Abstract
The Ser-Arg (SR)-related protein SRm160 is a coactivator of pre-mRNA splicing. It bridges splicing factors located at the 5′ splice site, branch site, and 3′ splice site. Recently, SRm160 has also been shown to be involved in mRNA export as part of an exon-junction complex. SRm160 is highly concentrated in splicing speckles but is also present in long branched intranuclear tracks connecting splicing speckles with sites at the nuclear lamina. In this study we identified domains of SRm160 important for spatial targeting within the nucleus and for binding to the nuclear matrix. Using a series of FLAG- and enhanced GFP-conjugated deletion mutants we found two contiguous sequences that independently target SRm160 to nuclear matrix sites at splicing speckled domains: amino acids 300–350 and 351–688. Constructs containing amino acids 300–350 were also targeted to sites peripheral to speckled domains where most mRNA originate subsequent to splicing. Sequences from the N-terminal domain localized proteins to the nuclear lamina near sites where mRNA leaves the nucleus.
Keywords: RNA splicing‖RNA export‖speckled domains‖SRm300
Since the discovery of RNA splicing (1), its mechanism has been elucidated by the clever use of in vitro assays (2). Simple precursor RNAs, usually with one small intron, are added to a nuclear extract. After the addition of ATP, spliceosomal complexes form, and introns are removed slowly. In marked contrast, native RNA splicing in cells is far more rapid and efficient, capable of processing more complicated substrates. Precursor RNAs as large as 80,780 bases with as many as 175 introns (3) are rapidly spliced, often in complicated but precise alternative patterns. The very rapid splicing seen in vivo likely reflects, in part, the accurate positioning of splicing substrates and factors by the highly ordered architecture of the nucleus. Most RNA splicing factors are concentrated in subnuclear structures that appear as speckled domains when visualized by immunofluorescence microscopy (4). When seen by electron microscopy, these correspond to interchromatin granule clusters (5) that are surrounded by regions rich in the perichromatin fibrils that contain many new transcripts (5, 6). A majority of these transcripts are spliced at or near speckled domains (7), and mechanisms have been described for recruiting splicing factors from these domains to newly activated genes (8, 9).
Evidence that the nuclear matrix has a critical role in RNA splicing has emerged from studies examining cells expressing a β-globin pre-mRNA splicing construct (10, 11). This precursor remains associated with the nuclear matrix after its isolation and is spliced rapidly after addition of the ATP (11). In contrast to conventional in vitro splicing reactions, splicing in situ on nuclear matrix preparations occurs without a lag period, indicating that spliceosomal commitment complexes are preassembled and fully functional.
Two strong candidates for factors that might couple splicing components are Ser-Arg (SR)-related matrix protein of 160 kDa (SRm160) and SR-related matrix protein of 300 kDa (SRm300), two high molecular mass SR-related proteins (11–15). These proteins are bound more tightly to the nuclear matrix than other SR proteins, are binding partners, and are constituents of in vitro-assembled spliceosomes. SRm160 serves as a coactivator of splicing, bridging the 5′ and 3′ splice sites by interacting with factors at those sites including U1 small nuclear ribonucleoprotein (RNP), U2 small nuclear RNP, SRp75, and SRp20 (13). In addition, SRm160/300 may serve to attach spliceosomes to the nuclear matrix. We suggest that SRm160 and SRm300 are key players in the recruitment of splicing factors to sites of splicing and in the assembly of presplicing complexes on the nuclear matrix, contributing to the greater efficiency of splicing in intact nuclei. Although both proteins participate in splicing reactions, SRm160 may be the more important for in vitro splicing, being required for the splicing of some RNA substrates (13, 14).
Most copies of SRm160 and SRm300 are concentrated in speckled domains. However, as visualized by immunoelectron microscopy, SRm160 but not SRm300 is also present in long intranuclear tracks that frequently connect to the interchromatin granule clusters (J.A.N., K. M. Wan, G. Krockmalnic, and S.W., unpublished data). These tracks suggest a role for SRm160 in intranuclear transport, perhaps of mRNA after splicing. This hypothesis is supported by work showing that in vitro, SRm160 remains bound to mRNAs after splicing (13) at sites 20–24 nucleotides upstream from exon–exon junctions in an exon-junction complex (EJC) also containing the mRNA export factors DEK, RNPS1, Y14, Aly/REF (16–18), and Magoh (19). SRm160 also stimulates 3′-end cleavage, a prerequisite for mRNA release to the cytoplasm (20). Its overexpression causes the premature release and cytoplasmic accumulation of intron-containing species (20).
In this study we have identified domains of SRm160 important for spatial targeting within the nucleus and for binding to the nuclear matrix. We find two contiguous sequences conferring SRm160s localization in splicing speckles and binding to the nuclear matrix. We have identified sequences that target fusion constructs to sites peripheral to speckled domains and sequences in the N-terminal domain that target SRm160 to sites at the nuclear lamina. These represent the origin and cytoplasmic transition point for most mRNA export from the nucleus.
Materials and Methods
Expression Vectors.
The FLAG epitope, MDYKDDDDK, was cloned between the HindIII and BamHI sites of the pCDNA 3 vector (Invitrogen). The cDNA coding for human SRm160 (13) was inserted in frame to FLAG between the BamHI and EcoRI sites. All FLAG-SRm160 and enhanced GFP (EGFP)-SRm160 constructs were derived from digested fragments of the original FLAG-SRm160 plasmid. For EGFP constructs either pEGFP-C1 or pEGFP-C3 vectors (CLONTECH) were used. For expression of VP22 fused to SRm160s nuclear matrix targeting signal (NMTS), the NMTS sequence coding for amino acids 300–688 was amplified by PCR (sequences for the primers are available upon request) and cloned in the KpnI and Xba sites of pVP22 (CLONTECH).
Cell Culture and Transfections.
HeLa cells were grown in DMEM (GIBCO/BRL) supplemented with 10% FBS to ≈60% confluence on 18-mm circular coverslips. Transfections were performed by using 1.5 μg of DNA and 7.5 μl of Superfect transfection reagent (Qiagen, Hilden, Germany) per coverslip.
Immunocytochemistry.
To prepare cells for immunocytochemistry, three different methods were applied.
Fixation/permeabilization.
Cells were washed in PBS before fixation with 4% formaldehyde in cytoskeletal buffer (CSK, 10 mM Pipes, pH 6.8/300 mM sucrose/100 mM NaCl/3 mM MgCl2/1 mM EGTA) for 15 min. To improve antibody penetration, the cells then were permeabilized by using 0.5% Triton X-100 in CSK for 5 min. All steps were performed at 4°C.
Permeabilization/fixation.
After washing in PBS, cells were incubated in 0.5% Triton X-100 in CSK for 10 min. This step removes soluble proteins, both cytoplasmic and nucleoplasmic. Then, cells were fixed in 4% formaldehyde in CSK. All steps were performed at 4°C.
Nuclear matrix preparation (21, 22).
After washing in PBS, soluble proteins were removed by extraction with 0.5% Triton in CSK containing 20 mM vanadyl riboside complex (5 Prime → 3 Prime) and 1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride (Roche, Indianapolis) for 5 min. After washing in CSK, the cells were incubated in extraction buffer [0.5% Triton X-100/10 mM Pipes, pH 6.8/250 mM ammonium sulfate/3 mM MgCl2/1 mM EGTA/20 mM vanadyl riboside complex/1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride] for 5 min. After another wash in CSK, DNA was extracted by incubation in digestion buffer [0.5% Triton X-100/10 mM Pipes, pH 6.8/300 mM sucrose/50 mM NaCl/3 mM MgCl2/1 mM EGTA/20 mM vanadyl riboside complex/1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride] containing 400 units/ml DNase I (RNase-free, Roche) at 30°C for 50 min. Except for DNase I digestion, all steps were performed at 4°C. Finally, the cells were fixed with 4% formaldehyde in CSK.
Antibody Staining.
Nonspecific binding sites were blocked with TBS-I [10 mM Tris, pH 7.7/150 mM NaCl/3 mM KCl/1.5 mM MgCl2, 0.05% (vol/vol) Tween 20/0.1% (wt/vol) BSA/0.2% (wt/vol) glycine] for at least 1 h. All antibodies were diluted in TBS-I and incubated at 4°C for at least 1 h. To wash unbound antibodies, cells were rinsed three times in PBS containing 0.05% (vol/vol) Tween 20. Antibody dilutions were as follows: BIC8 (mouse IgM), 1:20; B4A11 (mouse IgG), 1:20; Y12 (mouse IgG, kind gift of Ben Blencowe, University of Toronto, Toronto), and 1:50; lamin A/C (rabbit IgG, kind gift of Nilabh Chaudhary and Günther Blobel, The Rockefeller University, New York), 1:2,000. The secondary antibodies goat anti-mouse IgG (γ) + FITC or tetramethylrhodamine B isothiocyanate/goat anti-mouse IgM (μ) + FITC or tetramethylrhodamine B isothiocyanate were from Kirkegaard & Perry Laboratories. Coverslips were mounted with Prolong (Molecular Probes).
Results
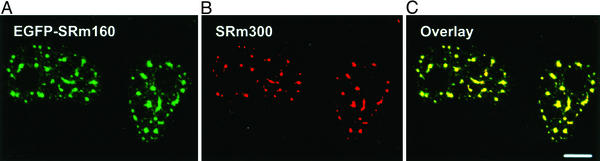
SRm160 has sequences responsible for spatial targeting to sites of RNA splicing and export as well as those responsible for its binding to the nuclear matrix. To identify these, we constructed a series of FLAG epitope-tagged deletion mutants and a corresponding set of sequence domains cloned as EGFP fusion proteins (Table 1). Ectopically expressed EGFP-SRm160 (Fig. 1A) was concentrated in splicing speckled domains where it colocalized with endogenous SRm160 (data not shown), with its binding partner SRm300 (Fig. 1 B and C), and with small nuclear RNPs detected with the Y12 antibody (data not shown). Thus, addition of EGFP to the N terminus of SRm160 did not alter its subnuclear localization.
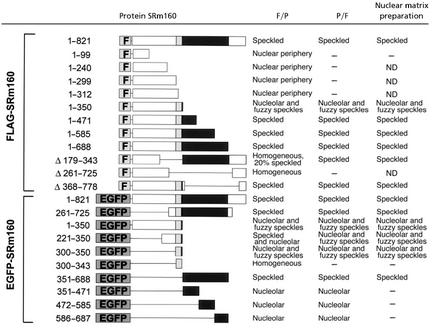
Table 1.
Amino acid domain analysis of SRm160 spatial targeting and nuclear matrix binding
A series of FLAG-SRm160 deletion mutants and a series of SRm160 domains fused to the C terminus of EGFP are presented. The localization of the protein expressed in HeLa cells is noted for cells that were fixed and then permeabilized (F/P), permeabilized and then fixed (P/F), or after a nuclear matrix preparation prior to fixation. ND, not determined.
Figure 1.
EGFP-SRm160 colocalized with endogenous SRm300 in speckled domains. (A) EGFP-SRm160 (red) was expressed in HeLa cells. Cells were permeabilized and fixed before immunofluorescent staining. (B) Endogenous SRm300 was localized with the B4A11 antibody. (C) EGFP-SRm160 (green) and SRm300 (red) were colocalized in speckled domains. (Scale bar, 10 μm.)
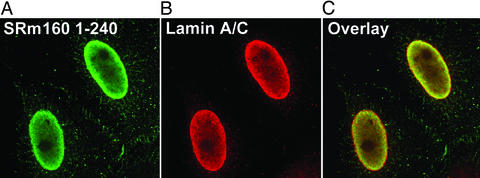
Surprisingly, all deletion mutants (Table 1) were located exclusively in the nucleus, suggesting the existence of multiple nuclear localization signals. Further, the nuclear localization of the N-terminal domain lacking any SR repeats (FLAG-SRm1601–99, see Table 1) strongly indicates that the nuclear retention of SRm160 does not depend on SR motifs. The SRm160 N terminus up to amino acid 299 (Table 1) did not target constructs to splicing speckles. Instead they colocalized with lamin A/C at the nuclear envelope as shown for FLAG-SRm1601–240 in Fig. 2. This attachment at the nuclear periphery was loose because permeabilization of cells with 0.5% Triton X-100 removed the signal (Table 1).
Figure 2.
FLAG-SRm1601–240 colocalized with lamin A/C at the nuclear periphery. (A) FLAG-SRm1601–240 was transiently expressed in HeLa cells. The cells were fixed and then permeabilized to permit antibody penetration. (B) Lamin A/C staining. (C) FLAG-SRm1601–240 and lamin A/C were colocalized at the nuclear periphery.
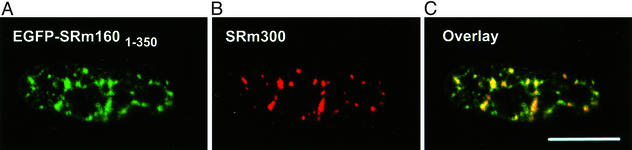
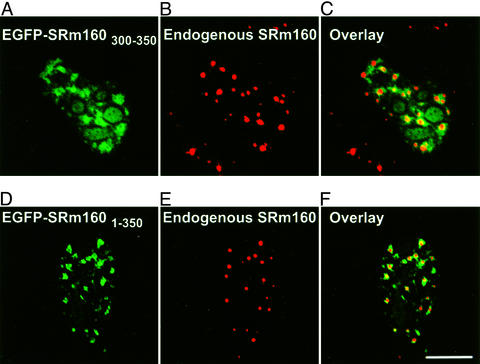
In contrast, all mutants containing at least the first 350 aa of SRm160 were found in splicing speckles (Table 1). This localization was unaltered after the removal of soluble proteins and chromatin in nuclear matrix preparations (Table 1). FLAG-SRm1601–350 (data not shown) and EGFP-SRm1601–350 (Fig. 3A) partially colocalized with the speckled domain protein SRm300 (Fig. 3 B and C), a binding partner of SRm160, suggesting that the N-terminal 350 aa of SRm160 are sufficient for speckle targeting. However, both FLAG-SRm1601–350 and EGFP-SRm1601–350 were also present in a region adjacent to splicing speckled domains (Fig. 3 A and C) that may correspond to the recently distinguished “paraspeckles” (23). Both were also detected in nucleoli in up to 50% of all transfected cells (data not shown). Dissecting the N-terminal domain further showed that SRm160300–350 was sufficient to target EGFP to the paraspeckle region adjacent to speckled domains (Fig. 4 A–C). After extraction to prepare the nuclear matrix (Fig. 4 D–F) there was a significant loss of fluorescence in nucleoli (compare Fig. 4 A and D). Although some EGFP-SRm160300–350 remained in the speckled domain itself (Fig. 4D), where it colocalized with endogenous SRm160 (Fig. 4E), most of the protein was concentrated in the peripheral paraspeckle region (Fig. 4 D and F).
Figure 3.
EGFP-SRm1601–350 colocalized with endogenous SRm300 at the nuclear matrix. (A) EGFP-SRm1601–350 was ectopically expressed in HeLa cells. (B) Soluble proteins and chromatin were removed (21) before detection of endogenous SRm300 with the B4A11 antibody. Both EGFP-SRm1601–350 and SRm300 were retained at sites on the nuclear matrix at sites of speckled domains (C), although the ratio of the two proteins varied between domains. (Scale bar, 10 μm.)
Figure 4.
Amino acids 300–350 targeted SRm160 to sites on the nuclear matrix and peripheral to speckled domains. EGFP-SRm160300–350 was transiently expressed in HeLa cells. EGFP-SRm160300–350 (A) in fixed cells was concentrated in and around speckled domains (overlay, C) as visualized by the B1C8 antibody for SRm160 (B) but was also visible in nucleoli. (D) EGFP-SRm300300–350 remained bound after the removal of soluble proteins and chromatin in a nuclear matrix preparation. The protein partially colocalized with endogenous SRm160 in speckled domains (E) but was more concentrated in regions adjacent to speckled domains (F). (Scale bar, 10 μm.)
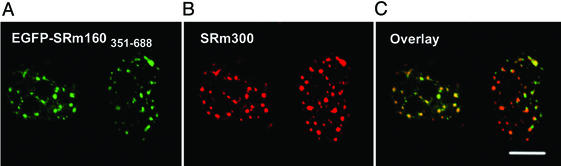
Interestingly, nuclear matrix binding was abolished completely for the slightly shorter EGFP-SRm160300–343 (Table 1), emphasizing the important role that amino acids 344–350 play in anchoring to the nuclear matrix. In summary, amino acids 300–350, although necessary for targeting the SRm160 N terminus to speckled domains, were inefficient in anchoring the EGFP fusion protein to speckled domains. However, we have identified a second sequence domain toward the C terminus of SRm160 that mediates localization to speckled domains and confers stronger binding to the nuclear matrix at those sites. Both EGFP-SRm160351–688 (Fig. 5A) and the internal deletion mutant FLAG-SRm160▵172–343 (Table 1) were localized to speckled domains where they colocalized with the endogenous SRm300 (Fig. 5 B and C). Because this sequence domain is rather large we sought a smaller sequence within this region sufficient for binding to splicing speckles. However, SRm160 amino acids 351–471, 472–585, and 586–688 were not able to target EGFP to speckled domains but instead sent it to nucleoli (Table 1 and data not shown), which suggests that SRm160 amino acids 351–688 constitute a minimal sequence for binding to splicing speckles at the nuclear matrix. If there is a shorter domain responsible for matrix binding, it only works within the structural context of the whole SRm160351–688 domain. Taken together, these data show the existence of two contiguous sequence domains of SRm160 responsible for targeting to speckled domains.
Figure 5.
EGFP-SRm160351–688 colocalized with endogenous SRm300 in splicing speckles. EGFP-SRm160351–688 was expressed in HeLa cells. The cells were permeabilized before fixation. Endogenous SRm300 (B) colocalizes with EGFPSRm160351–688 (A), although the ratio of the two proteins varied between domains (overlay, C). EGFP-SRm160351–688 was nuclear matrix-associated and the entire amino acid 351–688 domain was required for this targeting. (Scale bar, 10 μm.)
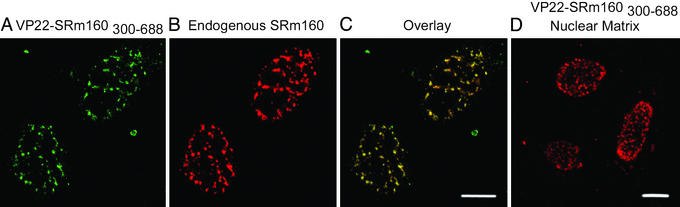
In additional experiments, the SRm160 nuclear matrix targeting sequences were fused to the herpes simplex virus protein VP22 also containing a myc tag. As expected, this fusion protein was readily observed in speckled domains (Fig. 6A), colocalized with endogenous SRm160 (Fig. 6 B and C) and bound tightly to the nuclear matrix (Fig. 6D). In contrast, the myc-tagged VP22 alone was homogeneously distributed in the nucleus but entirely lost when subjected to the salt-extraction step of a nuclear matrix preparation (data not shown). VP22 fusion proteins have been shown to penetrate cell membranes and move from transfected cells to the nuclei of nontransfected cells (24). Thus, we expected to find the fusion protein in all cells after transient transfection. However, no movement to untransfected cells was observed for VP22 fusions with the nuclear matrix targeting sequences of SRm160 (SRm160300–350 and SRm160351–688). Considering the strong nuclear matrix binding of the protein and the lack of nuclear-cytoplasmic shuttling of the full-length protein (J.A.N., K. M. Wan, G. Krockmalnic, and S.W., unpublished data), it is likely that the SRm160 nuclear matrix targeting sequences tether the fusion proteins and suppress the distinctive VP22 spreading to adjacent cells.
Figure 6.
The fusion protein VP22-SRm160300–688 was tightly bound to the nuclear matrix in speckled domains. (A) VP22-SRm160300–688, a fusion of the nuclear matrix targeting sequences of SRm160 with VP22, was expressed in HeLa cells. The cells were permeabilized before fixation. As shown in C, VP22-SRm160300–688 (A) was colocalized with the endogenous SRm160 (B) in speckled domains. (D) After a nuclear matrix preparation, VP22-SRm160300–688 remained with the nuclear matrix. VP22 alone did not localize to speckled domains and was not retained in nuclear matrix preparations (data not shown). (Scale bars, 10 μm.)
The availability of a draft human genome (3, 25) has enabled us to investigate the SRm160 genomic context. The gene for SRm160 is located on chromosome 1, spans a genomic sequence of ≈27 kb, and consists of 16 exons. Because functional domains often coincide with exons we looked for those exons encoding SRm160 speckle targeting and nuclear matrix binding sequences. Interestingly, the SRm160 weak nuclear matrix binding sequence almost exactly matches exon 7.
Discussion
We have identified the sequences within the splicing coactivator SRm160 that target it to the splicing factor-rich, speckled domains of the nucleus. These consist of two contiguous sequence domains of SRm160: amino acids 300–350 and 351–688. Both sequences are characterized by an unusual high content of arginine, proline, and serine: 88% for SRm160300–350 and 64% for SRm160351–688. SR domains have been reported to be sufficient for speckle localization for a subset of SR proteins (26–28). Although the SRm160 speckle targeting sequences contain some SR repeats, these are less clustered, more disperse, and scattered in smaller domains. Moreover, half of the SRm160 SR repeats, including the most prominent stretch from amino acids 277 to 292, are located in the N-terminal part of the protein, which is not involved in speckle binding. Thus within the context of SRm160, SR repeats alone may not be sufficient for speckle association, and other sequences besides the 5 SR repeats within SRm160300–351 and the 10 SR repeats within SRm160351–688 must support speckle targeting.
There have been other candidates for speckle domain targeting sequences. Examples are the RNA recognition motif (RRM) in the polypyrimidine track binding (PTB)-associated splicing factor (PSF) (29), the “ForkHead-associated” domain of the protein phosphatase-1 regulator NIPP1 (30), and the TP repeat domain of the splicing protein SF3b (155) (31). In contrast to other SR proteins, SRm160 does not contain an RRM. Neither SRm160 speckle targeting domain showed any sequence similarity to the ForkHead-associated domain. The SRm160 speckle targeting domains contain only three TP repeats compared with 27 for the speckle-directing motif of SF3b (155), and these were not sufficient to target the EGFP-SRm160472–585 fusion protein to splicing speckles. Taken together, the comparison with known speckle targeting sequences suggests that both SRm160 speckle determinants are previously uncharacterized motifs. SRm160 appearance in speckled domains correlated with binding to the nuclear matrix. Both speckle targeting sequences were also anchored to the nuclear matrix, whereas the SRm160 N-terminal domain, which did not localize to speckles, was solubilized by detergent permeabilization.
NMTSs have been described for the transcription factors AML-1 (32), YY1 (33), PIT-1 (34), and other nuclear proteins such as the kinase anchoring protein AKAP95 (35), the Epstein–Barr virus nuclear antigen leader protein (EBNA-LP) (36), and the glucocorticoid receptor (37, 38). No consensus sequence for these NMTSs has been found, and they have no significant similarity to the SRm160 NMTS. The lack of a consensus sequence for nuclear matrix binding may reflect the binding of these proteins to unique partners in different matrix-associated complexes.
Localization of sites of RNA splicing by fluorescence in situ hybridization, shows that a majority are clustered at or near speckled domains (7). It has been suggested that this splicing occurs in perichromatin fibrils that surround the interchromatin granule cluster lying at the heart of the speckled domain (39). Interestingly, all SRm160 deletion mutants containing only the weaker speckle targeting sequence (amino acids 300–350) were also found in regions adjacent to splicing speckles (Fig. 4). When fused to EGFP, this sequence directed the fusion protein to sites peripheral to speckled domains (Fig. 4). These correspond to sites enriched in perichromatin fibrils and with new transcripts and abundant RNA splicing. This amino acid domain of SRm160 would represent a targeting signal that is specific for this location in the nucleus, a site centrally important for gene expression. Similar regions peripheral to speckled domains have been found recently to contain three proteins, PSP1, PSP2, and p54/nrb, that traffic between these paraspeckles and the nucleolar periphery (23).
SRm160 remains preferentially and stably associated with the exon–exon product and not with the intron–lariat product after splicing (13). This has suggested a possible involvement of SRm160 in mRNA transport after the excision of introns. More recently, SRm160 has been found on spliced mRNAs at sites 20–24 nt upstream from exon–exon junctions in an EJC also containing the mRNA export factors DEK, RNPS1, Y14, Aly/REF (16–18), and Magoh (19). Y14 and the mRNA export factor REF continuously shuttle between nucleus and the cytoplasm (40, 41), whereas SRm160 is exclusively located in the nucleus (J.A.N., K. M. Wan, G. Krockmalnic, and S.W., unpublished data). The targeting of all N-terminal domains of SRm160 to the nuclear periphery suggests a possible role of the SRm160 N terminus in the subnuclear transport of spliced mRNA to specific sites near nuclear pores, where SRm160 would leave the complex. SRm160 is also present in long intranuclear tracks that are sometimes branched and frequently terminate near the nuclear pores (J.A.N., K. M. Wan, G. Krockmalnic, and S.W, unpublished data). Taking all these data into consideration, we suggest the following model: SRm160 is present in spliceosomal complexes, which are most often at or near speckled domains. After excision of introns, the complex disassembles, but SRm160 remains bound to the mRNA and recruits other RNA export factors to the EJC. The assembled complex moves along intranuclear tracks to the nuclear periphery where SRm160 detaches and returns to splicing speckled domains. The remaining messenger RNP complex containing Y14 and REF crosses to the cytoplasm through nuclear pores. After unloading the mRNA cargo, both RNA export factors shuttle back into the nucleus, ready to start another mRNA export cycle.
SR proteins including SRm160 and SRm300 are not detectable in nucleoli when visualized by immunofluorescence. Surprisingly, however, we observed nucleolar localization for a subset of truncated forms of SRm160. The SRm160 nucleolar localization might be suppressed or masked in the full-length protein because of stronger targeting signals such as speckle targeting sequences. Interestingly, the nucleolar RNA-binding protein NHPX undergoes a transient interaction with splicing speckles (42) before it irreversibly accumulates in nucleoli. It is unclear why SRm160 would have a nucleolar targeting signal or why SRm160 might traffic to the nucleolus, although the involvement of the nucleolus in mRNA export has been suggested (43).
Acknowledgments
We thank C. Stone for technical support. We are grateful to G. Bauren and P. Sharp (Massachusetts Institute of Technology) for their gift of an SRm160 cDNA, and we thank B. Blencowe (University of Toronto, Toronto), M. Mancini (Baylor College of Medicine, Houston), and J. Stein (University of Massachusetts) for helpful discussions and advice. This work was supported by American Cancer Society Grant RPG-99-262-01-GMC.
Abbreviations
- SR
Ser-Arg
- SRm160
SR-related matrix protein of 160 kDa
- SRm300
SR-related matrix protein of 300 kDa
- RNP
ribonucleoprotein
- EJC
exon-junction complex
- EGFP
enhanced GFP
- NMTS
nuclear matrix targeting signal
References
- 1.Berget S M, Moore C, Sharp P A. Proc Natl Acad Sci USA. 1977;74:3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharp P A. Cell. 1994;77:805–815. doi: 10.1016/0092-8674(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 3.Lander E S, Linton L M, Birren B, Nusbaum C, Zody M C, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Spector D L, Fu X D, Maniatis T. EMBO J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monneron A, Bernhard W. J Ultrastruct Res. 1969;27:266–288. doi: 10.1016/s0022-5320(69)80017-1. [DOI] [PubMed] [Google Scholar]
- 6.Puvion E, Puvion-Dutilleul F. Exp Cell Res. 1996;229:217–225. doi: 10.1006/excr.1996.0363. [DOI] [PubMed] [Google Scholar]
- 7.Smith K P, Moen P T, Wydner K L, Coleman J R, Lawrence J B. J Cell Biol. 1999;144:617–629. doi: 10.1083/jcb.144.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Misteli T, Caceres J F, Clement J Q, Krainer A R, Wilkinson M F, Spector D L. J Cell Biol. 1998;143:297–307. doi: 10.1083/jcb.143.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Misteli T, Caceres J F, Spector D L. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- 10.Zeitlin S, Parent A, Silverstein S, Efstratiadis A. Mol Cell Biol. 1987;7:111–120. doi: 10.1128/mcb.7.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeitlin S, Wilson R C, Efstratiadis A. J Cell Biol. 1989;108:765–777. doi: 10.1083/jcb.108.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blencowe B J, Nickerson J A, Issner R, Penman S, Sharp P A. J Cell Biol. 1994;127:593–607. doi: 10.1083/jcb.127.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blencowe B J, Issner R, Nickerson J A, Sharp P A. Genes Dev. 1998;12:996–1009. doi: 10.1101/gad.12.7.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blencowe B J, Bauren G, Eldridge A G, Issner R, Nickerson J A, Rosonina E, Sharp P A. RNA. 2000;6:111–120. doi: 10.1017/s1355838200991982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wan K M, Nickerson J A, Krockmalnic G, Penman S. Proc Natl Acad Sci USA. 1994;91:594–598. doi: 10.1073/pnas.91.2.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Hir H, Izaurralde E, Maquat L E, Moore M J. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Hir H, Moore M J, Maquat L E. Genes Dev. 2000;14:1098–1108. [PMC free article] [PubMed] [Google Scholar]
- 18.Le Hir H, Gatfield D, Izaurralde E, Moore M J. EMBO J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kataoka N, Diem M D, Kim V N, Yong J, Dreyfuss G. EMBO J. 2001;20:6424–6433. doi: 10.1093/emboj/20.22.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCracken S, Lambermon M, Blencowe B J. Mol Cell Biol. 2002;22:148–160. doi: 10.1128/MCB.22.1.148-160.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He D C, Nickerson J A, Penman S. J Cell Biol. 1990;110:569–580. doi: 10.1083/jcb.110.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nickerson J A, Krockmalnic G, Penman S. In: Cell Biology: A Laboratory Handbook. 2nd Ed. Celis J, editor. Boca Raton, FL: Academic; 1997. ,Vol. 2, pp. 184–189. [Google Scholar]
- 23.Fox A H, Lam Y W, Leung A K, Lyon C E, Andersen J, Mann M, Lamond A I. Curr Biol. 2002;12:13–25. doi: 10.1016/s0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- 24.Liu C S, Kong B, Xia H H, Ellem K A, Wei M Q. J Gene Med. 2001;3:145–152. doi: 10.1002/jgm.164. [DOI] [PubMed] [Google Scholar]
- 25.Venter J C, Adams M D, Myers E W, Li P W, Mural R J, Sutton G G, Smith H O, Yandell M, Evans C A, Holt R A, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 26.Gama-Carvalho M, Krauss R D, Chiang L, Valcarcel J, Green M R, Carmo-Fonseca M. J Cell Biol. 1997;137:975–987. doi: 10.1083/jcb.137.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Bingham P M. Cell. 1991;67:335–342. doi: 10.1016/0092-8674(91)90185-2. [DOI] [PubMed] [Google Scholar]
- 28.Hedley M L, Amrein H, Maniatis T. Proc Natl Acad Sci USA. 1995;92:11524–11528. doi: 10.1073/pnas.92.25.11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dye B T, Patton J G. Exp Cell Res. 2001;263:131–144. doi: 10.1006/excr.2000.5097. [DOI] [PubMed] [Google Scholar]
- 30.Jagiello I, Van Eynde A, Vulsteke V, Beullens M, Boudrez A, Keppens S, Stalmans W, Bollen M. J Cell Sci. 2000;113:3761–3768. doi: 10.1242/jcs.113.21.3761. [DOI] [PubMed] [Google Scholar]
- 31.Eilbracht J, Schmidt-Zachmann M S. Proc Natl Acad Sci USA. 2001;98:3849–3854. doi: 10.1073/pnas.071042298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng C, McNeil S, Pockwinse S, Nickerson J, Shopland L, Lawrence J B, Penman S, Hiebert S, Lian J B, van Wijnen A J, et al. Proc Natl Acad Sci USA. 1998;95:1585–1589. doi: 10.1073/pnas.95.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bushmeyer S M, Atchison M L. J Cell Biochem. 1998;68:484–499. [PubMed] [Google Scholar]
- 34.Mancini M G, Liu B, Sharp Z D, Mancini M A. J Cell Biochem. 1999;72:322–338. [PubMed] [Google Scholar]
- 35.Akileswaran L, Taraska J W, Sayer J A, Gettemy J M, Coghlan V M. J Biol Chem. 2001;276:17448–17454. doi: 10.1074/jbc.M101171200. [DOI] [PubMed] [Google Scholar]
- 36.Yokoyama A, Kawaguchi Y, Kitabayashi I, Ohki M, Hirai K. Virology. 2001;279:401–413. doi: 10.1006/viro.2000.0715. [DOI] [PubMed] [Google Scholar]
- 37.van Steensel B, Jenster G, Damm K, Brinkmann A O, van Driel R. J Cell Biochem. 1995;57:465–478. doi: 10.1002/jcb.240570312. [DOI] [PubMed] [Google Scholar]
- 38.Tang Y, Getzenberg R H, Vietmeier B N, Stallcup M R, Eggert M, Renkawitz R, DeFranco D B. Mol Endocrinol. 1998;12:1420–1431. doi: 10.1210/mend.12.9.0169. [DOI] [PubMed] [Google Scholar]
- 39.Huang S, Spector D L. J Cell Biochem. 1996;62:191–197. doi: 10.1002/(sici)1097-4644(199608)62:2<191::aid-jcb7>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 40.Kataoka N, Yong J, Kim V N, Velazquez F, Perkinson R A, Wang F, Dreyfuss G. Mol Cell. 2000;6:673–682. doi: 10.1016/s1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 41.Rodrigues J P, Rode M, Gatfield D, Blencowe B, Carmo-Fonseca M, Izaurralde E. Proc Natl Acad Sci USA. 2001;98:1030–1035. doi: 10.1073/pnas.031586198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leung A K, Lamond A I. J Cell Biol. 2002;157:615–629. doi: 10.1083/jcb.200201120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kadowaki T, Hitomi M, Chen S, Tartakoff A M. Mol Biol Cell. 1994;5:1253–1263. doi: 10.1091/mbc.5.11.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]