Abstract
Ste5, the prototypic mitogen-activated protein kinase (MAPK) scaffold protein, associates with plasma membrane-tethered Gβγ freed upon pheromone receptor occupancy, thereby initiating downstream signaling. We demonstrate that this interaction and membrane binding of an N-terminal amphipathic α-helix (PM motif) are not sufficient for Ste5 action. Rather, Ste5 contains a pleckstrin-homology (PH) domain (residues 388–518) that is essential for its membrane recruitment and function. Altering residues (R407S K411S) equivalent to those that mediate phosphoinositide binding in other PH domains abolishes Ste5 function. The isolated PH domain, but not a R407S K411S derivative, binds phosphoinositides in vitro. Ste5(R407S K411S) is expressed normally, retains Gβγ and Ste11 binding, and oligomerizes, yet is not recruited to the membrane in response to pheromone. Artificial membrane tethering of Ste5(R407S K411S) restores signaling. R407S K411S loss-of-function mutations abrogate the constitutive activity of gain-of-function Ste5 alleles, including one (P44L) that increases membrane affinity of the PM motif. Thus, the PH domain is essential for stable membrane recruitment of Ste5, and this association is critical for initiation of downstream signaling because it allows Ste5-bound Ste11 (MAPKKK) to be activated by membrane-bound Ste20 (MAPKKKK).
Keywords: Pheromone response, plasma membrane, mutants, yeast, Saccharomyces cerevisiae, Ste20
Yeast pheromone response links activation of a G-protein-coupled receptor to stimulation of an appropriate mitogen-activated protein kinase (MAPK) (for review, see Wang and Dohlman 2004). Ste5 protein (Elion 2001) is essential for this coupling: First, because it binds the MAPKKK (Ste11), the MAPKK (Ste7), and the MAPK (Fus3) of the MAPK cascade. Distinct regions of Ste5 interact with each kinase, as delimited by mutational analysis (Choi et al. 1994; Marcus et al. 1994; Printen and Sprague 1994; Inouye et al. 1997a); thus, Ste5 was the first recognized MAPK scaffold protein. Second, Ste5 delivers its kinase cargo to the plasma membrane, at least in part, via association with the Gβ subunit (Ste4) of the Gβγ complex released from the receptor-associated heterotrimeric G-protein (Whiteway et al. 1995; Pryciak and Huntress 1998). The region of Ste5 responsible for interaction with Gβγ includes a RING-H2 domain near its N terminus (Inouye et al. 1997b; Feng et al. 1998). Gβγ remains tethered at the plasma membrane via lipophilic modifications at the C-terminal end of Gγ (Ste18) (Hirschman and Jenness 1999). Gβγ-dependent docking of Ste5 at the plasma membrane permits efficient encounter of Ste5-bound Ste11 with its activator (MAPKKKK), Ste20 (Drogen et al. 2000), a so-called p21-activated protein kinase (PAK). The p21 that activates Ste20 is the small GTPase, Cdc42 (Lamson et al. 2002), which is also membrane-anchored via C-terminal lipophilic modifications (Ohya et al. 1993). Activated Ste20 is membrane-associated not only by binding of its N-terminal CRIB domain to Cdc42-GTP, but also because its C terminus contains a high-affinity binding site for Gβγ (Leeuw et al. 1998). Thus, like Ste5, Ste20 concentrates near activated pheromone receptors.
Ste5 self-associates, and this oligomerization is important for efficient signaling (Yablonski et al. 1996; Inouye et al. 1997b; Wang and Elion 2003). Ste5 undergoes constitutive nucleocytoplasmic shuttling in naïve cells, but accumulates stably at the cell cortex in the projection (“shmoo tip”) that forms on pheromone-treated cells (Pryciak and Huntress 1998; Mahanty et al. 1999), and reimport of Ste5 into the nucleus contributes to down-regulation of signaling (Künzler et al. 2001). To further delineate how Ste5 contributes to signal propagation, we isolated three mutations (P44L, C226Y, S770N) that constitutively activate Ste5 (Sette et al. 2000) by selecting for alterations of Ste5 that permit signaling even in cells lacking Gβγ. Although these alleles bypass the need for Gβγ, they do not bypass the need for the other known components of the pathway, most notably Ste20. Our ability to isolate such alleles raised a paradox, given the evidence that Ste5–Gβγ association is critical for membrane delivery of Ste11 and its activation by Ste20. This quandary suggested that the mutants unveil an alternative, relatively efficient, Gβγ-independent means by which Ste5 associates with the membrane.
Perhaps the mutants are secured at the cell cortex, even in the absence of Gβγ, because they enhance Ste5 association with other proteins that are themselves membrane-bound in a Gβγ-independent manner. Interactions of Ste5 with other plasma membrane-associated proteins, including Bem1 (Leeuw et al. 1995; Lyons et al. 1996) and, most recently, Cdc24 (Wang et al. 2005), have been reported. Bem1, an SH3 domain-containing adaptor protein, has an internal Phox-homology (PX) domain, and PX domains can bind phosphoinositides (Sato et al. 2001). Cdc24, the guanine nucleotide exchange factor (GEF) for Cdc42, has an internal pleckstrin-homology (PH) domain, and PH domains can also bind phosphoinositides (for review, see Lemmon and Keleti 2005).
Another possibility is that the constitutive alleles elevate some intrinsic ability of Ste5 to associate with membranes. Indeed, it has been demonstrated that P44L (and T52M, another constitutive Ste5 allele that we isolated by a different strategy [Hasson et al. 1994]) operate by enhancing the membrane-binding affinity of an N-terminal amphipathic α-helix in Ste5, dubbed the PM motif (Winters et al. 2005). However, some sequence analysis algorithms predict that Ste5 contains a PH domain. We suspected that the candidate PH domain might be highly relevant to Ste5 function because some PH domains can associate with both phosphoinositides and Gβγ (Lemmon and Keleti 2005). Recent genome-wide analysis of Saccharomyces cerevisiae PH domains relied on the SMART database, which did not pinpoint Ste5 (Yu et al. 2004). Hence, the properties and biological role of the presumptive PH domain in Ste5 have not been explored. Therefore, we applied genetic, biochemical, and cell biological approaches to assess the role of the predicted PH domain in Ste5. As documented here, this previously unrecognized and unstudied region of Ste5 is absolutely essential for the function of Ste5 in pheromone signaling because it is required to achieve stable membrane recruitment of Ste5.
Results
A candidate PH domain in Ste5
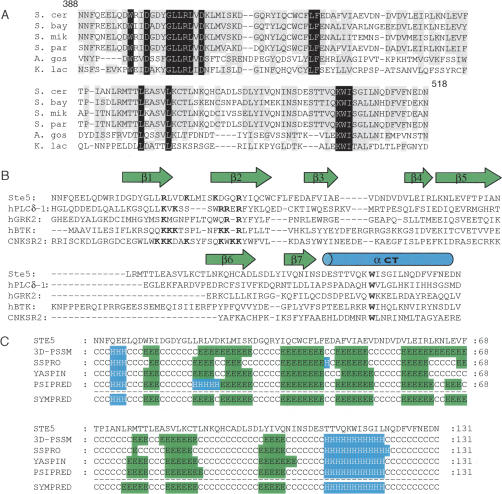
Both the PFAM (Bateman et al. 2004) and BLAST CDD (Marchler-Bauer et al. 2003) databases predict a PH domain (residues 400–512) in S. cerevisiae Ste5. This element lies in a slightly larger region (residues 388–518) that is highly conserved between S. cerevisiae Ste5 and its orthologs in other Saccharomyces species and even more distantly related yeasts (Fig. 1A). This segment possesses the sequence signatures (Fig. 1B) found in well characterized PH domains, even though PH domains in general can share as little as 10%–15% amino acid identity (Lemmon and Keleti 2005). In particular, the N-terminal portion of the presumptive PH domain in Ste5 contains basic residues situated at positions that are equivalent to those that mediate phosphoinositide binding and membrane targeting in crystal structures of other PH domains (Fig. 1B). Despite low sequence similarity, authentic PH domains all adopt a characteristic protein fold, consisting of a β-strand sandwich capped off by a C-terminal α-helix. Regardless of the algorithm used (Pollastri et al. 2002; Lin et al. 2005), the apparent PH domain in Ste5 is predicted to assume the same tertiary fold (Fig. 1C), in agreement with its alignment against other PH domains of known structure based on sequence similarity alone (Fig. 1B). Moreover, molecular modeling and threading programs (Kelley et al. 2000; McGuffin et al. 2000) predict that, compared with the database of known folds, all those closest to the predicted structure of this region of Ste5 are PH domains.
Figure 1.
Sequence alignment and analysis of the predicted S. cerevisiae Ste5 PH domain. (A) Alignment of the S. cerevisiae Ste5 PH domain (residues 388–518) with corresponding sequences in orthologs from Saccharomyces bayanus, Saccharomyces mikatae, Saccharomyces paradoxus, Ashbya gossypii, and Kluyveromyces lactis. (B) Alignment of the S. cerevisiae Ste5 PH domain with selected mammalian PH domains. Secondary structure elements (green arrows, β-strands; blue cylinder, α-helix) depict those in the crystal structure of the PLCδ1 PH domain (Lemmon and Keleti 2005). Basic residues in the β1–β2 loop and the conserved Trp in the C-terminal helix are highlighted (bold). (C) Secondary structure elements in the Ste5 PH domain predicted by the indicated algorithms: 3D-PSSM, SSPro, YAPSIN, PSIPRED, and SYMPRED. (Green/E) β-strand; (blue/H) α-helix; (C) random coil.
The predicted PH domain is essential for Ste5 function
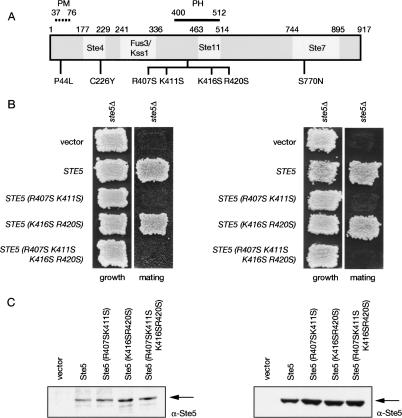
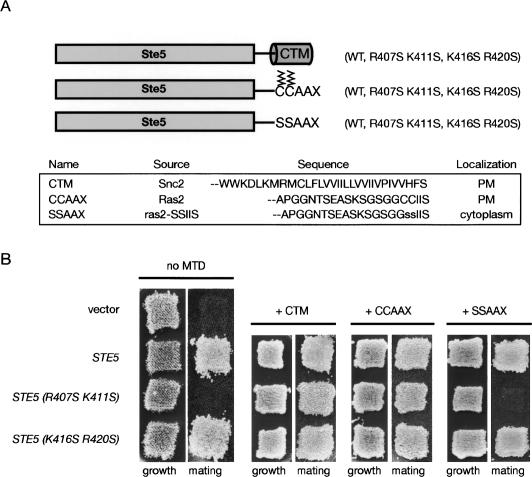
To investigate the role of this sequence in Ste5 function in vivo, three mutants were generated: Ste5(R407S K411S); Ste5(K416S R420S); and Ste5(R407S K411S K416S R420S) (Fig. 2A). These basic residues were selected because they lie at or near positions in PH domains of known structure that contact the phosphates on the head group of the phosphoinositides to which they bind. When expressed at near-endogenous level (from the native STE5 promoter on a CEN plasmid), Ste5(R407S K411S) failed to rescue the mating defect of a ste5Δ mutant, whereas Ste(K416S R420S) supported mating as well as wild-type Ste5 (Fig. 2B, left). The quadruple mutant displayed the same mating deficiency as Ste5(R407S K411S). The observed phenotypes were not due to any differences in expression or stability of these mutants, compared with each other or wild-type Ste5 (Fig. 2C, left). Nevertheless, partial loss-of-function mutations can sometimes be rescued by increased expression. Hence, as one means to assess the severity of the defect conferred by the R407S K411S mutations, the same test was performed with the same mutants overexpressed from the GAL1 promoter. Even when produced at a level 10–20 times above normal (Fig. 2C, right), both Ste5(R407S K411S) and the quadruple mutant were unable to support any detectable mating (Fig. 2B, right), indicating that the R407S K411S mutations confer a complete loss of function.
Figure 2.
The PH domain is essential for Ste5 function in vivo. (A) Ste5 primary structure. (White boxes) Regions implicated in binding the indicated proteins; (dotted line) location of the PM motif; (solid line) location of the predicted PH domain. Positions of previously characterized hyperactive alleles (P44L, C226Y, S770N) and basic residues mutated in this study (R407S K411S R416S R420S) are indicated. (B) Strain BYB69 (ste5Δ) was transformed with either an empty CEN vector or the same vector expressing wild-type Ste5, Ste5(R407S K411S), Ste5(K416S R420S), or Ste5(R407S K411S K416S R420S) from either the native STE5 promoter (left) or from the GAL1 promoter (right), and patch-mating assays were carried out as described in Materials and Methods. (C) Extracts of the same cultures as in B were prepared, resolved by SDS-PAGE, and analyzed by immunoblotting with either an α-Ste5 antiserum (gift of K. Benjamin) (left) or affinity-purified α-Ste5 IgG (Hasson et al. 1994) (right).
Ste5 PH domain binds phosphoinositides in vitro
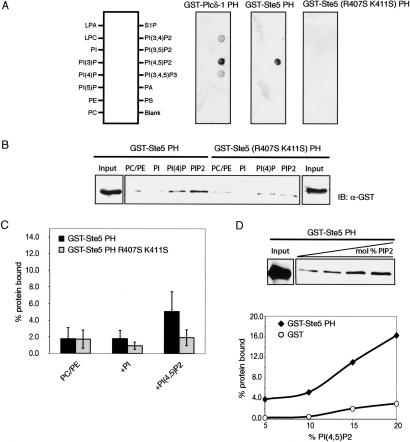
A hallmark of many PH domains (but far from all) is the ability to bind specific phosphoinositides in vitro (Lemmon and Keleti 2005). To determine whether the Ste5 PH domain binds phosphoinositides and whether the phenotypes of the mutants correlate with their phosphoinositide-binding capability, we used three independent methods to measure the association of the isolated Ste5 PH domain with these lipids. For this purpose, we constructed, expressed, and purified from bacterial cells a glutathione-S-transferase (GST)–Ste5(369–517) fusion protein, with or without the PH domain mutations. As a positive control, a GST fusion to the PH domain of mammalian phospholipase Cδ1 (PLCδ1), which binds well to PtdIns(4,5)P2, was expressed and purified in a similar manner. Using an overlay assay in which the GST–PH domain fusions were incubated with various lipids immobilized on nitrocellulose membranes (Yu et al. 2004), we found that the Ste5 PH domain bound to PtdIns(4,5)P2 and did so, in most experiments, at least as specifically as the PLCδ1 PH domain (Fig. 3A). Most importantly, no detectable binding of the PH domain of the mating-defective mutant, Ste5(R407S K411S), was ever observed by this method (Fig. 3A), whereas the nondefective Ste5(K416S K420S) mutant still displayed detectable binding (data not shown).
Figure 3.
The Ste5 PH domain binds PtdIns(4,5)P2. (A) Overlay on immobilized lipids. The indicated purified GST fusions (1 μM) were incubated with commercial filter strips (three right-most panels) on which the indicated phospholipids and related compounds were spotted in the pattern shown (left). (B) Binding to liposomes. Samples (1 μM) of the indicated purified GST fusions (input) were incubated with synthetic vesicles of the following compositions: PC/PE (77% DOPC, 22% DOPE); PI (52% DOPC, 22% DOPE, 10% DOPS, 5% DOPA, 10% PI); PI(4)P; and PIP2 [same as PI vesicles, except 5% PI and 5% PI(4)P or 5% PI(4,5)P2, respectively], each marked with 1% Texas Red-PE. (C) Summary of liposome-binding assays. Values, obtained as in B, represent the amount of vesicle-bound protein (expressed as percent of input) averaged over three to four independent trials. Bars indicate standard error. (D) Effect of vesicle content of PtdIns(4,5)P2. Binding assays were performed with GST–Ste5 PH domain or GST alone, as a control, as in B with liposomes containing the indicated fraction of PI(4,5)P2 (when PIP2 exceeded 10%, DOPC was decreased correspondingly).
To confirm the reliability of these results, binding of the same Ste5 PH domain constructs to phospholipid vesicles in solution was also examined. In this assay, artificial PtdCho/PtdEth liposomes without or with an increasing mole fraction of different phosphoinositides (and marked with Texas Red to follow the efficiency of vesicle recovery) were incubated with the GST–PH domain fusions and separated from excess unbound protein by flotation through a sucrose gradient. In agreement with the overlay assay, the Ste5 PH domain bound best to liposomes containing PtdIns(4,5)P2, and did so distinctly more avidly than the Ste5(R407S K411S) mutant (Fig. 3B). Indeed, in multiple trials, the R407S K411S mutations decreased PtdIns(4,5)P2 binding of the Ste5 PH domain to the background level (Fig. 3C). Based on the percent recovery at equivalent input concentrations, the Ste5 PH domain bound significantly more weakly than did the PLCδ1 PH domain (data not shown). However, as expected for authentic binding, the amount of vesicle-bound GST–Ste5 PH domain (but not GST alone) increased commensurately with an increase in the PtdIns(4,5)P2 content of the liposomes (Fig. 3D) or with an increase in the amount of the GST–Ste5 PH protein added (data not shown).
The same trends were observed using a third method, surface plasmon resonance, in which the PH domain constructs were flowed over vesicles covalently immobilized (via phytosphingosine) to the derivatized gold-plated surface of the detector cell: The Ste5 PH domain bound to PtdIns(4,5)P2-containing liposomes, and bound markedly better than the R407S K411S mutant over the range of protein concentrations tested (data not shown). We conclude that, like certain other isolated S. cerevisiae PH domains examined previously (Yu et al. 2004), the Ste5 PH domain binds PtdIns(4,5)P2 directly and reasonably specifically, but with relatively modest affinity. Nonetheless, taken together with the mating results, these findings suggested that the PH domain could have a direct and critical role in membrane recruitment of Ste5 and that the mating-defective behavior of the R407S K411S mutant might be explained by its inability to be targeted to and stably associate with the plasma membrane.
The PH domain is required for membrane association of Ste5
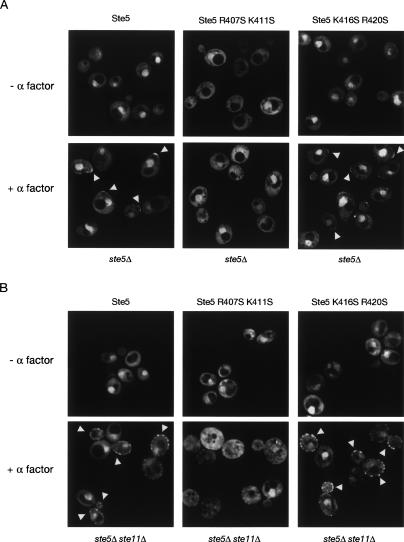
To examine membrane recruitment in live cells in real time, Ste5-GFP fusions with or without the PH domain mutations were constructed and expressed as the sole source of Ste5 in MATa haploids. After exposure to pheromone (α-factor), cells expressing either wild-type Ste5 or the functional Ste5(K416S R420S) mutant formed the typical “shmoo” shape, and a significant fraction of both proteins was rapidly relocalized to the cortex at the elongating cell pole (Fig. 4A, left and right), as observed for Ste5 by us and others previously (Pryciak and Huntress 1998; Mahanty et al. 1999; Sette et al. 2000). Ste5 and Ste5(K416S R420S) remained stably associated with the shmoo tip throughout the time period examined (up to 1 h). In contrast, cells expressing the nonfunctional Ste5(R407S K411S) mutant did not form shmoos and the protein was not recruited to the plasma membrane, even though the cells contain Gβγ, previously thought to be the major recruiter of Ste5 to the cell cortex (Fig. 4A, middle). We noted that Ste5(R407S K411S) also displayed elevated cytoplasmic fluorescence compared with wild-type Ste5; this increase was not due to any defect in nuclear import because Ste5(R407S K411S) accumulated in the nucleus to the same extent as wild-type Ste5 in an msn5Δ mutant in which pheromone-induced nuclear export is blocked (L.S. Garrenton, unpubl.). Unlike Ste5(R407S K411S), other Ste5 derivatives that exhibit an increased cytoplasmic level (e.g., Ste5–GST fusions) show an increase in membrane recruitment and constitutive activation of the mating pathway (Inouye et al. 1997b; Wang and Elion 2003).
Figure 4.
The PH domain is required for pheromone-induced membrane recruitment of Ste5. Strain BYB84 (ste5Δ) (A) or strain YAS1 (ste5Δ ste11Δ) (B) transformed with CEN plasmids expressing from the GAL1 promoter wild-type Ste5-GFP (pCJ80), Ste5(R407S K411S)-GFP (pLG35), or Ste5(K416S R420S)-GFP (pLG36) were grown, induced with galactose, treated with pheromone for 1 h, and viewed by deconvolution microscopy, as indicated in Materials and Methods. Panels depict a collage of representative images; arrowheads indicate Ste5-GFP accumulated at the cell cortex.
Given that Ste5(R407S K411S)-GFP is signaling-defective, its failure to be recruited to the plasma membrane at the shmoo tip might arise simply because the cells do not undergo pheromone-induced morphogenesis. To rule out this possibility, we stimulated Gβγ release in cells (ste11Δ) in which downstream MAPK signaling was blocked, thereby preventing subsequent shmoo formation (for review, see Knaus et al. 2005). Under these conditions, wild-type Ste5 and Ste5(K416S R420S) were recruited to numerous disperse membrane-associated puncta (Fig. 4B, left and right), whereas Ste5(R407S K411S) remained in the cytosol (Fig. 4B, middle). Thus, under all conditions tested, and in agreement with its inability to bind to phosphoinositide-containing vesicles in vitro, Ste5(R407S K411S) was completely defective in membrane recruitment in response to pheromone, indicating that the PH domain plays an essential role in Ste5 association with the plasma membrane in vivo. Moreover, this role of the PH domain lies upstream of the MAPK cascade and is required even when Gβγ is present.
Membrane targeting rescues the mating defectof the Ste5 PH domain mutant
If the sole role of the PH domain in Ste5 function is to help anchor this scaffold protein to the plasma membrane, then the signaling defect of the Ste5(R407S K411S) mutant should be overcome completely by tethering the protein to the plasma membrane by other means. To test this prediction, two different membrane targeting elements—the C-terminal transmembrane segment (CTM) of yeast Snc2 (a synaptobrevin-like R-type v-SNARE) and the S-palmitoylated and S-farnesylated C-terminal CCAAX box of yeast Ras2—were fused in-frame to the C terminus of Ste5 lacking or containing the PH domain mutations (Fig. 5A). As an additional control, we also used a C318S C319S derivative of the Ras2 CCAAX box that cannot be modified by any lipophilic substituent and consequently cannot mediate membrane association (Mitchell and Deschenes 1995). Prior work has shown that fusion of the Snc2 CTM and the Ras2 CCAAX box to wild-type Ste5 does not perturb its function, but rather bypasses the need for Gβγ and constitutively activates downstream signaling (Pryciak and Huntress 1998). Indeed, we found that attachment of the CTM and the CCAAX box permitted both normal Ste5 and the functional Ste5(K416S R20S) mutant to fully rescue the mating defect of ste5Δ cells (Fig. 5B, top and bottom). Consistent with the conclusion that the sole defect conferred by the nonfunctional PH domain mutant is a lack of membrane association, attachment of the CTM and the CCAAX box (but not the mutant SSAAX box) also fully restored mating in ste5Δ cells expressing Ste5(R407S K411S) (Fig. 5B, middle). Furthermore, as assessed by the level of expression of a pheromone-responsive reporter gene, FUS1∷HIS3, the Ste5(R407S K411S) mutant is fully capable of propagating a signal from a dominant allele (STE11-4), which produces a constitutively active kinase, Ste11(T596I), that bypasses the need for its activation at the membrane by Ste20 (Supplementary Fig. S1; Stevenson et al. 1992). Thus, the Ste5 PH domain mutations prevent Ste5 from functioning only when signaling is initiated at the plasma membrane, but not when the need for membrane recruitment is bypassed by artificial targeting or by an activated allele of Ste11. Together, these data argue strongly that the only defect of Ste5(R407S K411S) is its demonstrated inability to be recruited to the plasma membrane and, therefore, that the PH domain is essential for Ste5 interaction with the membrane.
Figure 5.
Forced membrane anchoring rescues the Ste5 PH domain mutant. (A) Membrane targeting elements. CTM (cylinder), C-terminal α-helical transmembrane segment of Snc2; S-palmitoylated and S-farnesylated (zig-zags) C-terminal segment of wild-type (CCAAX) and unmodified mutant (SSAAX) Ras2. (B) Mating proficiency of BYB69 (ste5Δ) cells expressing either empty vector, wild-type Ste5, or the indicated Ste5 mutants, not fused to any membrane targeting domain (no MTD) or fused to the elements shown in A, was analyzed using a patch-mating assay as in Figure 2B.
Ste5 PH domain mutant binds MAPKKK Ste11
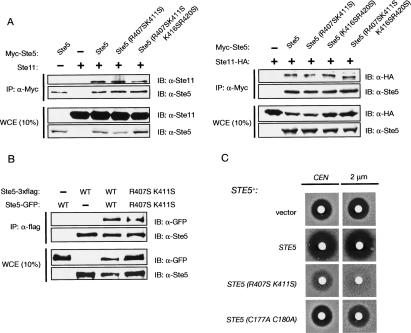
The predicted PH domain (400–512) partially overlaps with the region (residues 463–514) previously implicated in binding of Ste11 to Ste5 (Fig. 2A). Indeed, it was concluded that Ste11 binding is required for Ste5 membrane recruitment because mutations in this region [e.g., Ste5(Δ474–487)] prevented its association with both Ste11 and the plasma membrane (Wang and Elion 2003). However, the altered region corresponds to the C-terminal portion of the PH domain. In contrast to alterations of the C-terminal end of the PH domain, the nonfunctional Ste5(R407S K411S) and Ste5(R407S K411S K416S R420S) mutants associate with Ste11 indistinguishably from wild-type Ste5 or the functional Ste5(K416S R420S) mutant, as judged by coimmunoprecipitation from cell extracts, regardless of whether Ste11 was overexpressed (Fig. 6A, left) or expressed at its endogenous level from its chromosomal locus (Fig. 6A, right). Thus, the R407S K411S mutations that abrogate the function of the Ste5 PH domain do not alter the affinity of Ste5 for Ste11.
Ste5 PH domain mutant oligomerizes
Prior work indicates that Ste5 self-associates and that this oligomerization is important for its signaling function (Yablonski et al. 1996; Inouye et al. 1997b; Sette et al. 2000; Wang and Elion 2003). The PH domain lies within one of two regions in Ste5 (residues 335–586) that have been implicated in mediating its oligomerization. To determine whether the signaling defect of Ste5(R407S K411S) could be explained by an inability to self-associate, three independent and complementary approaches were taken. First, we coexpressed in MATa ste4Δ ste5Δ cells two differentially tagged versions of Ste5(R407S K411S), one fused to GFP and the other containing a 3xFlag epitope. Both derivatives of Ste5(R407S K411S) were coimmunoprecipitated (using anti-Flag antibody-coated beads) just as efficiently as the same derivatives of wild-type Ste5 (Fig. 6B). Second, when expressed in wild-type cells, Ste5(R407S K411S) acts as a potent dominant-negative (Fig. 6C). By contrast, overexpression of an oligomerization-defective mutant, Ste5(C177A C180A), has a significantly weaker effect. These results indicate that the observed inhibition by the Ste5 PH domain mutant cannot be due merely to titration of signaling components, but rather to formation of mixed oligomers with reduced capacity for membrane binding. Third, the mating defect of the Ste5(R407S K411S) mutant was not rescued by forced dimerization with a heterologous dimerization domain, GST (Maru et al. 1996), which as we previously have shown restores signaling function to known oligomerization-defective mutants of Ste5 (Supplementary Fig. S2). Hence, the inability of Ste5(R407S K411S) to function in vivo reflects a role for the PH domain in a property of Ste5 different from oligomerization, consistent with the other evidence presented for its essential role in membrane recruitment of Ste5.
The PH domain is not required for Gβγ binding to Ste5
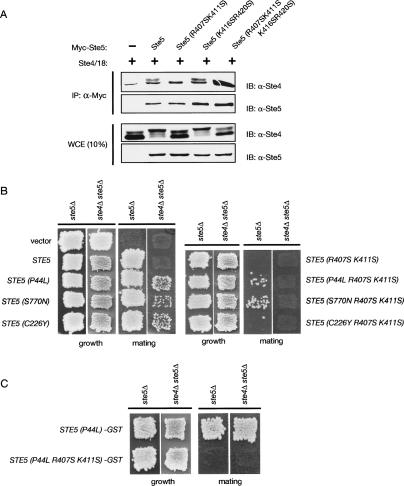
Interaction of Ste5 with membrane-anchored Gβγ freed upon exposure of cells to pheromone is an early, crucial, and well-established event in this signaling pathway (for review, see Wang and Dohlman 2004). Given the inability of Ste5(R407S K411S) to be recruited to the membrane in response to pheromone, it was possible that this PH domain mutant was unable to interact with Gβγ, in addition to its defect in binding to membrane phosphoinositides revealed by our in vitro experiments. However, as judged by coimmunoprecipitation from cell extracts, the nonfunctional mutants, Ste5(R407S K411S) and Ste5(R407S K411S K416S R420S), showed no detectable diminution in their ability to associate with Ste4 (Gβ), compared with wild-type Ste5 or the functional mutant, Ste5(K416S R420S), expressed under the same conditions (Fig. 7A).
Figure 7.
The Ste5 PH domain acts separately from, but synergistically with, Gβγ. (A) The Ste5 PH domain mutant retains Gβγ binding. Strain BYB84 (ste5Δ)—expressing from the GAL1 promoter either a vector control (−) or the c-Myc epitope-tagged Ste5 derivatives described in Figure 5A and coexpressing STE4 and STE18 under the control of the divergent GAL1–GAL10—was lysed, subjected to immunoprecipitation, and analyzed by immunoblotting as in Figure 6A using α-Ste5 antisera and α-Ste4 antisera. A trace of Ste4 associates with antibody-coated beads nonspecifically in the absence of Ste5, as we have observed previously (Inouye et al. 1997a). (B) Loss of PH domain function abrogates the constitutive activity of hyperactive Ste5 alleles. Strains BYB69 (ste5Δ) or BYB88 (ste4Δ ste5Δ) were transformed with empty 2μm-DNA vector or the same vector expressing from the GAL1 promoter either wild-type Ste5 or Ste5(R407S K411S), Ste5(P44L) or Ste5(P44L R407S K411S), Ste5(S770N) or Ste5(R407S K411S S770N), or Ste5(C226Y) or Ste5(C226Y R407S K411S), as indicated, and patch-mating assays were done as in Figure 2B. (C) Loss of PH domain function still abrogates the enhanced activity of the PM motif mutant, Ste5(P44L)–GST. Strain BYB88 (ste4Δ ste5Δ) was transformed with a CEN vector expressing from the GAL1 promoter either Ste5(P44L)–GST or Ste5(P44L R407S K411S)–GST, and patch-mating assays were performed.
Elevated expression of Ste5 leads to pathway activation, resulting in hyperphosphorylation of Ste4 (Cole and Reed 1991). Indeed, hyperphosphorylated Ste4 was the primary species present in extracts of cells expressing wild-type Ste5 or the functional Ste5(K416S R420S) mutant and coimmunoprecipitated with both of these proteins, whereas hypophosphorylated Ste4 was the major species present in extracts of cells expressing the nonfunctional Ste5(R407S K411S) or Ste5(R407S K411S K416S R420S) mutants and coimmunoprecipitated with those proteins (Fig. 7A). Thus, as judged by this additional indicator of early events in the response pathway, the PH domain mutations render Ste5 incompetent to transmit a pheromone-induced signal, and this defect is not due to an inability to associate with Gβγ. The above observations, taken together with the results of our analysis of subcellular localization (Fig. 4), make it clear that, in the absence of a functional PH domain, interaction with Gβγ is not sufficient for membrane recruitment of Ste5 in response to pheromone.
Hyperactive Ste5 alleles do not functionin the absence of an intact PH domain
Our initial impetus for examining mechanisms for membrane targeting of Ste5 other than by its interaction with Gβγ was the fact that we were able to generate hyperactive Ste5 alleles that promote signaling even when Gβγ is absent (Hasson et al. 1994; Sette et al. 2000). We reasoned that the activating mutations must enhance the ability of Ste5 to associate with the membrane. Indeed, the apparent explanation for two such alleles (P44L and T52M) is that they install markedly more hydrophobic residues into an N-terminal basic α-helix (PM motif), thereby elevating its intrinsic propensity to associate with acidic phospholipids (Winters et al. 2005). We reasoned further, however, that such an interaction may not be sufficient and that effective membrane recruitment of Ste5, even by these hyperactive alleles, may depend on synergy between binding of the PH domain to phosphoinositides and binding of the mutant PM motif to the membrane.
To test this possibility, we introduced the PH domain loss-of-function mutations, R407S K411S, into Ste5 mutants containing each of three different gain-of-function mutations: P44L, C226Y, and S770N. As observed before, wild-type Ste5 and all three hyperactive alleles (P44L, C226Y, and S770N) promoted robust mating in ste5Δ cells, whereas only the hyperactive alleles supported detectable mating in a ste4Δ ste5Δ double mutant (Fig. 7B, left). However, when the same hyperactive mutants were combined with the R407S K411S PH domain mutations, no mating occurred in the cells lacking Gβγ (ste4Δ ste5Δ strain) (Fig. 7B, right). Moreover, introduction of the R407S K411S mutations completely blocked signaling even when Ste5(P44L) was fused to GST, which greatly enhances the potency of the P44L allele in promoting mating (Fig. 7C), in agreement with prior work (Sette et al. 2000; Winters et al. 2005). Interestingly, in cells containing Gβγ (ste5Δ strain), two of the hyperactive alleles supported a detectable degree of mating, even when the PH domain was ablated (Fig. 7B, right).
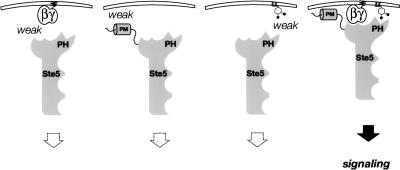
These observations indicate, first, that the hyperactive mutants cannot bypass the need for Gβγ when the PH domain is not functional. Second, the fact that the PH domain is essential for the hyperactive alleles to function in cells (ste4Δ ste5Δ) that completely lack Gβγ confirms that the PH domain contributes to membrane recruitment in a manner that does not depend on its direct interaction with Gβγ. Third, the fact that the presence of Gβγ partially suppressed the mating debility of the PH domain-defective hyperactive alleles provides further support for the conclusion that the PH domain works independently from, but synergistically with, Gβγ in recruitment of Ste5 to the membrane. Collectively, these findings show that no single mode of membrane interaction (via the PH domain, the mutant PM motif, or Gβγ binding) is sufficient for signaling, whereas inefficient signaling can be mounted by any combination of two of these modes, and that maximal signaling requires all three (Fig. 8).
Discussion
Our results demonstrate that a previously uncharacterized PH domain in Ste5 has an essential role in the function of this archetypal MAPK scaffold protein. Alteration of two basic residues in this domain abogrates the ability of Ste5 to promote signaling in response to pheromone, prevents Ste5 binding to a specific phosphoinositide in vitro, and eliminates membrane recruitment of Ste5 in vivo. These mutations do not affect any of the other functions of Ste5 attributed to this region of the protein (such as binding of Ste11 or oligomerization) or Gβγ binding. Thus, interaction of Ste5 with Gβγ is not sufficient for membrane tethering when the PH domain is nonfunctional. Fusion of the Ste5 PH domain mutant to the CTM domain of Snc2 or the CCAAX box of Ras2 efficiently ameliorates its signaling defect; thus, alternative means of membrane binding bypass the need for the PH domain. Even when the membrane-associating propensity of Ste5 is elevated by means intrinsic to Ste5 (such as mutations, like P44L, that increase the membrane affinity of the N-terminal PM motif) and Gβγ is available, signaling is still very inefficient when the PH domain is not functional. Conversely, the PH domain is not necessary to propagate a signal initiated by Ste11(T596I), a constitutively active mutant of the MAPKKK that bypasses the need for Ste20-initiated signaling at the plasma membrane. Hence, it seems clear that optimal function of Ste5 normally requires stable membrane recruitment via concerted action of the PH domain, the PM motif, and binding to Gβγ (Fig. 8).
At present, based on estimates from our in vitro phospholipid-binding studies (L.S. Garrenton, unpubl.), interaction of the Ste5 PH domain with PtdIns(4,5)P2 has a Kd = ∼5–10 μM, which is significantly weaker than that of the well characterized PtdIns(4,5)P2-specific PLCδ1 PH domain (Kd = 200 nM) (Lemmon and Keleti 2005). However, this property is not uncommon among PH domains. Only a small minority of known PH domains binds their cognate phosphoinositide with high affinity and specificity, whereas the large majority binds phosphoinositides less selectively and rather weakly (Kd = 10–20 μM or greater) (Yu et al. 2004). However, the specific nature of the Ste5 PH domain may be physiologically important because it may set a threshold that prevents inadvertent association of Ste5 with the membrane in the absence of authentic pheromone stimulation. Moreover, it may explain, at least in part, the apparently vital role of oligomerization in Ste5 function (Yablonski et al. 1996; Inouye et al. 1997b), given that multimerization of PH domains and other membrane-binding motifs greatly increases their membrane-binding avidity (Stefan et al. 2002; Winters et al. 2005). For example, the PH domain of mammalian dynamin-1 requires oligomerization for significant membrane association, and, when overexpressed in vivo, PH domain-defective dynamin-1 mutants act as dominant-negative inhibitors (Lemmon and Keleti 2005), as we have observed for Ste5(R407S K411S). In addition, the relatively modest affinity of the PH domain for membrane phosphoinositide may explain why it must act in cooperation with Gβγ binding and the PM motif to achieve effective membrane association. Multiple, but low-affinity, membrane targeting domains make pathway activation dependent on coincident and highly cooperative detection of both receptor occupancy and a receptive membrane milieu. Multiple sites of interaction, once joined, also provide relatively stable membrane localization, imposing spatial selectivity and thus a mechanism to ensure that only Ste5-bound Ste11 becomes activated.
Coordinate action of a PH domain with other membrane interaction and/or protein interaction motifs to drive membrane association is not unique to Ste5. The PH domain of the β-adrenergic receptor kinase, Grk2, binds both phosphoinositides and Gβγ, yet neither interaction alone supports efficient membrane recruitment (Lemmon and Keleti 2005). Likewise, the PH domain in a yeast PAK, Cla4 (Wild et al. 2004), and PH domains in PLCγ1 and in a Rho- and Rac GEF, Tiam1 (Lemmon and Keleti 2005), all act in conjunction with additional domains to achieve proper membrane recruitment and localization. Most tellingly, in S. cerevisiae pheromone response, another scaffold protein, Far1, serves to deliver the Cdc42 GEF, Cdc24, to the plasma membrane. Sequence alignments show that Far1 shares two regions of strong similarity with Ste5: an N-terminal RING-H2 domain (residues 202–251), which interacts with Gβγ (Butty et al. 1998), just like its counterpart in Ste5 (Inouye et al. 1997b; Feng et al. 1998); and a more internal segment (residues 416–550), corresponding to the region in Ste5 that we have demonstrated here contains its functional PH domain. Despite the presence of its Gβγ-binding RING-H2 domain, deletion of the PH domain-like element in Far1 prevents its recruitment to the shmoo tip during pheromone response (Wiget et al. 2004).
In this same regard, prior work has suggested that interaction of Ste5 with other proteins, specifically Bem1 and Cdc24, contributes to recruiting Ste5 to the shmoo tip to promote pheromone-induced signaling (Leeuw et al. 1995; Lyons et al. 1996; Wang et al. 2005). Although the evidence for these interactions is quite compelling, they are not essential for Ste5 function when Gβγ is present, in marked contrast to what we have demonstrated for the PH domain. For example, an allele (cdc24-4) of Cdc24 that cannot interact with Ste5 does not exhibit any detectable mating defect (Shimada et al. 2004), and even a null mutation (bem1Δ) does not eliminate mating (Lyons et al. 1996). In contrast, when Ste5 carries the PH domain mutations, it is unable to support any detectable mating. Although Ste5 interaction with Bem1 and/or Cdc24 may contribute to Ste5 function when it is at the plasma membrane, our results show that the PH domain makes an independent and more crucial contribution in the recruitment of Ste5 to the plasma membrane per se.
It has been suggested (Wang and Elion 2003) that a Leu zipper-like sequence (residues 428–455), which resides within the PH domain in Ste5, may mediate its oligomerization. However, all other structurally characterized Leu zippers that form coiled-coils lie in bona fide 4–3 hydrophobic repeats in sequences that adopt an α-helical conformation, whereas all of the algorithms we applied indicate that this segment of Ste5 will adopt a β-strand conformation, more compatible with the β-sheets found in PH domains. In this same study, the purported Leu zipper itself was not mutagenized; but, deletion (Δ474–487) or alteration (L482A L485A) of residues downstream of the proposed Leu zipper, which lie within the C-terminal half of the PH domain, prevented membrane recruitment and signaling, yet did not interfere with oligomerization, just as we have observed for the R407S K411S mutations near the N-terminal end of the PH domain. However, unlike Ste5(R407S K411S), the Δ474–487 deletion eliminated, and the L482A L485A mutations greatly reduced, Ste11 binding. Hence, the signaling defect of these alterations was attributed to some as yet undefined role for Ste11 in mediating membrane association of Ste5. However, the R407S K411S mutant, which binds Ste11 normally, clearly separates the role of the PH domain in membrane binding from Ste11 docking. Also, as we have shown here, wild-type Ste5, but not the PH domain mutant, is recruited to the membrane, even when Ste11 is absent, corroborating a Ste11-independent role for the PH domain.
The PH domain is a critical mediator of the stable association of Ste5 with the plasma membrane. In vitro the Ste5 PH domain binds preferentially to PtdIns(4,5)P2. Use of fluorescent reporter proteins and other means have shown that PtdIns(4,5)P2 is located predominantly, if not exclusively, in the plasma membrane in S. cerevisiae (Stefan et al. 2002). Thus, the PH domain presumably assists in membrane anchoring of Ste5 via its direct association with PtdIns(4,5)P2 in the plasma membrane. Given that the PH domain is critical for membrane binding, what does interaction of Ste5 with Gβγ contribute? We believe that the most likely explanation for why elimination of the PH domain ablates the ability of Gβγ to promote membrane recruitment is that a significant aspect of the physiological function of Gβγ binding to Ste5 is not to help anchor Ste5 to the membrane per se, but rather to cause a conformational change that exposes the PH domain so that it is accessible to interact with the membrane. We have at least some evidence that the PH domain may be masked in unstimulated Ste5. Only the isolated GST–PH domain bound to phospholipids in vitro; full-length Ste5 (purified from Escherichia coli as a fusion to MalE) did not bind to phospholipids in either the filter overlay or liposome-binding assays (L.S. Garrenton, unpubl.). Therefore, it is important to determine whether Gβγ-induced conformational changes and/or oligomerization may regulate the ability of the PH domain to drive membrane recruitment of Ste5.
Materials and methods
Yeast strains and media
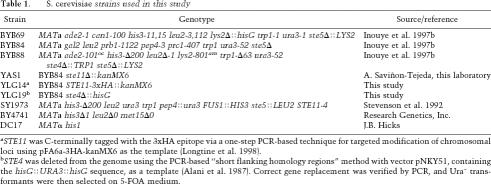
Cultivation of strains (Table 1) was at 30°C in standard rich (YP) or defined minimal (SC) media (Sherman et al. 1986) containing 2% glucose (Glc), 2% raffinose with 0.2% sucrose (Raf/Suc), or 2% galactose (Gal) and supplemented with appropriate nutrients to maintain selection for plasmids. Gene induction from the GAL1 promoter was performed as in Inouye et al. (1997a). Solid medium supplemented with 30 mM 3-aminotriazole (3-AT) was used, where indicated. Standard yeast genetic techniques were according to Sherman et al. (1986). Mating proficiency of MATa cells was assessed as described by Sprague (1991) using strain DC17 as the MATα tester.
aSTE11 was C-terminally tagged with the 3xHA epitope via a one-step PCR-based technique for targeted modification of chromosomal loci using pFA6a-3HA-kanMX6 as the template (Longtine et al. 1998).
bSTE4 was deleted from the genome using the PCR-based “short flanking homology regions” method with vector pNKY51, containing the hisG∷URA3∷hisG sequence, as a template (Alani et al. 1987). Correct gene replacement was verified by PCR, and Ura− transformants were then selected on 5-FOA medium.
Plasmids and recombinant DNA methods
Plasmids (Supplementary Table S1) were constructed and propagated in E. coli using standard recombinant DNA methods (Sambrook et al. 1989). Fidelity of all constructs was verified by nucleotide sequence analysis. All PCR utilized Turbo Pfu DNA polymerase (Stratagene). Ste5 PH domain mutants were generated by site-directed mutagenesis using appropriate mismatch primers.
Purification of GST fusions and overlay assay
To construct GST–Ste5 PH domain fusions with and without the R407S K411S mutations, an ∼400-base-pair (bp) fragment (corresponding to Ste5 residues 369–517) was amplified by PCR using pCJ117 and pLG21 as the templates, respectively, and ligated into pGEX-4T (Amersham-Pharmacia), yielding pLG49 and pLG50, respectively. A protease-deficient E. coli strain [BL21-CodonPlus(DE3)-RIL; Stratagene] carrying either pLG49 or pLG50 was grown to A600nm = 0.2, and expression of GST–Ste5–PH or GST–Ste5–PH(R407S K411S) was induced by addition of isopropyl-β-D-thiogalacto-pyranoside (0.4 mM final concentration). After vigorous aeration for 3 h at room temperature, cells were harvested and GST fusions were purified by column chromatography on glutathione-agarose using standard procedures.
To measure lipid binding, commercially prepared filters with immobilized phospholipids (PIP Strips, Eschelon Biosciences, Inc.) were blocked for 1 h at room temperature in Odyssey Blocking Solution (Li-Cor). The filters were then incubated at room temperature with purified GST fusions (0.5 μg/mL) in Odyssey Blocking Solution for 3 h, washed three times in Tris-buffered (pH 7.5) saline containing 0.1% Tween-20, and bound GST fusions were detected by incubation—first for ∼1.5 h with monoclonal α-GST antibodies, and then for 60 min with an appropriate secondary antibody conjugated to an infrared fluorophore—and visualized using an infrared imaging system (Odyssey, Li-Cor).
Binding to liposomes
Phospholipids and derivatives in chloroform (Avanti Polar Lipids) and Texas Red-DHPE (Molecular Probes) were mixed, evaporated, and hydrated at a final concentration of 1 mg/mL total phospholipid in HKME buffer (20 mM HEPES-KOH at pH 7.0, 160 mM KOAc, 1 mM MgCl2, and 0.1 mM EGTA) at room temperature with occasional vortexing. The resulting suspension of multilamellar vesicles was extruded through a polycarbonate filter (400-nm pore size). A sample (12 μL) of the resulting unilamellar liposomes was mixed with 48 μL HKME containing GST–Ste5–PH or GST–Ste5–PH(R407S K411S) (0.5–5.0 μM), and flotation was conducted as described by Matsuoka et al. (1998). Liposome-bound proteins were resolved by SDS-PAGE and analyzed by immunoblotting with monoclonal α-GST antibodies, as described above. Surface plasmon resonance measurements were made in a device from Reichert Instruments, Inc.
Subcellular localization by fluorescence microscopy
Transformants expressing GFP fusions were grown, treated with α-factor, and prepared for microscopic examination as described previously (Sette et al. 2000). Cells were viewed under a 100× objective using a Delta-Vision Spectris DV4 deconvolution microscope (Applied Precision LCC). Images were collected and processed using API SoftWoRx imaging software and Photoshop (Adobe Systems, Inc.).
Preparation of cell extracts, immunoprecipitation, and immunoblotting
Yeast cells were grown, harvested, and lysed; cell-free extracts were prepared; and final protein concentration was determined as described previously (Inouye et al. 1997a). For immunoprecipitation, samples (1 mg total protein) were diluted to a final volume of 300 μL in L buffer (Inouye et al. 1997a) containing 1% bovine serum albumin. Subsequent preclearing, immunoprecipitation, washing, and elution were also carried out according to Inouye et al. (1997a), with the exception that the Ste5-3xFlag derivatives were immunoprecipitated using anti-Flag M2 affinity gel (Sigma Chemical Corp.) according to the manufacturer’s instructions. The eluted immune complexes, solubilized in SDS-PAGE sample buffer, were boiled for 5 min, clarified by brief centifugation, resolved by 8%–12% SDS-PAGE, and analyzed by immunoblotting. Proteins resolved in slab gels (either cell extract or solubilized immune complexes) were transferred to nitrocellulose, incubated with appropriate primary antibodies, washed, incubated with appropriate secondary antibodies conjugated to infrared fluorophores, and visualized using an infrared imaging system. Ste5 was detected using either affinity-purified polyclonal rabbit IgG (Hasson et al. 1994) or another polyclonal rabbit anti-Ste5 antiserum (gift of Kirsten Benjamin, Molecular Sciences Institute, Berkeley, CA). Other antibodies were as follows: polyclonal rabbit anti-Ste4 (Hirschman et al. 1997), polyclonal goat anti-Ste11 (Santa Cruz Biotechnology, Inc.), mouse monoclonal anti-HA epitope (Covance), mouse monoclonal anti-c-Myc epitope (Evan et al. 1985), mouse monoclonal anti-GFP (Roche Diagnostics, Inc.), mouse monoclonal anti-GST (Sigma Chemical Corp.), Alexa Fluor 680-conjugated goat anti-rabbit IgG and donkey anti-goat IgG (Molecular Probes, Inc.), and IRDye800-conjugated goat anti-mouse IgG and donkey anti-rabbit IgG (Rockland Immunochemicals, Inc.).
Acknowledgments
We thank D. Ballon, E. Futai, R. Schekman, D. Jenness, M.A. Lemmon, K. Benjamin, and P. Pryciak for advice, reagents, and/or communication of unpublished results, and T.E. Ryan of Reichert Instruments, Inc., for help with SPR analysis. This work was supported by NIH Predoctoral Traineeships GM07232 (to L.S.G.) and GM07048 (to S.L.Y.), by NCI Predoctoral Traineeship CA09041 (to L.S.G.), by NIH Research Grant GM21841 (to J.T.), and by facilities provided by the Cancer Research Laboratory of the University of California, Berkeley.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1413706
References
- Alani E., Cao L., Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butty A.C., Pryciak P.M., Huang L.S., Herskowitz I., Peter M. The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science. 1998;282:1511–1516. doi: 10.1126/science.282.5393.1511. [DOI] [PubMed] [Google Scholar]
- Choi K.Y., Satterberg B., Lyons D.M., Elion E.A. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- Cole G.M., Reed S.I. Pheromone-induced phosphorylation of a G protein β subunit in S. cerevisiae is associated with an adaptive response to mating pheromone. Cell. 1991;64:703–716. doi: 10.1016/0092-8674(91)90500-x. [DOI] [PubMed] [Google Scholar]
- Drogen F., O’Rourke S.M., Stucke V.M., Jaquenoud M., Neiman A.M., Peter M. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr. Biol. 2000;10:630–639. doi: 10.1016/s0960-9822(00)00511-x. [DOI] [PubMed] [Google Scholar]
- Elion E.A. The Ste5p scaffold. J. Cell Sci. 2001;114:3967–3978. doi: 10.1242/jcs.114.22.3967. [DOI] [PubMed] [Google Scholar]
- Evan G.I., Lewis G.K., Ramsay G., Bishop J.M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Song L.Y., Kincaid E., Mahanty S.K., Elion E.A. Functional binding between Gβ and the LIM domain of Ste5 is required to activate the MEKK Ste11. Curr. Biol. 1998;8:267–278. doi: 10.1016/s0960-9822(98)70108-3. [DOI] [PubMed] [Google Scholar]
- Hasson M.S., Blinder D., Thorner J., Jenness D.D. Mutational activation of the STE5 gene product bypasses the requirement for G protein β and γ subunits in the yeast pheromone response pathway. Mol. Cell. Biol. 1994;14:1054–1065. doi: 10.1128/mcb.14.2.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschman J.E., Jenness D.D. Dual lipid modification of the yeast gγ subunit Ste18p determines membrane localization of Gβγ. Mol. Cell. Biol. 1999;19:7705–7711. doi: 10.1128/mcb.19.11.7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschman J.E., De Zutter G.S., Simonds W.F., Jenness D.D. The Gβγ complex of the yeast pheromone response pathway. Subcellular fractionation and protein–protein interactions. J. Biol. Chem. 1997;272:240–248. doi: 10.1074/jbc.272.1.240. [DOI] [PubMed] [Google Scholar]
- Inouye C., Dhillon N., Durfee T., Zambryski P.C., Thorner J. Mutational analysis of STE5 in the yeast Saccharomyces cerevisiae: Application of a differential interaction trap assay for examining protein–protein interactions. Genetics. 1997a;147:479–492. doi: 10.1093/genetics/147.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye C., Dhillon N., Thorner J. Ste5 RING-H2 domain: Role in Ste4-promoted oligomerization for yeast pheromone signaling. Science. 1997b;278:103–106. doi: 10.1126/science.278.5335.103. [DOI] [PubMed] [Google Scholar]
- Kelley L.A., MacCallum R.M., Sternberg M.J. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- Knaus M., Wiget P., Shimada Y., Peter M. Control of cell polarity in response to intra- and extracellular signals in budding yeast. Novartis Found. Symp. 2005;269:47–58. [PubMed] [Google Scholar]
- Künzler M., Trueheart J., Sette C., Hurt E., Thorner J. Mutations in the YRB1 gene encoding yeast ran-binding-protein-1 that impair nucleocytoplasmic transport and suppress yeast mating defects. Genetics. 2001;157:1089–1105. doi: 10.1093/genetics/157.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamson R.E., Winters M.J., Pryciak P.M. Cdc42 regulation of kinase activity and signaling by the yeast p21-activated kinase Ste20. Mol. Cell. Biol. 2002;22:2939–2951. doi: 10.1128/MCB.22.9.2939-2951.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeuw T., Fourest-Lieuvin A., Wu C., Chenevert J., Clark K., Whiteway M., Thomas D.Y., Leberer E. Pheromone response in yeast: Association of Bem1p with proteins of the MAP kinase cascade and actin. Science. 1995;270:1210–1213. doi: 10.1126/science.270.5239.1210. [DOI] [PubMed] [Google Scholar]
- Leeuw T., Wu C., Schrag J.D., Whiteway M., Thomas D.Y., Leberer E. Interaction of a G-protein β-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature. 1998;391:191–195. doi: 10.1038/34448. [DOI] [PubMed] [Google Scholar]
- Lemmon M.A., Keleti D. PH domains. In: Cesareni G., et al., editors. Modular protein domains. Wiley-VCH Verlag GmbH & Co.; Weinheim, Germany: 2005. pp. 337–363. [Google Scholar]
- Lin K., Simossis V.A., Taylor W.R., Heringa J. A simple and fast secondary structure prediction method using hidden neural networks. Bioinformatics. 2005;21:152–159. doi: 10.1093/bioinformatics/bth487. [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lyons D.M., Mahanty S.K., Choi K.Y., Manandhar M., Elion E.A. The SH3-domain protein Bem1 coordinates mitogen-activated protein kinase cascade activation with cell cycle control in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:4095–4106. doi: 10.1128/mcb.16.8.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahanty S.K., Wang Y., Farley F.W., Elion E.A. Nuclear shuttling of yeast scaffold Ste5 is required for its recruitment to the plasma membrane and activation of the mating MAPK cascade. Cell. 1999;98:501–512. doi: 10.1016/s0092-8674(00)81978-9. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Anderson J.B., DeWeese-Scott C., Fedorova N.D., Geer L.Y., He S., Hurwitz D.I., Jackson J.D., Jacobs A.R., Lanczycki C.J., et al. CDD: A curated Entrez database of conserved domain alignments. Nucleic Acids Res. 2003;31:383–387. doi: 10.1093/nar/gkg087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus A., Polverino A., Barr M., Wigler M. Complexes between STE5 and components of the pheromone-responsive mitogen-activated protein kinase module. Proc. Natl. Acad. Sci. 1994;91:7762–7766. doi: 10.1073/pnas.91.16.7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maru Y., Afar D.E., Witte O.N., Shibuya M. The dimerization property of glutathione S-transferase partially reactivates Bcr-Abl lacking the oligomerization domain. J. Biol. Chem. 1996;271:15353–15357. doi: 10.1074/jbc.271.26.15353. [DOI] [PubMed] [Google Scholar]
- Matsuoka K., Orci L., Amherdt M., Bednarek S.Y., Hamamoto S., Schekman R., Yeung T. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- McGuffin L.J., Bryson K., Jones D.T. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Mitchell D.A., Deschenes R.J. Characterization of protein prenylation in Saccharomyces cerevisiae. Methods Enzymol. 1995;250:68–78. doi: 10.1016/0076-6879(95)50063-4. [DOI] [PubMed] [Google Scholar]
- Ohya Y., Qadota H., Anraku Y., Pringle J.R., Botstein D. Suppression of yeast geranylgeranyl transferase I defect by alternative prenylation of two target GTPases, Rho1p and Cdc42p. Mol. Biol. Cell. 1993;4:1017–1025. doi: 10.1091/mbc.4.10.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollastri G., Przybylski D., Rost B., Baldi P. Improving the prediction of protein secondary structure in three and eight classes using recurrent neural networks and profiles. Proteins. 2002;47:228–235. doi: 10.1002/prot.10082. [DOI] [PubMed] [Google Scholar]
- Printen J.A., Sprague G.F., Jr. Protein–protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics. 1994;138:609–619. doi: 10.1093/genetics/138.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryciak P.M., Huntress F.A. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gβγ complex underlies activation of the yeast pheromone response pathway. Genes & Dev. 1998;12:2684–2697. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Sato T.K., Overduin M., Emr S.D. Location, location, location: Membrane targeting directed by PX domains. Science. 2001;294:1881–1885. doi: 10.1126/science.1065763. [DOI] [PubMed] [Google Scholar]
- Sette C., Inouye C.J., Stroschein S.L., Iaquinta P.J., Thorner J. Mutational analysis suggests that activation of the yeast pheromone response mitogen-activated protein kinase pathway involves conformational changes in the Ste5 scaffold protein. Mol. Biol. Cell. 2000;11:4033–4049. doi: 10.1091/mbc.11.11.4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink G.R., Hicks J.B. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1986. [Google Scholar]
- Shimada Y., Wiget P., Gulli M.P., Bi E., Peter M. The nucleotide exchange factor Cdc24p may be regulated by auto-inhibition. EMBO J. 2004;23:1051–1062. doi: 10.1038/sj.emboj.7600124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague G.F., Jr. Assay of yeast mating reaction. Methods Enzymol. 1991;194:77–93. doi: 10.1016/0076-6879(91)94008-z. [DOI] [PubMed] [Google Scholar]
- Stefan C.J., Audhya A., Emr S.D. The yeast synaptojanin-like proteins control the cellular distribution of phosphatidylinositol (4,5)-bisphosphate. Mol. Biol. Cell. 2002;13:542–557. doi: 10.1091/mbc.01-10-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson B.J., Rhodes N., Errede B., Sprague G.F., Jr. Constitutive mutants of the protein kinase STE11 activate the yeast pheromone response pathway in the absence of the G protein. Genes & Dev. 1992;6:1293–1304. doi: 10.1101/gad.6.7.1293. [DOI] [PubMed] [Google Scholar]
- Wang Y., Dohlman H.G. Pheromone signaling mechanisms in yeast: A prototypical sex machine. Science. 2004;306:1508–1509. doi: 10.1126/science.1104568. [DOI] [PubMed] [Google Scholar]
- Wang Y., Elion E.A. Nuclear export and plasma membrane recruitment of the Ste5 scaffold are coordinated with oligomerization and association with signal transduction components. Mol. Biol. Cell. 2003;14:2543–2558. doi: 10.1091/mbc.E02-10-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Chen W., Simpson D.M., Elion E.A. Cdc24 regulates nuclear shuttling and recruitment of the Ste5 scaffold to a heterotrimeric G protein in S. cerevisiae. J. Biol. Chem. 2005;280:13084–13096. doi: 10.1074/jbc.M410461200. [DOI] [PubMed] [Google Scholar]
- Whiteway M.S., Wu C., Leeuw T., Clark K., Fourest-Lieuvin A., Thomas D.Y., Leberer E. Association of the yeast pheromone response G protein βγ subunits with the MAP kinase scaffold Ste5p. Science. 1995;269:1572–1575. doi: 10.1126/science.7667635. [DOI] [PubMed] [Google Scholar]
- Wiget P., Shimada Y., Butty A.C., Bi E., Peter M. Site-specific regulation of the GEF Cdc24p by the scaffold protein Far1p during yeast mating. EMBO J. 2004;23:1063–1074. doi: 10.1038/sj.emboj.7600123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild A.C., Yu J.W., Lemmon M.A., Blumer K.J. The p21-activated protein kinase-related kinase Cla4 is a coincidence detector of signaling by Cdc42 and phosphatidylinositol 4-phosphate. J. Biol. Chem. 2004;279:17101–17110. doi: 10.1074/jbc.M314035200. [DOI] [PubMed] [Google Scholar]
- Winters M.J., Lamson R.E., Nakanishi H., Neiman A.M., Pryciak P.M. A membrane binding domain in the ste5 scaffold synergizes with gβγ binding to control localization and signaling in pheromone response. Mol. Cell. 2005;20:21–32. doi: 10.1016/j.molcel.2005.08.020. [DOI] [PubMed] [Google Scholar]
- Yablonski D., Marbach I., Levitzki A. Dimerization of Ste5, a mitogen-activated protein kinase cascade scaffold protein, is required for signal transduction. Proc. Natl. Acad. Sci. 1996;93:13864–13869. doi: 10.1073/pnas.93.24.13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.W., Mendrola J.M., Audhya A., Singh S., Keleti D., DeWald D.B., Murray D., Emr S.D., Lemmon M.A. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol. Cell. 2004;13:677–688. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]