Abstract
The importance in downstream target regulation of tertiary structure and DNA binding specificity of the protein encoded by the vnd/NK-2 homeobox gene is analyzed. The ectopic expression patterns of WT and four mutant vnd/NK-2 genes are analyzed together with expression of two downstream target genes, ind and msh, which are down-regulated by vnd/NK-2. Three mutants are deletions of conserved regions (i.e., tinman motif, acidic motif, and NK-2 box), and the fourth, Y54M vnd/NK-2, corresponds to a single amino acid residue replacement in the homeodomain. Of the four ectopically expressed mutant genes examined, only the Y54M mutation inactivates the ability of the vnd/NK-2 homeodomain protein to repress ind and msh. The acidic motif deletion mutant slightly reduced the ability of the protein to repress ind and msh. By contrast, both tinman and NK-2 box deletion mutants behaved as functional vnd/NK-2 genes in their ability to repress ind and msh. The NMR-determined tertiary structures of the Y54M vnd/NK-2 homeodomain, both free and bound to DNA, are compared with the WT analog. The only structural difference observed for the mutant homeodomain is in the complex with DNA and involved closer interaction of the methionine-54 with A2, rather than with C3 of the (−) strand of the DNA. This subtle change in the homeodomain–DNA complex resulted in modifications of binding affinities to DNA. These changes resulting from a single amino acid residue replacement constitute the molecular basis for the phenotypic alterations observed on ectopic expression of the Y54M vnd/NK-2 gene during embryogenesis.
Keywords: Drosophila‖neurobiology‖protein–DNA binding‖nuclear magnetic resonance
Homeobox-containing genes are of widespread interest because they encode transcription factors that regulate cell identity decisions in all eukaryotic species (1–3). Furthermore, mutations in these genes have been identified with various congenital anomalies and altered embryonic development or lethality in many species. The proteins encoded by these genes always contain a highly conserved 60-aa domain (i.e., the homeodomain) encoded by the 180-bp homeobox. In addition, other amino acid sequences conserved during evolution are also present in many homeodomain-containing proteins. Although it is now well established that some amino acid residues in specific positions are critical to function, the roles of these various conserved motifs are not fully understood, nor is the tertiary structural basis of their specificity of action well characterized. The homeodomain contains a helix–turn–helix motif and binds to specific DNA sequences. Proper regulation of transcription depends on this homeodomain–DNA interaction as well as on other protein–protein interactions, such as ones involving nuclear protein kinases and Groucho proteins (4). The specificity and strength of the homeodomain–DNA interaction are determined, at least in part, by particular amino acid residues found in selected positions in both the DNA-recognition helix and in the N-terminal arm of the homeodomain. Determination of the functional role of these conserved amino acid sequences and characterization of the structural behavior of critical amino acid residues that leads to functional specificity are fundamental to understanding how homeodomain proteins regulate transcription.
Among the homeobox genes, members of the NK family are of particular interest because they are expressed in neuroectodermal cells (5) and are associated with the development of vital organs (6, 7) such as the heart, lung, prostate, or thyroid, or with the formation of part of the CNS (8). Within the NK family of homeobox genes, the NK-2 class, whose founding member is vnd/NK-2 from Drosophila melanogaster (8–10), is defined by the presence of tyrosine in position 54 of the homeodomain (11, 12). This tyrosine is the major determinant of the nucleotide-binding specificity (13, 14) to a sequence of DNA that contains 5′-CAAGTG-3′ as its core (15–18) through direct interaction between the side chain of Y54 and the cytosine that complements the guanine (in bold) of the consensus DNA (14). This core DNA sequence is recognized by all members of the Nirenberg and Kim (NK)-2 class of homeodomains (15–18). The vnd/NK-2 promoter has been identified and contains several copies of this consensus sequence (19).
NK-2 class homeodomain proteins contain an evolutionarily conserved EH1, or tinman motif, on the N-terminal side of the homeodomain, as well as an NK-2-specific domain (NK-2 SD) on the C-terminal side of the homeodomain (20). The NK-2 SD is separated from the C terminus of the homeodomain by a short linker of 8–31 amino acid residues. In addition, an acidic domain on the N-terminal side of and near the homeodomain has been conserved during evolution in some, but not all, members of the NK-2 class of homeodomain proteins. The roles of these conserved amino acid residue segments are not fully understood, although the NK-2 SD has been shown to be an intramolecular inhibitor of a transcriptional activator domain (20).
Many homeobox genes in the NK-2 class are essential for proper development during embryogenesis (6–8, 21–23) for many species, including humans. The vnd/NK-2 gene of Drosophila is expressed in ventral neuroectodermal cells and then in ventral neuroblasts whose progeny give rise to part of the ventral nerve cord (8, 24). Neural pathways of development are initiated independently in three anterior to posterior stripes of neuroectodermal cells on each side of the embryo. The ventral nervous system defective (vnd)/NK-2 pattern initiates neural development in the ventromedial neuroectoderm (25), and the intermediate neuroblast defective (ind) homeodomain protein initiates neural development in the intermediate column of neuroectoderm (26–28). The mechanism of initiating neural development in the lateral neuroectoderm is unknown; however, muscle segment homeodomain (msh) is expressed in the lateral column of neuroectoderm (24, 26) and is required for the specification of some lateral neuroblasts. The vnd/NK-2 homeobox gene has been shown to mediate repression of the ind (25, 26) and msh (25, 27–29) homeobox genes. Gel shift and footprinting results indicate that the vnd/NK-2 protein represses ind through binding to regulatory sites that contain the nucleotide sequence 5′-CAAGTG-3′ (26). Important questions relate to the role of the entire vnd/NK-2 protein, including the other conserved segments, in addition to that of the homeodomain, in regulation of ind and msh. Our approach to this question involved deletions of each of the three conserved parts of the gene as well as alteration of the individual codon in the homeobox to replace tyrosine-54 by methionine in the homeodomain. The ectopic expression of the WT and four mutant vnd/NK-2 genes in Drosophila embryos was examined together with the effects on ind and msh expression. The alteration of a single amino acid residue in the homeodomain was designed to permit us to correlate functional changes with structural alterations in a region of the protein that is critical for sequence-specific binding to DNA.
Materials and Methods
DNA Constructs and Transgenic Flies.
The WT and mutant vnd/NK-2 genes were expressed ectopically throughout the embryo to examine the effect of the modifications on embryogenesis. The upstream activating sequence (UAS)-Gal4 system was used to overexpress these genes (30, 31). The UAS-vnd/NK-2WT construct was made by subcloning vnd/NK-2 cDNA into the EcoRI site of pUAST (30). To generate the UAS-vnd/NK-2 variants, the vnd/NK-2 cDNA was subcloned into the EcoRI site of a modified pBluescript (Stratagene) that had the cloning sites HpaI, NotI, EcoRI, and SfiI replaced. Alterations of the vnd/NK-2 cDNA were performed by PCR mutagenesis. The PCR primers used to make the various vnd/NK-2X constructs were as follows: a, GCCCTGCCATTGGGCGATAGTTCCAAACTGGGA; b, GCCACCCATCCGCATCACCCCAGTGCCCTGCCATTGGGCGATAGT; c, CATCCTGGTCTACTGCACGGCCATGCCACCCATCCGCATCACCCC; d, CACTAGTATTGATTTTTATTTAGAAGG; e, CTGCGAGAACCAACGCTCTGGCTCTGAGCTGAAGAATGCAGCA; f, GAAGATCTTCACCTGGGTCGGCGTCAGGCGGA; g, CTCCATCGATCCTCTGGTAATGGAAGTGCC; h, CACTGCCGTGACCACTGCCTGCTCCGTTGATCCCCT; i, TTGCATCCCCGCCGCCACTGCCGTGACCACTGCCTG; j, CGTCGGATCCATTTGCATCCCCGCCGCCACTGC; k, GAAGATCTGGTTTCAAAACCATCGCATGAAGACGAAGCGGGCGCAAAACGAGAA; and l, CACTAGTATTGATTTTTATTTAGAAGG.
UAS-vnd/NK-2Y54M Construct.
PCR was carried out with primers k/l to make a point mutation from TAC (the codon for Y54) to ATG (the codon for M54) at base pairs 2145–2147. The PCR product then was inserted into the BglI/SpeI-digested vnd/NK-2 cDNA.
UAS-vnd/NK-2Δacidic Construct.
A BamHI/BamHI fragment was replaced with a PCR product at the same location, but with one that lacks the acidic motif that corresponds to base pairs 1876–1935. The first round of PCR generated a DNA fragment that corresponds to base pairs 1740–1944 with the acidic box deletion. The second and third rounds of PCR were performed to extend this DNA fragment in the 3′ direction to the second BamHI site (base pair 1962). The combinations of primers are g/h, g/l, and g/j.
UAS-vnd/NK-2ΔtinmanConstruct.
a BSTx1/BGIII fragment was replaced with a PCR product lacking the tinman motif that also corresponds to base pairs 949–981 (primers e/f).
UAS-vnd/NK-2ΔNK-2 boxConstruct.
An AccI/SpeI fragment of the vnd/NK-2 cDNA was replaced by a PCR product corresponding to the same region of the cDNA, but lacking the NK-2 box (base pairs 2244–2294). To make this deletion fragment, three rounds of PCR were performed. The first round PCR (primers a/d) was designed to generate a 223- to 2838-bp fragment with the NK-2 box deletion. To extend this DNA fragment to the 5′ direction by the AccI site (base pair 2195), the second (primers b/d) and third round (primers c/d) of PCR were carried out. The final PCR product then was digested with AccI/SpeI and inserted into the vnd/NK-2 cDNA.
After modification, each of the vnd/NK-2 cDNAs was subcloned into the EcoRI site of pUAST. DNA (1 μg/μl) was injected into the Drosophila embryos of Δ2–3 Sb/Ser flies to generate the transgenic fly lines. Most P-element insertions were mapped to a specific chromosome.
Crosses.
Each of the UAS-vnd/NK-2X variants was crossed with the Kruppel (Kr) Gal4 driver lineKr-Gal4/TM3 Sb, which expresses the Gal4 transcription activator protein in a Kruppel expression pattern (32). The crosses were as follows: UAS-vnd/NK-2X/UAS-vnd/NK-2X × Kr(DNA regulatory region)Gal4/TM3 Sb. Thus, 50% of the embryo progeny contained the genotype UAS-vnd/NK-2/Kr(DNA regulatory region)Gal4 and ectopically expressed WT or mutant vnd/NK-2. The remaining 50% of the embryo progeny contained the genotype UAS-vnd/NK-2X/TM3 Sb and did not express ectopic vnd/NK-2. Embryos were collected and incubated to allow development to proceed to stage 5, 8, or 10. The embryos were then fixed and subjected to in situ hybridization (described below) with vnd/NK-2, ind, or msh probes.
In Situ Hybridization.
A 1.6-kb BamHI/EcoRI vnd/NK-2 fragment was subcloned into pBluescript to make the vnd/NK-2 antisense probe catalyzed by RNA polymerase (27). A PCR fragment (primers m/n) that corresponds base pairs 467–958 of the ind cDNA was cloned into pBluescript. The ind antisense RNA probe was then synthesized by using T7 RNA polymerase. A PCR product (primers o/p) that corresponds to base pairs 1470–1871 was cloned into the pCR4-TOPO (Invitrogen) to make the msh antisense probe by using T7 RNA polymerase.
The PCR primers used to make the antisense probes were as follows: m, ATAAGAATGCGGCCGCTCTACGGATCTCCAGTTGTGGGCGG; n, AAAACTGCAGACGCCTCAACCTTCAATTCGTGGAC; o, AAGTGCAACCTGCGGAAGCACAAGCCCAAC; and p, AGGATGAAGATGATCGTGGGCACTGTGGCC.
In situ hybridization experiments were carried out according to the 96-well In Situ Hybridization Protocol [Berkeley Drosophila Genome Project (BDGP); www.fruitfly.org] with a minor modification: 1.5-ml tubes were used for each reaction instead of the 96-well plates.
NMR Spectroscopy.
The protocols used to acquire the NMR data were described (14). The uniformly 13C/15N doubly labeled and 15N singly labeled Y54M mutant vnd/NK-2 homeodomain proteins were expressed from the pET15b vector in Escherichia coli grown in Bioexpress (Cambridge Isotopes Laboratories, Cambridge, MA) medium and purified as described (33). All NMR spectra were obtained on a Bruker Instruments (Billerica, MA) Avance 600 or an Avance 800 NMR spectrometer at 35°C. Both heteronuclear single quantum coherence (HSQC) and NOESY-HSQC experiments were obtained on the Y54M mutant and the spectra were compared with the spectra of the WT vnd/NK-2 homeodomain–DNA complex obtained (14). In addition, a 3D 13C-edited NOESY-HMQC experiment on the doubly labeled complex in 2H2O was performed to examine short distances between the M54 side chain and the DNA. A typical mixing time, τmix, for the NOESY experiments was 100 ms (14).
Electrophoretic Mobility-Shift Assays (EMSAs).
Complementary oligonucleotides that contain the vnd/NK-2 DNA consensus binding sequence were used as described (33). Apparent Kd values were obtained by quantitating the intensities of the EMSA bands and plotting the percentage of the total DNA that is bound to protein against the concentration of protein.
Mutant Selection.
To study vnd/NK-2-mediated repression of the ind or msh genes, 16 lines of transgenic flies (Table 1) were generated that ectopically express WT or mutant vnd/NK-2 proteins. An EcoRI vnd/NK-2 cDNA fragment that encodes the full-length WT vnd/NK-2 homeodomain protein was inserted in the polylinker of the P-element [Pp (UAST)] with a 5′-flanking hsp70 promoter and five binding sites for Gal-4 protein (five UASs) to make UAS-vnd/NK-2WT. In addition, P-elements with four mutant vnd/NK-2 cDNA constructs were prepared (27). One construct, the UAS-vnd/NK-2ΔNK-2 box, lacks the NK-2 box; i.e., the region of the vnd/NK-2 cDNA that encodes the NK-2 SD. The NK-2 SD is a 17-aa sequence that has been conserved during evolution and is separated from the C terminus of the homeodomain by a short linker. Another construct was the UAS-vnd/NK-2Δtinman, which contains a deletion of vnd/NK-2 cDNA that encodes the EF1 or tinman motif, another amino acid sequence that has been conserved during evolution. A third construct was UAS-vnd/NK-2Δacidic, which lacks the region of the gene that encodes an acidic domain found on the N-terminal side of the homeodomain. The fourth construct was UAS-vnd/NK-2Y54M, in which the codon for tyrosine-54 of the homeodomain, TAC, was replaced by the codon for methionine, ATG.
Table 1.
Transgenic lines and P-element locations
| Construct | Transformants
|
|
|---|---|---|
| P-element location | No. | |
| UAS-vnd/NK-2WT | 2nd chromosome | 3 |
| Unknown | 1 | |
| UAS-vnd/NK-2ΔNK-2 box | 2nd chromosome | 3 |
| UAS-vnd/NK-2Δtinman | 2nd chromosome | 2 |
| UAS-vnd/NK-2Δacidic | 2nd chromosome | 1 |
| Unknown | 2 | |
| UAS-vnd/NK-2Y54M | 2nd chromosome | 2 |
| X chromosome | 1 | |
| Unknown | 1 | |
Results and Discussion
Transgenic Lines.
A total of 16 transgenic lines of flies were obtained from injection into Drosophila embryos of each of the five P-element constructs. As shown in Table 1, between two and four transgenic lines fly lines were obtained for each P-element construct. We were able to identify the chromosomal location for most of these P-element constructs (Table 1).
Ectopic vnd Expression.
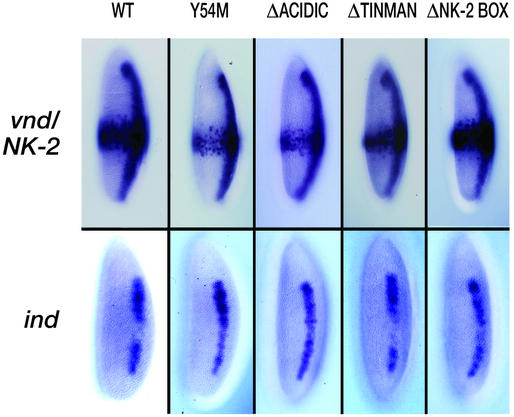
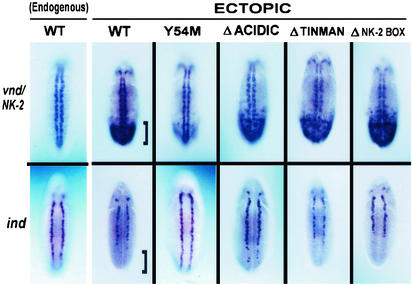
Ectopic expression of vnd/NK-2 mRNA was observed first at stage 5 as a broad band of mRNA in the middle of and encircling the embryo. At stage 8, the ectopic vnd/NK-2 mRNA band was found in the posterior ventral half of the embryo, and at stage 10, ectopic vnd/NK-2 mRNA was found in the posterior ventral quarter of the embryo. At stage 11, the broad areas of ectopically expressed vnd/NK-2 mRNA disappeared, and ectopic expression was found in minor populations of cells in various tissues. Fig. 1 shows the ectopic expression for the WT and mutant vnd/NK-2 genes in the Kruppel pattern (row 1) together with expression of ind at stage 5 of embryonic development (row 2). In the first row of Fig. 2, the endogenous expression for the WT vnd/NK-2 gene together with the ectopic expression patterns for the UAS-vnd/NK-2WT and vnd/NK-2 mutants, UAS-vnd/NK-2Y54M, UAS-vnd/NK-2Δacidic, UAS-vnd/NK-2Δtinman, and UAS-vnd/NK-2ΔNK-2 box are shown. The corresponding patterns for ind at stage 10 (or early stage 11) of embryonic development are shown in row 2 of Fig. 2. Fig. 3 shows msh expression patterns at stage 10 (or early stage 11) of embryonic development. The bracket is placed to indicate the region of high ectopic expression of the WT vnd/NK-2 mRNA (Fig. 2 Upper) and the repression of ind (Fig. 2 Lower).
Figure 1.
(Upper) Patterns of the ectopically expressed WT and mutant UAS-vnd/NK-2 genes at stage 5 of embryonic development. Embryos were stained by using a vnd/NK-2 or ind RNA probe as described in Materials and Methods. (Lower) The expression pattern at stage 5 of embryonic development of the ind gene in the presence of the ectopically expressed WT or mutant vnd/NK-2 genes.
Figure 2.
(Upper) Patterns of the endogenous and ectopically expressed WT and mutant UAS-vnd/NK-2 genes at stage 10 (or early stage 11) of embryonic development. Embryos were stained by using a vnd/NK-2 or ind mRNA probe as described in Materials and Methods. (Lower) The expression patterns of the ind genes in the presence of the endogenous WT (first column) or of the endogenous WT and ectopically expressed WT or mutant (second through sixth columns) vnd/NK-2 genes. Brackets indicate area of WT vnd/NK-2 expression and down-regulation of ind.
Figure 3.
Patterns of the expression of the msh gene in the presence of the ectopically expressed WT and mutant UAS-vnd/NK-2X genes at stage 10 (or early stage 11) of embryonic development. Embryos were stained by using a vnd/NK-2 or ind RNA probe as described in Materials and Methods.
For two of the mutant lines, UAS-vnd/NK-2Δtinman and UAS-vnd/NK-2ΔNK-2 box, no significant difference in the ectopic expression level of vnd/NK-2 relative to that of the WT UAS-vnd/NK-2WT transgene was seen at stages 5 or 10. Reductions in UAS-vnd/NK-2Y54M and UAS-vnd/NK-2Δacidic mRNA expression levels were seen in stage 5 (Fig. 1), stage 8 (data not shown), and stage 10 embryos (Figs. 2 and 3), with the greatest reduction being observed for the construct that codes for the single Y54M amino acid residue replacement in the vnd/NK-2 homeodomain, UAS-vnd/NK-2Y54M. This latter reduction in mRNA expression appears to take place where some neuroectodermal cells showed no mRNA expression, whereas other cells appeared to show relatively high levels of mRNA expression. The inability of the mutant protein encoded by UAS-vnd/NK-2Y54M to bind with the proper affinity to 5′-CAAGTG-3′ vnd/NK-2 binding sites is likely the origin of the observed overall lower mRNA expression. The vnd/nK-2 protein, directly or indirectly, is a feedback activator of the vnd/NK-2 gene expression (19, 27). In the case of UAS-vnd/NK-2Δacidic, the proximity of the negatively charged encoded protein to the homeodomain might decrease the affinity to the (negatively charged) DNA over that when the acidic domain is absent. For both the Y54M and Δacidic mutants, the most likely explanation for the reduction in mutant vnd/NK-2 mRNA is that these mutant proteins are less active autoregulators than the corresponding WT vnd/NK-2 proteins. Ectopic expression of WT or mutant vnd/NK-2 protein in all transgenic fly lines resulted in lethality.
ind Expression.
In previous studies, the vnd/NK-2 protein was shown to mediate repression of the ind gene (27). WT ind expression initiates at stage 5 and persists until the germ-band shortening stage (stage 11). In the neuroectoderm (stage 7), the expression of ind mRNA is characterized by a stripe on each side of the embryo and along the anterior/posterior axis, adjacent and lateral to the stripes of vnd/NK-2 mRNA. Ectopic expression of vnd/NK-2 mRNA from the WT UAS-vnd/NK-2WT transgene greatly reduced ind expression in the posterior part of stage 5, stage 8 (data not shown), and stage 10 embryos (Figs. 1 and 2). Expression of mutant vnd/NK-2 mRNA lacking the acidic region from the UAS-vnd/NK-2Δacidic transgene was less effective in down-regulating the ind gene than the expression of the WT UAS-vnd/NK-2WT transgene (Fig. 2). Ectopic expression of the mutant UAS-vnd/NK-2Y54M transgene, which has the homeodomain tyrosine-54 residue replaced by methionine, showed little or no activity as a repressor of the ind gene expression. However, expression of two of the deletion mutant vnd/NK-2 lines from the UAS-vnd/NK-2Δtinman or UAS-vnd/NK-2ΔNK-2box were as effective in down-regulating the ind gene as ectopic expression of WT vnd/NK-2 from the UAS-vnd/NK-2WT transgene. Similar results were obtained with three distinct UAS-vnd/NK-2Y54M transgenic fly lines and with two distinct UAS-vnd/NK-2Δacidic fly lines. Analogous results with all four mutant transgenes were likewise observed with embryos at other stages of development (data not shown).
msh Expression.
vnd/NK-2 also has been shown to down-regulate the msh homeobox gene (26, 31). msh is expressed initially in the lateral neuroectoderm at stage 6 as two anterior-posterior stripes of cells adjacent and lateral to the cells that express ind. The stripes of cells that express msh change to segmentally repeated clusters of cells that express msh at late stage 8 and, soon afterward, multiple clusters that express msh appear in each hemisegment (26, 34). Similar to the expression of ind, repression of msh was not found in the presence of the ectopically expressed UAS-vnd/NK-2Y54M mutant transgene (Fig. 3). The ectopic expression of WT vnd/NK-2 from the UAS-vnd/NK-2WT transgene results in repression of the msh gene in the posterior-ventrolateral portion of the embryo. Ectopic expression of mutant vnd/NK-2 mRNA lacking the acidic domain from the UAS-vnd/NK-2Δacidic transgene was less effective in down-regulating the msh gene relative to that of the WT vnd/NK-2 transgene.
Structures of the WT and Mutant Y54M Homeodomain–DNA Complexes.
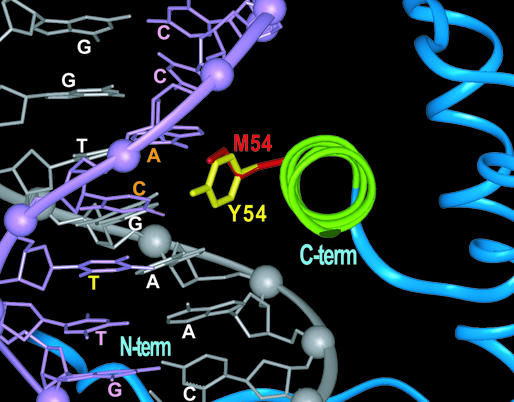
The inability of the Y54M vnd/NK-2 protein to repress expression of ind and msh suggests a critical role for the homeodomain–DNA interaction. This role may involve both specificity and affinity. To address this suggestion and to compare the conformations of the tyrosine and methionine side chains in position 54 of the vnd/NK-2 homeodomain both in the free state and bound to a 16-bp segment of DNA, structural studies were carried out by NMR spectroscopy. These studies were performed on an 80-aa protein that encompasses the Y54M vnd/NK-2 homeodomain (33). NMR was the technique of choice, because the homeodomain does not crystallize in the absence of DNA. NMR spectroscopy was used previously to generate the coordinates of the WT vnd/NK-2 homeodomain in both the free (35) and the DNA-bound states (14). The tertiary structure of the Y54M vnd/NK-2 homeodomain in the absence of DNA, as evidenced by the HSQC spectra (12, 35), is unchanged over that of the WT analog. Furthermore, this replacement of tyrosine-54 with methionine does not result in any measurable alteration in secondary structure or thermal stability of the free homeodomain in solution. Helix III of the homeodomain, the DNA recognition helix, is 11 residues in length and goes from P42 through H52 in the absence of DNA, as seen for the WT homeodomain. For the Y54M mutant homeodomain bound to DNA, complete assignment of the proton 15N, and 13C resonances showed no discernable differences between the WT and mutant homeodomains except around position 54. The implication of this result is that the global fold of the Y54M mutant homeodomain is identical to that of its WT analog. Two heteronuclear 3D NOESY experiments showed that the individual atom–atom distances between the mutant homeodomain and the 16-bp DNA fragment are essentially the same as those determined for the WT homeodomain (14, 33), except for local differences associated with the Y54 and M54 side chains. As in the case of the WT vnd/NK-2 homeodomain, the third helix of the Y54M mutant is inserted into the major groove of the DNA and makes the majority of specific intermolecular contacts (i.e., has short atom–atom distances) with the DNA bases. A superposition of the structures of the two homeodomains that shows the orientations of the WT Y54 and mutant M54 side chain in the DNA major groove is shown in Fig. 4. A notable feature of this superposition is the similarity of the conformations of the M54 side chain and the Y54 side chain including part of the tyrosine ring. The only apparent qualitative difference between the WT and M54 homeodomains is in the set of atom–atom interactions the tyrosine and methionine side chains experience with the DNA. The longer tyrosine side chain shows additional atom–atom interactions with the DNA, including those with the thymine that complements the adenine (bold) of 5′-CAAGTG-3′.
Figure 4.
Superimposed 3D structures of the WT and Y54M mutant vnd/NK-2 homeodomain–DNA complexes, where the region around helix III (green) and the DNA recognition helix are shown. The DNA bases (orange) indicate that both tyrosine-54 (yellow) and methionine-54 (red) are in close proximity (<6 Å). The base in yellow indicates that only tyrosine-54 (yellow) is in close proximity (<6 Å). The coding strand that contains the 5′-CAAGTG-3′ consensus sequence of the DNA is white and the noncoding strand is pink.
Homeodomain–DNA Binding.
The NMR studies have demonstrated that position 54 makes important contacts with the DNA, and these contacts have been shown to be critical to recognition of the unusual sequence of DNA (13, 33). Replacement of the tyrosine with methionine alters the strengths of the interactions with the various neighboring DNA bases. Whereas the structural studies show the importance of Y54 in sequence-specific DNA binding, the transgenic experiments demonstrate that this interaction of Y54 with the DNA is critical for proper function.
To quantify the interaction of the WT and Y54M vnd/NK-2 homeodomains with DNA, the binding affinities to selected 18-bp segments of DNA that contain the consensus sequence (Table 2) were determined. Because the structural studies showed that the side chain of residue 54 interacts with two variable positions, 4 and 5, of the DNA consensus sequence (i.e., 5′-CAAGTG-3′, where positions 4 and 5 are shown in bold), only these two bases were modified in this study to resemble the binding site (A18 and 5 in Table 2) for Antennapedia and ftz, which contain methionine in position 54 of their homeodomains (22). The apparent Kd values were determined from EMSAs. Although the apparent Kd values differ somewhat from those previously obtained (12, 18, 33), probably because of slightly different experimental conditions, the results (Table 2) show that the Y54M replacement reduces the affinity for the NK-2 binding site (18NK-2 no. 1) by ≈10-fold. Replacement of the G in position 4 by T (18NK-2 no. 3 and 18NK-2 no. 6) likewise reduces the Kd by ≈10-fold. The double replacement in the DNA of GT by TG (18NK-2 no. 6) suggests that there is a slight preference of the Y54M vnd/NK-2 homeodomain for DNA that contains G in position 5 rather than T (18NK-2 no. 4 and 18NK-2 no. 6). This alteration in specificity of the Y54M vnd/NK-2 homeodomain might be understandable, because the methionine side chain in position 54 is in close spatial proximity to the base in position 5 (i.e., 5′CAAGTG-3) of the DNA that complements the T (in bold), although admittedly it is difficult to quantify these interactions solely from the structural information. The structural alterations seen for the Y54M change in vnd/NK-2 are only local in nature involving residue 54 side-chain orientations, and the changes in DNA-binding specificity are subtler than those seen in an analogous assay on ftz-DNA binding (36, 37).
Table 2.
EMSA for binding of the WT and Y54M vnd/NK-2 homeodomain to a set of 18-bp DNA oligomers
| No. | Sequence |
Kd, nM*
|
Kd, relative
|
||
|---|---|---|---|---|---|
| WT | Y54M | WT | Y54M | ||
| 1 | TGTGTCAAGTGGCTGTAG | 13 | 135 | 1.0 | 10.3 |
| 3 | --------T—-------- | 136 | 160 | 10.4 | 12.2 |
| 4 | ---------G-------- | 138 | 130 | 10.6 | 10.0 |
| 6 | --------TG-------- | 211 | 122 | 16.1 | 9.3 |
| A18 | CTCTAATGGCTTTTTCTC | 313 | 260 | 23.9 | 19.9 |
| 5 | ---C-------------- | 284 | 301 | 21.7 | 22.9 |
The core sequence is bold. The underlined area indicates DNA bases whose complements on the (−) strand are close to Y54 or M54, respectively, of the WT or mutant vnd/NK-2 homeodomain. Hyphens indicate identity with the sequence above.
The measured affinities are apparent Kd values only.
One might suspect that the apparent inability of the Y54M mutant vnd/NK-2 protein to repress the ind and msh genes may be because of the decreased level of UAS-vnd/NK-2Y54M mRNA compared with the WT or other mutant vnd/NK-2 mRNA levels (18, 25) seen at stage 10 (Fig. 1). However, at stage 5, the mRNA level of the UAS-vnd/NK-2Y54M appears comparable to that of UAS-vnd/NK-2WT. Thus, at least at stage 5, the inability of the Y54M mutant protein to repress ind and msh is not because of a lower expression level.
The combined transgenic, structural, and binding data presented here demonstrate that the ability of vnd/NK-2 to repress the two downstream target genes, msh and ind, depends on a local very specific interaction between the homeodomain and its cognate DNA. The presence of tyrosine-54 results in the correct interaction required for autoregulation of vnd/NK-2, directly or indirectly, as well as for down-regulation of both ind and msh by the vnd/NK-2 homeoprotein. Having the structure of the vnd/NK-2–DNA complex was necessary to the interpretation of these transgenic results. Although the structural studies do not directly yield binding affinities, and because they have shown that the only change in the Y54M mutant homeodomain is in the side-chain interaction with the DNA, the inability of the mutant homeoprotein to function most likely results from the alteration in the affinity for the DNA. This reduction in the homeodomain–DNA binding affinity appears to result in complete elimination of the ability of the vnd/NK-2 gene to regulate its downstream targets. The results from such a diverse set of transgenic and biophysical experiments demonstrate not only the utility of the approach but also that control in embryonic development can be exquisitely sensitive to very small molecular alterations. The results described here demonstrate, through tertiary structural studies, the sensitivity of individual amino acid residue replacements in the alteration of the transcriptional regulation of downstream targets. A mutation of tyrosine-54 to cysteine in a highly sequentially and structurally homologous homeodomain, CSX or NKX-2.5, has been suggested to be an origin of atrial septum defect in the human heart (21). The heart alterations seen for this Y54C replacement were as pronounced as those observed where a stop codon occurred before the homeobox (21). The results described above indicate an analogous alteration in DNA-binding affinity because of the presence of cysteine-54 in CSX/NKX-2.5 together with no other structural modification. In addition, the results described in this study show a relation between structural and functional modularity. In cases where single residue mutations are related to genetic disease, further combined structural studies and transgenic investigations on this and other homeodomain proteins using appropriate animal models thus become critical.
Acknowledgments
We thank Drs. Alan Peterkofsky and Edward D. Korn for careful reading of the manuscript and Dr. Hyun Sook Lee for help with the homeodomain–DNA binding experiments. J.A.F. thanks Dr. Kae-Jung Hwang for help with the generation of Figs. 1–3 and preparation of the manuscript. J.W.M. and J.-H.J. acknowledge financial support from the Minority Biomedical Research Support program of the National Institute of General Medical Sciences, National Institutes of Health, Grant GM-08016 (to J.W.M.).
Abbreviations
- vnd
ventral nervous system defective
- NK
Nirenberg and Kim
- NK-2 SD
NK-2-specific domain
- ind
intermediate neuroblasts defective
- msh
muscle segment homeobox
- UAS
upstream activating sequence
References
- 1.Gehring W J. Science. 1987;230:1245–1252. doi: 10.1126/science.2884726. [DOI] [PubMed] [Google Scholar]
- 2.Scott M P, Tamkun J W, Hartzell G W., III Biochim Biophys Acta. 1989;989:25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- 3.Gehring W J, Affolter M, Burglin T. Annu Rev Biochem. 1994;63:487–526. doi: 10.1146/annurev.bi.63.070194.002415. [DOI] [PubMed] [Google Scholar]
- 4.Choi C Y, Kim Y H, Kwon H J, Kim Y. J Biol Chem. 1999;274:33194–33197. doi: 10.1074/jbc.274.47.33194. [DOI] [PubMed] [Google Scholar]
- 5.Bober E, Baum C, Braun T, Arnold H H. Dev Biol. 1994;162:288–303. doi: 10.1006/dbio.1994.1086. [DOI] [PubMed] [Google Scholar]
- 6.Lints T J, Parsons L M, Hartley L, Lyons I, Harvey R P. Development (Cambridge, UK) 1993;119:419–431. doi: 10.1242/dev.119.2.419. [DOI] [PubMed] [Google Scholar]
- 7.Kimura S, Hara Y, Pineau T, Fernandez-Salguero P, Fox C H, Ward J M, Gonzales F J. Genes Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- 8.Jimenez F, Martin-Morris L E, Valasco L, Chu H, Sierra J, Rosen D R, White K. EMBO J. 1995;14:3487–3495. doi: 10.1002/j.1460-2075.1995.tb07355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim Y, Nirenberg M. Proc Natl Acad Sci USA. 1989;86:7716–7720. doi: 10.1073/pnas.86.20.7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nirenberg M, Nakayama K, Nakayama Y, Kim Y S, Mellerick D, Wang L-H, Webber K, Lad R. Ann NY Acad Sci. 1995;758:224–242. doi: 10.1111/j.1749-6632.1995.tb24830.x. [DOI] [PubMed] [Google Scholar]
- 11.Guazzi S, Price M, De Felice M, Damante G, Mattei M G, Di Lauro R. EMBO J. 1990;9:3631–3639. doi: 10.1002/j.1460-2075.1990.tb07574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsao D H H, Gruschus J M, Wang L-H, Nirenberg M, Ferretti J A. Biochemistry. 1994;33:15053–15060. doi: 10.1021/bi00254a014. [DOI] [PubMed] [Google Scholar]
- 13.Gruschus J M, Tsao D H H, Wang L-H, Nirenberg M, Ferretti J A. Biochemistry. 1997;36:5372–5380. doi: 10.1021/bi9620060. [DOI] [PubMed] [Google Scholar]
- 14.Gruschus J M, Tsao D H H, Wang L-H, Nirenberg M, Ferretti J A. J Mol Biol. 1999;289:529–545. doi: 10.1006/jmbi.1999.2774. [DOI] [PubMed] [Google Scholar]
- 15.Harvey R P. Dev Biol. 1996;178:203–216. doi: 10.1006/dbio.1996.0212. [DOI] [PubMed] [Google Scholar]
- 16.Chen C Y, Schwartz R J. J Biol Chem. 1995;270:15628–15633. doi: 10.1074/jbc.270.26.15628. [DOI] [PubMed] [Google Scholar]
- 17.Damante G, Fabbro D, Pellizzari L, Civitareale D, Guazzi S, Polycarpou-Schwartz M, Cauci S, Quadrifoglio F, Formisano S, DiLauro R. Nucleic Acids Res. 1994;22:3075–3083. doi: 10.1093/nar/22.15.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L-H, Chmelik R, Nirenberg M. Proc Natl Acad Sci USA. 2002;99:12721–12726. doi: 10.1073/pnas.202461199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saunders H-M H, Koizumi K, Odenwald W, Nirenberg M. Proc Natl Acad Sci USA. 1998;95:8316–8321. doi: 10.1073/pnas.95.14.8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watada H, Mirmira R G, Kalamaras J, German M S. Proc Natl Acad Sci USA. 2000;97:9443–9448. doi: 10.1073/pnas.97.17.9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasahara H, Lee B, Shott J-J, Benson D W, Seidman J G, Seidman C E, Izumo S. J Clin Invest. 2000;106:299–308. doi: 10.1172/JCI9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krude H, Schutz B, Biebermann H, von Moers A, Schnabel D, Neitzel H, Tonnies H, Weise D, Lafferty A, Schwarz S, et al. J Clin Invest. 2002;109:475–480. doi: 10.1172/JCI14341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasahara H, Bartunkova S, Schinke M, Tanaka M, Izumo S. Circ Res. 1998;82:936–946. doi: 10.1161/01.res.82.9.936. [DOI] [PubMed] [Google Scholar]
- 24.Mellerick D M, Nirenberg M. Dev Biol. 1995;171:306–316. doi: 10.1006/dbio.1995.1283. [DOI] [PubMed] [Google Scholar]
- 25.Skeath J B, Parganiban G F, Carroll S B. Development (Cambridge, UK) 1994;120:1517–1524. doi: 10.1242/dev.120.6.1517. [DOI] [PubMed] [Google Scholar]
- 26.Weiss J B, Von Ohlen T, Mellerick D M, Dressler G, Doe C Q, Scott M P. Genes Dev. 1998;12:3591–3602. doi: 10.1101/gad.12.22.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu H, Parras C, White K, Jimenez F. Genes Dev. 1998;12:3613–3624. doi: 10.1101/gad.12.22.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDonald J A, Holbrook S, Isshiki T, Weiss J B, Doe C Q, Mellerick D M. Genes Dev. 1998;12:3603–3612. doi: 10.1101/gad.12.22.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Von Ohlen T, Doe C O. Dev Biol. 2000;224:362–372. doi: 10.1006/dbio.2000.9789. [DOI] [PubMed] [Google Scholar]
- 30.Brand A H, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 31.Mellerick D M, Modica V. J Neurobiol. 2002;50:118–136. doi: 10.1002/neu.10022. [DOI] [PubMed] [Google Scholar]
- 32.Giesen K, Hummel T, Stollewerk A, Harrison S, Travers A, Klambt C. Development (Cambridge, UK) 1997;118:2307–2316. doi: 10.1242/dev.124.12.2307. [DOI] [PubMed] [Google Scholar]
- 33.Weiler S, Gruschus J M, Tsao D H H, Yu L, Wang L-H, Nirenberg M, Ferretti J A. J Biol Chem. 1998;273:10994–11000. doi: 10.1074/jbc.273.18.10994. [DOI] [PubMed] [Google Scholar]
- 34.D'Alessio M, Frasch M. Mech Dev. 1996;58:217–231. doi: 10.1016/s0925-4773(96)00583-7. [DOI] [PubMed] [Google Scholar]
- 35.Tsao D H H, Gruschus J M, Wang L-H, Nirenberg M, Ferretti J A. J Mol Biol. 1995;251:297–307. doi: 10.1006/jmbi.1995.0435. [DOI] [PubMed] [Google Scholar]
- 36.Percival-Smith A, Müller M, Affolter M, Gehring W J. EMBO J. 1990;9:3967–3974. doi: 10.1002/j.1460-2075.1990.tb07617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schier A F, Gehring W J. Proc Natl Acad Sci USA. 1993;90:1450–1454. doi: 10.1073/pnas.90.4.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]