Abstract
The transcription factor absent, small, or homeotic discs 2 (ash2) gene is a member of the trithorax group of positive regulators of homeotic genes. Mutant alleles for ash2 are larval/pupal lethals and display imaginal disc and brain abnormalities. The allele used in this study is a true mutant for the trithorax function and lacks the longest transcript present in wild-type flies. In an attempt to identify gene targets of ash2, we have performed an expression analysis by using cDNA microarrays. Genes involved in cell cycle, cell proliferation, and cell adhesion are among these targets, and some of them are validated by functional and expression studies. Even though trithorax proteins act by modulating chromatin structure at particular chromosomal locations, evidence of physical aggregation of ash2-regulated genes has not been found. This work represents the first microarray analysis, to our knowledge, of a trithorax-group gene.
The trithorax group (trx-G) of activators and the Polycomb group (Pc-G) of repressors maintain the correct expression of several key developmental regulators, including the homeotic genes. Pc-G mutants exhibit posterior transformations in embryos and adults caused by derepression of homeotic loci in flies (1) and vertebrates (2). In contrast, proteins of the trx-G are required for the maintenance of activation of homeotic loci (3). Pc-G and trx-G proteins function in distinct multiprotein complexes that are believed to control transcription by changing the structure of chromatin, organizing it into either a “closed” or an “open” conformation (ref. 4 and references therein). It is thought that Pc-G and trx-G regulate many targets in addition to homeotic genes, indicating that epigenetic maintenance of activated or repressed states might represent a fundamental developmental mechanism (5).
The ash2 (absent, small, or homeotic discs 2) gene is a member of the trx-G discovered, together with ash1, in a screen for late larval/early pupal lethals that had imaginal discs abnormalities (6–9). The ASH2 protein has a proline-, glutamic acid-, serine-, and threonine-rich region sequence characteristic of short-lived proteins, a putative double zinc-finger domain, a bipartite nuclear localization signal, and a SPRY domain (10). Biochemical studies have shown that ASH1 and ASH2 are subunits of distinct protein complexes and that ASH2 elutes in fractions with an apparent native molecular mass of 500 kDa (11). More recently it has been reported that the Saccharomyces cerevisiae SET1 complex includes two putative ASH2 homologues as well as a protein (SET1) with high similarity to TRX. This complex methylates histone 3 lysine 4, reinforcing the notion that methylation is important for regulating the transcriptional accessibility of chromatin (12–14).
Mutations in ash2 cause the homeotic transformations expected for genes in this group in addition to a variety of additional pattern formation defects. ash2 mutant hemizygotes that are able to survive until eclosion include supernumerary legs, duplication of thoracic bristles, and transformation of campaniform sensilla to bristles (15). The line l(3)112411 was isolated from a collection of P-lacW element insertional mutagenesis in the third chromosome (16) and corresponds to a new ash2 allele. The few homozygous flies that reach the adult stage are sterile and display anomalous patterns of appendage differentiation. Clonal analysis in adult wings of homozygous cells for the stronger allele ash2I1 reveals a role in vein–intervein patterning, because a reduction of intervein tissue and an increase of vein tissue are observed autonomously and nonautonomously in the clones (17). Moreover, a failure to form joints or fusion of several fragments leads to shortened legs when big clones are generated. Taken together, the pleiotropic phenotypes observed could not be explained only by changes in homeotic gene expression; therefore, more genes should be responding to the loss of ash2 function.
In this work, we have applied cDNA microarray technology to analyze the transcription profile of ash2I1 mutant larvae in comparison with WT, in an attempt to delineate the transcriptional consequences of lack of ash2 function and to identify genes that may fulfill the criteria of ash2 targets. Microarrays have been used to study a variety of biological processes, from differential gene expression in yeast sporulation (18) to human tumors (19). In the case of Drosophila, they were initially applied to analyze development during metamorphosis (20) and more recently for analyzing patterns of transcription under different situations or mutant conditions (21–26). The microarray analysis presented here represents the first approach, to our knowledge, to monitoring the genome wide-expression profile from a mutant of the trx-G. The regulated genes have been automatically classified and clustered according to the functional criteria in the Gene Ontology (GO) database (27), with the aim of finding a differential distribution among the regulated genes.
Materials and Methods
Canton-S and ash2I1/TM6C strains were maintained on standard medium and experiments performed at 25°C. Details of mitotic clone generation, 5′-rapid amplification of cDNA ends, Northern blot, and RT-PCR are published as Supporting Materials and Methods on the PNAS web site, www.pnas.org.
Microarray Analysis.
One to three micrograms of poly(A) RNA from WT or mutant larvae were labeled by reverse transcription incorporation of Amino-allyl dUTP and coupling to cyanine dye (Cy3- or Cy5-NHS esters, Amersham Biosciences) and hybridized to cDNA microarrays constructed by using PCR products directly amplified from the drosophila gene collection 1.0 (www.fruitfly.org/dgc/index.html). genepix 3.0 (Axon Instruments, Foster City, CA), convertdata 3.33 (www.le.ac.uk/cmht/microarray_lab/microarray_softwares/microarray_softwares.htm) and Microsoft excel were used for acquiring and mining data. One Class Response Significance Analysis of Microarrays (sam) Analyses (28) were conducted with different fold-change thresholds. Automatic functional annotation of the regulated genes in our microarray experiments has been obtained by using the GO database (27). Complete information on the methods used and controls carried out is published as supporting information on the PNAS web site, which also contains a comprehensive description on how GO classification, chromosomal localization, and sequence analysis were performed. Additional data and detailed PERL scripts can be accessed at www.ub.es/epidd/arrays/index.htm.
Results and Discussion
The ash2I1 Mutant Lacks the trx-G Function.
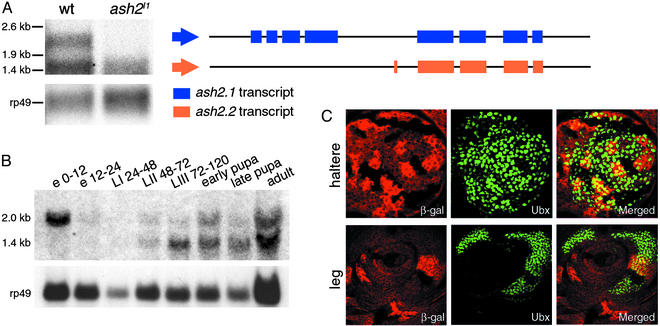
In the work presented here, we have used the allele ash2I1, obtained after excision of the P-lacW transposon present in line l(3)12411 (17). It is lethal in early pupa, and homozygous larvae have reduced and abnormal imaginal discs and brain (data not shown and Fig. 5A). The molecular alterations present in the ash2I1 allele are small changes (2-bp deletion and 5-bp insertion) in the fourth intron of the gene (17). Northern blot analysis of poly(A) RNA extracted from third instar larvae showed the presence of two transcripts (2 and 1.4 kb) with potential coding sequence in WT and only the small one in ash2I1 mutant flies (Fig. 1A). The longer transcript would account for the already described ASH2 protein (15), and 5′-rapid amplification of cDNA ends results supported by in silico predictions from geneid (29) and genscan (30) showed that the 1.4-kb transcript, identical in both WT and ash2I1, contains exons 5–8 present in the previously described ash2 transcript plus a novel 62-bp exon containing 28-bp encoding for amino acids (Fig. 1A). If translated, the resulting protein would be 350 aa in length and would lack the proline-, glutamic acid-, serine-, and threonine-rich region sequence and the putative double zinc-finger domain, also found in other trx-G proteins. Developmental Northern blot of WT flies showed that both transcripts are present at all stages of development except in early embryos, where only the long maternal transcript was detected (Fig. 1B). Because the insertion (TTAGG) detected in the fourth intron of the ash2I1 allele (17) creates a putative splicing acceptor site (31) that could generate a transcript containing a premature translation termination codon, it is tempting to speculate on a nonsense mediated decay of such RNA species (32). To confirm that our mutant behaves as a true trx-G mutant, we generated genetic mosaics in haltere and leg imaginal discs with the aid of the flipase–flipase recombination target (FLP-FRT) technique and analyzed the expression of ultrabithorax (UBX) by immunohistochemistry (see supporting information on the PNAS web site). The down-regulation of UBX accumulation in the homozygous ash2I1 tissue (Fig. 1C) proves this mutant behaves as a trithorax mutant regarding homeotic function.
Figure 5.
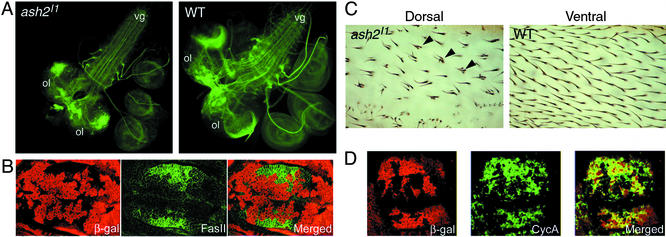
ash2 has a role in regulating cell adhesion, development of neural system, and cell cycle. (A) Third instar larval brains from ash2I1/ash2I1 and WT individuals stained with anti-FASII. ol, optic lob; vg, ventral ganglion. Note the disruption of the neural pattern in both the optic lobe and the comissures of the ventral ganglion. (B) FLP-FRT clones on Minute (M−) background of ash2I1 mutant cells from a wing imaginal disc tested for the up-regulated protein Fasciclin II (green). (C) Detail of a clone ash2I1/ash2I1 in the dorsal layer of cells of an adult wing (Left) and the same area (WT tissue) focused on the opposite wing side (Right). Note that in the WT condition each cell develops one hair, whereas in the dorsal mutant surface some cells develop multiple hairs (arrowheads). (D) Immunodetection of the down-regulated CycA (green) on M− background FLP-FRT clones. (B and D Left) Staining with anti-β-galactosidase antibody in red (see Fig. 1 legend).
Figure 1.
(A–C) Molecular characterization of WT and mutant ash2 mRNA. (A) Northern blot analysis of poly(A) RNA extracted from WT and ash2I1 homozygous third instar larvae. Structure of the 2- (ash2.1) and 1.4-kb (ash2.2) transcripts after performing rapid amplification of cDNA ends is shown. (B) Developmental Northern blot of ash2 expression. In A and B, rp49 was used as a loading control. e, embryo; L, larval stages; numbering indicates hours after egg laying. (C) Down-regulation of UBX protein accumulation in ash2I1 mutant clones generated in haltere and leg imaginal discs by FLP-FRT-induced mitotic recombination. (Left) Staining with anti-β-galactosidase antibody:WT cells (bright red), heterozygous cells (red), and homozygous ash2I1 mutant cells (lack of red staining). (Center) Staining with anti-UBX antibody (green). (Right) Merged images.
Transcription Profile of ash2I1 Homozygous Mutant Larvae.
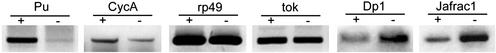
To identify downstream genes of ash2 function, we have compared the population of mRNA species isolated from homozygous ash2I1 third instar larvae with that of stage-matched WT. Four completely independent cDNA microarray experiments were carried out with poly(A) RNA isolated from separate extractions. The microarrays were constructed by using the ESTs from the drosophila gene collection 1.0, which contains about one-third of the Drosophila genes. This collection lacks some genes known to be regulated by ash2 such as Ubx. A total of 5,139 cDNAs with a different FlyBase identifier (33) were printed; 4,163 of them passed the quality filters in at least two of the experiments (see supporting information on the PNAS web site) and could be used in a One Class Response sam analysis (28). With a false discovery rate of ≈0.025 and a fold change threshold of 1.75, we identified 235 genes of the 4,163 (5.6%) in the sam input whose expression levels change significantly in the mutant, pointing to ash2 as a putative regulator of them (see Fig. 6, which is published as supporting information on the PNAS web site, and additional data at www.ub.es/epidd/arrays/index.htm). Before inputting data with sam, the mean Pearson's correlation coefficient among all replicate experiments for the 235 regulated genes was 0.88, indicating a good reproducibility of ratios. One hundred forty of these genes were positively regulated and 95 negatively regulated. Down-regulated genes include ash2 with a 1.76-fold change, a rather high value if we acknowledge the presence of the 1.4-kb transcript in the ash2I1 mutant. We verified the differential expression levels of candidate genes by performing semiquantitative RT-PCR analysis on selected genes (Fig. 2).
Figure 2.
Validation of the microarray analysis by RT-PCR of WT (+) and ash2I1 (−) mRNA samples on selected genes. Punch (Pu) and Cyclin A (CycA) are down-regulated genes; ribosomal protein rp49 and the endopeptidase tolkin (tok) are unaltered genes; Dodeca-satellite-binding protein 1 (Dp1) and thiredoxin peroxidase1 (Jafrac1) are up-regulated genes.
Functional Classification of Regulated Genes.
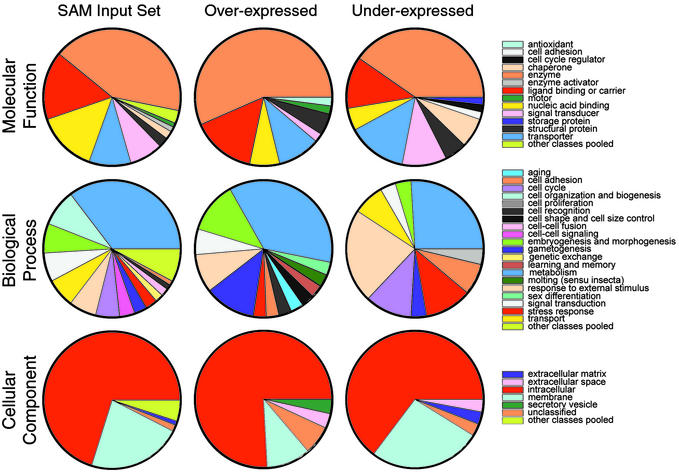
Functional annotation of the genes present in the sam 1.75 output has been performed aiming to discover features characteristic to the sets of up- and down-regulated genes. We classified the cDNAs present in our chips according to GO database (27). GO provides a controlled vocabulary organized in a hierarchical way that can be applied to describe gene products from eukaryotic organisms regarding: molecular function (MF), biological process (BP), and cellular component (CC). GO terms were found for 55% of the 5,139 unique genes. From the 4,163 genes given to sam as input, 56% had GO terms, and 62% of the 235 regulated genes also had them. The GO classifications of the sam input set, the set of all genes in the chip, and the set of all genes in FlyBase (additional data at www.ub.es/epidd/arrays/index.htm) were very similar for the different categories studied (MF levels 1 and 2, BP levels 1 and 2, and CC levels 1 and 2), strongly suggesting that the sam input set is a good representation of all known genes in Drosophila. To evaluate whether ash2 was acting predominantly on a certain group of functionally related genes, we compared the GO classification of the regulated genes (up or down) with that of the whole set of genes given as input to sam. Fig. 3 shows that the distribution of classes is quite different, both when comparing the regulated genes with the sam input genes and when comparing up- with down-regulated genes (Fig. 3 and additional data at www.ub.es/epidd/arrays/index.htm), strongly suggesting that the results of our microarray experiment are indeed capturing underlying biological phenomena and are not purely artifactual.
Figure 3.
Classification of regulated genes (up- and down-) according to GO (MF, level 1; CC, level 2; and BP, level 2) and comparison with the genes used for the study (SAM input set). To assess the statistical significance of differences between sets of genes, we have calculated pairwise Chi2. Chi2 P values are given in the following order for each of the classes: SAM input vs. whole fly, overexpressed vs. SAM input and underexpressed vs. SAM input. The P values are as follows: MF1, 0.820, 0.008, and 0.006; BP2, 0.845, 0.000, and 0.000; and CC2, 0.931, 0.002, and 0.384.
In addition to distributing the sam 1.75 set of genes among GO classes, we have hierarchically clustered them according to their GO terms (MF level 1 and BP level 2), so that genes having similar sets of GO terms are placed closer in Fig. 4. The position of the genes in this tabular representation is (broadly) associated with its biological function. Indeed, the more GO terms share two of them (or two groups of them), the more similar they are supposed to be and the closer they will be to one another in the figure. For example, the down-regulated heat-shock proteins and the up-regulated gene Frost have been grouped together in Fig. 4 and are included in the classes MF chaperones and BP response to external stimulus, two of the classes that stand out in the set of down-regulated genes in Fig. 3. We believe methods like this of presenting microarray results will become more useful as more genes from Drosophila and other organisms are given GO terms, and as more microarray data from different eukaryotic organisms become available. By clustering genes from different sources using GO terms and microarray data, we will be able to find functional homologies between genes and/or pathways from different organisms even when there is no conservation whatsoever at the sequence level.
Figure 4.
Genes (rows) differentially expressed in the mutant ash2I1 clustered according to their MF level 1, and BP level 2 GO terms (columns). MF and BP descriptors are also clustered. A given gene has a GO term if the intersecting cell is dark gray (up-regulated) or light gray (down-regulated). Genes (CG nos. and BcDNAs) presenting only one GO term have been excluded. “Additional Information” has been extracted from FlyBase, interactive fly, and references herein. A POSTSCRIPT image with tree branches can be found at www.ub.es/epidd/arrays/index.htm.
In Fig. 3, it is clear that regarding MF, nucleic acid-binding proteins are underrepresented in both sets of regulated genes and, whereas enzymes are overrepresented only in the set of up-regulated genes, so are chaperones in the down-regulated set. As to BP, GO classifications are quite characteristic of both up- and down-regulated genes. Genes related to cell organization and biogenesis are absent from the sets of regulated genes. The set of up-regulated genes is clearly enriched with genes involved in gametogenesis but lacks genes involved in cellular transport, whereas genes involved in responses to external stimulus, stress response, cell adhesion, and cell proliferation are overrepresented among down-regulated genes. Finally, concerning CC, regulated genes (both up- and down-) have an excess of genes acting in the extracellular space, whereas up-regulated genes show an excess of genes acting in the secretory vesicle and a deficiency of membrane proteins.
Because trx-G might be acting on chromatin structure, making DNA accessible to other proteins involved in transcriptional regulation, and some genes that have been found to be regulated by ash2 are located adjacent or very close to each other on the genome (i.e., Hsp23, Hsp26, Hsp27, and aay or Cyp6a8 and Cyp6a20), we mapped differentially expressed genes in the sam 1.75 output set onto the D. melanogaster genome sequence with the aim of investigating possible colocalization of coregulated genes. To assess the degree of aggregation for coregulated genes, we computed the Moran's index (34) to measure the correlation between the localization of the genes along the chromosomes and their level of expression (see supporting information on the PNAS web site). We found no evidence of physical aggregation of coregulated genes, because the value of the Moran's index (0.01) was not significant after 1,000 Monte Carlo tests with randomly shuffled data. The same results have been obtained in whole-genome expression analysis of snf/swi mutants of S. cerevisiae (35). More recently, it has been found that chromosomal position of signature genes is random when studying the transcript profiles in aging and calorically restricted flies (36), although evidence for large domains of similarly expressed genes in the Drosophila genome has also been reported (37).
Finally, for 165 genes (72 down- and 93 up-regulated), complete mRNA exonic coordinates were annotated, and the corresponding full-length cDNA sequences were obtained. A number of sequence characteristics were computed on these genes (G+C content, number of exons, length of transcripts) and compared with those of the whole set of full-length cDNAs from Drosophila (4,002 sequences). As expected, no striking differences were found among up- and down-regulated genes and the whole set of fly genes (data not shown).
Confirmation of Target Genes.
Genes involved in cell adhesion and/or development of the neural system (i.e., FasII, mfas, Ama, Lac, and shg) are two of the main classes regulated by ash2 (Figs. 3 and 4). We have focused in the up-regulated gene FasII as an example of those and performed clonal analysis on a Minute background, to assess whether the behavior of the FasII transcript observed with this ash2 mutant was also kept at the protein level in wing imaginal discs (Fig. 5B). Homozygous mutant cells show a clear up-regulation of FASII, mainly in the wing pouch area further away from the dorsoventral margin, where we have found FASII to be very slightly expressed in WT wing discs (data not shown). The up-regulation of FasII and other cell adhesion molecules like mfas, together with the up-regulation of the transcription factor vri, could explain some of the phenotypes previously found by clonal analysis (17), such as disruption of vein–intervein patterning, because it is known that preferential accumulation of specific adhesion molecules characterizes the final stages of vein differentiation (38). Furthermore, because FASII is involved in the development of the neural system, we have compared its pattern of expression in ash2I1 mutant brains with that of WT and have found that they present a distorted phenotype in the optic lobes as well as fasciculation defects in the ventral ganglion (Fig. 5A), a process in which FASII plays a central role (39, 40). A gain-of-function screen done by Kraut et al. (41) identified ash2 and FasII as genes involved in the development of the neural system, further supporting a relationship between them.
In a search for putative downstream genes for Set1, many genes involved in transcriptional regulation of growth and cell cycle control were found (12). According to our results, ash2 also seems to act on genes that fall into these classes (see Figs. 3 and 4). For example, Eip75B, vri, jing, or HmgD could be classified in the first class, whereas CyclinA (CycA), cutlet, Pu, mei-S332, or mus209 could do so in the second. We have confirmed, by clonal analysis in wing imaginal discs, that the protein product of the down-regulated gene CycA is present to a lesser extent in mutant clones (Fig. 5D). The down-regulation of CycA can play a role in the proliferation defects observed in mutant cells when clones are generated in the imaginal discs (17). Furthermore, in genetic mosaics, homozygous mutant ash2I1 cells show effects on both cell differentiation and cell size. Normal wing cells develop a single hair, whereas ash2I1 cells can develop either a single or multiple hairs. In addition, the spacing between ash2I1 mutant cells is increased compared with that of WT ones, suggesting that ash2I1 mutant cells are larger (Fig. 5C).
Preliminary promoter analysis of the regulated genes does not seem to reveal any defined and clear pattern (see supporting information on the PNAS web site). Although this needs further investigation, it may be difficult to find any consensus sequence, mainly because the regulated genes reflect all of the transcripts present at that moment and therefore a particular cellular status. Even though some authors (42, 43) have described cis-acting Trithorax Response Element sequences needed for some trx-G proteins to exert their function, it is not yet possible to discern between primary and secondary targets of ASH2 on the basis of this information. However, we can state that ASH2 function is required for a wide variety of biological processes during larval development.
Supplementary Material
Acknowledgments
We thank G. Hulburt for technical support, J. F. Abril for help with POSTSCRIPT, S. Vives for statistical advice, and J. Baguñà for helpful discussions. S.B. and E.B. are supported by fellowships from the Ministerio de Ciencia y Tecnología. Grants BMC2000-0766, GEN2001-4846-C05-02, and HI2001-0009 from Ministerio de Ciencia y Tecnología, Spain, have supported this work. We thank the Developmental Studies Hybridoma Bank, University of Iowa, for providing the anti-Fasciclin II antibody.
Abbreviations
- GO
Gene Ontology
- SAM
significance analysis of microarrays
- MF
molecular function
- BP
biological process
- CC
cellular component
- trx-G
trithorax group
- UBX
ultrabithorax
- FLP–FRT
flipase–flipase recombination target
References
- 1.Simon J. Curr Opin Cell Biol. 1995;7:376–385. doi: 10.1016/0955-0674(95)80093-x. [DOI] [PubMed] [Google Scholar]
- 2.Schumacher A, Magnuson T. Trends Genet. 1997;13:167–170. [PubMed] [Google Scholar]
- 3.Kennison J A. Annu Rev Genet. 1995;29:289–303. doi: 10.1146/annurev.ge.29.120195.001445. [DOI] [PubMed] [Google Scholar]
- 4.Mahmoudi T, Verrijzer C P. Oncogene. 2001;20:3055–3066. doi: 10.1038/sj.onc.1204330. [DOI] [PubMed] [Google Scholar]
- 5.Francis N J, Kingston R E. Nat Rev Mol Cell Biol. 2001;2:409–421. doi: 10.1038/35073039. [DOI] [PubMed] [Google Scholar]
- 6.Shearn A, Rice T, Garen A, Gehring W. Proc Natl Acad Sci USA. 1971;68:2695–2698. doi: 10.1073/pnas.68.10.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shearn A, Hersperger E, Hersperger G. Roux's Arch Dev Biol. 1987;196:231–242. doi: 10.1007/BF00376347. [DOI] [PubMed] [Google Scholar]
- 8.Shearn A. Genetics. 1989;121:517–525. doi: 10.1093/genetics/121.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LaJeunesse D, Shearn A. Mech Dev. 1995;53:123–139. doi: 10.1016/0925-4773(95)00430-0. [DOI] [PubMed] [Google Scholar]
- 10.Ponting C, Schultz J, Bork P. Trends Biochem Sci. 1997;22:193–194. doi: 10.1016/s0968-0004(97)01049-9. [DOI] [PubMed] [Google Scholar]
- 11.Papoulas O, Beek S J, Moseley S L, McCallum C M, Sarte M, Shearn A, Tamkun J W. Development (Cambridge, UK) 1998;125:3955–3966. doi: 10.1242/dev.125.20.3955. [DOI] [PubMed] [Google Scholar]
- 12.Nislow C, Ray E, Pillus L. Mol Biol Cell. 1997;8:2421–2436. doi: 10.1091/mbc.8.12.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roguev A, Schaft D, Shevchenko A, Pijnappel W W, Wilm M, Aasland R, Stewart A F. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagy P L, Griesenbeck J, Kornberg R D, Cleary M L. Proc Natl Acad Sci USA. 2002;99:90–94. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adamson A L, Shearn A. Genetics. 1996;144:621–633. doi: 10.1093/genetics/144.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deak P, Omar M M, Saunders R D, Pal M, Komonyi O, Szidonya J, Maroy P, Zhang Y, Ashburner M, Benos P, et al. Genetics. 1997;147:1697–1722. doi: 10.1093/genetics/147.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amorós M, Corominas M, Deak P, Serras F. Int J Dev Biol. 2002;46:321–324. [PubMed] [Google Scholar]
- 18.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown P O, Herskowitz I. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 19.Perou C M, Sorlie T, Eisen M B, van de Rijn M, Jeffrey S S, Rees C A, Pollack J R, Ross D T, Johnsen H, Akslen L A, et al. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 20.White K P, Rifkin S A, Hurban P, Hogness D S. Science. 1999;286:2179–2184. doi: 10.1126/science.286.5447.2179. [DOI] [PubMed] [Google Scholar]
- 21.Leemans R, Egger B, Loop T, Kammermeier L, He H, Hartmann B, Certa U, Hirth F, Reichert H. Proc Natl Acad Sci USA. 2000;97:12138–12143. doi: 10.1073/pnas.210066997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furlong E E, Andersen E C, Null B, White K P, Scott M P. Science. 2001;293:1629–1633. doi: 10.1126/science.1062660. [DOI] [PubMed] [Google Scholar]
- 23.De Gregorio E, Spellman P T, Rubin G M, Lemaitre B. Proc Natl Acad Sci USA. 2001;98:12590–12595. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irving P, Troxler L, Heuer T S, Belvin M, Kopczynski C, Reichhart J M, Hoffmann J A, Hetru C. Proc Natl Acad Sci USA. 2001;98:15119–15124. doi: 10.1073/pnas.261573998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Churchill G A, Oliver B. Nat Genet. 2001;29:355–356. doi: 10.1038/ng1201-355. [DOI] [PubMed] [Google Scholar]
- 26.Arbeitman M N, Furlong E E, Imam F, Johnson E, Null B H, Baker B S, Krasnow M A, Scott M P, Davis R W, White K P. Science. 2002;297:2270–2275. doi: 10.1126/science.1072152. [DOI] [PubMed] [Google Scholar]
- 27.Ashburner M, Ball C A, Blake J A, Botstein D, Butler H, Cherry J M, Davis A P, Dolinski K, Dwight S S, Eppig J T, et al. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tusher V G, Tibshirani R, Chu G. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parra G, Blanco E, Guigó R. Genome Res. 2000;10:511–515. doi: 10.1101/gr.10.4.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burge C, Karlin S. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 31.Pertea M, Lin X, Salzberg S L. Nucleic Acids Res. 2001;29:1185–1190. doi: 10.1093/nar/29.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hentze M W, Kulozik A E. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 33.Anonymous. Nucleic Acids Res. 1999;27:85–88. doi: 10.1093/nar/27.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moran P A P. Biometrika. 1950;37:17–23. [PubMed] [Google Scholar]
- 35.Sudarsanam P, Iyer V R, Brown P O, Winston F. Proc Natl Acad Sci USA. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pletcher S D, Macdonald S J, Marguerie R, Certa U, Stearns S C, Goldstein D B, Partridge L. Curr Biol. 2002;12:712–723. doi: 10.1016/s0960-9822(02)00808-4. [DOI] [PubMed] [Google Scholar]
- 37.Spellman P T, Rubin G M. J Biol. 2002;1:5.1–5.8. doi: 10.1186/1475-4924-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Celis J F. Int J Dev Biol. 1998;42:335–343. [PubMed] [Google Scholar]
- 39.Lin D M, Goodman C S. Neuron. 1994;13:507–523. doi: 10.1016/0896-6273(94)90022-1. [DOI] [PubMed] [Google Scholar]
- 40.Lin D M, Fetter R D, Kopczynski C, Grenningloh G, Goodman C S. Neuron. 1994;13:1055–1069. doi: 10.1016/0896-6273(94)90045-0. [DOI] [PubMed] [Google Scholar]
- 41.Kraut R, Menon K, Zinn K. Curr Biol. 2001;11:417–430. doi: 10.1016/s0960-9822(01)00124-5. [DOI] [PubMed] [Google Scholar]
- 42.Chang Y L, King B O, O'Connor M, Mazo A, Huang D H. Mol Cell Biol. 1995;15:6601–6612. doi: 10.1128/mcb.15.12.6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tillib S, Petruk S, Sedkov Y, Kuzin A, Fujioka M, Goto T, Mazo A. Mol Cell Biol. 1999;19:5189–5202. doi: 10.1128/mcb.19.7.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.