Abstract
Genetic alterations in tumor cells often lead to the emergence of growth-stimulatory autocrine and paracrine signals, involving overexpression of secreted peptide growth factors, cytokines, and hormones. Increased levels of these soluble proteins may be exploited for cancer diagnosis and management or as points of therapeutic intervention. Here, we combined the use of controlled vocabulary terms and sequence-based algorithms to predict genes encoding secreted proteins from among ≈12,500 sequences represented on oligonucleotide microarrays. Expression of these genes was queried in 150 carcinomas from 10 anatomic sites of origin and compared with 46 normal tissues derived from the corresponding sites of tumor origin and other body tissues and organs. Of 74 different genes identified as overexpressed in cancer tissues, several encode proteins with demonstrated clinical diagnostic application, such as α-fetoprotein in liver carcinoma, and kallikreins 6 and 10 in ovarian cancer, or therapeutic utility, such as gastrin-releasing peptide/bombesin in lung carcinomas. We show that several of the other candidate genes encode proteins with high levels of tumor-associated expression by immunohistochemistry on tissue microarrays and further demonstrate significantly elevated levels of another novel candidate protein, macrophage inhibitory cytokine 1, a distant member of the tranforming growth factor-β superfamily, in the serum of patients with metastatic prostate, breast, and colorectal carcinomas. Our results suggest that the combination of annotation/protein sequence analysis, transcript profiling, immunohistochemistry, and immunoassay is a powerful approach for delineating candidate biomarkers with potential clinical significance and may be broadly applicable to other human diseases.
Keywords: gene expression‖microarray‖genome ontology‖sequence analysis‖ immunohistochemistry
Many of the genetic alterations selected during malignant transformation disrupt paracrine signaling networks, resulting in the release of tumor cells from normal growth constraints or the establishment of new signaling pathways that confer growth and survival advantages on cancer cells (1). The emergence of cancer-specific autocrine and paracrine signals is often facilitated by the inappropriate, supraphysiological expression of secreted proteins or their receptors (2). Many small peptide hormones have been identified as highly expressed in cancer, with potential clinical utility for therapeutic intervention or as diagnostic or prognostic biomarkers. Recent genomic analyses have identified an increasingly large number of genes overexpressed in cancers, of which several encode secreted proteins (e.g., see refs. 3–5). Although the functional impact of these overexpressed proteins is unclear, their clinical utility is emerging. For example, cDNA microarray analysis has revealed significantly elevated expression of the prostasin and osteopontin genes in ovarian cancers, and correspondingly high levels of these proteins have subsequently been detected in the serum of ovarian cancer patients (6, 7). Bioinformatic analyses have also revealed the emergence of autocrine loops in hematopoietic and solid tumors by correlating the expression of genes encoding known ligands and their cognate receptors in cancer tissue samples (8).
Here, we sought global methods to identify genes encoding secreted proteins in common human carcinomas from among thousands of genes represented on commercially available microarrays. The approaches described herein led to the identification of 2,300 genes inferred to be extracellular through annotation- and sequence analysis-based approaches, of which 74 were overexpressed in one or more carcinoma types relative to their physiologically normal counterpart tissues. The overexpression of several of these genes was confirmed at the protein level, as well as in the serum of cancer patients. Our results suggest that many of these proteins may be clinically relevant to cancers of multiple anatomic origins.
Materials and Methods
Tissue and Serum Samples.
Carcinoma samples used for RNA extraction and GeneChip hybridization have been described (4, 5, 9). The set of normal counterpart tissues comprised two breast, four colon, two gastric, three kidney, four liver, four lung, four ovary, four pancreas, and nine prostate samples. The University of Virginia Human Investigation Committee approved the use of the human malignant and normal counterpart tissue samples obtained at the university. To estimate the normal range of serum protein, 260 normal serum samples were obtained from the Red Cross Blood Bank (Adelaide, Australia) as described (10). Patient serum samples were obtained from subjects who had stage IV disease according to the 1997 Union Internationale Contre le Cancer/American Joint Committee on Cancer classification. Samples from 26 patients were obtained, eight with colorectal cancer, nine with breast cancer, and nine with prostate cancer. All patients were being treated at St. Vincent's Hospital (Sydney) and the serum samples were obtained with the informed consent of the patient.
RNA Extraction and GeneChip Hybridization.
Each of the normal and carcinoma samples was processed as described (9). The GeneAtlas set of normal human tissues represents a defined subset of samples recently described in detail (see ref. 11; http://expression.gnf.org). RNA was extracted, and cRNA was prepared and hybridized on oligonucleotide microarrays (U95a GeneChip, Affymetrix, Santa Clara, CA) exactly as described (4, 5, 9). All average difference values <20, including negative average difference values, were raised to a value of 20.
Mining for Genes Encoding Secreted Proteins.
Two approaches were used in parallel to mine for genes encoding secreted proteins. First, we used the Genome Ontology (GO) consortium annotations (12) to identify genes associated with keywords implying extracellularity (www.geneontology.org). Specifically, Affymetrix probe-set identification numbers were used to retrieve gene-associated sequences from Affymetrix. blast comparisons of the Affymetrix sequences to the UniGene database (www.ncbi.nlm.nih.gov/UniGene) gave rise to exemplar sequences, which were used to query the LocusLink database (www.ncbi.nlm.nih.gov/LocusLink). GO terms associated with each gene were retrieved and searched for each of the following 30 terms: blood coagulation, blood coagulation factor, cell–cell signaling, cell communication, complement activation, complement component, diuretic hormone, ephrin, extracellular, extracellular matrix, extracellular matrix glycoprotein, extracellular matrix structural protein, extracellular space, hormone, insulin-like growth factor receptor ligand, IL-12 receptor ligand, IL-2 receptor ligand, IL-4 receptor ligand, IL-5 receptor ligand, IL-6 receptor ligand, IL-7 receptor ligand, IL-8 receptor ligand, leukemia inhibitor factor receptor ligand, ligand, neuropeptide hormone, opsonin, protein secretion, secreted phospholipase A2, tissue kallikrein, and vascular endothelial growth factor receptor ligand.
In a second approach, predicted amino acid sequences for each gene on the U95a array were obtained from RefSeq (13) and analyzed with the emboss programs tmap (14, 15) and sigcleave (16). Both programs accept arbitrary sequences of amino acids as arguments; sigcleave estimates the likelihood of an authentic signal peptide cleavage site, and tmap predicts the number of transmembrane regions. For sigcleave, we chose a high-confidence threshold (>3.5), which corresponds to 95% specificity and 95% sensitivity of detection. We noted that several authentic signal peptide sequences were counted as transmembrane regions by tmap and therefore chose to admit proteins having as many as one transmembrane domain. This approach is likely to identify proteins that are initially anchored to the plasma membrane before secretion [e.g., CA15–3 (17), CD44 (18), and CD27 (19)].
Differential Gene Expression.
Genes were selected as “differentially expressed” if the measured expression levels were at least 3-fold greater in one or more tumors from a specific anatomic site versus their correspondingly normal tissue counterpart, and at least 2-fold greater than the levels of expression in any of the GeneAtlas tissues (11). Data were processed in cluster and visualized in treeview (20).
Validation of Gene Expression by RT-PCR.
RT-PCR was carried out exactly as described (4). Primers for the relaxin-1 gene were: forward, 5′-GCGGCATGAGCACCTGGAG-3′ and reverse, 5′-GATTGCTGTCTGCGGCTTCACTT-3′.
Tissue Microarrays (TMAs) and Immunohistochemistry (IHC).
Two TMAs containing 0.6-mm cores were constructed from zinc formalin-fixed, paraffin-embedded specimens by using a Tissue Microarrayer (Beecher Instruments, Silver Spring, MD). Each microarray contained one core of each neoplasm whose transcripts were profiled in the study (with the exception of the ovarian serous papillary carcinomas that had been profiled as described in ref. 4). An independent set of 17 serous papillary carcinomas was included in the TMAs, as well as cores of selected normal tissues. For IHC performed on the TMAs and selected whole tissue sections, the avidin-biotin immunoperoxidase method was used. Slides of deparaffinized tissue sections were placed in citrate buffer and treated with microwave heat for 20 min. Primary antibodies used included monoclonal anti-MUC-2 (clone CCP58; 1:40 dilution; BioGenex); monoclonal anti-maspin (clone EAW24; 1:50 dilution; Novocastra, Newcastle, U.K.); and rabbit polyclonal anti-neuropeptide Y (NPY) (1:2,000 dilution; Research Diagnostics, Flanders, NJ). After the addition of the biotinylated secondary antibody, avidin-biotin immunoperoxidase was applied. Diaminobenzidine was used as the chromogen. Sections were counterstained with hematoxylin. Staining for NPY was scored as negative or positive, whereas for MUC-2, tumors were classified as negative, 1+ (<10% positive cells), 2+ (10–50% cells), or 3+ (>50% cells). For maspin, tumors were scored as negative, 1+ (<50% positive cells and weak intensity), 2+ (<50% and moderate or >50% positive and weak intensity), 3+ (<50% and strong), 4+ (>50% and moderate), or 5+ (>50% and strong).
Macrophage Inhibitory Cytokine 1 (MIC-1) ELISA.
The MIC-1 sandwich ELISA used for the quantification of MIC-1 in serum was performed as described (10, 21) by using a mouse monoclonal anti-hMIC-1 antibody, 26G6H6, for antigen capture and sheep anti-MIC-1 antibody 233B-P for detection. Three human serum samples, which had undergone multiple serum MIC-1 determinations in the formatted assay, were used as controls. Sample diluent was used as a background control. All samples were assayed in triplicate.
Results
Identification of Genes Encoding Secreted Proteins.
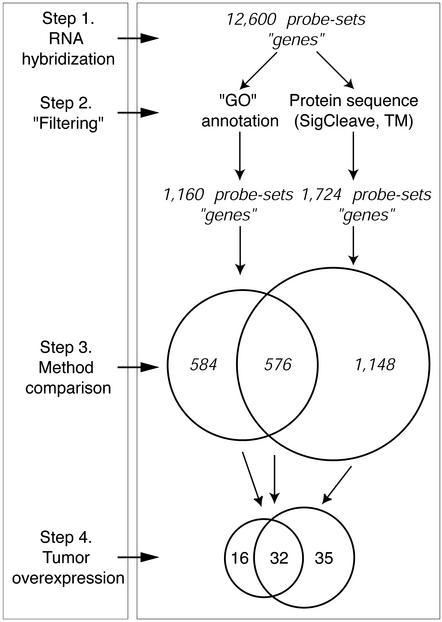
Two approaches were used to select for genes on the Affymetrix U95a GeneChip array that were likely to encode secreted proteins (Fig. 1). First, we asked whether the annotation(s) associated with each gene implied the secretion of the encoded protein into the extracellular space. Specifically, we mapped probe-sets from the Affymetrix U95a GeneChip to annotations provided by the GO consortium via the National Center for Biotechnology Information's LocusLink database (see Materials and Methods), which provides a controlled vocabulary related to a protein's molecular function, biological process, and cellular component (12). Of the annotations associated with genes on the U95a GeneChip, we focused on 30 of these terms (see Materials and Methods), identifying 1,160 genes. In a parallel approach, we looked for genes whose protein sequence features implied the presence of a signal peptide cleavage site, as well as the absence of transmembrane domains, thus suggesting a protein product that would be secreted through a membrane. Using conservative thresholds for the algorithms that we used (tmap and sigcleave, see Materials and Methods), we identified a set of 1,724 genes that potentially encoded extracellular proteins. Together, these two methods identified a nonredundant subset of 2,308 genes, of which 576 were obtained with both methods (Fig. 1).
Figure 1.
Mining for genes that encode secreted proteins. Oligonucleotide probe-sets were filtered for candidate genes encoding secreted proteins by two distinct approaches (step 2, filtering). Affymetrix gene probe-sets were mapped to GO Consortium annotations, and those with evidence suggesting secretion of the encoded protein were identified (1,160 total). Protein sequences of the genes represented on the oligonucleotide microarray were interrogated by using two sequence-based algorithms, sigcleave, which estimates the likelihood of an authentic signal peptide cleavage site in arbitrary amino acid sequence data, and tmap (TM), which predicts transmembrane regions in proteins. A series of 1,724 probe-sets (genes) met the criteria imposed by both sequence algorithms. Probe sets selected by each method were then compared for the degree of overlap (step 3, method comparison), and, finally, their overexpression in carcinomas of different anatomic origin (step 4, tumor differential expression).
Candidate Genes Encoding Secreted Proteins Are Overexpressed in Tumors of Diverse Anatomic Origin.
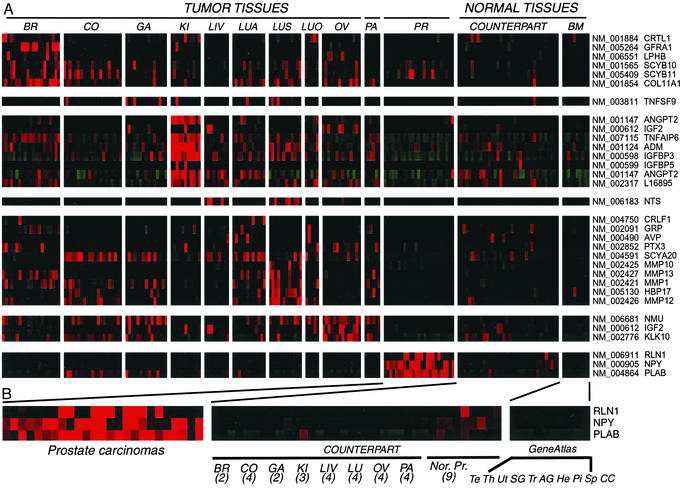
We next examined the expression of these 2,308 genes in a series of 150 carcinomas representing samples from 10 distinct anatomic origins, 46 normal tissues from the corresponding anatomic sites, and nine other anatomic sites not represented in our “tumor/normal” collection (11) (Fig. 2). We sought genes whose expression was high in tumors of one or more sites of origin, with correspondingly low or absent expression in other normal body tissues (see Materials and Methods). Eighty-three of the 2,308 (3.6%) probe-sets met these criteria (Fig. 1; Table 1, which is published as supporting information on the PNAS web site, www.pnas.org), representing 74 different genes. Of the 32 probe-sets (30 distinct genes) identified by both annotation- and sequence-based approaches (designated as GO and TMSC in Table 1; Fig. 2A), we found strong evidence that almost all of them encode secreted proteins; only 3/30 genes (the RET coreceptor, GFRA-1; member 9 of the tumor necrosis factor superfamily, TNFR9; and the cytokine receptor-like factor 1, CRLF-1) are unlikely to encode such proteins. For the 16 probe-sets (15 different genes) identified by annotation alone (designated as GO only in Table 1), we found clear evidence that 11/15 were secreted. However, only 9/29 unique genes selected by features within their amino acid sequences alone had evidence of secretion (designated as TMSC only in Table 1). The majority of these genes were found to be glycosylphosphatidylinositol-anchored or integral membrane proteins.
Figure 2.
Candidate secreted biomarkers elevated in multiple cancer types. (A) Thirty-two genes encoding secreted proteins selected by annotation- and sequence-based analyses had significant overexpression in at least one tumor-normal counterpart tissue pair (>3-fold), and significant overexpression in tumors compared with any other normal tissue (>2-fold). Counterpart tissues are tissues at the sites of tumor origin; GeneAtlas tissues are those described in ref. 11. BR, breast (ER+, ER−); CO, colorectal; GA, gastric/esophagus adenocarcinoma; KI, kidney; LI, liver; LUA, adenocarcinoma of the lung; LUS, squamous carcinoma of the lung; LUO, lung other, small cell lung carcinomas, large cell undifferentiated carcinomas of the lung; OV, ovary; PA, pancreas; PR, prostate. Gene symbols are depicted to the right. (B) An expanded view of genes preferentially up-regulated in carcinomas of the prostate. Numbers of tissue samples from each counterpart site are given in parentheses. GeneAtlas tissues are: Te, testis; Th, thyroid; Ut, uterus; SG, salivary gland; Tr, trachea; AG, adrenal gland; He, heart; Pi, pituitary gland; Sp, spinal cord; CC, cerebral cortex; Nor. Pr, normal prostate. Transcript levels were normalized in cluster and visualized in treeview (20).
Validation of Candidate Gene and Encoded Protein Overexpression in Tumor Samples.
Significant validation of our approach comes from the observation that many of the genes identified here encode secreted proteins previously shown to be dysregulated in cancer tissue (e.g., by other transcript-based approaches or IHC), or shown to be elevated in the serum from cancer patients compared with matched controls. The latter include gastrin-releasing peptide (GRP/bombesin) in lung carcinomas (22), kallikreins 6 and 10 (KLK6, KLK10) in ovarian carcinomas (23, 24), α-fetoprotein (AFP) in liver carcinomas (25), and mammaglobin A (MGBA) in breast carcinomas (26).
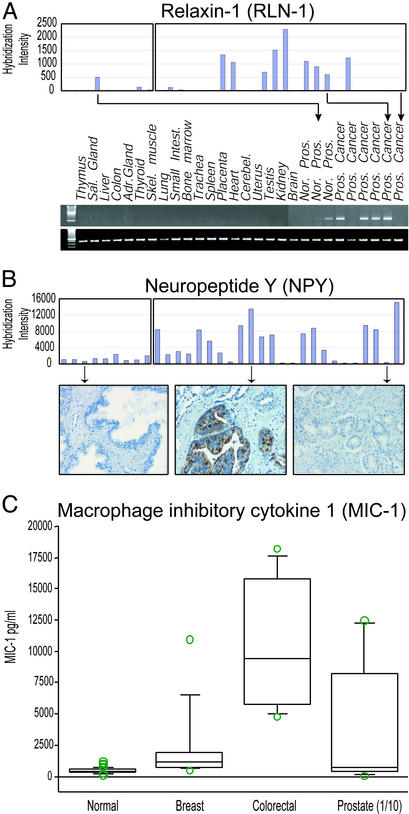
For many of the 74 genes that we discovered with this approach, we validated tumor overexpression by using several independent methods. For example, in prostate carcinomas (highlighted in Fig. 2B), we confirmed the highly tissue-selective expression of relaxin-1 (RLN1), a small peptide hormone of the insulin family involved in remodeling the birth canal (27), by semiquantitative RT-PCR in nine prostate and 19 nonprostatic tissues. Expression levels of RLN-1 determined by PCR were entirely consistent with the results obtained by microarray hybridization (Fig. 3A).
Figure 3.
Validation of microarray gene expression by RT-PCR, IHC, and ELISA. (A) RNAs from multiple different human tissues. Three normal and six primary prostate carcinomas were reverse-transcribed and amplified under standard conditions by using primers directed toward relaxin-1 (4). (Upper) The primary microarray data are shown (hybridization intensity on the y axis; samples on the x axis). (Lower) A representative PCR is shown. Primers specific for 18S were used to control for the amount of amplified cDNA. (B) IHC was performed with an anti-NPY antibody on whole tissue sections. (Upper) Primary microarray data are shown. (Lower) Examples of IHC staining in normal, microarray-positive, and microarray-negative prostate cancers. (C) Box plot showing MIC-1 serum levels in normal subjects (n = 260) and patients with breast (n = 10), colon (n = 8), and prostate (n = 9) carcinoma. P values were determined by comparison with normal by ANOVA. The prostate serum MIC-1 levels were divided by 10 (1/10) for graphical representation.
For NPY, which was highly expressed in 15/25 prostate carcinomas compared with normal prostate tissue, we stained TMAs containing 229 carcinomas and 36 normal tissue samples of diverse anatomic origin with a commercial anti-NPY antibody. In normal prostate tissues, we found staining in nerves and a few prostate secretory epithelial cells, whereas in prostate carcinomas that had high NPY gene expression, we found correspondingly high levels of protein expression (Fig. 3B). The anti-NPY antibody did not stain carcinomas of other anatomic sites on the TMAs, nor other non-neural or non-neuroendocine normal tissues that we included on these arrays. In carcinomas of other sites, we also obtained similarly consistent results by using antibodies directed against other candidate proteins. As expected from transcript analysis, antibodies specific for MUC-2 (in the group designated GO only in Table 1) showed selective expression in carcinomas of the colon, whereas antibodies specific for maspin showed high expression in carcinomas of the colon, gastroesophagous, lung, and pancreas compared with normal tissue sites (Fig. 4, which is published as supporting information on the PNAS web site). Although we did find evidence for decreased maspin expression in some ductal carcinomas of the breast, compared with normal ductal breast tissue, consistent with its purported role as a tumor suppressor in breast cancer (28), we also identified cases of breast carcinoma with significantly elevated levels of the maspin protein (Fig. 5, which is published as supporting information on the PNAS web site). In a small series of breast tumors, we found that maspin expression correlated with estrogen receptor status of the tumor, consistent with recent reports of maspin gene overexpression in estrogen receptor-negative carcinomas (29).
Candidate Genes Are Overexpressed in Independent Datasets.
Because of the relatively small number of anatomically distinct carcinoma samples that were used in our analysis, we queried candidate gene expression in other independent datasets. For example, publicly accessible data from Bhattacharjee et al. (30) enabled us to query the expression of 26 lung cancer candidate genes in 203 lung tissues, including 17 samples of normal lung, 127 adenocarcinomas, 21 squamous carcinomas, 20 pulmonary carcinoids, and six small cell undifferentiated carcinomas (Fig. 6, which is published as supporting information on the PNAS web site). This analysis demonstrated some striking correlates of gene expression with histological subtype. For example, GRP/bombesin was overexpressed predominantly in small cell lung carcinomas and carcinoids, consistent with published reports (31), and in only 2/127 adenocarcinomas; maspin, in contrast was expressed in almost all of the squamous cell carcinomas, but only in a minority of adenocarcinomas, and not at all in carcinoids. The specificity of these results is reflected by the relative lack of expression of relaxin-1 and NPY, which we included as controls based on their predominantly prostate cancer-specific expression. Compared with normal lung tissue, and based on their relative histological specificity, we suggest that proteins such as maspin, and others that remain to be validated (e.g., heparin-binding protein 17 and galectin 7) are worthy of further investigation as diagnostic biomarkers.
Serum Levels of MIC-1 Protein Are Elevated in Patients with Metastatic Cancer.
To begin to address the clinical significance of candidate genes for noninvasive diagnosis of cancer, we used a recently described ELISA (21) to evaluate the serum levels of the transforming growth factor-β superfamily cytokine, MIC-1 (32), the gene for which we found highly elevated levels of expression in prostate carcinomas and, to a lesser extent, carcinomas of the colon and breast. Using serum samples from 26 patients with metastatic breast, colorectal, and prostate carcinomas, we found significantly elevated levels of the MIC-1 protein compared with the mean basal level of the protein determined from 260 normal serum samples. Overall, we found elevated serum MIC-1 levels in 8/8, 6/10, and 8/9 patients with colorectal, breast, and prostate carcinomas, respectively. Notably, the levels of MIC-1 protein in the serum of patients with metastatic prostate carcinomas were significantly and markedly higher than those from patients with breast and colorectal carcinomas (P < 0.0001; ANOVA), consistent with the differences in the levels of gene expression that we observed in these carcinomas.
Discussion
The approach presented here was designed to exploit large cancer transcript databases to identify genes encoding secreted proteins with up-regulated expression in carcinomas relative to normal tissues. The two methods that we used, annotation-based searching and analysis of amino acid sequences for specific features, are complementary to the extent that they query fundamentally different aspects of biological knowledge. We initially focused on gene annotation alone to identify candidate genes of interest. However, the genes represented on commercially available arrays (e.g., the Affymetrix U95a GeneChip) are mostly well characterized, with defined biological roles. With the completion of the genome sequence, and the construction of arrays based increasingly on transcripts of unknown function or transcripts based on in silico predictions, annotation-based approaches will become less useful, and one must begin to rely on other computational approaches to define genes of interest. In that regard, we evaluated multiple methods to interrogate predicted amino acid sequences for relevant features; in this case, features that imply secretion of the encoded protein into the extracellular space. The two algorithms used here, sigcleave and tmap, were found to perform well and were typically easy to implement.
A significant fraction of the 74 genes identified in this study have been described by others as dysregulated in cancer (at the RNA and/or protein level) or shown to have clinical utility. Some of these show diagnostically elevated levels of secreted protein in the serum of cancer patients, and others have been shown to lead to the emergence of autocrine loops that can, or have been, targeted therapeutically. We have also shown significantly elevated levels of several of the proteins encoded by these genes in cancer tissues, as well as highly elevated levels of a candidate protein, MIC-1, by ELISA in the serum of patients with metastatic prostate, breast, and colorectal carcinomas, demonstrating the potential clinical utility of the current approach for the discovery of tissue and serum biomarkers.
A small, but growing, number of examples of successfully translating microarray-based observations of gene overexpression into candidate serum biomarkers now exist. These include osteopontin and prostasin in ovarian carcinomas (6, 7), TIMP-1 in pancreatic adenocarcinoma (33), and MIC-1 in metastatic prostate, breast, and colorectal carcinomas described here. Based on these data, we suggest many of the other proteins described herein, such as relaxin and NPY in prostate carcinomas, neuromedin U in ovarian carcinomas, Gal7 and heparin-binding protein 17 in squamous carcinomas of the lung, among others, may have similar utility (see Table 1). Interestingly, proteins such as maspin, which has to date been considered a suppressor protein (28), may also find utility as a biomarker of several cancers, most particularly for squamous carcinomas of the lung. In this tumor, we identified strong, almost uniform IHC staining, as well as high expression in an expanded set of pulmonary carcinomas (30).
Up-regulation of several of the secreted proteins described here may also result in growth stimulatory autocrine loops. Overexpression of GRP/bombesin in several cancers can stimulate growth via cognate GRP receptors, specific antagonists of which can reverse this effect (34). Our data suggest that this loop may be particularly important in carcinoid tumors of the lung, as well as small cell undifferentiated carcinomas, as described (22). The detection of elevated levels of GRP in the serum of cancer patients may help stratify those who would most benefit from anti-GRP-based therapies (if they are proven beneficial). The limitation of our approach toward identifying autocrine loops is that we have not systematically evaluated the expression of the cognate receptors, which, in many cases are not known. Graeber and Eisenberg (8) have described methods based on correlation of gene expression levels to identify ligand–receptor pairs concomitantly up-regulated in solid and hematological cancers (8). Although their approach is based on a priori knowledge of the ligand–receptor pair, we envision an approach in which one may be able to correlate ligand gene expression (i.e., the gene expression levels of the secreted proteins identified here) to the transcript levels of genes predicted to encode receptors, thereby driving discovery of novel autocrine loops in cancer.
In summary, we describe and apply an approach for identifying genes that encode secreted proteins overexpressed in human carcinomas from multiple anatomic sites. Because several of the genes are known to be dysregulated in cancer, and are diagnostically or therapeutically useful, we suggest that many of the other genes presented here, some of which we have validated, may find similar utility. Interrogation of the entire transcriptome in human cancers and normal tissues, coupled with refined methods for sequence motif searching, and biological methods for enrichment (35) are likely to yield many more proteins of importance secreted by these malignancies.
Supplementary Material
Acknowledgments
We are grateful to our colleagues at the University of Virginia and the Genomics Institute for valuable discussions, Mr. Chris Benner for critical evaluation of the methods used, and Drs. Quinn Deveraux and John Hogenesch for critical evaluation of the manuscript.
Abbreviations
- GO
Genome Ontology
- IHC
immunohistochemistry
- TMA
tissue microarray
- MIC-1
macrophage inhibitory cytokine 1
- NPY
neuropeptide Y
- GRP
gastrin-releasing peptide
References
- 1.Hanahan D, Weinberg R A. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Sporn M B, Roberts A B. Nature. 1985;313:745–747. doi: 10.1038/313745a0. [DOI] [PubMed] [Google Scholar]
- 3.Buckhaults P, Rago C, St. Croix B, Romans K E, Saha S, Zhang L, Vogelstein B, Kinzler K W. Cancer Res. 2001;61:6996–7001. [PubMed] [Google Scholar]
- 4.Welsh J B, Zarrinkar P P, Sapinoso L M, Kern S G, Behling C A, Monk B J, Lockhart D J, Burger R A, Hampton G M. Proc Natl Acad Sci USA. 2001;98:1176–1181. doi: 10.1073/pnas.98.3.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Welsh J B, Sapinoso L M, Su A I, Kern S G, Wang-Rodriguez J, Moskaluk C A, Frierson H F, Jr, Hampton G M. Cancer Res. 2001;61:5974–5978. [PubMed] [Google Scholar]
- 6.Mok S C, Chao J, Skates S, Wong K, Yiu G K, Muto M G, Berkowitz R S, Cramer D W. J Natl Cancer Inst. 2001;93:1458–1464. doi: 10.1093/jnci/93.19.1458. [DOI] [PubMed] [Google Scholar]
- 7.Kim J H, Skates S J, Uede T, Wong K K, Schorge J O, Feltmate C M, Berkowitz R S, Cramer D W, Mok S C. J Am Med Assoc. 2002;3:1671–1679. doi: 10.1001/jama.287.13.1671. [DOI] [PubMed] [Google Scholar]
- 8.Graeber T G, Eisenberg D. Nat Genet. 2001;29:295–300. doi: 10.1038/ng755. [DOI] [PubMed] [Google Scholar]
- 9.Su A I, Welsh J B, Sapinoso L M, Kern S G, Dimitrov P, Lapp H, Schultz P G, Powell S M, Moskaluk C A, Frierson H F, Jr, Hampton G M. Cancer Res. 2001;61:7388–7393. [PubMed] [Google Scholar]
- 10.Brown D A, Bauskin A R, Fairlie W D, Smith M D, Liu T, Xu N, Breit S N. BioTechniques. 2002;33:118–124. doi: 10.2144/02331rr03. [DOI] [PubMed] [Google Scholar]
- 11.Su A I, Cooke M P, Ching K A, Hakak Y, Walker J R, Wiltshire T, Orth A P, Vega R G, Sapinoso L M, Moqrich A, et al. Proc Natl Acad Sci USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashburner M, Ball C A, Blake J A, Botstein D, Butler H, Cherry J M, Davis A P, Dolinski K, Dwight S S, Eppig J T, et al. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pruitt K D, Maglott D R. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Persson B, Argos P. J Protein Chem. 1997;16:453–457. doi: 10.1023/a:1026353225758. [DOI] [PubMed] [Google Scholar]
- 15.Milpetz F, Argos P, Persson B. Trends Biochem Sci. 1995;20:204–205. doi: 10.1016/s0968-0004(00)89009-x. [DOI] [PubMed] [Google Scholar]
- 16.von Heijne G. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duffy M J, Shering S, Sherry F, McDermott E, O'Higgins N. Int J Biol Markers. 2000;15:330–333. doi: 10.1177/172460080001500410. [DOI] [PubMed] [Google Scholar]
- 18.Lockhart M S, Waldner C, Mongini C, Gravisaco M J, Casanova S, Alvarez E, Hajos S. Oncol Rep. 1999;6:1129–1133. doi: 10.3892/or.6.5.1129. [DOI] [PubMed] [Google Scholar]
- 19.Kersten M J, Evers L M, Dellemijn P L, van den Berg H, Portegies P, Hintzen R Q, van Lier R A, von dem Borne A E, van Oers R H. Blood. 1996;87:1985–1989. [PubMed] [Google Scholar]
- 20.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore A G, Brown D A, Fairlie W D, Bauskin A R, Brown P K, Munier M L, Russell P K, Salamonsen L A, Wallace E M, Breit S N. J Clin Endocrinol Metab. 2000;85:4781–4788. doi: 10.1210/jcem.85.12.7007. [DOI] [PubMed] [Google Scholar]
- 22.Heasley L E. Oncogene. 2001;20:1563–1569. doi: 10.1038/sj.onc.1204183. [DOI] [PubMed] [Google Scholar]
- 23.Diamandis E P, Yousef G M, Soosaipillai A R, Bunting P. Clin Biochem. 2000;33:579–583. doi: 10.1016/s0009-9120(00)00182-x. [DOI] [PubMed] [Google Scholar]
- 24.Luo L Y, Bunting P, Scorilas A, Diamandis E P. Clin Chim Acta. 2001;306:111–118. doi: 10.1016/s0009-8981(01)00401-6. [DOI] [PubMed] [Google Scholar]
- 25.Johnson P J. Clin Liver Dis. 2001;5:145–159. doi: 10.1016/s1089-3261(05)70158-6. [DOI] [PubMed] [Google Scholar]
- 26.Fleming T P, Watson M A. Ann NY Acad Sci. 2000;923:78–89. doi: 10.1111/j.1749-6632.2000.tb05521.x. [DOI] [PubMed] [Google Scholar]
- 27.Bani D. Gen Pharmacol. 1997;28:13–22. doi: 10.1016/s0306-3623(96)00171-1. [DOI] [PubMed] [Google Scholar]
- 28.Sager R, Sheng S, Pemberton P, Hendrix M J. Adv Exp Med Biol. 1997;425:77–88. [PubMed] [Google Scholar]
- 29.Martin K J, Kritzman B M, Price L M, Koh B, Kwan C P, Zhang X, Mackay A, O'Hare M J, Kaelin C M, Mutter G L, et al. Cancer Res. 2000;60:2232–2238. [PubMed] [Google Scholar]
- 30.Bhattacharjee A, Richards W G, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et al. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sunday M E, Choi N, Spindel E R, Chin W W, Mark E J. Hum Pathol. 1991;22:1030–1039. doi: 10.1016/0046-8177(91)90011-d. [DOI] [PubMed] [Google Scholar]
- 32.Bootcov M R, Bauskin A R, Valenzuela S M, Moore A G, Bansal M, He X Y, Zhang H P, Donnellan M, Mahler S, Pryor K, et al. Proc Natl Acad Sci USA. 1997;94:11514–11519. doi: 10.1073/pnas.94.21.11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou W, Sokoll L J, Bruzek D J, Zhang L, Velculescu V E, Goldin S B, Hruban R H, Kern S E, Hamilton S R, Chan D W, et al. Cancer Epidemiol Biomarkers Prev. 1998;7:109–112. [PubMed] [Google Scholar]
- 34.de Castiglione R, Gozzini L. Crit Rev Oncol Hematol. 1996;24:117–151. doi: 10.1016/1040-8428(96)00220-x. [DOI] [PubMed] [Google Scholar]
- 35.Diehn M, Eisen M B, Botstein D, Brown P O. Nat Genet. 2000;25:58–62. doi: 10.1038/75603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.