Abstract
ARC1 is a novel U-box protein required in the Brassica pistil for the rejection of self-incompatible pollen; it functions downstream of the S receptor kinase (SRK). Here, we show that ARC1 has E3 ubiquitin ligase activity and contains several motifs that influence its subcellular localization. ARC1 can shuttle between the nucleus, cytosol, and proteasome/COP9 signalosome (CSN) when expressed in tobacco BY-2 suspension-cultured cells. However, ARC1 localization to the proteasome/CSN occurs only in the presence of an active SRK. In the pistil, ubiquitinated protein levels increase specifically with incompatible pollinations, but they do not change in ARC1 antisense-suppressed pistils. In addition, inhibition of the proteasomal proteolytic activity disrupts the self-incompatibility response. We propose that ARC1 promotes the ubiquitination and proteasomal degradation of compatibility factors in the pistil, which in turn leads to pollen rejection.
INTRODUCTION
Pollination and fertilization in flowering plants involve a complex series of events with tightly regulated cell–cell interactions and signaling between the pollen and the pistil. This process typically starts with pollen recognition and adhesion to the stigmatic surface of the pistil, followed by pollen hydration, germination, and pollen tube growth through the pistil to the ovules, where fertilization occurs (reviewed by Lord and Russell, 2002). In many flowering plants, there is an added layer of complexity in which mechanisms exist to prevent self-fertilization and avoid inbreeding. One such mechanism is self-incompatibility, which enables the plant to discriminate between “self” and “nonself” pollen, with the subsequent rejection of self pollen (reviewed by Stone and Goring, 2001; Wheeler et al., 2001).
In Brassica, the self-incompatibility response causes the inhibition of self pollen early after pollen contact with the pistil; consequently, pollen tube growth beyond the pistil surface is prevented. The genetic system that regulates this form of self-incompatibility is multi-allelic and requires that both the pistil and the pollen-producing anther share the same self-incompatibility alleles for pollen rejection. There are two tightly linked multi-allelic genes that act as the primary determinants of Brassica self-incompatibility and encode components of a ligand–receptor kinase complex (reviewed by Silva and Goring, 2001; Kachroo et al., 2002). The male determinant of self-incompatibility is the SCR/SP11 ligand, a small pollen coat protein present on the surface of the pollen grain (Schopfer et al., 1999; Takayama et al., 2000). The female determinant of self-incompatibility is the S receptor kinase (SRK), a receptor Ser/Thr kinase, which is localized to the plasma membrane of the stigmatic papillae on the surface of the pistil (Giranton et al., 2000; Takasaki et al., 2000; Silva et al., 2001). An allele-specific interaction between SCR/SP11 and SRK has been demonstrated and proposed to cause a signaling cascade in the pistil that leads to pollen rejection (Kachroo et al., 2001; Takayama et al., 2001). How the SCR/SP11 interaction with SRK leads to receptor activation and induces the self-incompatible response is not well characterized.
We previously conducted a yeast two-hybrid screen for SRK kinase domain–interacting proteins to begin to dissect the SRK signaling pathway. Two of three interactors isolated, THL1 and THL2, encoded members of the thioredoxin h family and subsequently were shown to act as inhibitors of SRK in the absence of SCR/SP11 ligand (Bower et al., 1996; Cabrillac et al., 2001). The binding of the ligand to SRK results in the alleviation of THL1's inhibition of SRK and the subsequent activation of the kinase domain (Cabrillac et al., 2001). The third interacting protein isolated, ARC1, interacts with the kinase domain of SRK in a phosphorylation-dependent manner via a C-terminal arm repeat domain and is phosphorylated in turn by SRK (Gu et al., 1998). In addition, ARC1 is known to be a positive regulator of the self-incompatibility system, because suppression of ARC1 transcripts in the pistils of self-incompatible Brassica napus W1 plants resulted in a partial breakdown in the self-incompatibility response (Stone et al., 1999).
However, questions remain regarding the cellular function of ARC1 and the mechanisms by which it participates in the SRK signaling pathway to bring about self pollen inhibition. To begin to address these issues, we used a transient expression assay to characterize several different protein motifs within ARC1 and their roles in mediating the intracellular localization of wild-type and mutant forms of ARC1 under various conditions. In addition, we examined the involvement of the ubiquitin/proteasome system in the Brassica self-incompatibility response. Our results indicate that during self-incompatibility, SRK uses, through ARC1, ubiquitin-dependent proteasomal protein degradation to promote pollen rejection; this also illustrates a novel mechanism for receptor kinase signaling.
RESULTS
The Subcellular Localization of ARC1 Is Altered in the Presence of an Active SRK
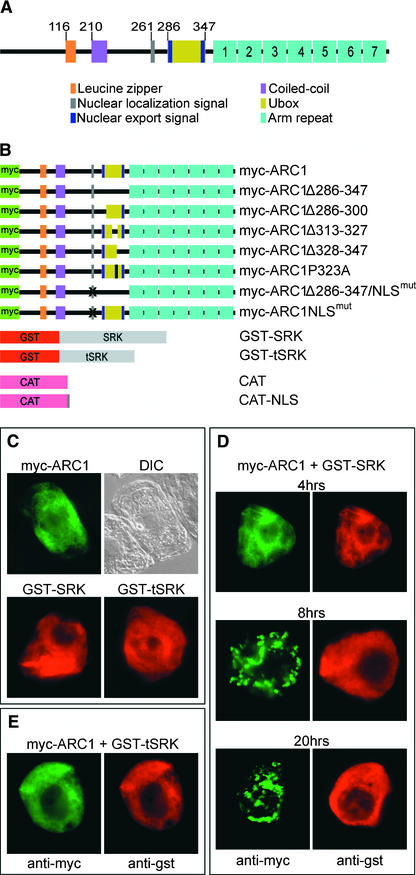
As a first step in understanding how ARC1 causes pollen rejection, we searched for protein motifs that might define its cellular function. Previously, ARC1 was found to contain a C-terminal arm repeat domain and a U-box motif that is a modified ring finger (Gu et al., 1998; Aravind and Koonin, 2000) (Figure 1A). ARC1 also contains several other putative protein motifs, including a Leu zipper, a coiled-coil domain, a nuclear localization signal (NLS), and two nuclear export signals (NESs) (Figure 1A). To determine if these various motifs play roles in defining the intracellular localization of ARC1, we used tobacco BY-2 suspension-cultured cells as a transient expression system followed by immunofluorescence microscopy. The BY-2 cell culture system is well characterized (Nagata et al., 1992); compared with time-consuming and more difficult Brassica transformation methods, BY-2 cells are a far more viable system for testing a variety of ARC1 constructs modified by site-directed mutagenesis. In addition, the Brassica stigmatic papillae contain a large central vacuole that compresses the cytosol and organellar compartments against the plasma membrane and cell wall, making microscopic analyses and subcellular localization studies difficult.
Figure 1.
ARC1 and SRK Constructs Tested in Tobacco BY-2 Cells, and the Subcellular Localization of myc-ARC1 and GST-SRK.
(A) Predicted protein motifs in ARC1. Numerals at top indicate the positions of the motifs in the amino acids sequence.
(B) Schemes of the various ARC1 and SRK fusion proteins examined in BY-2 cells.
(C) Subcellular localization of transiently expressed myc-ARC1, GST-SRK, and GST-tSRK in BY-2 cells at 20 h after biolistic bombardment. Cytosolic localization is observed for all proteins expressed individually. A differential interference contrast (DIC) image of the myc-ARC1–expressing cell is shown.
(D) Time-course study of BY-2 cells cobombarded with myc-ARC1 and GST-SRK and allowed to express proteins for 4, 8, and 20 h. myc-ARC1 is localized to the cytosol at 4 h and then relocalizes to several distinct subcellular sites by 8 h and remains there at 20 h. Cytosolic localization is observed for GST-SRK in all cells.
(E) Control cotransformation of BY-2 cells with myc-ARC1 and GST-tSRK at 20 h. Cytosolic localization is observed for both myc-ARC1 and the inactive form of GST-SRK.
BY-2 cells were transformed by biolistic bombardment with constructs containing either wild-type or mutated forms of ARC1 under the control of the 35S promoter of Cauliflower mosaic virus (Figure 1B). The myc epitope tag was fused to the N terminus of ARC1 (myc-ARC) to allow immunodetection of the protein and to avoid any potential cross-hybridization with a large family of ARC1-related proteins in plants (Azevedo et al., 2001). Immunofluorescence microscopic analyses of all myc-ARC1 proteins were performed at 20 h after bombardment unless stated otherwise. To ensure that the intracellular location(s) of ARC1 was not affected by the addition of the myc epitope, transient transformations also were performed using either N-terminal hemagglutinin epitope–tagged ARC1 or a chloramphenicol acetyltransferase (CAT)–ARC1 fusion protein, and in all cases, results similar to those presented for myc-ARC1 were obtained (data not shown).
Figure 1C shows that at 20 h after biolistic bombardment, transiently expressed myc-ARC1 was distributed throughout the cytosol of an individual BY-2 cell (Figure 1C). However, the subcellular localization of the protein was altered dramatically when a glutathione S-transferase (GST)–SRK fusion protein was coexpressed in the same cell. GST-SRK consists of the kinase domain from SRKA14, starting just after the transmembrane domain, and is constitutively active and capable of interacting with and phosphorylating ARC1 in vitro (Gu et al., 1998). A time course study of myc-ARC1/GST-SRK–cotransformed cells revealed that myc-ARC1 was present throughout the cytosol at 4 h after bombardment but was localized to another distinct subcellular site by 8 h and remained so at 20 h after bombardment (Figure 1D). The redistribution of ARC1 in BY-2 cells that coexpressed GST-SRK required SRK kinase activity, because cotransformations with a truncated and inactive SRK, GST-tSRK, did not affect the cytosolic localization of myc-ARC1 (Figure 1E). Both GST-SRK and GST-tSRK were observed in all cells to be distributed exclusively throughout the cytosol (Figures 1C to 1E).
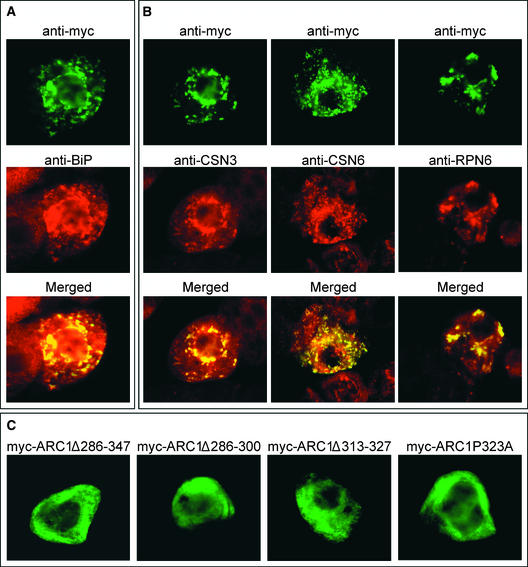
ARC1 Colocalizes with an Endoplasmic Reticulum Protein and Subunits of the Proteasome and COP9 Signalosome in the Presence of an Active SRK, and This Localization Is U-Box Dependent
To identify the subcellular structures that myc-ARC1 relocalized to from the cytosol in GST-SRK–coexpressing cells, we first compared the immunofluorescence staining patterns of myc-ARC1 with those of several resident enzymes/proteins that specifically mark various plant cell organelles. No apparent colocalization was observed between myc-ARC1 and marker proteins for peroxisomes, mitochondria, or Golgi (data not shown). However, the fluorescence attributable to myc-ARC1 in the presence of GST-SRK resembled the fluorescence attributable to the lumenal binding protein (BiP), a well-established resident of the endoplasmic reticulum (ER) (Munro and Pelham, 1986) (Figure 2A). Interestingly, the pattern of BiP fluorescence in transformed cells appeared to be altered from that of the surrounding untransformed cells, suggesting that BiP was recruited to myc-ARC1–containing structures. The localization of endogenous BiP was not altered in either BY-2 cells cobombarded with myc-ARC1 and GST-tSRK or in various control bombardments, including mock transformations (data not shown). Likewise, the omission of primary antibodies did not yield any fluorescence, as expected (data not shown).
Figure 2.
myc-ARC1 in BY-2 Cells Coexpressing GST-SRK Colocalizes with an ER Protein and Subunits of the Proteasome/CSN.
(A) After cotransformation with GST-SRK, myc-ARC1 colocalizes with the endogenous ER marker protein BiP, as illustrated by the merged image.
(B) After cotransformation with GST-SRK, myc-ARC1 colocalizes with the endogenous subunits of the CSN, CSN3 and CSN6, and with the 26S proteasomal subunit, RPN6, as illustrated by the merged images.
(C) The ARC1 U-box is required for the localization of myc-ARC1 to the proteasome/CSN. After cotransformation with GST-SRK, myc-ARC1Δ286-347, myc-ARC1Δ286-300, myc-ARC1Δ313-327, and myc-ARC1P323A remain cytosolic and do not sort to the proteasome/CSN, as seen for myc-ARC1. GST-SRK is localized to the cytosol in all transformed cells (data not shown).
The presence of a U-box in ARC1 and the fact that the cytosolic face of ER membranes is a major site for proteasomal protein degradation (Sommer et al., 2001) led us to postulate that ARC1 may participate in the ubiquitin/proteasome pathway and that the distinct subcellular sites at which myc-ARC1 localizes represent proteasomes adjacent to the regions of the ER at which lumenal BiP accumulates (Enenkel et al., 1998). To examine this possibility, BY-2 cells were cotransformed with myc-ARC1 and GST-SRK, and the localization of myc-ARC1 was compared with that of the endogenous proteasomal proteins RPN6, CSN3, and CSN6. RPN6 is a subunit of the 19S regulatory particle or “lid” of the 26S proteasome (Ferrell et al., 2000), whereas CSN3 and CSN6 are subunits of the COP9 signalosome (CSN), a multiprotein complex with similarity to the 26S proteasome lid (Peng et al., 2001a, 2001b). Figure 2B shows that the immunofluorescence patterns attributable to myc-ARC1 and CSN3, CSN6, or RPN6 were identical, indicating that myc-ARC1 was localized to the proteasome/CSN. As with BiP, the CSN3, CSN6, and RPN6 staining patterns in myc-ARC1/GST-SRK–coexpressing cells were altered compared with the staining in adjacent untransformed cells, suggesting that there is an enrichment of proteasomes/CSNs at certain sites of the ER where lumenal BiP also accumulated. Because of the lack of an ER targeting signal in ARC1, it is unlikely that the protein is in the ER lumen; it is more likely located on the outside of the ER with the proteasome/CSN. In addition, myc-ARC1 did not appear to be degraded by the proteasome/CSN, because myc-ARC1 fluorescence remained unchanged when examined at 48 h after bombardment (data not shown).
U-box domains are highly conserved among fungi, plants, and animals and are required for the E3 ubiquitin ligase activity of U-box proteins (Aravind and Koonin, 2000; Patterson, 2002). Therefore, we investigated the role of ARC1's U-box in mediating the proteasome/CSN localization of myc-ARC1 in the presence of the active SRK. Deletions in myc-ARC1 of either the entire U-box (myc-ARC1Δ286-347) or smaller portions of the U-box (myc-ARC1Δ286-300 and myc-ARC1Δ313-327) as well as the substitution (with Ala) of a conserved Pro residue in the U-box (myc-ARC1ΔP323A) all resulted in the cytosolic localization of altered myc-ARC1 when coexpressed with GST-SRK (Figure 2C). Overall, these results indicate that an intact U-box motif is required for the localization of myc-ARC1 from the cytosol to proteasomes/CSNs at ER membranes and that the phosphorylation of myc-ARC1 by GST-SRK likely is involved in this sorting process.
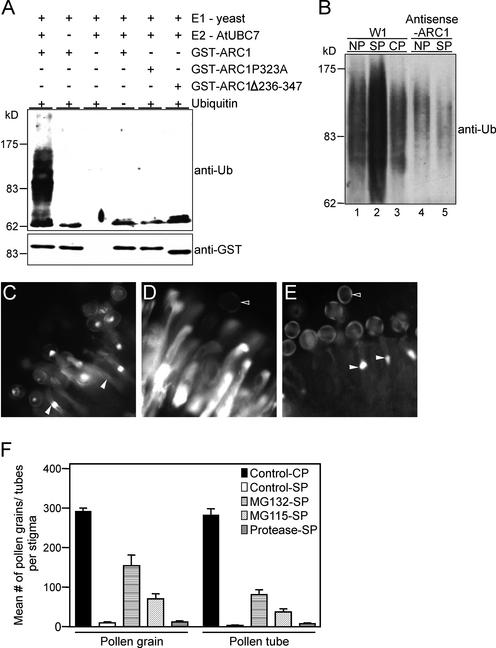
ARC1 Mediates Protein Ubiquitination, and the Proteasomal Function Is Required for Self-Incompatibility
Protein ubiquitination generally involves three enzymes: the ubiquitin-activating enzyme (E1), the ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3) (Sommer et al., 2001). To determine whether ARC1 functions as an E3 ligase and can mediate protein ubiquitination, in vitro ubiquitination assays were performed using a commercially available yeast E1 enzyme, a His-tagged Arabidopsis E2 enzyme, UBC7 (van Nocker et al., 1996), and a GST-tagged ARC1. The bacterial proteins present in the unpurified E2 enzyme extracts serve as substrates for ubiquitination in this assay (Hatakeyama et al., 1997; Lorick et al., 1999). Under these in vitro conditions, E3 ligases are able to use bacterial proteins as substrates, and any potential for cross-contamination is removed because Escherichia coli does not contain a ubiquitination system (Hatakeyama et al., 1997; Lorick et al., 1999). Figure 3A shows that GST-ARC1 combined with the E1 and E2 enzymes mediated the polyubiquitination of proteins, as evident by the multiple high molecular mass proteins detected with anti-ubiquitin antibodies. However, ARC1 does not appear to be polyubiquitinated, because only a single band of the expected size for GST-ARC1 was detected (Figure 3A). Omission of any of the components, as well as deletion of the ARC1 U-box motif or mutation of the conserved Pro, resulted in a loss of protein ubiquitination (Figure 3A). Thus, ARC1 possesses U-box–dependent E3 ubiquitin ligase activity.
Figure 3.
ARC1 Has E3 Ubiquitin Ligase Activity and Mediates Ubiquitination in the Self-Incompatibility Response.
(A) ARC1 has U-box–dependent E3 ligase activity in vitro. For the in vitro ubiquitination assay, GST-ARC1 was tested with the yeast E1 ubiquitin-activating enzyme and the Arabidopsis E2 ubiquitin-conjugating enzyme (UBC7). The presence of all three enzymes leads to the polyubiquitination of bacterial proteins (detected with the anti-ubiquitin antibody [anti-Ub]). No ubiquitination is observed with the ARC1 U-box mutant constructs GST-ARC1Δ286-347 and GST-ARC1P323A. GST-ARC1 is not polyubiquitinated, because only a single polypeptide is detectable with the anti-GST antibodies.
(B) Ubiquitinated protein levels increase in B. napus pistils after self-incompatible pollination. Self-incompatible W1 pollination of W1 pistils (SP) increases the levels of ubiquitinated proteins compared with that of nonpollinated (NP) and compatible Westar pollinated (CP) W1 pistils. In ARC1 antisense-suppressed W1 pistils, the levels of ubiquitinated proteins are not affected significantly after self-pollination (SP). Total protein was extracted from pollinated and nonpollinated stigmas and used in protein gel blot analysis with anti-ubiquitin antibodies. The Bio-Rad protein reagent assay and Coomassie blue gel staining were used to determine protein concentrations for equal loading in each lane.
(C) to (E) Proteasomal inhibitors disrupt the self-incompatible response. Fluorescence microscopic analyses of pollen germination and tube growth in compatible pollination of a W1 pistil with Westar pollen (C), self-incompatible pollination of a W1 pistil with W1 pollen (D), and self-incompatible pollination of a W1 pistil treated with the proteasomal inhibitor MG132 before pollination with untreated W1 pollen (E). Closed arrowheads indicate growing pollen tubes that successfully penetrate the stigmatic surface. Open arrowheads indicate inhibited pollen grains.
(F) Mean number of pollen grains adhering to the stigmatic papillae and pollen tubes growing beyond the stigmatic surface after treatment of pistils with proteasomal inhibitors. Controls include compatible pollination of water/DMSO-treated W1 pistils with Westar pollen (CP) and self-incompatible W1 pollination of water/DMSO-treated W1 pistils (SP). W1 pistils were treated with a proteasomal inhibitor, either MG132 or MG115, before self-incompatible pollination with untreated W1 pollen (MG132-SP and MG115-SP). As a control for any potential nonspecific effects, W1 pistils also were treated with a cocktail of general protease inhibitors before self-incompatible pollination with untreated W1 pollen (Protease-SP). Both MG132 and MG115 cause significant breakdown in the rejection of self-incompatible pollen.
We next examined the levels of ubiquitinated proteins in pollinated and nonpollinated pistils from wild-type B. napus W1 and ARC1 antisense-suppressed W1 plants (Figure 3B). W1 pistils were pollinated with either self-incompatible pollen or compatible pollen from self-compatible B. napus cv Westar plants. A strong increase in the levels of ubiquitinated proteins was observed in self-incompatible pollinated pistils compared with that in the nonpollinated or compatible pollinated W1 pistils (Figure 3B). However, an increase in the levels of ubiquitinated proteins was not observed with the ARC1 antisense-suppressed W1 pistils after self-pollination (Figure 3B).
To determine if the ubiquitination-mediated protein degradation by the proteasome was required for self-incompatibility, we studied the effects of two proteasomal inhibitors, Cbz-leu-leul-leucinal (MG132) and Cbz-leu-leul-norvalinal (MG115), on pollinated Brassica napus pistils. Both MG132 and MG115 effectively inhibit the proteolytic activity of the 26S proteasome (Rock et al., 1994). In control compatible pollinations, W1 pistils contained, on the stigmatic surface, a large number of pollen grains that had germinated and produced pollen tubes growing down the pistil (Figures 3C and 3F). These events were inhibited in the control W1 pistils after self-incompatible W1 pollination; very few pollen grains adhered to the stigmatic surface, and any resulting pollen tubes were unable to grow beyond the stigma surface (Figures 3D and 3F). However, treatment of W1 pistils with MG132 followed by pollination with untreated self-incompatible W1 pollen resulted in a breakdown in the self-incompatibility response; approximately 15 times more pollen grains adhered to the MG132-treated stigmatic surface compared with the control self-incompatible pollination (Figures 3E and 3F). Of the pollen grains that adhered to the MG132-treated W1 pistils, ∼50 to 60% germinated and produced a pollen tube that grew beyond the stigmatic surface (Figure 3F). A similar but less pronounced effect was observed with MG115 (Figure 3F). This effect was specific to proteasomal inhibitors, because self-incompatibility remained intact in pistils treated with a cocktail of general protease inhibitors (Figure 3F). In addition, the effect of MG132 and MG115 was specific to the self-incompatibility response and did not affect compatible pollinations (data not shown). Together, these results indicate that proteasomal protein degradation is involved in the rejection of self-incompatible pollen on Brassica pistils.
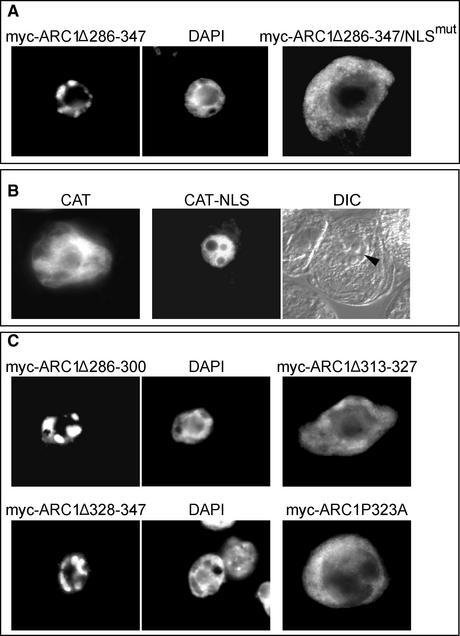
Nuclear Localization and Export of ARC1
During the examination of the localization of the various myc-ARC1 U-box deletion constructs, we observed that under certain conditions, myc-ARC1 localized to the nucleus rather than the cytosol. For instance, when myc-ARC1 with its entire U-box deleted (ARC1Δ286-347) was either transformed alone (Figure 4A) or cotransformed with the inactive GST-tSRK (data not shown), the modified ARC1 protein localized entirely to the nucleus. This observed nuclear localization of myc-ARC1Δ286-347 was not entirely unexpected, because ARC1 contains a putative NLS (260KKqRKR266) that is similar to the monopartite SV40-type NLS (Bogerd et al., 1996). Indeed, direct evidence that the ARC1 NLS can mediate nuclear localization was obtained when substitution of the Lys residues with Ala residues within the NLS of the U-box–deleted myc-ARC1 (myc-ARC1Δ286-347/NLSmut) abolished nuclear localization (Figure 4A). In addition, fusion of the ARC1 NLS to the C terminus of CAT was sufficient to redirect the reporter protein from the cytosol to the nucleus (Figure 4B).
Figure 4.
Nuclear Localization and Export Signals Affect the Subcellular Location of ARC1.
(A) Nuclear localization of the ARC1 U-box deletion mutant myc-ARC1Δ286-347. The nucleus of the myc-ARC1Δ286-347–expressing cell is shown in the corresponding 4′,6-diamidino-2-phenylindole (DAPI)–stained image. myc-ARC1Δ286-347/NLSmut, in which the U-box is deleted and the NLS is mutated, is localized exclusively to the cytosol.
(B) The NLS of ARC1 is capable of redirecting the reporter protein CAT from the cytosol to the nucleus. CAT alone is localized predominantly in the cytosol, whereas the fusion protein CAT-NLS is localized exclusively in the nucleus. The position of the nucleus (arrowhead) in the CAT-NLS–expressing cell is apparent in the corresponding differential interference contrast (DIC) image of the same cell.
(C) Deletion of different portions of the ARC1 U-box–containing putative NESs results in the nuclear accumulation of both myc-ARC1Δ286-300 and myc-ARC1Δ328-347. The DAPI-stained nucleus is shown at right of each cell. Cytosolic localization is observed when another section of the ARC1 U-box, myc-ARC1Δ313-327, is deleted or a conserved Pro in the U-box, myc-ARC1ΔP323A, is mutated.
Because myc-ARC1 with its U-box deleted was localized to the nucleus, the predominantly cytosolic localization observed for intact myc-ARC1 (Figure 1C) could be attributable in part to topogenic signals that are located in the U-box region and that mediate nuclear export and/or cytosolic retention. Upon inspection of the amino acid sequence in ARC1's U-box, we identified two Leu/Ile-rich regions that may function as NESs. One of these two amino acid sequences, 332LSFVSNLAL340, matches well with the consensus Rev/Rex-type NES [L-X(2-3)-L/P/I/V/M-X(2-3)-L-X-L/I] (Bogerd et al., 1996). Figure 4C shows that when these two regions were deleted independently, the resulting myc-ARC1 mutant proteins (myc-ARC1Δ286-300 and myc-ARC1Δ328-347) accumulated exclusively in the nucleus. However, when other portions of the ARC1 U-box were modified so that the two putative NESs remained intact, the resulting myc-ARC1 mutants (myc-ARC1Δ313-327 and myc-ARC1P323A) were localized to the cytosol (Figure 4C). These results indicate that the two regions within the ARC1 U-box predicted to contain NESs, amino acids 286 to 300 and 328 to 347, both are required for the cytosolic localization of ARC1 and that the disruption of either region redirects ARC1 to the nucleus.
Cytosolic Localization of ARC1 Is Maintained by the XPO1 Nuclear Export Pathway
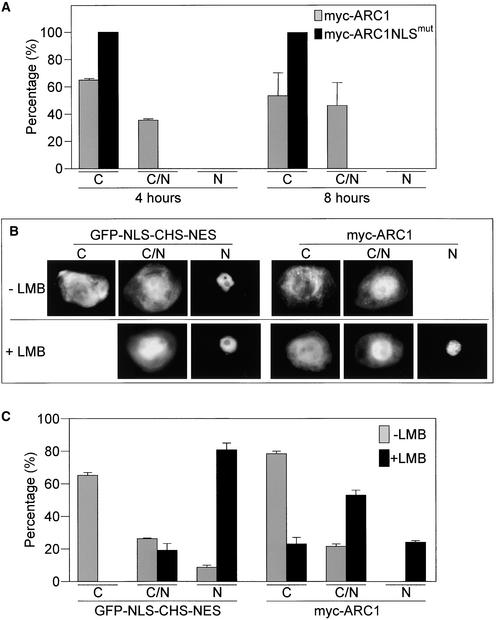
Given that ARC1 contains topogenic signals for both nuclear localization and export, we reexamined the distribution of intact myc-ARC1 at earlier times after biolistic bombardment to determine if ARC1 actually entered the nucleus. Compared with 20 h after bombardment, when myc-ARC1 was localized exclusively to the cytosol (Figure 1C), at 4 and 8 h after bombardment, myc-ARC1 was distributed in both the cytosol and the nucleus in ∼35 to 50% of the transformed cells (Figure 5A). However, myc-ARC1NLSmut was cytosolic in all transformed cells examined (Figure 5A). These data indicate that myc-ARC1 does enter the nucleus, but because the protein tends to accumulate over time predominantly in the cytosol, it may be actively excluded from the nucleus.
Figure 5.
Nuclear Localization of ARC1.
(A) myc-ARC1 accumulates in the nucleus in a percentage of transformed cells at 4 and 8 h after bombardment. No nuclear localization is observed for myc-ARC1NLSmut. C, cytosolic localization; C/N, cytosolic and nuclear localization; N, nuclear localization.
(B) and (C) Treatment with the nuclear export inhibitor LMB promotes the accumulation of myc-ARC1 in the nucleus.
(B) BY-2 cells were transformed with myc-ARC1 and then incubated with either LMB (dissolved in ethanol) or with ethanol alone. Cells were analyzed at 4 h after biolistic bombardment. The fusion protein GFP-NLS-CHS-NES served as a positive control.
(C) Percentage of transformed BY-2 cells at 4 h after bombardment in the presence or absence of LMB displaying localization of either myc-ARC1 or GFP-NLS-CHS-NES in the cytosol only (C), in the cytosol and the nucleus (C/N), or in the nucleus only (N).
A number of NES-containing proteins in yeast, plants, and mammals undergo nuclear export via the XPO1-dependent nuclear export pathway (Ossareh-Nazari et al., 1997; Stade et al., 1997; Haasen et al., 1999). To determine if the nuclear export of ARC1 uses this pathway, we evaluated the localization of myc-ARC1 in the presence of leptomycin B (LMB), a specific inhibitor of the XPO1 nuclear export receptor, Exportin1/Crm1p (Ossareh-Nazari et al., 1997). The fusion protein green fluorescent protein (GFP)-NLS-chalcone synthase (CHS)-NES, which contains the SV40 large T antigen NLS and the HIV-1 protein Rev NES, was used as a positive control in these experiments (Haasen et al., 1999). Figures 5B and 5C show that transiently expressed GFP-NLS-CHS-NES, in the absence of LMB, was localized, as expected, to the cytosol in the majority (70%) of transformed cells. However, when the XPO1 nuclear export pathway was blocked with LMB, GFP-NLS-CHS-NES relocalized to the nucleus in most (80%) of the transformed cells (Figures 5B and 5C). Similarly, treatment with LMB altered the subcellular localization pattern of myc-ARC1 from being predominantly cytosolic to nuclear. That is, >75% of the transformed cells showed myc-ARC1 either partially or completely localized to the nucleus in the presence of LMB (Figures 5B and 5C).
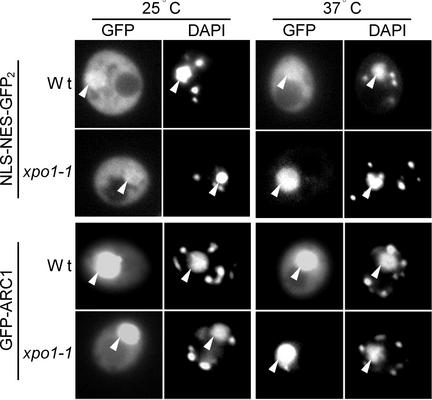
XPO1-mediated nuclear export of ARC1 also was studied in yeast cells containing a xpo1-1 temperature-sensitive mutation (Stade et al., 1997). GFP-tagged ARC1 as well as a NLS-NES-GFP2 control construct were transformed independently into both wild-type and xpo1-1 temperature-sensitive yeast. Figure 6 shows that NLS-NES-GFP2 was localized primarily to the cytosol in wild-type and xpo1-1 cells at the permissive temperature of 25°C. At the nonpermissive temperature of 37°C, NLS-NES-GFP2 remained mainly cytosolic in wild-type cells but was localized almost exclusively to the nucleus in xpo1-1 cells (Figure 6). At 25°C, GFP-ARC1 was distributed between the cytosol and the nucleus, with greater accumulation of the protein in the nucleus in both wild-type and xpo1-1 cells. The increased nuclear accumulation of GFP-ARC1 in yeast cells is in contrast to the predominantly cytosolic localization observed for either myc-ARC1 (Figures 1C and 5C) or GFP-ARC1 (data not shown) in BY-2 cells and suggests that the ARC1 NLS is more active than the NESs in yeast. Nevertheless, at 37°C, the pattern of GFP-ARC1 localization was unchanged compared with that at 25°C in wild-type cells; however, it was exclusively nuclear in xpo1-1 cells (Figure 6), confirming that ARC1 is actively excluded from the nucleus via the XPO1 nuclear export pathway.
Figure 6.
ARC1 Is Exported from the Nucleus in Yeast via the XPO1 Nuclear Export Pathway.
The reporter constructs NLS-NES-GFP2 and GFP-ARC1 were expressed individually in either wild-type (Wt) or temperature-sensitive xpo1-1 yeast cells and grown at either 25°C or shifted to the nonpermissive temperature of 37°C. Both NLS-NES-GFP2 and GFP-ARC1 are localized strictly to the nucleus in xpo1-1 yeast grown at 37°C, at which temperature XPO1 nuclear export is blocked. Nuclei are visualized by 4′,6-diamidino-2-phenylindole (DAPI) staining. Note that DAPI stains both nuclear and mitochondrial DNA in living yeast cells. Arrows indicate the colocalization of the GFP reporter protein and DAP1 staining in the nucleus of the same cell.
DISCUSSION
Receptor kinase signaling is emerging as essential in many aspects of plant growth and development. Several hundred putative receptor kinase genes have been identified in the Arabidopsis genome, all with Ser/Thr kinase domains, and are grouped into several different subclasses based on their predicted extracellular domains (Shiu and Bleecker, 2001). However, only a few receptor kinases, such as CLV1, BRI1, FLS2, and SRK, have been well studied with ligands and some downstream signaling components identified (Becraft, 2002; Gomez-Gomez and Boller, 2002; Kachroo et al., 2002).
ARC1 was isolated previously through an interaction screen in which it was found to bind to the phosphorylated form of SRK and to function downstream of SRK to cause self-incompatible pollen rejection (Gu et al., 1998; Stone et al., 1999). However, the manner in which ARC1 promotes the self-incompatible response was not known. In this study, we found that ARC1 contains several different motifs that, depending on the presence or absence of an active SRK, influence the subcellular localization of the protein. For instance, in the absence of an active SRK, ARC1 enters the nucleus, likely via an NLS, but the protein is predominantly cytosolic as a result of the action of at least two NESs and the XPO1 nuclear export pathway. On the other hand, modified ARC1 lacking its NESs is localized in the cytosol when coexpressed with an active SRK. This finding suggests that the phosphorylation of ARC1 by SRK inhibits the targeting function of the NLS. The presence of an active SRK, as well as the U-box in ARC1, is important for the localization of ARC1 to the 26S proteasome/CSN. Again, phosphorylation of ARC1 by SRK may be responsible for this sorting. Overall, the varied localization of ARC1 raises the question of what function(s) the protein serves at each of these different subcellular sites with respect to the self-incompatibility response.
The purpose of ARC1 being redirected from the cytosol to the proteasome/CSN in the presence of an active SRK is not to degrade ARC1 itself, because there was no evidence that the fluorescence attributable to proteasome/CSN-localized myc-ARC1 was diminished (Figure 2B). In addition, previous work with ARC1 antisense-suppressing plants (Stone et al., 1999) revealed that the loss of ARC1 causes a breakdown in self-incompatibility, which is contrary to what would be predicted if ARC1 was being degraded. Given that ARC1 has E3 ubiquitin ligase activity, the more plausible explanation is that the protein carries ubiquitinated substrates to the proteasome/CSN for their subsequent degradation. This explanation is consistent with the increased levels of ubiquitinated proteins detected after self-incompatible pollinations and with the concomitant decrease of ubiquitinated proteins in ARC1 antisense-suppressed pistils.
The multiple ubiquitinated bands detected after self-incompatible pollination do not necessarily reflect the number of substrates being ubiquitinated, because individual proteins typically are modified with varying numbers of ubiquitin molecules to produce a range of higher molecular mass proteins (Yokouchi et al., 2001). However, the large increase in the level of ubiquitinated proteins suggests that more than one substrate is targeted for ubiquitination, in accordance with self-incompatible pollen being blocked at multiple stages. For example, the substrates normally may promote the fertilization process by releasing water for pollen hydration or by loosening the stigmatic papillar cell walls for pollen tube growth; they also may be involved in pollen tube guidance. Degradation of any of these proteins would block the fertilization process. Moreover, the fact that there was a breakdown of self-incompatibility after the inhibition of proteasomal proteolytic function by the application of either MG132 or MG115 indicates that protein degradation is directly required for self-incompatibility. Finally, the self-incompatible response is thought to be restricted to the site of pollen contact with the stigmatic papilla (Dickinson, 1995; Lolle and Pruitt, 1999). Thus, targeted protein degradation also would need to be restricted to this region to allow for the localized inhibition of self-incompatible pollen.
Given the role for ARC1 in mediating protein degradation at the proteasome/CSN during the self-incompatibility response, the question arises of what function ARC1 serves in being shuttled into and out of the nucleus. One possibility is that there is an indirect relationship between ARC1's nuclear localization and its self-incompatibility function(s). For example, ARC1 may shuttle to the nucleus to interact with its substrate, as shown for the hRPF1/Nedd4 E3 ligase, which is involved in the degradation of a nuclear protein despite being localized predominantly to the cytosol (Hamilton et al., 2001). Alternatively, nucleocytosolic partitioning may regulate the function of ARC1 either through potential modifications or interactions with nuclear proteins. This latter role for nuclear shuttling has been found to be important in the function of the yeast Ste5p mitogen-activated protein kinase cascade scaffold protein, which ultimately is localized to the plasma membrane (Elion, 2001). A second possibility is a role for ARC1 in the nucleus as an E3 ligase or some other function unrelated to self-incompatibility. For example, the mammalian c-Cbl protein is involved in a number of signaling pathways, yet it can function independently as an E3 ligase or as a multi-adaptor protein (Thien and Langdon, 2001).
Although we have no evidence for a biological role for ARC1 other than in self-incompatibility, this could be attributable simply to functional redundancy. Arabidopsis contains a large family of ARC1-related proteins, with 42 members containing a U-box domain followed by an arm repeat domain (Azevedo et al., 2001; J. Salt, S.L. Stone, and D.R. Goring, unpublished data). In addition, several ARC1-related sequences can be found by Basic Local Alignment Search Tool (BLAST) searches of partial Brassica genomic DNA sequences (D.R. Goring, unpublished data). At least one of these ARC1-related members in potato, PHOR1, was found to localize to the nucleus after treatment with the gibberellin hormone and appears to play a role in gibberellin signal transduction (Amador et al., 2001). A similar test with ARC1 showed no effects of gibberellin on the subcellular localization of myc-ARC1 (data not shown). Clearly, more work is required to understand the importance of ARC1 nucleocytosolic partitioning.
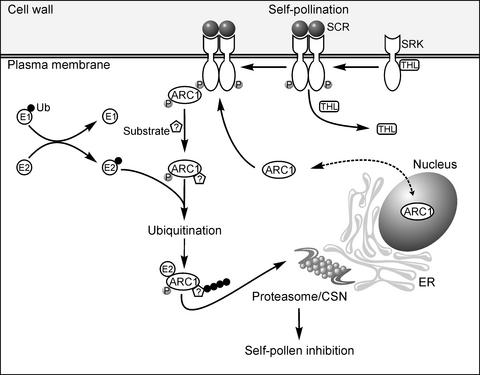
Based on this and other work with ARC1 and previous knowledge of SRK signaling, we propose the following model for the Brassica self-incompatibility response (Figure 7). In the absence of pollen, ARC1 shuttles between the nucleus and the cytosol. Activation of SRK by self-pollination then results in the phosphorylation of ARC1 and the relocalization of the protein to assembled proteasomes/CSNs situated on the cytosolic face of the ER membranes. Although not presented in the model shown, our data indicate also that at least one reticuloplasmin, BiP, is recruited to specific regions in the ER lumen adjacent to the site where proteasomes/CSNs and ARC1 accumulate on the cytosolic side of the ER membranes. ARC1 may recruit its substrate(s) from the nucleus, the cytosol, and/or the ER via retrograde protein transport (Sommer et al., 2001). The proposed ARC1 phosphorylation may influence ARC1's subcellular localization, interactions with substrate(s), or ubiquitin ligase activity. For example, the unphosphorylated form of the mammalian c-Cbl E3 ligase can mediate the in vitro ubiquitination of its substrate, c-Src; however, increased c-Src ubiquitination was observed in the presence of phosphorylated c-Cbl (Yokouchi et al., 2001). ARC1 has at least two putative protein–protein interaction domains in its N-terminal region, a Leu zipper and a coiled-coil domain, both of which may facilitate substrate binding while associating through its U-box motif with the E2 ubiquitin-conjugating enzyme. Evidence for this latter step comes from another study in which the U-box motif was shown to facilitate an interaction between the UIP5 U-box protein and the UbcM4 E2 enzyme (Pringa et al., 2001). ARC1-bound substrates then would be ubiquitinated and targeted for degradation by the proteasome/CSN.
Figure 7.
Model for the Role of ARC1 in Protein Degradation in the Self-Incompatibility Response.
In the absence of self-incompatible pollen, SRK is inhibited by THL1 and ARC1 shuttles actively between the nucleus and the cytosol, although it is localized primarily in the cytosol as a result of NESs. Activation of SRK by the pollen ligand SCR alleviates SRK inhibition by THL1 and promotes SRK autophosphorylation. This results in the recruitment and binding of cytosolic ARC1 to phosphorylated SRK. ARC1, in turn, is phosphorylated and relocalizes to proteasomes/CSNs assembling at the cytosolic face of the ER membranes. ARC1 may recruit substrate(s) in the nucleus, cytosol, and/or exported from the ER through its N-terminal Leu zipper and/or its coiled-coil domain and interacts through its U-box with a cognate E2 enzyme that is linked to ubiquitin (Ub). At the proteasome/CSN, ARC1 then mediates the ubiquitination and proteasomal degradation of its substrate(s). Although these ARC1 substrates are unknown at present, they may represent pistil factors that normally promote pollen hydration, germination, and pollen tube growth.
The colocalization of ARC1 with both subunits of the CSN and the lid particle of the 26S proteasome (Figure 2B) indicates that ARC1-mediated protein degradation may use both multiprotein complexes. The CSN is thought to function as a scaffold, linking signal transduction and ubiquitination with the 26S proteasome (Schwechheimer and Deng, 2001). E3 ligases also are known to interact with subunits of the CSN as well as the 26S proteasome, likely to direct ubiquitinated substrate(s) for degradation (Xie and Varshavsky, 2000; Schwechheimer et al., 2001). In plants, proteasomal protein degradation has been well studied in the auxin-dependent degradation of the Aux/IAA proteins and in the regulation of photomorphogenesis by the CSN (Schwechheimer and Deng, 2001; Hellmann and Estelle, 2002). However, these systems represent only a fraction of what likely occurs in plants in terms of regulation by ubiquitination and proteasomal protein degradation. As one indication of this, large families of putative F-box and ring-finger proteins, both of which are key regulatory factors in proteasome/CSN protein degradation processes, have been annotated in the Arabidopsis genome (Gagne et al., 2002; Kosarev et al., 2002). Finally, the SRK-ARC1 signaling pathway described here may be a more broadly used pathway, based on the large number of SRK-related receptor kinases and ARC1-related U-box proteins present in Arabidopsis (Azevedo et al., 2001; Shiu and Bleecker, 2001).
METHODS
Plasmid Construction
The plant expression vector pRTL2 was used for all tobacco (Nicotiana tabacum) BY-2 cell transformations (Lee et al., 1997). All ARC1 constructs were subcloned into pRTL2 so that they were in frame with a 5′ myc epitope tag. myc-ARC1Δ286-347 was generated using PCR to delete sequences that encode the entire U-box. Site-directed mutagenesis (Stratagene Quickchange) was used to introduce the substitutions K261A, K263A, and K264A into myc-ARC1 and myc-ARC1Δ286-347, yielding myc-ARC1NLSmut and myc-ARC1Δ286-347/NLSmut, respectively. Site-directed mutagenesis also was used to generate myc-ARC1Δ286-300, myc-ARC1Δ313-327, myc-ARC1Δ328-347, and myc-ARC1P323A. Chloramphenicol acetyltransferase–nuclear localization signal (CAT-NLS) was generated by subcloning an annealed synthetic oligonucleotide encoding ARC1's putative NLS followed by a stop codon in frame with the 3′ end of CAT in pRTL2 (Lee et al., 1997). The plasmid GFP-NLS-CHS-NES was described previously (Haasen et al., 1999). To construct GST-SRK, the GST-tagged SRKA14 kinase domain was amplified by PCR from pGEX-3X (Gu et al., 1998) and subcloned into pRTL2. To construct GST-tSRK, two NsiI sites located toward the 3′ end of SRKA14 were used to remove essential portions of the kinase domain before subcloning GST-SRKA14 into pRTL2. pYes2/GFP-ARC1 was constructed by fusing GFP in frame to the N terminus of full-length ARC1. pYes2/GFP is a high-copy (2μ) vector that contains the GAL1-inducible promoter and the URA3-selectable marker. The plasmid pPS1372 (2μ; URA3) contains the ADHI constitutive promoter and sequences that encode two copies of the GFP fused to both the SV40 large-T NLS and the protein kinase inhibitor NES (Taura et al., 1998).
Tobacco Cell Transformation and Immunofluorescence Microscopy
Tobacco BY-2 suspension-cultured cells were grown and transformed as described previously (Lee et al., 1997). Transformations were performed with a biolistic particle delivery system (Bio-Rad) using either 10 μg of plasmid DNA or, for cotransformations, 5 μg of each plasmid DNA. After bombardment, cells were left in the dark for the appropriate time, fixed with paraformaldehyde, permeabilized with Triton X-100, and then processed for immunofluorescence microscopy. For leptomycin B (LMB) treatments, LMB was dissolved in ethanol (10 μg/mL) and used at a final concentration of 2 μM (Haasen et al., 1999). After transformation, cells were treated with LMB or ethanol (in controls) for 4 h, fixed, permeabilized, and processed for immunofluorescence microscopy as described above.
Sources and concentrations for the primary and secondary antibodies used were as follows: mouse anti-c-myc (1:500) was from Bio/Can Scientific (Mississauga, ON, Canada); rabbit anti-GST (1:500) was from Santa Cruz Biotechnology (Santa Cruz, CA); rabbit anti-maize BiP (1:500) (Fontes et al., 1991); rabbit anti-CSN3, anti-CSN6, and anti-RPN6 (all used at 1:200 dilutions) (Peng et al., 2001a, 2001b); mouse anti-CAT (undiluted hybridoma medium) was provided by S. Subramani (University of California, San Diego); goat anti-mouse Alexa Fluor 488 (1:1000) was from Cedar Lane Laboratories (Hornby, ON, Canada); and goat anti-rabbit rhodamine red-X (1:1000) was from Bio/Can Scientific.
Stained BY-2 cells were mounted on slides in 90% glycerol with 1 mg/mL n-propyl gallate (Sigma-Aldrich). Yeast cells were immobilized on glass slides coated with 1% poly-l-Ly (Sigma-Aldrich). Images of cells were acquired using a Zeiss Axioskop 2 MOT epifluorescence microscope (Carl Zeiss, Thornwood, NY) with either a Zeiss ×100 Plan Apochromat oil-immersion objective for yeast cells or a Zeiss ×63 Plan Apochromat oil-immersion objective for BY-2 cells and a Retiga 1300 charge-coupled device camera (Qimaging, Burnaby, BC, Canada).
Recombinant Protein Purification and in Vitro Ubiquitination Assay
The full-length cDNA of Arabidopsis thaliana E2, UBC7 (van Nocker et al., 1996), was amplified by reverse transcriptase–mediated PCR from total leaf mRNA using gene-specific primers and subcloned into the His tag vector pET15b. Wild-type and mutated ARC1 cDNAs were cloned into the pGEX 4T-1 vector. All fusion proteins were expressed and purified from Escherichia coli strain BL21(DE3)pLysS as described by Hardtke et al. (2002) and Hatakeyama et al. (2001).
In vitro ubiquitination assays were performed as described by Hatakeyama et al. (2001). Reaction mixtures (25 μL) contained 0.5 μg of yeast E1 (BostonBiochem, Cambridge, MA), 1 μg of purified E2 or 2 μL of lysate containing E2, 1 μg of E3, 25 mM Tris-HCl, pH 7.5, 1 mM MgCl2, 120 mM NaCl, 2 mM ATP, 0.3 mM DTT, 1 μg of ubiquitin, 1 mM creatine phosphate, and 1 unit of phosphocreatine kinase (Sigma-Aldrich). Reactions were incubated at 30°C for 2 h and terminated by adding 2× SDS sample buffer and heating at 95°C for 5 min. The samples were resolved on an 8% SDS-PAGE gel followed by protein gel blot analysis with rabbit anti-ubiquitin antibodies (Sigma-Aldrich).
Brassica Pollination and Protein Extraction
One day before anthesis, buds from Brassica napus W1 and ARC1 antisense plants were emasculated and left covered overnight. Pistils were pollinated for 1 h, and then the stigmas were collected and stored at −80°C. Total protein extracts were prepared by grinding the stigmas in liquid nitrogen followed by extraction buffer containing 50 mM Tris-HCl, 50 mM NaCl, 10 mM MgCl2, 5 mM EDTA, 5 mM DTT, and 10% glycerol (v/v) supplemented with plant protease inhibitor cocktail (Sigma-Aldrich). The Bio-Rad protein reagent assay and Coomassie Brilliant Blue R250 staining of proteins separated by SDS-PAGE gel were used to determine protein concentrations for equal loading. Ten micrograms of protein was separated on an 8% SDS-PAGE gel followed by protein blot analysis with rabbit anti-ubiquitin antibodies (Sigma-Aldrich).
Proteasome Inhibitor Studies
B. napus W1 buds were collected 1 day before anthesis and incubated overnight with their pedicels submerged in 50 μM of the proteasome inhibitor MG132 or MG115 (10 mM stock in DMSO; Sigma-Aldrich). For controls, the Sigma-Aldrich plant protease inhibitor cocktail was used at the recommended concentration, or DMSO only was added. The pistils then were pollinated with untreated pollen, left for 3 h, and analyzed as described previously (Stone et al., 1999).
Yeast Strains, Growth Conditions, and Transformation
The wild-type and xpo1-1 Saccharomyces cerevisiae strains (W303; Mat a/α, ade2-1 ura 3-1, his3-11,15 tip 1-1 leu2-3, 112 can 1-100) were described previously (Stade et al., 1997; Neville and Rosbash, 1999). Untransformed yeast cells were maintained on YPAD plates (1% w/v yeast extract, 2% w/v peptone, 2% w/v dextrose, and 0.04% w/v adenine) and grown at either 25°C (xpo1-1) or 30°C (wild type). Yeast were transformed with plasmid DNA using the lithium acetate method (Gietz and Woods, 1994) and plated on synthetic dextrose plates (2% w/v dextrose, 0.67% yeast nitrogen base without amino acids) supplemented with appropriate amino acids (Bufferad; Difco). Liquid culturing of transformed cells was performed essentially as described by Stade et al. (1997). Briefly, cells containing either pYes2.1/GFP-ARC1 or pPS1372 (NLS-NES-GFP2) were grown overnight at 25°C in synthetic dextrose medium containing appropriate amino acid supplements. The next day, cultures containing pYes2.1/GFP-ARC1 were diluted to 0.2 OD600 in synthetic galactose medium (2% w/v galactose, 0.67% yeast nitrogen base without amino acids, plus amino acid supplements) and grown at 25°C until mid log phase. Portions of all cultures were shifted to the restrictive temperature (37°C) for 5 min, and GFP signals were analyzed via fluorescence microscopy.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We are especially grateful to Z. Peng and X.-W. Deng (Yale University, New Haven, CT) for providing the rabbit anti-CSN3, anti-CSN6, and anti-RPN6 polyclonal antibodies and for helpful suggestions; to M. Yoshida (University of Tokyo) for leptomycin B; to R. Boston (North Carolina State University, Raleigh) for providing the anti-maize BiP antibody; to T. Merkle (University of Freiburg, Germany) for pGFP-NLS-CHS-NES; to D. Mangroo (University of Guelph, Canada) for pYes2/GFP and helpful suggestions; to P. Silver (Harvard Medical School, Boston, MA) for pPS1372; to M. Rosbash (Brandeis University, Waltham, MA) for the xpo1-1 yeast mutant; and to I. Smith (University of Guelph) for assistance with Figure 7. This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC) to D.R.G. and R.T.M., by Premier's Research Excellence Awards to D.R.G. and R.T.M., and by a NSERC graduate scholarship to S.L.S.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.009845.
References
- Amador, V., Monte, E., Garcia-Martinez, J.L., and Prat, S. (2001). Gibberellins signal nuclear import of PHOR1, a photoperiod-responsive protein with homology to Drosophila armadillo. Cell 106, 343–354. [DOI] [PubMed] [Google Scholar]
- Aravind, L., and Koonin, E.V. (2000). The U-box is a RING finger: A common domain in ubiquitination. Curr. Biol. 10, 132–134. [DOI] [PubMed] [Google Scholar]
- Azevedo, C., Santos-Rosa, M.J., and Shirasu, K. (2001). The U-box protein family in plants. Trends Plant Sci. 6, 354–358. [DOI] [PubMed] [Google Scholar]
- Becraft, P.W. (2002). Receptor kinase signaling in plant development. Annu. Rev. Cell Dev. Biol. 18, 163–192. [DOI] [PubMed] [Google Scholar]
- Bogerd, H.P., Fridell, R.A., Benson, R.E., Hua, J., and Cullen, B.R. (1996). Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol. Cell. Biol. 16, 4207–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower, M.S., Matis, D.D., Fernandes-Carvalho, E., Mazzurco, M., Gu, T., Rothstein, S.J., and Goring, D.R. (1996). Two members of the thioredoxin-h family interacts with the kinase domain of a Brassica S locus receptor kinase. Plant Cell 8, 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrillac, D., Cock, M., Dumas, C., and Gaude, T. (2001). The S-locus receptor kinase is inhibited by thioredoxins and activated by pollen coat proteins. Nature 410, 220–223. [DOI] [PubMed] [Google Scholar]
- Dickinson, H. (1995). Dry stigmas water and self-incompatibility in Brassica. Sex. Plant Reprod. 8, 1–10. [Google Scholar]
- Elion, E.A. (2001). The Ste5p scaffold. J. Cell Sci. 114, 3967–3978. [DOI] [PubMed] [Google Scholar]
- Enenkel, C., Lehmann, A., and Kloetzel, P.-M. (1998). Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelope-ER network in yeast. EMBO J. 17, 6144–6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell, K., Wilkinson, C.R.M., Dubiel, W., and Gordon, C. (2000). Regulatory subunit interactions of the 26S proteasome, a complex problem. Trends Biochem. Sci. 25, 83–88. [DOI] [PubMed] [Google Scholar]
- Fontes, E.B., Shank, B.B., Wrobel, R.L., Moose, S.P., O'Brian, G.R., Wurtzel, E.T., and Boston, R.S. (1991). Characterization of an immunoglobulin binding protein homolog in the maize floury-2 endosperm mutant. Plant Cell 3, 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne, J.M., Downes, B.P., Shiu, S.H., Durski, A.M., and Vierstra, R.D. (2002). The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 11519–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R.D., and Woods, R.A. (1994). High efficiency transformation in yeast. In Molecular Genetics of Yeast: Practical Approaches, J.A. Johnston, ed (New York: Oxford University Press), pp. 121–134.
- Giranton, J.-L., Duma, C., Cock, M., and Gaude, T. (2000). The integral membrane S-locus receptor kinase of Brassica has serine/threonine kinase activity in a membranous environment and spontaneously forms oligomers in planta. Proc. Natl. Acad. Sci. USA 97, 3759–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gomez, L., and Boller, T. (2002). Flagellin perception: A paradigm for innate immunity. Trends Plant Sci. 7, 251–256. [DOI] [PubMed] [Google Scholar]
- Gu, T., Mazzurco, M., Sulaman, W., Matias, D.D., and Goring, D.R. (1998). Binding of a novel arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc. Natl. Acad. Sci. USA 95, 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasen, D., Kohler, C., Neuhaus, G., and Merkle, T. (1999). Nuclear export of proteins in plants: AtXPO1 is the export receptor for the leucine-rich export signals in Arabidopsis thaliana. Plant J. 20, 695–705. [DOI] [PubMed] [Google Scholar]
- Hamilton, M.H., Tcherepanova, I., Huibregtse, J.N., and McDonnell, D.P. (2001). Nuclear import/export of hRPF1/Nedd4 regulates the ubiquitin-dependent degradation of its nuclear substrate. J. Biol. Chem. 276, 26324–26331. [DOI] [PubMed] [Google Scholar]
- Hardtke, C.S., Okamoto, H., Stoop-Myer, C., and Deng, X.W. (2002). Biochemical evidence for ubiquitin ligase activity of the Arabidopsis COP1 interacting protein 8 (CIP8). Plant J. 30, 385–394. [DOI] [PubMed] [Google Scholar]
- Hatakeyama, S., Jensen, J.P., and Weissman, A.M. (1997). Subcellular localization and ubiquitin-conjugating enzyme (E2) interactions of mammalian HECT family ubiquitin protein ligases. J. Biol. Chem. 272, 15085–15092. [DOI] [PubMed] [Google Scholar]
- Hatakeyama, S., Yada, M., Matsumoto, M., Ishida, N., and Nakayama, K. (2001). U-box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 276, 33111–33120. [DOI] [PubMed] [Google Scholar]
- Hellmann, H., and Estelle, M. (2002). Plant development: Regulation by protein degradation. Science 297, 793–797. [DOI] [PubMed] [Google Scholar]
- Kachroo, A., Nasrallah, M.E., and Nasrallah, J.B. (2002). Self-incompatibility in the Brassicaceae: Receptor-ligand signaling and cell-to-cell communication. Plant Cell 14 (suppl.), S227.–S238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo, A., Schopfer, C.R., Nasrallah, M.E., and Nasrallah, J.B. (2001). Allele-specific receptor ligand interactions in Brassica self-incompatibility. Science 293, 1824–1826. [DOI] [PubMed] [Google Scholar]
- Kosarev, P., Mayer, K.F., and Hardtke, C.S. (2002). Evaluation and classification of RING-finger domains encoded by the Arabidopsis genome. Genome Biol. 3, RESEARCH0016. [DOI] [PMC free article] [PubMed]
- Lee, M.S., Mullen, R.T., and Trelease, R.N. (1997). Oilseed isocitrate lyases lacking their essential type 1 peroxisomal targeting signal are piggybacked to glyoxysomes. Plant Cell 9, 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolle, S.J., and Pruitt, R.E. (1999). Epidermal cell interactions: A case of local talk. Trends Plant Sci. 4, 14–20. [DOI] [PubMed] [Google Scholar]
- Lord, E.M., and Russell, S.D. (2002). The mechanisms of pollination and fertilization in plants. Annu. Rev. Cell Dev. Biol. 18, 81–105. [DOI] [PubMed] [Google Scholar]
- Lorick, K.L., Jensen, J.P., Fang, S., Ong, A.M., Hatakeyama, S., and Weissman, A.M. (1999). RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 96, 11364–11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro, S., and Pelham, H.R. (1986). An Hsp70-like protein in the ER: Identity with the 78 kd glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell 46, 291–300. [DOI] [PubMed] [Google Scholar]
- Nagata, T., Nemoto, Y., and Hasezawa, S. (1992). Tobacco BY-2 cell line as the “Hela” cell in the cell biology of higher plants. Int. Rev. Cytol. 132, 1–30. [Google Scholar]
- Neville, M., and Rosbash, M. (1999). The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 18, 3746–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossareh-Nazari, B., Bachelerie, F., and Dragemont, C. (1997). Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science 278, 141–144. [DOI] [PubMed] [Google Scholar]
- Patterson, C. (2002). A new gun in town: The U-box is a ubiquitin ligase domain. Sci. STKE 22, PE4.. [DOI] [PubMed] [Google Scholar]
- Peng, Z., Serino, G.F., and Deng, X.-W. (2001. a). A role for Arabidopsis CSN in multifaceted developmental processes revealed by the characterization of its subunit 3. Development 128, 4277–4288. [DOI] [PubMed] [Google Scholar]
- Peng, Z., Serino, G.F., and Deng, X.-W. (2001. b). Molecular characterization of subunit 6 of the COP9 signalosome and its role in multifaceted developmental processes in Arabidopsis. Plant Cell 13, 2393–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringa, E., Martinez-Noel, G., Muller, U., and Harbers, K. (2001). Interaction of the RING finger-related U-box motif of a nuclear dot protein with ubiquitin-conjugating enzymes. J. Biol. Chem. 276, 19617– 19623. [DOI] [PubMed] [Google Scholar]
- Rock, K.L., Gramm, C., Rothstein, L., Clark, K., Stein, R., Dick, L., Hwang, D., and Goldberg, A.L. (1994). Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78, 761–771. [DOI] [PubMed] [Google Scholar]
- Schopfer, C.R., Nasrallah, M.E., and Nasrallah, J.B. (1999). The male determinant of self-incompatibility in Brassica. Science 286, 1697–1700. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., and Deng, X.-W. (2001). CSN revisited: A novel mediator of protein degradation. Trends Cell Biol. 11, 420–426. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., Serino, G., Callis, J., Crosby, W.L., Laypina, S., Deshaies, R.J., Gray, W.M., Estelle, M., and Deng, X.-W. (2001). Interactions of the CSN with the E3 ubiquitin ligase SCFTIR1 in mediating auxin response. Science 292, 1370–1382. [DOI] [PubMed] [Google Scholar]
- Shiu, S.H., and Bleecker, A.B. (2001). Plant receptor-like kinase gene family: Diversity, function and signaling. Sci. STKE. 18, RE22.. [DOI] [PubMed] [Google Scholar]
- Silva, N.F., and Goring, D.R. (2001). Mechanisms of self-incompatibility in flowering plants. Cell. Mol. Life Sci. 58, 1988–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, N.F., Stone, S.L., Christe, L.N., Sulaman, W., Nazarain, K.A.P., Burnett, M., Arnoldo, M.A., Rothstein, S.J., and Goring, D.R. (2001). Expression of the S receptor kinase in self-compatible Brassica napus cv. Westar leads to the allele-specific rejection of self-incompatible Brassica napus pollen. Mol. Gen. Genet. 265, 552–559. [DOI] [PubMed] [Google Scholar]
- Sommer, T., Jarosch, E., and Lenk, U. (2001). Compartment-specific functions of the ubiquitin-proteasome pathway. Rev. Physiol. Biochem. Pharmacol. 142, 97–160. [DOI] [PubMed] [Google Scholar]
- Stade, K., Ford, C.S., Guthrie, C., and Weis, K. (1997). Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90, 1041–1050. [DOI] [PubMed] [Google Scholar]
- Stone, S.L., Arnoldo, M., and Goring, D.R. (1999). A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science 286, 1729–1731. [DOI] [PubMed] [Google Scholar]
- Stone, S.L., and Goring, D.R. (2001). The molecular biology of self-incompatibility in flowering plants. Plant Cell Tissue Organ Cult. 67, 93–114. [Google Scholar]
- Takasaki, T., Hatakeyama, K., Suzuki, G., Watanabe, M., Isogal, A., and Hinata, K. (2000). The S receptor kinase determines self-incompatibility in Brassica stigma. Nature 403, 913–916. [DOI] [PubMed] [Google Scholar]
- Takayama, S., Shiba, H., Iwano, M., Shimosato, H., Che, F.S., Kai, N., Watanabe, M., Suzuki, G., Hinata, K., and Isogai, A. (2000). The pollen determinant of self-incompatibility in Brassica campestris. Proc. Natl. Acad. Sci. USA 97, 1920–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama, S., Shimosato, H., Shiba, H., Funato, M., Che, F.S., Watanabe, M., Iwano, M., and Isogai, A. (2001). Direct ligand-receptor complex interaction controls Brassica self-incompatibility. Nature 413, 534–538. [DOI] [PubMed] [Google Scholar]
- Taura, T., Krebber, H., and Silver, P.A. (1998). A member of the Ran-binding protein family, Yrb2p, is involved in nuclear protein export. Proc. Natl. Acad. Sci. USA 95, 7427–7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thien, C.B., and Langdon, W.Y. (2001). Cbl: Many adaptations to regulate protein tyrosine kinases. Nat. Rev. Mol. Cell. Biol. 2, 294–307. [DOI] [PubMed] [Google Scholar]
- van Nocker, S., Walker, J.M., and Vierstra, R.D. (1996). The Arabidopsis thaliana UBC7/13/14 genes encode a family of multiubiquitin chain-forming E2 enzymes. J. Biol. Chem. 271, 12150–12158. [DOI] [PubMed] [Google Scholar]
- Wheeler, M.J., Franklin-Tong, V.E., and Franklin, F.C.H. (2001). The molecular and genetic basis of pollen-pistil interactions. New Phytol. 151, 565–584. [DOI] [PubMed] [Google Scholar]
- Xie, Y., and Varshavsky, A. (2000). Physical association of ubiquitin ligases and the 26S proteasome. Proc. Natl. Acad. Sci. USA 97, 2497–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokouchi, M., Kondo, T., Sanjay, A., Houghton, A., Yoshimura, A., Komiya, S., Zhang, H., and Baron, R. (2001). Src-catalyzed phosphorylation of c-Cbl leads to the interdependent ubiquitination of both proteins. J. Biol. Chem. 276, 35185–35193. [DOI] [PubMed] [Google Scholar]