraries, respectively. The smaller pie charts in each panel represent the TUGs containing ESTs from both libraries. The three sections of each smaller pie chart represent TUGs with one EST from both libraries (“SS → C”), one EST from one library, and multiple ESTs from the second library (“SC → C”), and multiple ESTs from both libraries (“CC → C”). In A, 1-mm tassel primordia (library 946) is compared with 0.5- to 2.0-cm tassels encompassing stages of organ specification and early differentiation (library 618). In B, library 618 is compared with 1- to 2-cm immature ear (library 606) containing similar stages of organ differentiation. In C, library 606 is compared with developing endosperm 10 to 14 d postpollination (library 605).
As shown in Figure 1A, TUGs from tassel primordia before organ differentiation (project 946) share only 14% of TUGs with tassels after organ differentiation (project 618). Tassel and ear at the same stage of spikelet differentiation show only 10% overlap (Fig. 1B). The extent of overlap between these inflorescence projects is similar to the extent of overlap between 10- to 14-d-old endosperm (i.e. endosperm at the stage when storage protein genes are first transcribed) and immature ear (Fig. 1C). Because our micro-array analysis was focused on the comparison of endosperm and immature ear, additional detail is worth noting in this comparison. The 3,113 endosperm TUGs (from 6,109 ESTs) and 2,595 ear TUGs (from 4,845 ESTs) overlap by only 12%, and the common sequences are drawn approximately equally from the S + S, S + C, and C + C classes. Of 2,163 singlets in 605 and 1,850 singlets in 606, only 181 contigs formed when the two groups of singlets were combined. In these two projects, >95% of the sequences are from the 5′ end of each cDNA clone.
To assess the significance of the overlap percentage, we randomly halved two of the largest EST projects, 614 and 946, and calculated the degree of overlap between the two halves of each project. Because these comparisons are between samples of ESTs from the same source, once robust sampling is completed such that all ESTs are recovered multiple times, 100% overlap is expected; in less complete samples, singlets can only be represented in one half or the other. Based on six repeated random assignments, the range of overlap was found to be 41% to 44% for 614 and 32% to 34% for 946, with about equal fractions of the S + S, S + C, and C + C classes. Thus, we conclude that incomplete EST sampling is only one of the factors contributing to the low overlap between different sources.
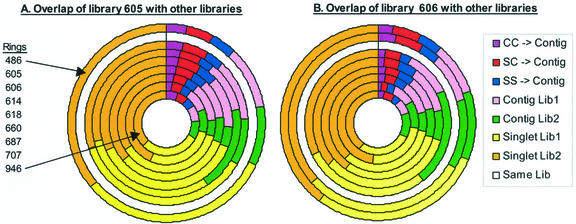
To compare the overlap of the other projects with projects 605 and 606, doughnut charts were constructed as shown in Figure 2. Each concentric ring in a doughnut chart represents the TUG overlap between two projects, one of which is kept constant across all comparisons (the white ring). The first three elements of each ring (starting at the top and moving clockwise) represent the percent of TUGs containing at least one EST from each of the two projects being compared. This is equivalent to the secondary pie charts in Figure 1, and the same color scheme is used. These shared sequences represent 9.7% to17.2% of the total TUGs from endosperm (Fig. 2A) and ear (Fig. 2B) and the other projects, respectively. Note that endosperm and ear typically share different contigs with the other projects. The next two elements of each ring are contigs comprised of ESTs from only one of the libraries in the comparison. As mentioned for the pie charts, the largest elements in the pair-wise analysis of libraries are the singlets in each library. These singlet classes comprise 27.6% to 37.5% of the total TUGs for endosperm and 25.0% to 34.5% for immature ear in comparisons to all eight other libraries.
Figure 2.
Multiple pair-wise comparisons of TUG overlap between one EST library and each of the eight other EST libraries listed in Table I. A, Comparison of the endosperm library (605) with the other libraries with each ring representing a comparison between library 605 and one other library. The order of the library comparisons is listed in the figure, starting with the outermost ring (library 486). B, Comparison of the immature ear library (606) with the other libraries. The first three sections of each ring (starting at the top and moving clockwise) correspond in color and meaning to the sections of the smaller pie charts in Figure 1. As in Figure 1, these three sections represent TUG overlap between the two libraries. The next four sections correspond in color and meaning to the sections of the larger pie charts in Figure 1.
Pair-wise comparisons for the other libraries gave similar results. Additional pie and doughnut charts are displayed at http://zmdb.iastate.edu/zmdb/publications/Fetal01-sm.html. The general conclusion is that at this level of EST sampling of developmentally staged organs or organ mixtures, distinctive suites of genes are detected with low overlap among organs. Of the EST projects examined, mixed adult organs (707) show the largest number of distinctive contigs that do not match either 605 or 606. Mixed adult organs (707) and embryos (687) have the highest percent of distinct singlets in pair-wise comparisons. Because EST sampling was not exhaustive, many expressed genes may have been overlooked. In particular, genes with low constitutive expression or moderate expression but in very limited domains within organs may not be defined by an EST.
Abundant ESTs
Up to 5% of the ESTs sequenced from a library assembled into a contig with five or more ESTs solely from that library as outlined in Table III. These contigs presumably represent genes that are highly expressed in particular tissues. Knowledge of EST representation provides candidate genes for recovery of promoters conferring stage or organ-specific expression. Compilation of maize contigs with a user-specified percent representation in a specific organ can be generated at http://www.tigr.org/tdb/zmgi/.
Table III.
Potential source-specific, highly expressed genes identified by EST profiles
| Source (Project) | No. of Contigs | No. of ESTs | Example of Gene Products |
|---|---|---|---|
| Leaf primordia (486) | 7 | 47 | Thioredoxin peroxidase (13 ESTs), cystathionine gamma-synthase (eight ESTs) |
| 10- to 14-d-old Endosperm (605) | 24 | 311 | Zein (two TUCs, 83 ESTs total) |
| 1- to 2-cm Immature ear (606) | 4 | 25 | MADS box protein (seven ESTs) |
| 4-d-old Root (614) | 46 | 307 | Glutamic acid-rich protein precursor (14 ESTs) |
| <2-cm Tassel (618) | 1 | 8 | No significant match |
| Mixed tassel stages (660) | 15 | 112 | Dihydroflavonol reductase (16 ESTs), expansin 1 (11 ESTs) |
| Mixed embryo stages (687) | 10 | 103 | Early embryogenesis protein (35 ESTs), peroxiredoxin (13 ESTs) |
| Mixed adult organs (707) | 30 | 213 | Photosystem II 10-kD polypeptide (16 ESTs), Cys proteinase precursor (15 ESTs) |
| 1-mm Tassel (946) | 4 | 21 | Histone H4 (six ESTs), MADS transcription factor (five ESTs) |
Columns two and three give the no. of contigs and total no. of ESTs in these contigs for contigs with at least five ESTs solely from the source indicated in column one. Column four gives one or two examples of corresponding putative gene products, inferred on the basis of high sequence similarity, for contigs with the highest number of ESTs.
Highly sampled contigs are listed at http://zmdb.iastate.edu/zmdb/publications/Fetal01-sm.html. All are found in at least two libraries, and nine were represented in all libraries. Five of the 30 most abundant EST clusters have no significant match at GenBank and could represent novel maize genes or possibly genes from fungi, insects, or other organisms associated with maize. It is surprising that although retrotransposons make up about two-thirds of the maize genome (SanMiguel et al., 1996), only 0.25% of maize ESTs correspond to these elements (Vicient et al., 2001; data not shown).
Ubiquitously expressed genes are represented by 26 contigs found in all nine of the EST projects. As shown in Table V, available at http://zmdb.iastate.edu/zmdb/publications/Fetal01-sm.html, all but three of these 26 contigs have more than one EST from each of the nine libraries. Based on similarity to known maize genes or high similarity to genes of known function in other organisms, nearly all of these widely expressed ESTs represent “housekeeping” functions. Despite their ubiquity, the majority of these widely expressed genes exhibit a skewed distribution among libraries, with abundant representation in only one or a few libraries. For example, of the 172 α-tubulin1 ESTs, 118 were recovered from immature ear, and of the 46 glutathione S-transferase1 representatives, 20 were from seedling root. A nonuniform distribution of EST number per library is seen in over two-thirds of the contigs containing ESTs from four or more libraries. These results suggest that quantitative differences in gene expression, even of widely expressed genes, often occur. The EST contig with the most uniform expression pattern is elongation factor 1α (EF-1α; TUC02-01-02-1471.1), which is represented by three to 11 ESTs per library and 69 ESTs altogether (http://zmdb.iastate.edu/zmdb/publications/Fetal01-sm.html). The EF-1α gene family is complex in maize and is estimated to contain 10 to 15 members based on DNA-blot hybridization (Carneiro et al., 1999); the family is represented by nearly 500 ESTs that assemble into 15 contigs and eight singlets, with two of the contigs ubiquitously expressed. The seven sequenced representatives of the family are 90% to 98% identical in the reading frames, and analysis of gene-specific expression requires probes from the 3′-untranslated regions (Carneiro et al., 1999). Although TUC02-01-02-1471.1 is not one of the fully characterized EF-1α genes, it is likely that the 3′ region of the corresponding gene would be a good internal loading control for RNA-blot hybridization assays in which diverse RNA sources are present.
Duplicate Genes with Quantitatively Different Expression Patterns
For pairs of closely related contigs, we asked if there were examples of high representation in one library combined with absence in other libraries. TUC01-12-19-1991.1 and TUC01-09-30-4459.2 share >99% sequence similarity to maize cytosolic glyceraldehyde-3-phosphate dehydrogenase genes Gpc3 and Gpc4, respectively. Eleven ESTs derived from the 660 library were from Gpc4 (TUC01-09-30-4459.2), but none were found for Gpc3 (TUC01-12-19-1991.1). TUC01-12-19-4269.1 and TUC05-31-1869.1 both share approximately 95% nucleotide sequence and high overall similarity to maize zein protein. TUC01-12-19-4269.1 is expressed in early embryo as judged by the presence of 15 ESTs derived from the early embryo library (687), although TUC05-31-1869.1 has no early embryo matches. In contrast, TUC05-31-1869.1 contains ESTs expressed in the endosperm library (605), while TUC01-12-19-4269.1 lacks contributions from the endosperm EST group. Contamination of embryo tissue samples by endosperm could explain these results, although we would have expected recovery of multiple types of zein ESTs in that case. A third example of possible tissue-specific expression is provided by TUC01-26-861.2 and TUC01-12-19-3881.1, which share approximately 89% sequence similarity to a subunit of the vacuolar proton ATPase. TUC01-12-19-3881.1 is expressed in root as judged by the contribution of 23 ESTs from library 614, but there were no representatives from other libraries. The root ESTs do not contribute to TUC01-26-861.2. Considering all aspects of EST analysis, the organs and tissues sampled show distinct gene expression profiles, suggesting that detailed analysis of EST organ distribution among gene families will provide hypotheses for tissue-specific expression that can be tested further.
Micro-Array Hybridization Analysis of Gene Expression
The EST analysis indicates low TUG overlap between libraries, despite the availability of relatively long ESTs representing 42,824 5′ and 19,687 3′ sequences from a total of 51,665 plasmids (some of which were sequenced multiple times) analyzed from the nine major libraries. This leads to the important question as to whether frequency distributions of ESTs within libraries adequately represent mRNA abundances within the organs and tissues examined. As an alternative means to assay mRNA expression patterns, we examined micro-array hybridization profiles from the 605 endosperm and 606 ear micro-arrays.
Micro-arrays of the 605 and 606 projects were separately printed on glass slides. The 605 micro-arrays were printed in two formats: as a single array with adjacent duplicate elements (605.04) and as two adjacent arrays (605.03). A panel of control elements was spotted as duplicate elements at the top and bottom of the single array (605.04) or as single elements at the top and bottom of each replicate array (605.03). The 606 micro-arrays were printed as a single array with adjacent triplicate elements; controls were spotted as triplicate elements at the top and bottom of the array. Array formats are described in more detail in “Materials and Methods” and at http://zmdb.iastate.edu/zmdb/microarray/arrays-info.html. Controls include individual clones, specifically selected for this purpose, as well as ESTs identified through data mining of the sequences that were present within the individual libraries. A description of the controls used on the micro-arrays can be found at http://zmdb.iastate.edu/zmdb/microarray/controls.html.
To evaluate the reproducibility of hybridization signals obtained from the micro-arrays, data from several different types of experiments were used. These include experiments in which: (a) only one labeled RNA was used in the hybridization, (b) a mixture of the same RNA labeled separately with Cy3 or Cy5 was used, or (c) a mixture of RNA from two different tissues labeled separately with Cy3 or Cy5 was used in reciprocal pair-wise hybridizations (dye reversal experiments). To compare gene expression patterns of endosperm and ear tissues, poly(A+) mRNA was prepared from 10- to 14-d-old endosperm and 1- to 2-cm ear primordia, at similar stages and of the same genotype as those used for library construction. Details of the labeling and hybridization protocols are provided in “Materials and Methods” and at http://zmdb.iastate.edu/zmdb/microarray/protocols.html. Descriptions of experiments, hybridization images, and original data sets are available at http://zmdb.iastate.edu/zmdb/microarray/data.html. Simple linear correlation analysis was used in pair-wise comparisons to evaluate variation within and between micro-arrays and within and between slides. The Pearson correlation coefficient (R) was computed as a means to quantitatively describe the strength of the relationship between replicates.
Micro-Array Reproducibility within Single Glass Slides
In this set of experiments, we employed the 605.04 slides containing a single micro-array with duplicate adjacent array elements. Correlation analysis was performed between the adjacent elements comparing the absolute signal intensities of the two replicates. We also employed the 606 micro-arrays containing a single micro-array, but with triplicate, adjacent array elements. In this case, the signal intensities for the first and third spot were compared (comparable correlation coefficients were observed for the other two possible pair-wise comparisons). If two labels were applied to the same slide, comparisons of signal intensities from each channel, as well as the ratio of signal intensities, are reported. For those within slide, highly local comparisons (over distances of approximately 200 μm) R values ranged from 0.92 to 0.98 for the individual channel intensity values (five and eight separate hybridizations of the 605 and 606 micro-arrays, respectively). Comparison of the ratio of signals (log10 transformed) between replicates (two separate hybridizations of each micro-array type) yielded R values ranging from 0.77 to 0.95. Because the ratio values combine the variation of two individual signal measurements, they would be expected to be more variable.
In the next experiment, we employed the 605.03 slides, containing duplicate adjacent micro-arrays on the same glass slide, to compare the reproducibility of hybridization within the same slides but over larger distances (18 mm). The signal intensities produced by each array element in one micro-array were compared with the corresponding element in the second array on the same slide. Correlation analysis yielded R values ranging from 0.95 to 0.97 in two separate hybridizations. We also examined reproducibility of hybridization within a glass slide when a mixture of two different preparations of the same RNA was applied. In this experiment, we used total RNA from ear separately labeled with Cy3 or Cy5 and applied the mixture to two 606 slides. The signal intensities produced by each array element in one channel were compared with the corresponding element in the second channel on the same slide. Correlation analysis resulted in R values of 0.99 in two separate hybridizations.
Between Micro-Array Reproducibility
In the next series of experiments, we compared the signal intensities of hybridization produced by each array element on one slide with the corresponding element on a second slide. In these experiments, the mean signal intensities for replicate elements in the Cy3 channel on one slide were compared with mean signal intensities in the Cy5 channel on a second slide. In this dye reversal experiment, R values ranged from 0.61 to 0.92 for all comparisons involving two and three separate hybridizations of the 605 and 606 micro-arrays, respectively. Hybridization in the dye reversal experiment was more variable between replicate glass slides than between replicate elements within the same slide.
Micro-Array-Based Analysis of Gene Expression in Different Tissues
After establishing the reproducibility of hybridization, micro-arrays were used to examine the patterns of hybridization between tissues, with the goal of estimating shared expression. Mixtures of labeled targets from endosperm and ear RNA were applied to 606 ear and 605 endosperm micro-arrays in dye-reversal experiments. To normalize signal intensities, we applied rank correlation analysis to identify a subset of the plant control genes whose expression patterns are similar among tissues (as described in “Materials and Methods” and more fully at http://zmdb.iastate.edu/zmdb/publications/Fetal01-sm.html). After two iterations of correlation analysis of hybridization signals and exclusion of outliers, the slope of the resulting trend line was used to normalize signal intensities between channels. Control genes that were consistently expressed more highly in endosperm than ear tissue included vacuolar ATPase and EFs 1α and TU. Control genes that were consistently more highly expressed in ear as compared with endosperm tissue included MNB1b binding protein, an HMG like protein, and one of the Histone 2A gene family members. As expected, control genes that are not highly expressed in endosperm or ear tissue, such as those required for photosynthesis and anthocyanin production, had low signal intensities in both channels.
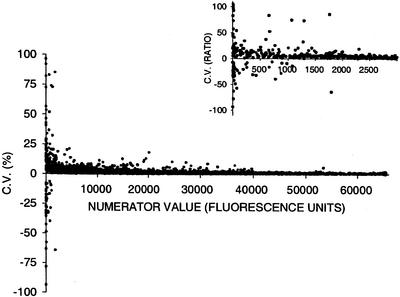
The mean of the observed signal intensities in each channel and the coefficient of variation for the ratio of signals were calculated from the replicate elements on each slide. A comparison of the coefficient of variation of the signal ratios versus signal intensity indicates that noise in the signal ratios diminishes as signal intensity increases (Fig. 3); this pattern did not differ among slides. Typically, at a signal intensity of 2,000 units (about 3% of the maximum signal intensity), the amplitude of the coefficient of variation in hybridization signal ratios is nearly constant; we used this threshold to identify ESTs with specific hybridization signals above background noise.
Figure 3.
Analysis of the reproducibility of measurement of fluorescence ratio values using the 606 ear tissue micro-array, as a function of the absolute values of the ratio numerator. Reproducibility is expressed in terms of the coefficient of variation of the three ratio values measured for each replicated array element.
Based on the comparison of TUGs identified in the EST sampling, we found only 12% of TUGs in common, using the criterion of 95% match over 40 bases. Micro-array hybridization is conducted at a stringency at which 90% matching over 60 bases should suffice to form a stable hybrid; therefore, we can calculate an expectation for the percent “overlap” between endosperm and ear by conducting a new EST assembly of the elements printed on the 605 (endosperm) and 606 (ear) micro-arrays. This new estimate is conservative because experimental data indicate that cross hybridization occurs on micro-arrays when individual gene targets retain 80% to 85% similarity to one or more of 142 Arabidopsis cytochrome P450 genes (Xu et al., 2001). At the 90% match criterion, there is a significant reduction in the number of TUGs on each array because singlets and contigs coalesce. After a 19.8% reduction in total TUGs for the endosperm array, there are 1,640 singlets and 605 clusters. A 21% reduction in TUGs for the ear array leaves 1,519 singlets and 493 clusters. After reassembling the ESTs represented on the arrays at this 90% criterion, 40% of the 605 array elements assembled with a printed 606 EST and 46% of the 606 elements assembled with an array element on the endosperm slides. As a consequence, the expected heterologous overlap is much higher than the 12% calculated at the stringent (95%) criterion of TUG assembly (Fig. 1C).
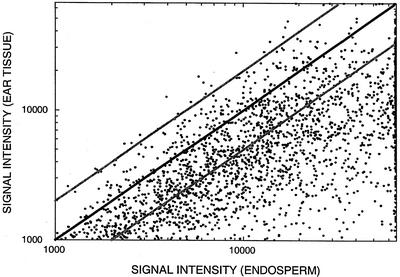
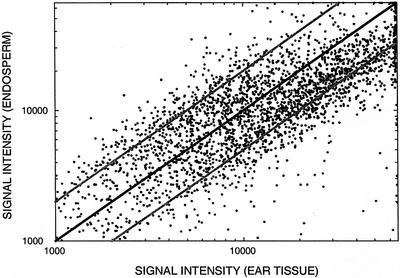
For experimental comparison of TUG expression patterns between tissues, the signal intensity of a contig (composed of multiple ESTs) was represented by the maximum signal intensity of all ESTs in that contig. Figures 4 and 5 compare the hybridization signal intensity of endosperm and ear targets for each TUG on an endosperm and ear micro-array, respectively. Data are presented for only one experiment of each micro-array type; patterns of hybridization did not differ between the replicate micro-arrays of each type nor in a subsequent replicate of the entire experiment with new RNA samples hybridized to pairs of ear and endosperm slides (data not shown). On the endosperm micro-array (Fig. 4), the signal intensity for most TUGs is greater for target derived from endosperm than from ear over the entire signal intensity range. This pattern differs from that observed on the ear micro-array (Fig. 5), where the signal intensity for most TUGs is similar for targets from both tissues.
Figure 4.
Hybridization signal intensity (fluorescence units) of endosperm and ear RNA to each TUG on an endosperm micro-array. For contigs, which are comprised of two or more ESTs, hybridization signal intensity is represented by the maximum signal intensity of all ESTs in the contig. The black line indicates equivalent signal intensities. Gray lines indicate 2-fold differences in signal intensity. Data for TUGs with hybridization signals less than 1,000 fluorescence units are omitted.
Figure 5.
Hybridization signal intensity (fluorescence units) of ear and endosperm RNA to each TUG on an ear micro-array. For contigs, hybridization signal intensity is represented by the maximum signal intensity of all ESTs in the contig. The black line indicates equivalent signal intensities. Gray lines indicate 2-fold differences in signal intensity. Data for TUGs with hybridization signals less than 1,000 fluorescence units are omitted.
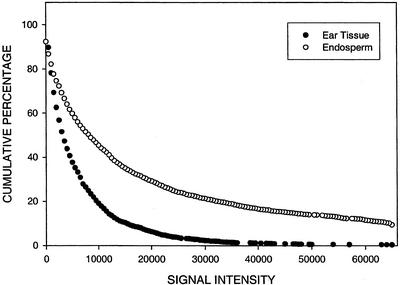
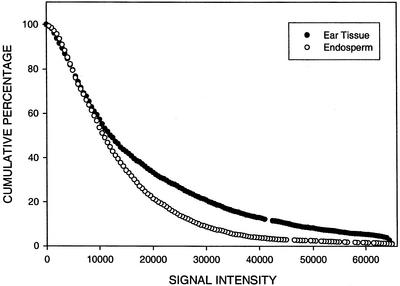
Figures 6 and 7 show the cumulative percentage of TUGs that hybridize with endosperm and ear targets as a function of hybridization signal intensity on an endosperm and ear micro-array, respectively. Mixtures of labeled targets from endosperm and ear RNA were applied to two 606 ear and two 605 endosperm micro-arrays in dye reversal experiments. The entire experiment was repeated with independent RNA preparations. On endosperm micro-arrays, at a conservative threshold of 2,000 units, an average of 83% of the TUGs hybridize with endosperm target and 57% hybridize with ear target. Approximately 10% of the TUGs that hybridize with endosperm target have signal intensity values that saturate, whereas fewer than 1% of that hybridize with ear target have values this high. As would be expected from the scatter plot (Fig. 4), a larger percentage of TUGs hybridize with endosperm, compared with ear target, over the entire range of signal intensity values on an endosperm micro-array (Fig. 6). This pattern differs from that observed on the ear micro-array where the average from four experiments is that 94% of the TUGs hybridize with targets from both tissues; data from one experiment are shown in Figure 7. Approximately the same percentage of TUGs (1%) hybridize with ear and endosperm targets at maximum signal intensity. Over the entire range of signal intensity values, the percentage of TUGs that hybridize with ear target is only slightly greater (<15%) than with endosperm target on the ear micro-array (Fig. 7).
Figure 6.
The cumulative percentage of TUGs that hybridize with endosperm and ear RNA as a function of hybridization signal intensity (fluorescence units) on an endosperm micro-array. For contigs (comprised of two or more ESTs), hybridization signal intensity is represented by the maximum signal intensity of all ESTs in the contig. The total number of TUGs is 2,800.
Figure 7.
The cumulative percentage of TUGs that hybridize with ear and endosperm RNA as a function of hybridization signal intensity (fluorescence units) on an ear micro-array. For contigs (comprised of two or more ESTs) hybridization signal intensity is represented by the maximum signal intensity of all ESTs in the contig. The total number of TUGs is 2,553.
Our general conclusions are that the TUGs contained within the endosperm project are typically more endosperm specific, are generally expressed at a higher level in endosperm, and that 10% of them had saturating signal intensities. In contrast, the TUGs contained within the ear project are more equally expressed in these two tissues. Furthermore, the micro-array experiments demonstrate that overlap is significantly greater than would be predicted by EST sampling or even by a 90% criterion of EST assembly: Approximately 57% and 94% of the TUGs hybridized above the 2,000 unit threshold with the heterologous target on the endosperm and ear micro-arrays, respectively.
DISCUSSION
ESTs are a quick and economical method for discovery of genes with moderate to abundant transcript levels. By sampling diverse organs at discrete developmental stages, high-quality ESTs assembled at stringent criteria can provide information on which genes in a gene family are expressed at quantitatively higher levels at specific stages in the plant life cycle. There are currently more than 106,000 maize ESTs in the public databases. A central annotation problem for EST collections is to estimate redundancy and to cluster ESTs into contigs that represent unique gene fragments. Such analysis is periodically performed at the ZmDB maize genome database (http://zmdb.iastate.edu). The most recent assembly of September 30, 2001, resulted in 28,220 TUGs composed of 15,095 contigs and 13,125 singlets. The number of TUGs is typically overestimated because ESTs from different parts of the same transcript type are reported separately until an EST overlap is generated.
EST assembly that clusters sequences based on a minimum criterion of 95% sequence identity in a region of >40 bases (criteria used in the ZmDB assemblies) should separate loci that are derived from the two progenitor species of modern maize provided sequence polymorphisms accumulate at a rate greater than one base change per 40 bases every 20 MYA (Gaut and Doebley, 1997). In fact, even alleles of most loci contain numerous nucleotide polymorphisms (Alfenito et al., 1998; Selinger and Chandler, 1999). The currently available ESTs are drawn mainly from modern inbred lines of maize (B73, W23, and Oh43) that are not closely related to each other. Even with this very narrow view of allelic diversity, base polymorphisms are approximately 1% (V. Walbot, unpublished data), predicting that many ancient duplications should be readily identified as different genes during EST assembly. More recent duplications complicate the picture and orthologues cannot be distinguished from paralogues (Doyle and Davis, 1998) without knowledge of the genomic location of the genes corresponding to the ESTs. Individual loci diverge at different rates, and base pair changes accumulate more quickly in regions of protein-coding genes with fewer functional constraints (Sanderson, 1998). Paralogous pairs, arising from local gene duplication, could diverge more or less quickly, or at the same rate, as orthologous gene copies within the same gene family. It is also plausible that coding regions under few restraints will have diverged >5%, whereas those regions under selection may have diverged very little, if any. Given the incomplete information on gene sequence provided by EST sampling, detecting rate variation is difficult.
The most striking feature of the EST collection for maize is that relatively few ubiquitously expressed genes were identified. By RNA-excess DNA-RNA hybridization analysis, it was estimated that approximately 5,000 genes were expressed in common among the major organ systems of tobacco (Nicotiana tabacum; Kamalay and Goldberg, 1980, 1984), but we found only 26 TUGs that were universally expressed in nine EST collections. EST sequencing can more readily distinguish members of gene families than can RNA hybridization analysis; however, EST sampling is highly sensitive to transcript representation. It is likely that only TUGs with moderate to high expression would be detected by EST sampling. In contrast, hybridization analysis can detect transcripts over several orders of magnitude of RNA representation, but is unlikely to distinguish closely related members of gene families. As a consequence, there may well be thousands of ubiquitously expressed types of genes with different family members varying qualitatively and quantitatively in their expression. EST recovery can highlight significant quantitative differences in representation as was found for Gpc3/Gpc4, zeins, and a likely vacuolar proton ATPase in which some TUGs of the gene family are recovered only in specific cDNA projects.
The second major finding is the low extent of overlap of TUGs between EST projects. These results indicate that for the readily recovered transcript classes, each tissue and developmental stage sampled has a relatively distinctive suite of moderately to highly expressed genes, often specific members of gene families. It seems likely that individual members of gene families are, in general, expressed in distinct patterns within maize. Ear (606) and tassel primordia (618) at the same stage of development, just after specification of floral organs, exhibit just 10% TUG overlap (Fig. 1B). Tassels at three stages of development—before organogenesis (946), after organogenesis but before cell expansion (618), and mixed stages of maturing tassels (660)—collectively display TUG overlap of only 4.0% (293/7,365). The estimates of overlap from EST sampling represent a lower bound because with more extensive EST sequencing more genes expressed in common are likely to be identified, including transcripts expressed at low levels in one or both organs.
Because of the limitation of EST sampling to fully define mRNA representation, we turned to micro-array analysis to test the conclusion that tissues and organs express largely discrete suites of genes. Micro-arrays of ESTs or other representations of genes are a powerful tool for identifying genes that are coordinately expressed during a particular environmental treatment, in a specific genetic background, or at a defined developmental stage. Array analysis, which examines the covariance of expression patterns, can identify genes with similar quantitative and qualitative aspects of expression. Confidence in interpreting the results comes from identifying previously well-studied genes in the expected patterns such as during acquisition of systemic acquired resistance (Maleck et al., 2000), under light or circadian regulation (Schaffer et al., 2001), after drought and cold (Seki et al., 2001), or after salt treatment (Kawasaki et al., 2001). Additional genes with the same expression patterns as the known examples are then candidates implicated in the same process.
In our study, we used ESTs as candidates of tissue specificity and then used micro-array experiments to ask how well such preliminary evidence on expression predicted hybridization behavior with a second tissue. Confidence in the results provided by micro-arrays is enhanced if it can be demonstrated that the process of hybridization is intrinsically accurate. Our results indicate very high within-slide reproducibility, with somewhat lower reproducibility between slides. These results are similar to other published data, in most cases exceeding the corresponding values reported (Girke et al., 2000; Kawasaki et al., 2001; Xu et al., 2001). In terms of sensitivity, micro-arrays appear to be slightly less sensitive than RNA-blot hybridization (Brown et al., 2001; Taniguchi et al., 2001).
In experiments with differentially labeled endosperm and ear targets, we detected extensive hybridization to both the 605 (endosperm) and 606 (ear) micro-arrays: 57% of the ear targets hybridized to the endosperm TUGs and 94% of the endosperm targets hybridized to the ear micro-arrays. These results set an upper bound of the overlap in gene expression between endosperm and immature ear because cross hybridization among closely related gene family members has undoubtedly occurred. Reassembly of contigs at the 90% match criterion reduced the proportion of unique TUGs within each array type to just 30% of elements printed and greatly increased the size of some contigs as multiple gene family members co-assembled. Relevant to the experimental results, the 90% match criterion predicted 40% endosperm target and 46% ear target hybridization to the heterologous array, based solely on the types of ESTs recovered in each project; this calculation cannot take into account shallow sampling within an EST project. At an even lower criterion, an even larger fraction of the ESTs within each array would assemble into contigs, and there would be an even higher prediction of overlap between ear and endosperm RNA samples.
The true extent of overlap in gene expression between ear and endosperm most likely lies between the estimates based on ESTs and micro-array hybridization. EST sampling is clearly incomplete, resulting in an under-estimate. Second, hybridization can occur among members of gene families, overestimating the number of expressed genes in common using micro-array profiling. A recent study using micro-arrays fabricated with seed-derived ESTs of Arabidopsis similarly found 60% to 77% overlap in expression with heterologous organs (Girke et al., 2000). These high percentages agree with the prior estimates based on solution hybridization (Kamalay and Goldberg, 1980), indicating that most organs express similar types of genes.
Because maize gene families have diverged during the approximately 20 MYA since the separation of the two species that later formed the allotetraploid leading up to modern maize (Gaut and Doebley, 1997), we would expect many families to contain members with extensive similarity in some portion of the transcribed region. As a consequence, the micro-array experiments demonstrate that one or more gene family members corresponding to a particular printed EST are present in an RNA sample without defining precisely which family member(s) are transcribed. To attain the precision in gene identification afforded by deep sampling of libraries producing long, high-quality EST sequences, micro-arrays must be fabricated that can distinguish loci. This could be accomplished either by using the most polymorphic portions of genes, such as the 3′-untranslated regions, or by designing gene-specific oligonucleotide probes more sensitive to mismatches over a shorter length.
MATERIALS AND METHODS
EST Sequencing
cDNA libraries were prepared from nine different tissue sources. EST collections are identified by three-digit project numbers assigned at the Stanford Genome Technology Center (Palo Alto, CA; Table I). Detailed descriptions of the tissue samples used for cDNA library construction are provided at http://zmdb.iastate.edu/zmdb/EST/libraries.html (for the analysis reported here, data from EST projects 707 and 945 were combined and listed as project 707 because these cDNAs were derived from the same library). Bacterial colonies containing cloned cDNAs were transferred into 96 deep-well blocks containing Terrific Broth (1.2 mL well−1) supplemented with the appropriate antibiotic. Cultures were grown overnight and harvested by centrifugation (1,000g). Plasmid DNA was isolated using the Qiagen Rapid Extraction Alkaline Lysis Prep 96 Plasmid Kit (product catalog no. 26173, QIAGEN, Valencia, CA) according to the manufacturer's specifications. Sequencing utilized ABI PRISM BigDye Primer CycleSequencing Kits (Applied Biosystems, Foster City, CA) as recommended by the manufacturer (http://www.appliedbiosystems.com/products/productdetail.cfm?ID=82). After sequencing at the Stanford Genome Technology Center, DNA was transferred into 96-well plates, stored at 4°C, and shipped frozen to the University of Arizona (Tucson).
Except for library 486, which was sequenced in part using gel-based equipment (ABI377, Applied Biosystems), sequencing was performed with MegaBACE capillary sequencers (Molecular Dynamics, Sunnyvale, CA). Base calling and quality assessment were evaluated using Phred (Ewing and Green, 1998). Phred scores report the confidence in base calling by indicating the expected error frequency, a function of sequencing technology and chemistry. For example, a Phred score of 30 indicates a probability of one base calling error in 1,000 bases. In general, a Phred score of y indicates a probability of one base calling error in 10 (y/10) bases. ESTs from a single library were assembled using Phrap (Gordon et al., 1998) to determine the rate of new gene discovery within a project. Some plates were sequenced multiple times while optimizing sequencing conditions; however, only a single sequence was included in the count of EST recovery from each well from each library plate. EST sequences of 100 nucleotides or greater and with a Phred score >15 were reported to GenBank in batch mode within 24 h of retrieval from the MegaBACE raw data files.
EST Assembly
EST contig assembly of all maize (Zea mays) ESTs in GenBank is periodically performed at ZmDB using the ZmDBAssembler protocol (http://zmdb.iastate.edu/zmdb/EST/assembly.html). ZmDBAssembler provides the logical flow between several third party programs used in the protocol, including BLAST (Altschul et al., 1997) and CAP3 (Huang and Madan, 1999). BLAST is used to indicate preliminary sequence clustering based on similarity. CAP3 is used for the ultimate assembly and derivation of consensus sequences. We used strict assembly criteria for CAP3, requiring that two ESTs overlap for at least 40 bases with at least 95% sequence identity to be put into the same contig. These criteria minimized the creation of chimeric contigs and assembly of ESTs from ancient duplicated genes into a single contig. On the other hand, ESTs from the same gene may not be clustered together because there are nonoverlapping clones from the 5′ and 3′ ends of long cDNAs, extensive sequence polymorphisms, or alternative pre-mRNA processing.
The assembly creates two sequence classes: TUCs and TUSs. Contigs are EST clusters with two or more member ESTs. Singlets are ESTs that are not significantly similar to any other ESTs. The combined TUCs and TUSs represent an approximate set of TUGs. “Tentative” indicates that all classifications are subject to constant and frequent changes as new ESTs are added to the assembly. New assemblies of available ESTs are compiled approximately every 4 months and reported at ZmDB. The analysis in this paper is based on the assembly of September 7, 2000. TUC names are assigned based on the last assembly date that changed a given TUC. For example, TUC09-07-5391.1 is a contig assembled on September 7, 2000. The number 5,391 reflects the contig number in that particular assembly. The terminal digit would be different from 1 only if a preliminary contig was split up by refined analysis with CAP3. The history of TUC names over successive assemblies can be traced at ZmDB.
Project Overlap
The degree of overlap between two EST projects was assessed by the fraction of TUGs from the two projects that comprise common contigs using all items available in GenBank. This procedure estimates the fraction of genes expressed in both conditions represented by the two projects. Precisely, for each TUG, we derived the count of member ESTs from each cDNA library. Then, for each pair of projects, contigs were placed into one of the first five categories below, whereas singlets were assigned to one of the last two categories: contig/contig → contig, more than one EST from each of the two libraries; singlet/contig → contig, one EST from one library and two or more ESTs from the second library; singlet/singlet → contig, a single EST from each of the two libraries; contig NNN, more than one EST but all from the first library (NNN); contig MMM, more than one EST but all from the second library (MMM); singlet NNN, a singlet from the first library; and singlet MMM, a singlet from the second library.
The fraction of TUGs placed in the first three categories gives the degree of overlap. Because EST sampling was incomplete, actual overlap will be higher than calculated.
Micro-Array Fabrication
A detailed description of micro-array construction, data sets, and hybridization methods can be found at http://zmdb.iastate.edu/zmdb/microarray. For this study, micro-arrays representing the 605 and 606 EST projects were separately printed on glass slides. Copies of the slides can be ordered online at http://zmdb.iastate.edu/zmdb/microarray/ordering.html.
The amplified inserts of all clones from the 605 EST project were printed on the “605 endosperm micro-arrays,” generating arrays with considerable internal redundancy of EST representation for some genes. In contrast, before PCR amplification and printing on the “606 ear micro-arrays,” the 606 EST project was consolidated by removal of 1,932 clones for which no sequence data was available. Information on ESTs contained within each project and their locations on the micro-array slides are provided at http://zmdb.iastate.edu/zmdb/microarray/libraries.html. In addition to the project maize ESTs, a panel of controls was selected for printing on all micro-arrays (see http://zmdb.iastate.edu/zmdb/microarray/controls.html). A liquid-handling robot (Beckman-Coulter Biomek 2000, Fullerton, CA) was used to remove 5-μL aliquots (1/10 volume) of the selected amplicons to a new plate. Each amplicon was diluted with 15 μL of sterile water and stored at −20°C. PCR reactions were carried out in duplicate 75-μL reactions in a 96-well format, and the products were combined. The final reaction concentrations were as follows: 0.2 mm dNTPs, 0.2 μm primers, 2.25 units of Taq Polymerase (product D1806, Sigma, St. Louis), and dilution of 10× reaction buffer (100 mm Tris-HCl, pH 8.3; 500 mm KCl; and 15 mm MgCl2) to 1× with sterile water. The primers used for all libraries were 5′-C6 amino modified (Bio-Synthesis, Inc., Lewisville, TX). Primer sequences employed for amplification of the 605 EST clones were: forward 5′ CTG CAG TAA TAC GAC TCA CTA TAG 3′ and reverse 5′ CTA TTC GAT GAT GAA GAT ACC 3′. Primer sequences for amplification of the 606 EST clones were: forward 5′ GTA ATA CGA CTC ACT ATA GGG C 3′ and reverse 5′ AAT TAA CCC TCA CTA AAG GG 3′. Aliquots (1–2 μL) of template DNA were added to the 96-well PCR plate containing reaction mix using a sterile 96-well replicating tool. Reaction conditions were as follows: 94°C for 2 min; 40 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min; then 72°C for 10 min and hold at 4°C. All products were analyzed by electrophoresis of a small aliquot (2 μL) of the amplicons using 1.2% (w/v) agarose gels prepared in Tris acetate EDTA buffer (Sambrook et al., 1989). Sample clean up was done using Multiscreen96 PCR Plates (product MANU03050, Millipore, Bedford, MA) according to the manufacturer's instructions. Purified amplicons were stored in V-Bottom 96-well plates (product 651-180, Greiner, Lake Mary, FL). Plates were dried at 60°C for 2 h using a Speedvac Concentrator (Thermo Savant, Holbrook, NY) and stored at −20°C. For printing, samples were dissolved by addition of 15 μL of 2× SSC buffer (Sambrook et al., 1989). Before printing, the plates were incubated at room temperature for at least 1 h. After each printing was completed, the plates were dried (60°C for 20 min) using a Speedvac concentrator, and returned to storage at −20°C. For subsequent printing, samples were resuspended in 15 μL of sterile distilled water and then incubated at room temperature for at least 1 h. Micro-array slides were printed using an OmniGridder printer (GeneMachines, San Carlos, CA) equipped with eight printing pins (product no. 11077-1 Rev A MicroQuill 2000 Microarray Printing Tip, Majer Precision Engineering, Tempe, AZ). Micro-arrays were printed on Sigma Screen Silane Slides (product no. S7934-50EA). One hundred micro-array slides were produced in each printing; finished slides were stored in slide containers in the dark at room temperature.
The 605 and 606 projects (plus associated controls) were printed on separate glass slides. The 605 micro-arrays were printed in two formats: as a single array with adjacent duplicate elements (605.04) and as two adjacent arrays (605.03). The 606 micro-arrays were printed as a single array with three replicate spots for each element. The control plates were printed twice (at the start and end of each print). The 605 micro-arrays were produced from 84 sample plates (8,064 elements), and 93 different controls, to give a total of 16,500 array elements, with a center-to-center spacing of 190 μm. The elements that were of sufficient sequence length and quality were reported to GenBank (5,534); these ESTs represented approximately 2,800 TUGs. The 606 micro-arrays were produced from 52 sample plates (4,980 elements) and two control plates (113 controls) for a total of 15,618 array elements, with 195-μm spacing; these ESTs represented approximately 2,550 TUGs. A more detailed description of the format used to print each micro-array can be found at http://zmdb.iastate.edu/zmdb/microarray/arrays-info.html.
Preparation of RNA Samples and Micro-Array Hybridization
RNA samples were prepared from endosperm 10 to 14 d after pollination and from 1- to 2-cm immature ears of self-pollinated OH43 inbred plants grown in spring 2000 in Tucson, AZ. RNA was purified from tissue samples pulverized in liquid nitrogen. Total RNA was isolated using TRIzol (GibcoBRL Life Technologies, Rockville, MD) and poly(A+) mRNA was purified using DynaBeads Oligo (dT)25 (Dynal A.S., Oslo) according to manufacturers' instructions. For each sample, either 200 μg of total RNA or 4 μg of poly(A+) mRNA was labeled using either Cy3- or Cy5-dUTP (products PA53022 and PA55022, Amersham Pharmacia, Piscataway, NJ). Sigma's AMV-RT Kit (product STR1-KT) was used for the labeling reaction according to manufacturer's instructions. A more detailed description of RNA isolation and labeling protocols can be found at http://zmdb.iastate.edu/zmdb/microarray/protocols.html. Before hybridization, slides were held face down over a 42°C water bath for 5 to 10 s to rehydrate the array elements and then snap dried on a 70°C to 80°C heat block for 3 to 10 s. DNA was cross-linked to the glass slide using 65 mJ of 254-nm UV-C radiation (FB UVXL-1000 UV Cross Linker set to 650 × 100 μJ/cm2, Fisher Scientific, Pittsburgh). Slides were then washed for 2 min in 1% (w/v) SDS on an orbital shaker, washed for 2 min in 95°C water, rinsed by plunging rapidly 10 to 20 times in a 100% (w/v) ethanol bath at room temperature, and immediately dried by centrifugation at 50g to 100g for 2 to 5 min. Dry arrays were used immediately or were stored, for less than 7 d, at room temperature.
Hybridization followed the slide manufacturer's recommended protocol with slight modifications (Sigma Technical Bulletin MB-745). The hybridization mixture consisted of 4 μg of each labeled mRNA target or, if total RNA was used, 200 μg of each labeled target; 2 μL of Liquid Block (Amersham product RP3601); 4 μL of 20× SSC buffer; and 1 μL of 2% (w/v) SDS, in a final volume of 30 μL. The mixture was denatured at 95°C for 2 min and then transferred to ice. The hybridization mixture was applied to a micro-array slide preheated to 65°C on a heat block, and quickly covered with a coverslip (Sigma Hybrislips Z36 591-2). The micro-array was then immediately transferred to a prewarmed hybridization chamber (50-mL plastic screw-top tube containing a paper towel moistened with 3× SSC) and then incubated overnight (8–12 h) at 62°C. To terminate hybridization, the slide was processed by successive 5-min washes in 2× SSC and 0.5% (w/v) SDS at 62°C, and in 0.5× SSC and then 0.05× SSC at room temperature. Slides were immediately dried by centrifugation at 50g to100g for several minutes. A more detailed description of DNA immobilization and hybridization can be found at http://zmdb.iastate.edu/zmdb/microarray/protocols.html.
Micro-Array Data Acquisition and Analysis
Slides were scanned within 8 h of hybridization using a GSI Lumonics ScanArray 3000 (Packard BioChip Technologies, Billerica, MA). Spot finding and analysis of signal intensity were carried out using ImaGene software (BioDiscovery, Los Angeles).
For each element on the micro-array, net signal intensity was computed from the median signal minus the median value of the local background. Local background is calculated from the median signal in the ring-shaped area surrounding the element, 40 microns from the element and 70 microns in width. To normalize the signal intensities between different tissues, we applied rank correlation to the signal intensity values of 78 elements to identify control genes whose gene expression patterns do not vary between tissues. The 78 elements are comprised of the two replicates of 39 separate maize controls (http://gremlin3.zool.iastate.edu/zmdb/microarray/controls.html). Those elements that differed in signal intensity rank by 20% of the total number of ranks (16 ranks) were excluded from the normalization because the expression of these genes is not similar among tissues. For the remaining control genes, the mean signal intensities for replicate elements in one channel (tissue 1) were compared with mean signal intensities in the second channel (tissue 2) on same slide. The slope of the trend line was used to normalize the signal intensity values of the control elements. We found that after normalization, some of the control elements with high ranks in both channels still had signal ratios that were >2 or <0.5, suggesting tissue specific expression patterns. These were excluded and the normalization process was reiterated. The slope of the subsequent trend line was used for the normalization of all elements on the slide. The genes contributing to normalization are listed at http://gremlin3.zool.iastate.edu/zmbd/microarray/605+606controls.html. Further details of normalization can be found at http://gremlin3.zool.iastate.edu/zmbd/microarray/furtherdetails.html. Ratios of signal intensities were calculated by dividing the signal intensity from the experimental condition (e.g. RNA from the heterologous tissue) by that from the control condition (e.g. RNA from the same tissue used to make the micro-array elements). The mean and coefficients of variation for the observed signal intensities in each channel and the ratio of signals were calculated from the two replicate elements (605 endosperm micro-array) and three replicate elements (606 ear micro-array) on each slide. Simple linear correlation analysis was used in pair-wise comparisons to evaluate variation within and between micro-arrays and within and between slides. The Pearson correlation coefficient (R) was computed as a means to quantitatively describe the strength of the relationship between replicates.
In comparison of TUG expression patterns between tissues, signal intensity for a contig was represented by the maximum signal intensity among all member ESTs of the contig. Signal intensity of the contig was determined for each channel separately; this means that the maximum signal in each channel does not necessarily come from the same EST. More information on data analysis can be found at http://gremlin3.zool.iastate.edu/zmdb/microarray/protocols.html.
ACKNOWLEDGMENTS
We thank Brian Nakao, Gurpreet Randhawa, Bret Schneider, and Khaled Sarsour of the Stanford Genome Technology Center for their work in EST production sequencing and members of the maize community for supplying cDNA libraries, as listed at http://zmdb.iastate. edu/give. Liqun Xing made substantial contributions to the data analysis in the early stages of this work while he was at Iowa State University. We thank Dominic DeCianne (University of Arizona) for assistance with PCR amplification of ESTs, gel electrophoresis, and micro-array printing.
Footnotes
This work was supported by the National Science Foundation Plant Genome Research Program as part of the Maize Gene Discovery, DNA Sequencing, and Phenotypic Analysis project (grant no. DBI–9872657).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010681.
LITERATURE CITED
- Alfenito MR, Souer E, Buell R, Koes R, Mol J, Walbot V. Functional complementation of anthocyanin sequestration in the vacuole by widely divergent glutathione S-transferases. Plant Cell. 1998;10:1135–1149. doi: 10.1105/tpc.10.7.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Brown AJP, Planta RJ, Restuhadi F, Bailey DA, Butler PR, Cadahia JL, Cerdan ME, DeJonge M, Gardner DCJ, Gent ME et al. Transcript analysis of 1003 novel yeast genes using high-throughput northern hybridizations. EMBO J. 2001;20:3177–3186. doi: 10.1093/emboj/20.12.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce W, Folkerts O, Garnaat C, Crasta O, Roth B, Bowen B. Expression profiling of the maize flavonoid pathway genes controlled by estradiol-inducible transcription factors CRC and P. Plant Cell. 2000;12:65–79. doi: 10.1105/tpc.12.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro NP, Hughes PA, Larkins BA. The eEF1A gene family is differentially expressed in maize endosperm. Plant Mol Biol. 1999;41:801–813. doi: 10.1023/a:1006391207980. [DOI] [PubMed] [Google Scholar]
- Chandler VL, Radicella JP, Robbins TP, Chen J, Turks D. Two regulatory genes of the maize anthocyanin pathway are homologous: isolation of B utilizing R genomic sequences. Plant Cell. 1989;1:1175–1183. doi: 10.1105/tpc.1.12.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Walbot V. Computational methods for gene annotation: the Arabidopsis genome. Curr Opin Biotechnol. 2001;12:126–130. doi: 10.1016/s0958-1669(00)00185-3. [DOI] [PubMed] [Google Scholar]
- Coe EH, McCormick SM, Modena SA. White pollen in maize. J Hered. 1981;72:318–320. [Google Scholar]
- Covitz PA, Smith LS, Long SR. Expressed sequence tags from a root-hair-enriched Medicago truncatula cDNA library. Plant Physiol. 1998;117:1325–1332. doi: 10.1104/pp.117.4.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delseny M, Cooke R, Raynal M, Grellet F. The Arabidopsis thaliana cDNA sequencing projects. FEBS Lett. 1997;405:129–132. doi: 10.1016/s0014-5793(97)00184-1. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Davis JI. Homology in molecular phylogenetics: a parsimony perspective. In: Soltis DE, Soltis PA, Doyle JJ, editors. Molecular Systematics of Plants. Boston: Kluwer Academic Publishers; 1998. pp. 101–131. [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequences traces using PHRED: II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing RM, Kahla AB, Poirot O, Lopez F, Audic S, Claverie JM. Large-scale statistical analyses of rice ESTs reveal correlated patterns of gene expression. Genome Res. 1999;9:950–959. doi: 10.1101/gr.9.10.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken P, Niesbach-Klosgen U, Weydemann U, Marechald-Rouard L, Saedler H, Wienand U. ). EMBO J 10: 2605–2612. 1991. The duplicated chalcone synthease genes C2 and Whp (white pollen) of Zea mays are independently regulated: evidence for translational control of WHP expression by the anthocyanin intensifying gene ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai XW, Lal S, Xing LQ, Brendel V, Walbot V. Gene discovery using the maize genome database ZmDB. Nucleic Acids Res. 2000;28:94–96. doi: 10.1093/nar/28.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Doebley JF. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci USA. 1997;94:6809–6814. doi: 10.1073/pnas.94.13.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke T, Todd J, White J, Benning C, Ohlrogge J. Microarray analysis of developing Arabidopsis seeds. Plant Physiol. 2000;124:1570–1581. doi: 10.1104/pp.124.4.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. CONSED: a graphical tool for sequencing finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamalay JC, Goldberg RB. Regulation of structural gene-expression in tobacco. Cell. 1980;19:935–946. doi: 10.1016/0092-8674(80)90085-9. [DOI] [PubMed] [Google Scholar]
- Kamalay JC, Goldberg RB. Organ-specific nuclear RNAs in tobacco. Proc Natl Acad Sci USA. 1984;81:2801–2805. doi: 10.1073/pnas.81.9.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki S, Borchert C, Deyholos M, Wang H, Brazille S, Kawai K, Galbraith DW, Bohnert HJ. Gene expression profiles during the initial phase of salt stress in rice (Oryza sativa L.) Plant Cell. 2001;13:889–906. doi: 10.1105/tpc.13.4.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CO, Kim HY, Kim MG, Lee SI, Chung WS, Park SH, Hwang I, Cho MJ. Expressed sequence tags of Chinese cabbage flower bud cDNA. Plant Physiol. 1996;111:577–588. doi: 10.1104/pp.111.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA, Dangl JL, Dietrich RA. The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat Gen. 2000;26:403–410. doi: 10.1038/82521. [DOI] [PubMed] [Google Scholar]
- Matsumura H, Nirasawa S, Terauchi R. Technical advances: transcript profiling in rice (Oryza saliva L.) seedlings using serial analysis of gene expression (SAGE) Plant J. 1999;20:719–726. doi: 10.1046/j.1365-313x.1999.00640.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Sanderson MJ. Estimating rate and time in molecular phylogenies: beyond the molecular clock? In: Soltis DE, Soltis PA, Doyle JJ, editors. Molecular Systematics of Plants. Boston: Kluwer Academic Publishers; 1998. pp. 242–264. [Google Scholar]
- SanMiguel P, Tikhonov A, Jin Y-K, Motochoulskaia N, Zakharvo D, Melake-Berhan A, Springer PC, Edwards KJ, Lee M, Avramova Z et al. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E. Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell. 2001;13:113–123. doi: 10.1105/tpc.13.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitroing of gene-expression patterns with a complementary-DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K. Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell. 2001;13:61–72. doi: 10.1105/tpc.13.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinger DA, Chandler VL. Major recent and independent changes in levels and patterns of expression have occurred at the b gene, a regulatory locus in maize. Proc Natl Acad Sci USA. 1999;96:15007–15012. doi: 10.1073/pnas.96.26.15007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterky F, Regan S, Karlsson J, Hertzberg M, Rohde A, Holmberg A, Amini B, Bhalerao R, Larsson M, Villarroel R et al. Gene discovery in the wood-forming tissues of poplar: analysis of 5,692 expressed sequence tags. Proc Natl Acad Sci USA. 1998;95:13330–13335. doi: 10.1073/pnas.95.22.13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe JG, Foye PE, Erlander MG, Hilbush BS, Bodzin LJ, Durham JT, Hasel KW. TOGA: an automated parsing technology for analyzing expression of nearly all genes. Proc Natl Acad Sci USA. 2000;97:1976–1981. doi: 10.1073/pnas.040537997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi M, Miura K, Iwao H, Yamanaka S. Quantitative assessment of DNA microarrays: comparison with Northern blot analyses. Genomics. 2001;71:34–39. doi: 10.1006/geno.2000.6427. [DOI] [PubMed] [Google Scholar]
- Vicient CM, Jääskeläinen MJ, Kalendar R, Schulman AH. Active retrotransposons are a common feature of grass genomes. Plant Phyiol. 2001;125:1283–1292. doi: 10.1104/pp.125.3.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290:2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- Wilson WA, Harrington SE, Woodman WL, Lee M, Sorrells ME, McCouch SR. Inferences on the genome structure of progenitor maize through comparative analysis of rice, maize and the domesticated panicoids. Genetics. 1999;153:453–473. doi: 10.1093/genetics/153.1.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Bak S, Decker A, Paquette SM, Feyereisen R, Galbraith DW. Microarray-based analysis of gene expression in very large gene families: the cytochrome P450 gene superfamily of Arabidopsis thaliana. Gene. 2001;272:61–74. doi: 10.1016/s0378-1119(01)00516-9. [DOI] [PubMed] [Google Scholar]