Abstract
Posttranslational modification of histones has emerged as a key regulatory signal in eukaryotic gene expression. Recent genetic and biochemical studies link H3-lysine 9 (H3-K9) methylation to HP1-mediated heterochromatin formation and gene silencing. However, the mechanisms that target and coordinate these activities to specific genes is poorly understood. Here we report that the KAP-1 corepressor for the KRAB-ZFP superfamily of transcriptional silencers binds to SETDB1, a novel SET domain protein with histone H3-K9-specific methyltransferase activity. Although acetylation and phosphorylation of the H3 N-terminal tail profoundly affect the efficiency of H3-K9 methylation by SETDB1, we found that methylation of H3-K4 does not affect SETDB1-mediated methylation of H3-K9. In vitro methylation of the N-terminal tail of histone H3 by SETDB1 is sufficient to enhance the binding of HP1 proteins, which requires both an intact chromodomain and chromoshadow domain. Indirect immunofluoresence staining of interphase nuclei localized SETDB1 predominantly in euchromatic regions that overlap with HP1 staining in nonpericentromeric regions of chromatin. Moreover, KAP-1, SETDB1, H3-MeK9, and HP1 are enriched at promoter sequences of a euchromatic gene silenced by the KRAB–KAP-1 repression system. Thus, KAP-1 is a molecular scaffold that is targeted by KRAB-ZFPs to specific loci and coordinates both histone methylation and the deposition of HP1 proteins to silence gene expression.
Keywords: Histone methylation, SET domain, chromatin, KRAB domain
Macromolecular protein complexes containing enzymatic activities that modify the N-terminal tails of the core histones have emerged as key regulators of gene expression in eukaryotes. The constellation of histone modifications, including acetylation, phosphorylation, ubiquination, and methylation, create both synergistic and antagonistic signals that correlate with the transcriptional activity of a gene. This emerging histone code is hypothesized to create an architecture in chromatin that is recognized by nonhistone chromosomal proteins, which then effect the dynamic transition between transcriptionally active versus transcriptionally silent chromatin domains (Jenuwein and Allis 2001). Moreover, the combinatorial nature of these histone modifications and the chromatin-associated proteins that recognize these signals may represent an epigenetic marking system responsible for setting and maintaining heritable programs of gene expression during cellular differentiation and organism development.
The role of histone acetylation and phosphorylation in regulation of transcription has been extensively characterized (Cheung et al. 2000; Strahl and Allis 2000; Berger 2001). Although it is well established that arginine and lysine methylation of histones occurs in vivo, the function of these specific modifications remains to be fully described (Strahl et al. 1999). Similar to the discovery of histone acetyltransferases (HATs) and deacetylases (HDACs), the role of histone methylation in the regulation of chromatin structure and gene expression has been greatly facilitated by the identification of the responsible enzymes. The discovery that the nuclear receptor co-activator associated protein CARM1 is an H3-specific arginine methyltransferase and that the mammalian homologs of the Drosophila melanogaster heterochromatin protein Su(var)3-9 are H3-specific lysine methyltransferases significantly supported the involvement of histone methylation in gene regulation (Chen et al. 1999; Rea et al. 2000). In the later case, methylation was highly selective for Lys 9, with the methyltransferase function mapping to the evolutionarily conserved SET domain homology (Rea et al. 2000). The H3-K9 methylation (H3-MeK9) mark establishes a high-affinity binding site for the recruitment of the HP1 family of heterochromatin proteins through its chromodomain (Jacobs et al. 2001; Jacobs and Khorasanizadeh 2002). Furthermore, genetic experiments link the localization of Swi6 and HP1 at condensed chromatin sequences to the histone methyltransferase activity of Clr4 and SUVAR39H1, respectively (Bannister et al. 2001; Lachner et al. 2001; Nakayama et al. 2001). Thus, substantial genetic, biochemical, and cytological evidence links the selective methylation of histone H3-K9 and the deposition of HP1 proteins to chromatin sequences whose transcription is epigenetically silenced. Additional studies have shown that the H3-MeK9 epitope globally distinguishes transcriptionally silent from active chromatin domains in vivo, and methylation of histone H3 Lys 9 represents an early molecular mark on the X chromosome during X inactivation (Heard et al. 2001; Litt et al. 2001; Noma et al. 2001; Boggs et al. 2002). However, the mechanisms that target H3-K9 methylation and coordinate HP1 deposition to specific cis regulatory sequences in vivo remain to be fully defined.
A prime candidate for a molecule that could coordinate these activities is the KAP-1 corepressor. This protein serves as a universal, obligatory corepressor for the >220 KRAB domain zinc-finger proteins (ZFPs) that are encoded by the human genome (Friedman et al. 1996). Each KRAB-ZFP contains an N-terminal 75-amino-acid KRAB box that binds directly to KAP-1, and a C-terminal array of C2H2 zinc fingers, which mediate sequence-specific DNA binding (Mark et al. 1999). The tripartite RBCC region of KAP-1 functions as an integrated structural unit that is necessary and sufficient for oligomerization and KRAB binding (Fig. 1A; Peng et al. 2000a,b). The PHD finger and bromodomain of KAP-1 form a cooperative transcriptional repression unit that recruits the NuRD HDAC complex (Capili et al. 2001; Schultz et al. 2001). A separate repression domain in KAP-1 containing a core PxVxL motif binds directly to the chromoshadow domain of the HP1 protein family (Ryan et al. 1999; Lechner et al. 2000). From these data we postulate that KAP-1 functions as a scaffold that, in turn, coordinates the activities of large macromolecular complexes that modify chromatin structure to silence gene expression. Because the HP1 chromodomain can bind to the methylated Lys 9 of histone H3, we reasoned that KAP-1 repression complexes in vivo might contain histone H3-K9 methyltransferase activity, which would function to coordinate HP1-mediated repression of transcription at KRAB-ZFP target genes.
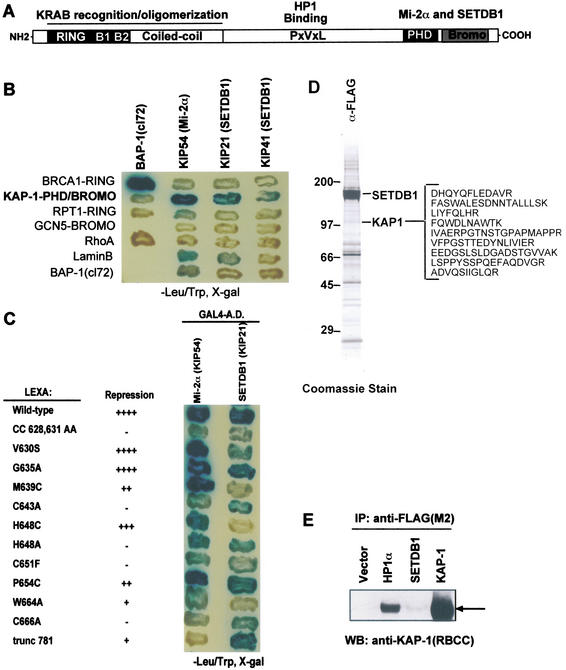
Figure 1.
The KAP-1 corepressor interacts with the putative histone methyltransferase SETDB1. (A) Schematic illustration of the KAP-1 corepressor. The oligomerization and KRAB-binding domain map to the RBCC region of KAP-1. The chromoshadow domain of the HP1 family of chromosomal proteins directly binds to a PxVxL motif in KAP-1. The PHD finger and bromodomain of KAP-1 form a cooperative repression domain that interacts with Mi-2α and SETDB1. (B) The KAP-1 PHD finger and bromodomain interact with Mi-2α (KIP54) and SETDB1 (KIP21 and KIP 41). (C) Mutations in the PHD finger and bromodomain that impair transcriptional repression by KAP-1 impair the association with SETDB1 (KIP21). (D) Coomassie blue staining of anti-Flag immunopurified SETDB1 from transfected HEK293 cells. MS/MS peptide identification, definitively identified 11 overlapping peptides of KAP-1, illustrated at the right in single-letter amino acid abbreviations. (E) Anti-KAP-1 Western blot of Flag immunoprecipitates from HEK293-transfected nuclear extracts.
Results
KAP-1 associates with a novel SET-domain protein, SETDB1
To identify effectors of KAP-1-directed transcriptional repression, we used the PHD finger and bromodomain of KAP-1 as bait in a two-hybrid screen. We previously reported the identification of a novel isoform of the Mi-2α subunit of the NuRD histone deacetylase complex that bound to this bipartite repression domain (Schultz et al. 2001). Here we report that KAP-1 specifically associated with two independent overlapping amino acid sequences (KIP21 and KIP41) that are encoded by the putative histone methyltransferase (HMTase) gene SETDB1 (Fig. 1B; Harte et al. 1999). As illustrated in Figure 1C, mutations deleterious to the repression activity of the KAP-1 PHD finger and bromodomain significantly impaired the association between KAP-1 and either SETDB1 or Mi-2α (Schultz et al. 2001). Note that some of the mutations differentially affect Mi-2α and SETDB1 binding, which raises the possibility that Mi-2α and SETDB1 may bind to different surfaces of the KAP-1 protein. To confirm the association between KAP-1 and SETDB1 in vivo, we generated a full-length Flag-epitope-tagged expression vector and immunopurified SETDB1 from transfected HEK293 cells. The spectrum of polypeptides was subjected to MS/MS peptide analyses, which definitively identified KAP-1 as a nonstoichiometric, associated polypeptide (Fig. 1D,E). Eleven KAP-1 polypeptides were identified spanning a significant portion of the KAP-1 open reading frame (amino acids 239–790). From these data we conclude that the KAP-1 corepressor interacts with the SETDB1 protein in vivo.
SETDB1 is a novel histone H3-specific methyltransferase
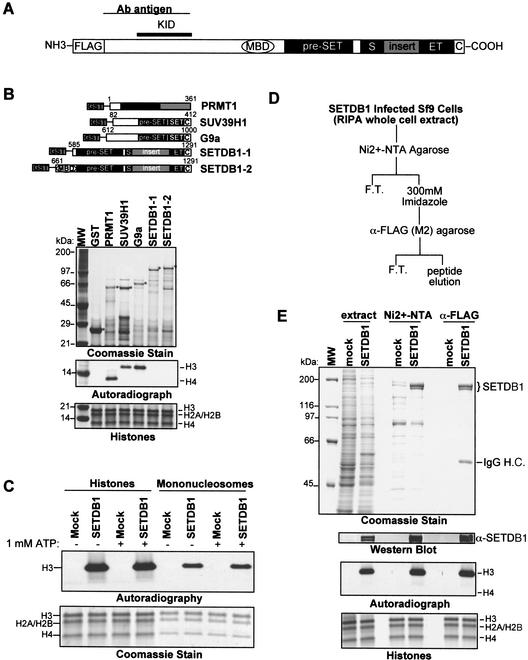
The primary amino acid sequence of SETDB1 revealed several interesting signature motifs including a CpG-DNA methyl binding domain of the MeCP2 family, and homology to the SET (SuVar3-9, Enhancer of Zeste, Trithorax) domain (Fig. 2A). Interestingly, the SET-domain homology of SETDB1 is interrupted by a 347-amino-acid insertion to create a bifurcated domain (Harte et al. 1999). This unique insertion is evolutionarily conserved from the human protein to lower eukaryotes, including Caenorhabditis elegans and D. melanogaster, suggesting that the SET domain may possess functionally separable domains. It has been previously shown that the SET-domain homology of SUV39H1 and the two adjacent cysteine-rich regions (pre-SET and post-SET) possess intrinsic histone methyltransferase (HMTase) activity that is dependent on the integrity of all three domains (Rea et al. 2000). To test whether SETDB1 possessed intrinsic HMTase activity, we expressed and purified two different recombinant GST–SETDB1 fusion proteins that encode the entire putative catalytic domain (amino acids 585–1291 and 661–1291, respectively). Unlike recombinant PRMT1, SUV39H1 and G9a proteins, the recombinant SETDB1 proteins failed to show any appreciable methylation of core histones (Fig. 2B). However, SETDB1 that was immunopurified from transiently transfected HEK293 cells (Fig. 1D) showed a robust histone H3-specific methyltransferase activity for core histones and mononucleosome substrates (Fig. 2C). Identical enzymatic activity was observed for a SETDB1 protein expressed and affinity-purified to near homogeneity from baculovirus-infected Sf9 cell extracts (Fig. 2E). Therefore, we postulate that SETDB1 may require posttranslational modification or a small molecular weight cellular cofactor(s) to function as a histone methylase.
Figure 2.
SETDB1 is a histone H3-specific methyltransferase. (A) Schematic illustration of the SETDB1 protein. The position of the pre-SET, SET, and post-SET (C) homologies at the C terminus are indicated. The 347-amino-acid insertion in the SET domain is indicated by the gray box. (MBD) A CpG DNA methyl-binding domain. The minimal KAP-1 interaction domain (KID) is defined by amino acids of SETDB1 present in two-hybrid clone KIP21. The region of SETDB1 (amino acids 1–377) used to raise a polyclonal antibody is illustrated. (B) Schematic illustration (top) of recombinant GST-PRMT1, GST-SUV39H1, GST-G9a, and GST-SETDB1 histone methyltransferase proteins and enzymatic activities (bottom) of affinity-purified proteins expressed in E. coli. Coomassie stain illustrates the affinity-purified GST–proteins. Autoradiograph shows [3H]methyl-labeled histone H3 and H4 from an in vitro HMTase assay with each respective methyltransferase. (Bottom panel) A Coomassie stain representing equal amounts of core histone octamers per each reaction, whose identities are labeled respectively. (C) Peptide eluate of anti-Flag immunopurified SETDB1 from transiently transfected HEK293 cells (Fig. 1D) revealed histone H3-specific methyltransferase activity in an in vitro HMTase assay with either core histones or chicken erythrocyte mononucleosomes as substrates. Coomassie blue stain shows the loading of histones. Autoradiograph shows corresponding [3H]methyl-labeled products. (D) Strategy to affinity-purify enzymatically active SETDB1 expressed in Sf9 baculovirus-infected cells. (E) Coomassie stain illustrates Ni2+-NTA and α-Flag M2 affinity-purified SETDB1 from baculovirus-infected Sf9 cells. Autoradiograph illustrates [3H]methyl-labeled histone H3. Western blot confirms identity of SETDB1 during the purification. (Bottom panel) A Coomassie stain representing equal amounts of core histone octamers per each reaction.
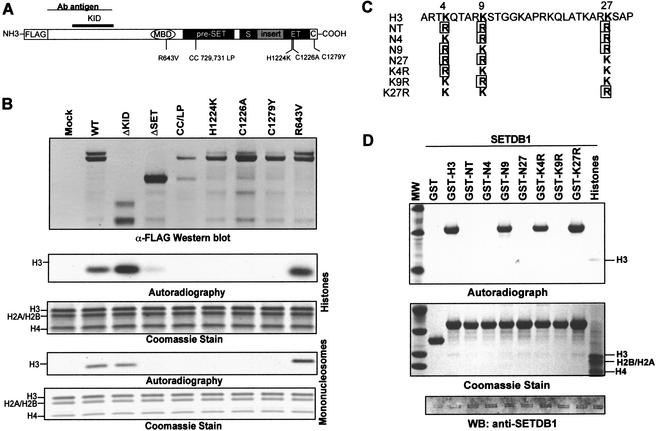
To evaluate the role of the pre-SET, SET, and post-SET domains in the HMTase activity, we engineered several mutants of SETDB1 (Fig. 3A). The HMTase activity of a protein containing a deletion of the post-SET domain and the C-terminal SET homology was significantly impaired, at least in this assay system. Furthermore, single amino acid substitutions at highly conserved residues in each of these subdomains reduced the methylase activity to undetectable levels. However, deletion of the putative KAP-1 interaction domain (KID) increased activity. A point mutation in the MBD homology had no effect on this enzymatic activity (Fig. 3B). Thus, similar to other members of the SET domain family of histone methyltransferases, SETDB1 requires the pre-SET, SET, and post-SET homologies for full enzymatic activity in vitro. Remarkably, the unique 347-amino-acid insertion in the SET domain appears to have no effect on the catalytic activity of SETDB1. These data support previous hypotheses that the SET domain structurally may be composed of two separable functional domains (Katsani et al. 2001). Furthermore, binding to KAP-1 is apparently not required for the methyltransferase activity. Although we cannot rule out the contributions of other endogenous polypeptides present in the immunopurified preparations of SETDB1, purification of SETDB1 to near homogeneity from Sf9 baculovirus-infected cell extracts indicates that it is sufficient to mediate histone methylation (Fig. 2E).
Figure 3.
SETDB1 selectively methylates Lys 9 of histone H3. (A) Schematic illustrating the relative position of single amino acid substitutions synthetically introduced into the MBD, pre-SET, SET, and post-SET domains as indicated. (B) Deletion of the post-SET and part of the SET homologies and single amino acid mutations at highly conserved residues within the catalytic domain (pre-SET, SET, and post-SET) impair the H3-methylase activity of SETDB1. The anti-Flag Western blot confirms the expression and Flag immunopurification of the indicated proteins. The ΔKID and ΔSET proteins correspond to amino acids 570–1291 and 1–951 of SETDB1, respectively. (C) Amino acid sequence of the N-terminal tail of histone H3 (1–30) is shown at the top with the K4, K9, and K27 residues highlighted. The various lysine to arginine mutations in K4, K9, and K27 derived to determine the substrate specificity of SETDB1 are indicated. All lysine (K) to arginine (R) mutations at K4, K9, and K27 are boxed. (D) The SETDB1 methyltransferase activity is highly specific for K9. Five micrograms of the corresponding GST–H3N protein was used as substrate in the in vitro HMTase assay with Flag-purified SETDB1 (Fig. 1D). Coomassie blue stain shows the purified GST–histone H3 substrates. Autoradiograph shows corresponding [3H]methyl-labeled products. Western blot confirms presence of Flag-SETDB1 in the HMTase reaction.
SETDB1 is a highly selective H3-K9 methyltransferase
To define the site specificity of H3 methylation by SETDB1, we used a series of purified, recombinant GST-histone tail proteins with several lysine-to-arginine substitutions as substrates (Fig. 3C; Tachibana et al. 2001). We found that H3 methylation by SETDB1 is highly selective for Lys 9 (Fig. 3D). A substrate (NT) in which K4, K9, and K27 were each mutated to arginine failed to be methylated. Substrates with double lysine-to-arginine mutations (N4, N9, N27) revealed methylation of a substrate with only K9 (N9) preserved. A substrate with a single arginine substitution at K9 (K9R) confirmed the specificity of SETDB1 for K9. These data confirm that additional posttranslational modifications (i.e., acetylation, phosphorylation, methylation) of the substrate are not required for H3-K9 methylation by SETDB1. Moreover, when K9 was mutated to arginine (K9R), SETDB1 did not change its specificity to K27, despite the fact that this residue lies in a strikingly similar amino acid sequence (TKxxARKS) as K9.
The histone code and H3-K9 methylation by SETDB1
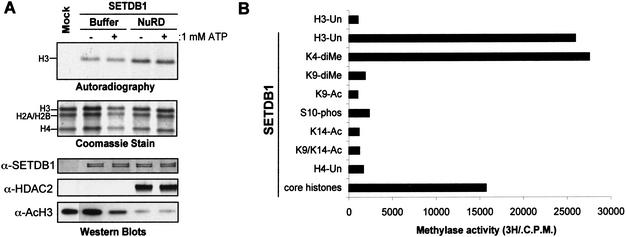
We next determined whether preexisting modifications to histone tails could influence the ability of SETDB1 to methylate its substrate. The core histones isolated from calf thymus were pretreated with catalytic amounts of a homogeneously pure preparation of the histone deacetylase complex NuRD (data not shown). We then evaluated the efficiency of SETDB1 to methylate this deacetylated core histone octamer substrate. As illustrated in Figure 4A, pretreatment of the histone substrate with the purified NuRD complex resulted in an approximately twofold enhancement in methylation, which was in agreement with a global decrease in H3 acetylation, suggesting that the site of methylation is either naturally acetylated, or that global acetylation of H3 interferes with the enzyme's recognition of the substrate.
Figure 4.
Dissecting the histone code and methylation by SETDB1. (A) Pretreatment of a core histone substrate with a homogenously pure histone deacetylase complex, NuRD, enhanced methylation of histone H3 in vitro, concomitantly with deacetylation of histone H3 (anti-AcH3 Western blot). Coomassie blue stain shows equal loading of histone proteins. Autoradiograph shows corresponding [3H]methyl-labeled products. Anti-SETDB1 and anti-HDAC2 Western blots show the presence of SETDB1 and HDACs in the corresponding HMTase reactions. (B) Effect of histone modifications on the enzymatic activity of SETDB1. One microgram of unmodified or acetylated (K9-Ac, K14-Ac, K9,K14-Ac), phosphorylated (S10-phos), or methylated (K4-diMe, K9-diMe) peptides corresponding to the N-terminal tail of histone H3 and H4 were used as substrates in the in vitro methylation assay with Flag-purified SETDB1. Methylation was quantified via a filter binding assay and represented as raw counts per minute (C.P.M.) incorporated.
To define which posttranslational modifications (i.e., acetylation, methylation, and phosphorylation) of histone H3 affect the SETDB1 methylase activity, we tested the activity of SETDB1 against a panel of peptide substrates possessing either an individual or a combination of modifications (Fig. 4B). Flag-purified SETDB1 robustly methylated the unmodified H3 substrate, but not an H4 peptide. Interestingly, a peptide substrate methylated at K4 had no apparent effect on this activity. As expected, any modification (methylation or acetylation) of H3-K9 inhibited SETDB1-mediated methylation. Furthermore, phosphorylation of S10 or acetylation of K14 also dramatically inhibited the methylation of the substrate. These observations are similar to that previously observed for the related K9-specific histone H3 methyltransferase SUV39H1, indicating that these proteins likely recognize the substrate in a similar fashion and possess a similar catalytic mechanism. We therefore conclude that in vivo the ability of SETDB1 to methylate histones within a target locus will likely require coordination with deacetylase complexes and putative histone phosphatases.
H3-K9 methylation by SETDB1 enhances HP1 binding
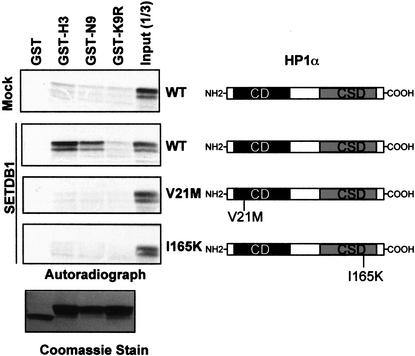
Because HP1 proteins bind methylated K9 histone peptides, we tested whether SETDB1 could stimulate HP1 binding to the N-terminal tails of histone H3. As illustrated in Figure 5, methylation of the GST-H3 and GST-N9 substrates by SETDB1 significantly enhanced the efficiency of HP1α binding to the N-terminal tail of histone H3. This binding activity was abolished by a mutation in the chromodomain (V21M) of HP1α. Furthermore, a mutation in the chromoshadow domain (I165K) that affects the dimerization of HP1α significantly impaired the HP1:histone interaction (Lechner et al. 2000). From this series of data we conclude that SETDB1 is a highly selective histone H3-K9 methylase fully capable of stimulating the binding of HP1 proteins to histone H3.
Figure 5.
SETDB1 methylation of histone H3-K9 enhances HP1 binding. In vitro GST-binding assay between HP1α and GST–H3N. GST–H3N substrates were premethylated with Flag-purified SETDB1 (Fig. 1D) and 15 μM S-adenosyl-L-methionine (Sigma). 35S-L-methinonine labeled in vitro translated HP1α proteins were incubated with the methylated GST–H3N proteins. HP1-histone complexes were eluted by denaturation and resolved on 10% SDS-PAGE gels, and bound HP1α was visualized by fluorography. Coomassie blue stain shows the purified, methylated GST–histone H3 substrates. Schematic diagram of HP1α to the right illustrates the domain organization (CD, chromodomain; CSD, chromoshadow domain) of this protein family and the relative position of the V21M and I165K mutations (Lechner et al. 2000).
Endogenous SETDB1 is a euchromatic H3-specific methyltransferase
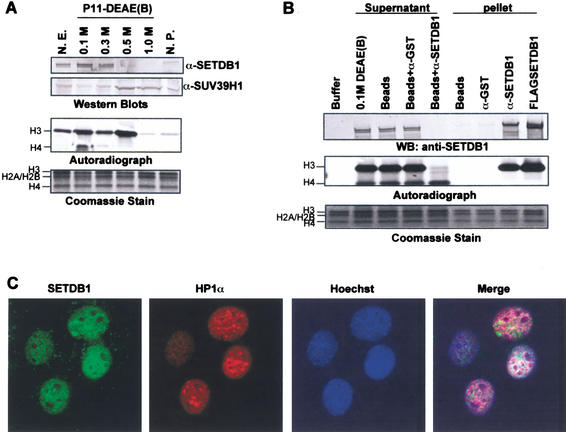
To confirm that endogenous SETDB1 possessed methyltransferase activity we produced a polyclonal antibody that specifically recognizes the protein (Fig. 2A). Western blot analysis of phosphocellulose-fractionated soluble HeLa nuclear extract revealed that SETDB1 primarily elutes in the 0.1 M and 0.3 M KCl elutions, whereas SUV39H1 is present in the 0.5 M and 1.0 M KCl elutions. The fractions containing either protein showed robust H3 methylase activity (Fig. 6A). Antibodies against SETDB1 efficiently immunodepleted nearly all the histone H3 methylase activity from the 0.1 M P11 extract without affecting the H4 activity (Fig. 6B). Moreover, the pellet of the SETDB1 immunoprecipitate retained a strong histone H3 activity that is comparable to that of Flag-purified SETDB1 (Fig. 1D). These observations strongly suggest that endogenous SETDB1 represents an abundant histone H3 methyltransferase. Indirect immunofluoresence staining of asynchronous populations of NIH/3T3 cells revealed that SETDB1 is localized predominantly in euchromatic regions of interphase nuclei and excluded from nucleoli and islands of condensed chromatin, as determined by Hoechst stain and immunostaining with a monoclonal antibody to HP1α (Fig. 6C). However, there is significant overlap between SETDB1 and HP1α in euchromatic regions of the nucleus. Therefore, we propose that SETDB1 functions independently of SUV39H1/H2 and is one cellular HMTase responsible for global euchromatic H3-K9 methylation maintained in the Suv39h double knockout mouse (Peters et al. 2001).
Figure 6.
Endogenous SETDB1 represents a major histone H3-specific methyltransferase. (A) Biochemical fractionation of H3-specific methlytransferases from HeLa nuclear extract. HeLa nuclear extract was fractionated by phosphocellulose (P11) chromatography as previously described (Bochar et al. 2000). HMTase activity was monitored by the in vitro methylation assay. Elution of SETDB1 and SUV39H1 from the P11 column was monitored by Western blot analysis. (B) Supernatants of SETDB1 immunodepleted nuclear extract were devoid of H3 HMTase activity. We incubated 150 μg of the 0.1 M P11 fractionated nuclear extract with protein A-agarose and either affinity-purified anti-GST or anti-SETDB1 IgG. Supernatants and pellets from these immunoprecipitates were assayed for HMTase activity. Coomassie blue stain shows equal amounts of core histone substrate in each reaction. Autoradiograph shows corresponding [3H]methyl-labeled products. Anti-SETDB1 Western blot shows efficient immunodepletion of SETDB1 from the 0.1 M P11 extract. (C) Indirect immunofluoresence of interphase nuclei of NIH/3T3 cells. Affinity-purified polyclonal SETDB1-specific IgG globally stained euchromatic nuclear territories of interphase nuclei (FITC) with little overlap in A-T-rich condensed chromatin domains visualized by Hoechst stain and monoclonal HP1α IgG (Texas Red).
SETDB1 enhances H3-K9 methylation and HP1 at an endogenous locus stably silenced by the KRAB–KAP-1 repression system
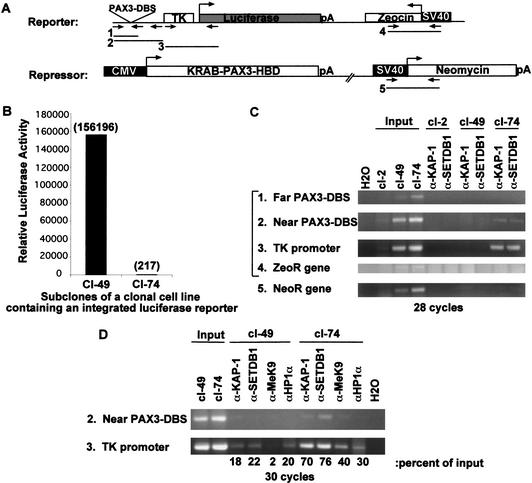
The above results suggest that SETDB1 functions to methylate histone H3-K9 in euchromatic territories of the nucleus to facilitate HP1 deposition. Furthermore, our biochemical data suggest that the KRAB-ZFP–KAP-1 repression complex could target H3-K9 methylation of endogenous gene promoters by SETDB1 facilitating the deposition of HP1 proteins to silence gene expression. To test this hypothesis, chromatin immunoprecipitation (ChIP) experiments were done with a cell line that contains a stably integrated, euchromatic luciferase transgene that is subject to KRAB-mediated repression (Fig. 7A). This two-plasmid system is based on a hormone-regulatable DNA-binding KRAB domain fusion (Ayyanathan et al. 2000) and a TK-luciferase reporter transgene as its target. The fusion protein is fully capable of forming a ternary complex with KAP-1 and HP1 (data not shown). A comprehensive analysis of clonal NIH3T3 cell lines that contain these two plasmids has shown the following: (1) The KRAB domain is a strong repressor of an integrated target gene; (2) repression is relatively short-range, and accompanied by localized chromatin compaction in the promoter region and spatial recruitment to subnuclear territories enriched in condensed chromatin; (3) the KAP-1 corepressor and HP1α/γ proteins are physically associated with the repressed gene in a highly localized manner as judged by ChIP assays. Most remarkable, a large fraction of cells from a hormone-treated population can be isolated that maintain a silent expression state of the transgene for >40 population doublings in the absence of hormone, suggesting that the silenced state is mitotically heritable.
Figure 7.
The KRAB–KAP-1 repression system targets SETDB1 and enhances H3-K9 methylation and HP1 recruitment to promoters of transcriptionally silenced genes. (A) Schematic representation of a two-plasmid system used to create a stably integrated luciferase transgene in NIH/3T3 cells that is regulated by a heterologous KRAB repressor protein. Numbered arrow sets represent the relative position of PCR primers used for PCR amplification of DNA retained by ChIP. (B) Two single-cell subclones containing the heterologous KRAB–PAX3–HBD transcriptional repressor and the integrated luciferase transgene, which is either expressed (cl-49) or stably silenced (cl-74) following hormone treatment. Luciferase activities were measured in subconfluent populations of cells and reported as relative light units per milligram of protein. (C) ChIP experiments showing the colocalization of KAP-1 and SETDB1 at the TK promoter region of the luciferase transgene in the cells where transcription of the luciferase gene has been stably silenced (cl-74). Formaldehyde cross-linked chromatin from cl-49 and cl-74 cells was immunoprecipitated with either affinity-purified KAP-1 or SETDB1 IgG. An equal amount of promoter sequence in cl-49 and cl-74 nucleosomal preparations was determined by PCR from 1% of the input chromatin. PCR-amplified DNA fragments are illustrated in A. cl-2 represents a negative control cell line. (D) ChIPs of cross-linked chromatin with KAP-1, SETDB1, HP1α, and MeK9 antiserum as in C. Bold numbers below each lane represent quantitation of amplified DNA, expressed as percentage of signal intensity for the amplified input DNA.
This system provides a model for HP1-dependent silencing and variegation of a euchromatic gene expression in a mammalian cell line (K. Ayyanathan and F.J. Rauscher III, in prep.), and we have used it as described below to evaluate the role of SETDB1 and histone H3 MeK9 in the stable silencing of the luciferase transgene. We used clonal cell lines that showed either robust expression of luciferase (cl-49) or nearly complete silencing of the luciferase transgene (cl-74; Fig. 7B). In cells containing a silenced luciferase transgene (cl-74), the ChIP experiments indicated that both KAP-1 and SETDB1 readily cross-linked to the luciferase transgene and were significantly colocalized around the TK promoter region of the integrated reporter. In contrast, KAP-1 and SETDB1 were undetectable by ChIP analysis at the TK promoter region in cells (cl-49) showing significant luciferase activity (Fig. 7C). Furthermore, we observed little binding of these proteins to the promoter region of the linked Zeocin resistance locus, which is nearly 3.0 kb downstream of the TK promoter, or at the unlinked Neomycin resistance gene present in the same cells (Fig. 7C). Clonal populations of cells containing only the integrated luciferase transgene failed to show any localization of KAP-1 or SETDB1 to the TK promoter region, suggesting that a DNA-bound KRAB repression module was required for KAP-1 and SETDB1 recruitment (data not shown). We next determined whether the localization of SETBD1 to the TK promoter region enhanced H3-K9 methylation and the recruitment of HP1. As illustrated in Figure 7D, a comparison between these two cell lines has revealed that HP1α and its chromatin ligand, H3-MeK9, are enriched in chromatin containing the TK promoter sequences in cells with a hormone-induced, stably silenced transgene. These data suggest that the KAP-1 corepressor functions as a molecular platform that coordinates the sequential recruitment of histone methyltransferases and the deposition of HP1 at a euchromatic locus to stably silence gene expression that is mitotically heritable.
Discussion
The role of histone modifications in the regulation of chromatin structure and subsequent regulation of eukaryotic gene expression is rapidly being defined (Strahl and Allis 2000; Jenuwein and Allis 2001). The hypothesis that is emerging from these studies is that the combination of modifications on a histone or perhaps other histones within the same nucleosome creates an indexing system, which formats chromatin to properly express the genetic information of the genome, and facilitates the establishment of specialized nuclear structures. However, key questions in understanding all of these histone modifications include the following: (1) How are they targeted to specific gene regulatory elements? (2) What non-histone chromosomal proteins interpret this histone code? (3) How is recognition of this code translated into a change in gene activity?
Here we report that the KAP-1 corepressor of the KRAB-ZFP family of sequence-specific transcriptional repressors associates with SETDB1, a histone H3 Lys 9 methyltransferase, and targets it to gene promoters transcriptionally silenced by a model KRAB repressor protein. Mutation of conserved amino acid residues in the catalytic domain, including the pre-SET, SET, and post-SET domains, significantly impairs the enzymatic activity of SETDB1. Although acetylation and phosphorylation of the H3 N-terminal tail profoundly affect the efficiency of H3-K9 methylation by SETDB1, we found that a substrate methylated at H3-K4 does not affect methylation of H3-K9 by SETDB1. In vitro methylation of the N-terminal tails of histone H3 by SETDB1 is sufficient to enhance the binding of HP1 proteins. Surprisingly, SETDB1 localizes exclusively in euchromatic territories of interphase nuclei and overlaps with HP1 staining in nonpericentromeric regions of chromatin. Furthermore, we show that SETDB1 can be targeted to a stably silenced euchromatic gene via the KAP-1 corepressor. Moreover, recruitment of SETDB1 enhances H3-K9 and HP1 localization to cis-regulatory sequences of a gene silenced by the KRAB–KAP-1 repression system. We propose that KRAB-ZFPs bind to its cognate recognition sequence and then recruits KAP-1 to form a scaffold that coordinates the assembly histone deacetylases, histone methylases, and the deposition of HP1 proteins to silence gene expression by forming a facultative heterochromatin environment.
SETDB1 mediates K9 methylation
The role of histone methylation in the emerging histone code has been revolutionized by the discovery that proteins with the highly conserved SET domain function as lysine-specific histone methyltransferases (Rea et al. 2000). Analysis of the primary amino acid sequence of SETDB1 revealed the presence of amino acid modules highly homologous to the pre-SET, SET, and post-SET domains of the SUV39H1 protein family (data not shown). Interestingly, the SET-domain homology of SETDB1 is interrupted by a 347-amino-acid insertion that is evolutionarily conserved in homologous proteins in lower eukaryotes. Although a recombinant SETDB1 composed of the minimal catalytic domain (pre-SET, SET, and post-SET domains) was not active in our methyltransferase assay, SETDB1 purified from eukaryotic cell extracts showed robust H3-specific HMTase activity using calf thymus core histones, chicken mononucleosomes, or H3 N-terminal peptides as substrates. It is not clear why the recombinant SETDB1 proteins are not active when compared with other histone methyltransferases that are active as recombinant proteins (SUV39H1, G9a, PRMT1). Although we cannot rule out a small-molecular-weight cofactor such as NAD or FAD, we favor a hypothesis that SETDB1 must require posttranslational modification(s) for activity over a cellular protein cofactor, because SETDB1 expressed in baculovirus-infected Sf9 cells and affinity-purified to near homogeneity was sufficient to show identical enzymatic activity as endogenous sources of SETDB1. During our affinity purification of enzymatically active SETDB1, we consistently observed two forms. Moreover, we noticed a tight correlation between the slower-migrating protein species by SDS-PAGE/Western blotting and the enzymatic activity of SETDB1 (Fig. 3B). It is possible that the faster-migrating species is a degradation product. However, peptide-mapping experiments revealed changes in the peptide profiles between the two species that are consistent with small-molecular-weight modifications. In this regard, we found that SETDB1 is constitutively phosphorylated in asynchronous populations of cells, a finding consistent with other members of the SUV39H1 family (D.C. Schultz and F.J. Rauscher III, unpubl.). Nonetheless, the exact types and extent of posttranslational modification of SETDB1 will need to be determined to fully assess the role they have in its enzymatic activity.
Mechanism of K9 methylation
Although methylation of histones has long been established, only recently have the functional consequences of site-specific methylation in modulation of chromatin structure and gene transcription begun to be elucidated. The recent discovery that the H3-MeK9 mark establishes a high-affinity ligand for binding the HP1 family of heterochromatin proteins and targeting its proper subcellular location has provided one of the first links between a histone modification and a repressive chromatin environment for gene expression (Bannister et al. 2001; Jacobs et al. 2001; Lachner et al. 2001; Nakayama et al. 2001; Jacobs and Khorasanizadeh 2002). Here we show that de novo methylation of recombinant histone H3 N-terminal tails by SETDB1 is sufficient to enhance the affinity of HP1 binding. Moreover, the recruitment of SETDB1 to a chromatinized locus targeted for repression by the KRAB–KAP-1 repression system coincides with significant enrichment of H3-K9 methylation and HP1 deposition. In both the in vitro and in vivo experiments, enhanced HP1 binding was completely dependent on H3-K9 methylation. As expected, HP1 binding to the methylated histone tail in vitro required a functional chromodomain. Furthermore, we found that optimal HP1 binding also required a functional chromoshadow domain, as a mutation that disrupts the dimerization of the HP1 protein significantly impaired this association. Thus, HP1 binding to methylated H3-K9 is not exclusively chromodomain-dependent. The later observation implies a potential role for any of the chromoshadow domain binding partners (i.e., KAP-1, CAF150, SP100) in stabilizing of the H3-MeK9–HP1 interaction. Addition of recombinant KAP-1 did not stimulate or enhance HP1 binding to the methylated H3 tail in an in vitro solution-binding assay (D.C. Schultz, unpubl.). One important question that remains to be answered about the HP1–H3-MeK9 interaction is whether the chromodomains of a single HP1 molecule bind to the methylated H3-K9 tails of a single nucleosome or two adjacent nucleosomes. Because KAP-1 exists as a homotrimer in solution, it is possible that KAP-1 may increase the stoichiometry of HP1–H3-MeK9 interactions at a targeted locus by functioning as a nucleosome cross-linking agent.
A possible interplay between H3-K4 and H3-K9 methylation
A more global analysis of H3-K9 methylation at specific loci in Schizosaccharomyces pombe and higher eukaryotes found this chromatin mark to be preferentially associated with transcriptionally inactive chromatin. In contrast, histone H3 Lys 4 methylation has been shown to correlate with histone H3 acetylation and chromatin regions permissive to gene transcription. Moreover, these data indicate that H3-K4 and H3-K9 methylation are mutually exclusive at these two specific loci (Heard et al. 2001; Litt et al. 2001; Noma et al. 2001; Boggs et al. 2002). Furthermore, H3-K4 methylation has been exclusively associated with transcriptionally active macronuclei, but not inactive micronuclei, of Tetrahymena (Strahl et al. 1999). Interestingly, we found that a substrate methylated at H3-K4 had little effect on the ability of SETDB1 to methylate H3-K9. This observation appears to be counterintuitive with the mutual exclusive hypothesis for these two modifications. Furthermore, this observation is in stark contrast to findings for SUV39H1, whose enzymatic activity for H3-K9 is significantly reduced by a methylated H3-K4 substrate (Nishioka et al. 2002). One explanation for such differences might be explained by a histone code hypothesis. In this particular case the pattern of histone modifications on the same histone appear to independently regulate the enzymatic activity of two different enzymes that modify the same residue. In this regard, SETDB1 may be instrumental in maintaining H3-K9 methylation at boundary elements and, thus, prevent the spreading of transcriptionally active chromatin domains from juxtaposed regions enriched in H3-K4 methylation. Alternatively, SETDB1 methylation of H3-K9 may function in concert with H3-K4 to repress transcription in highly specialized chromatin territories or structures. Consistent with this hypothesis, H3-K4 methylation by the Saccharomyces cerevisae protein Set1, the predominant H3-K4 methyltransferase, is required for repression of RNA polymerase II transcription within rDNA, telomeres, and the silent mating-type locus (Nislow et al. 1997; Briggs et al. 2001; Miller et al. 2001; Roguev et al. 2001). Characterization of the H3-K4 methylation status at our engineered transgene and further identification of endogenous loci regulated by SETDB1 will be required to define more accurately the role SETDB1 has in regulation of chromatin structure containing both H3-K4 and H3-K9 methylation.
Histone methylation and DNA methylation
Interestingly, SETDB1 is a unique member of a subclass of SET proteins that possess a canonical CpG DNA methyl binding domain (MBD), which other proteins use to bind methylated DNA (Wade and Wolffe 2001). One model would predict that SETDB1 binds directly to hypoacetylated regions (imprinting centers, inactive X chromosome, transposons, etc.) of methylated DNA and subsequently methylates the corresponding histones of surrounding nucleosomes. Alternatively, it may be that H3-K9 methylation by SETDB1 plays a role in gene-specific targeting of DNA methylation, based on a recent genetic study in Neurospora crassa that has linked maintenance of genomic DNA methylation to histone H3-K9 methylation (Tamaru and Selker 2001). Preliminary results suggest that modifying agents of DNA methylation can alter the silenced state of the integrated transgene (K. Ayyanathan and F.J. Rauscher III, unpubl.). The KRAB–KAP-1 repression system may therefore provide a useful tool to dissect the interplay between histone methylation and DNA methylation in establishing epigenetic states of gene silencing.
Role of KRAB-ZFPs in establishing facultative heterochromatin
These observations strongly suggest a role for the KRAB-ZFP superfamily of transcriptional repressors in sequence-specific establishment of gene silencing. KAP-1 appears to have the capacity to coordinate the biochemical activities required to induce and maintain the assembly of higher-order chromatin structure. We propose that KRAB-ZFPs selectively bind to cognate cis-regulatory elements and recruit the KAP-1 corepressor to the targeted locus. Because KAP-1 is obligatory for KRAB-mediated repression, it appears that the effector molecules of silencing are likely from the network of proteins that interact with KAP-1. Through our studies, we define KAP-1 as a molecular scaffold that coordinates at least four activities necessary for gene specific silencing: (1) targeting to specific promoters (via the >220 KRAB zinc-finger proteins in the human genome); (2) histone deacetylation via the NuRD/HDAC complex; (3) H3-K9 methylation via SETDB1; and (4) deposition of HP1 protein, which collectively facilitate the nucleation of facultative heterochromatin to silence gene expression. To date, Rb is the only other corepressor protein that seems to coordinate similar activities in the repression of E2F target genes (Nielsen et al. 2001). Thus, the KRAB–KAP-1 repression system is one of the best-characterized mammalian systems for gene-specific silencing of euchromatic genes by targeting HP1 proteins. Moreover, the abundance of the KRAB domain zinc-finger proteins in the human proteome and the potentially diverse array of DNA sequences they recognize potentially make this family of gene-specific silencers a master regulator in establishing epigenetic programs of gene silencing during cellular differentiation and organism development.
Materials and methods
Plasmids
Full-length human SETDB1 (KIAA0067) was obtained from the Kazasu DNA Research Institute. Coding sequences for SETDB1 were subcloned NotI/BamHI into pCMV2 (Sigma) to create the CMV-driven Flag-tagged SETDB1 mammalian expression vector. The ΔKID (amino acids 570–1291) expression construct was created by subcloning a HindIII/BamHI fragment into pCMV2. The ΔSET (amino acids 1–951) expression construct was created by subcloning a NotI/BglII fragment into pCMV2. Amino acid substitutions in SETDB1 (R643V, CC 729, 731 LP, H1224K, C1226A, C1279Y) were created using Quick Change PCR mutagenesis strategies (Stratagene). For protein expression in Escherichia coli, a 2.2-kb BamHI fragment encoding amino acids 661–1291 of human SETDB1 was subcloned into pGEX-5X-1. Similarly, a 2.6-kb XhoI/SalI fragment encoding amino acids 585–1291 was subcloned into pGEX-5X-1. For antigen production, a 1.4-kb XhoI fragment encoding amino acids 1–377 from pACT-KIP41 was subcloned into pGEX-4T-1. Previously described PHD finger and bromodomain mutations in KAP-1 were subcloned into the SmaI site of pBTM116 (Capili et al. 2001; Schultz et al. 2001). The GST–histone H3 and GST–G9a bacterial expression plasmids were previously described (Tachibana et al. 2001). Bacterial protein expression and in vitro GST-binding assays were performed as described previously (Ryan et al. 1999; Lechner et al. 2000). Appropriate reading frame fusions and integrity of flanking sequences for all constructs created by PCR were confirmed by DNA sequence analysis of both strands.
Baculovirus SETDB1
The full-length coding sequence for SETDB1 was subcloned via EcoRI/BamHI into pAcHLTa (Pharmingen) to create a tandem 6His/Flag-tagged SETDB1 baculovirus transfer vector. Recombinant baculovirus-infected Sf9 cells (mock or SETDB1) were harvested 48 h postinfection by lysing in RIPA buffer (50 mM Tris at pH 7.9, 150 mM NaCl, 0.5% deoxycholate, 0.1% SDS, 1 mM PMSF, 10 μg/mL pepstatin, 10 μg/mL aprotonin, 10 μg/mL leupeptin, 1 mM Benzamidine, 50 mM NaF, 10 mM NaOV4). Whole-cell extracts were clarified at 100,000g, and supernatants were incubated in batch with Ni2+-NTA agarose (QIAGEN), washed with 20 column volumes of RIPA buffer, followed by 10 column volumes of BC100 (20 mM Tris-HCl at pH 8.0, 100 mM NaCl, 0.2 mM EDTA, 10% glycerol, 0.2 mM PMSF, 0.2% Tween 20). Bound proteins were step-eluted with BC100 supplemented with 300 mM imidazole. Ni2+-NTA eluates were immediately incubated with anti-Flag M2-conjugated agarose (Sigma) at 4°C for 2 h, washed with 20 column volumes of BC1000 (1 M NaCl), followed by 10 column volumes of BC100. Bound proteins were eluted twice in batch with 2 column volumes of BC100 supplemented with 400 μg/mL Flag M2 peptide at 4°C for 1 h each.
Yeast two-hybrid system
The yeast two-hybrid system as modified by Stan Hollenberg was used for all yeast experiments. A human oligo(dT)-primed B-cell cDNA library was screened as described previously (Jensen et al. 1998).
Immunoprecipitation
HEK293 cells were transiently transfected with lipofectamine, and nuclear extracts were prepared 36–48 h posttransfection, as previously described (Ryan et al. 1999). Then 5–10 mg of nuclear extract adjusted to 100 mM NaCl was incubated with 100 μg of anti-Flag M2 (Sigma) at 4°C for 2–4 h. Immune complexes were washed three times with BC500 (20 mM Tris-HCl at pH 8.0, 500 mM NaCl, 0.2 mM EDTA, 10% glycerol, 0.2 mM PMSF, 0.2% Tween 20), once with BC100 (20 mM Tris-HCl at pH 8.0, 100 mM NaCl, 0.2 mM EDTA, 10% glycerol, 0.2 mM PMSF, 0.2% Tween 20), and eluted with 400 μg/mL Flag M2 peptide. Eluted proteins were resolved on a 4%–12% NuPAGE gel in MOPS running buffer (Invitrogen). Proteins were visualized by silver staining or Western blotting to PVDF as previously described (Ryan et al. 1999). For endogenous SETDB1 immunoprecipitation studies, 100 μg of a DEAE-bound, 0.1 M phosphocellulose elution of HeLa nuclear extract was incubated with 5 μg of affinity-purified SETDB1 antibody and 5 μL of protein G-Sepharose (Pharmacia) at 4°C for 2 h. Bound immune complexes were washed three times with BC100 and twice with HMTase buffer prior to assaying for HMTase activity. MS/MS peptide identification was performed at The Wistar Institute Protein Microchemistry Facility, using microcapillary reverse phase HPLC nanospray tandem mass spectrometry on a Finnigan LCQ quadrupole ion trap mass spectrometer.
In vitro histone methyltransferase reactions
In a 40-μL reaction volume, enzyme, 5 μg of core histones (Roche Biochemicals), 2 μg of chicken erythrocyte mononucleosomes, or 5 μg of GST–H3N, and 500 nCi of S-adenosyl-[3H-methyl]-L-methionine (3H-AdoMet; 72 Ci/mmole; NEN Life Science Products) were incubated at 37°C for 1 h in 50 mM Tris (pH 8.5), 20 mM KCl, 10 mM MgCl2, 10 mM β-mercaptoethanol, and 250 mM sucrose. Reactions were terminated by the addition of 5× SDS-buffer. Histones were resolved on 4%–12% NuPage gels in MES running buffer and visualized by Coomassie blue R250 stain. [3H]Methyl labeling was detected by fluorography in 22% PPO solution. Dried gels were exposed to Kodak MRX film. Western Blotting was done as previously described (Ryan et al. 1999).
Immunofluoresence
NIH3T3 cells were grown on glass coverslips in DMEM medium containing 10% calf serum and immunostained as previously described (Maul et al. 1998). The murine SETDB1 protein was visualized by indirect immunofluoresence with an antigen-purified rabbit polyclonal antibody diluted 1:400. DNA was counterstained with Hoechst 33258 (Sigma), and coverslips were mounted with Fluoromount G (Fisher Scientific). Cells were visualized with an inverted light microscope (Leica).
Chromatin immunoprecipitation
The chromatin immunoprecipitation (ChIP) experiments were done essentially as previously described with some modifications (Orlando et al. 1997). Cells were cross-linked with 1% formaldehyde at 37°C for 20 min. The cross-linking reaction was quenched by washing the cells several times with cold TBS (50 mM Tris at pH 8.0, 200 mM NaCl). Cells were scraped into cold TBS supplemented with 5 mM butyric acid. Chromatin was enriched for by washing the cells once in 20 mM Tris HCl (pH 8.0), 0.25% Triton X-100, 200 mM NaCl, 10 mM EDTA, 0.5 mM EGTA, 1 μg/mL aprotonin, leupetitn, pepstatin, 1 mM Benzamidine, 50 mM NaF, 10 mM NaOV3, and 5 mM butyric acid. The cells were centrifuged, resuspended in IP buffer (20 mM NaCl, 0.05% DOC, 0.5% Triton X-100, 0.5% NP-40, 200 mM NaCl, 5 mM butyrate, and protease inhibitors), and sonicated to an average fragment size of 300–500 bp. Solubilized chromatin was clarified by centrifugation at 12,000g, and the supernatant was preincubated for 2 h with protein A agarose beads blocked with salmon sperm DNA and BSA. Precleared chromatin was incubated with 5–10 μg of anti-KAP-1 (Schultz et al. 2001), anti-SETDB1, anti-H3 MeK9 (Upstate Biotechnology), and anti-HP1α (D.C. Schultz, unpubl.) at 4°C for 12–16 h. Immune complexes were bound to protein A agarose beads at 4°C for an additional 2–3 h. The beads were washed four times with IP buffer, two times with high salt buffer (IP buffer with 0.4 M NaCl), once with LiCl buffer (10 mM Tris at pH 8.0, 250 mM LiCl, 0.5% NP40, 1% Triton X-100, 0.1% DOC, 5 mM EDTA, and protease inhibitors), and two times with TE. The DNA–protein complexes were eluted from the protein A beads with 50 mM Tris (pH 8.0), 200 mM NaCl, 5 mM EDTA, and 1% SDS at room temperature for 1 h. The supernatant was transferred to a fresh tube and cross-links were reversed at 65°C for 6–12 h. Samples were treated with 30 μg of Proteinase K (Roche Biochemicals) at 55°C for 2 h, extracted once with phenol, and the DNA was precipitated with 2.5 volumes of ethanol plus 20 μg of glycogen as carrier. Precipitated DNA was pelleted, washed once with 70% ethanol, dried, and resuspended in 25 μL of water. The DNA was analyzed by PCR using specific primer pairs to promoter sequences of the integrated plasmids.
Acknowledgments
We thank Brian D. Strahl and C. David Allis for anti-H3 MeK9 IgG, chicken mononucleosomes, and histone H3 MeK4, MeK9 peptides. We thank Yoichi Shinkai for GST–H3N and mutant plasmids. We thank David Speicher and The Wistar Institute Cancer Center Core Protein Microchemistry Facility for the MS/MS analysis. We thank William Wunner and the Wistar Institute Cancer Center Core Protein Expression Facility for production of baculovirus-expressed SETDB1. D.C.S. was supported by DAMD 17-98-1-8269 and The Wistar Institute Cancer Training Grant CA 09171. G.M. is supported by AI41136. F.J.R. is supported in part by National Institutes of Health grants CA 92088, Core grant CA 10815, GM 54220, DAMD 17-96-1-6141, ACS NP-954, the Irving A. Hansen Memorial Foundation, and The Susan G. Komen Breast Cancer Foundation. Early parts of this work were supported by the Pew Scholars Program in the Biomedical Sciences (F.J.R.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL dcs17@po.cwru.edu; FAX (216) 368-3395.
E-MAIL Rauscher@wistar.upenn.edu; FAX (215) 898-3929.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.973302.
References
- Ayyanathan K, Fredericks WJ, Berking C, Herlyn M, Balakrishnan C, Gunther E, Rauscher FJ., III Hormone-dependent tumor regression in vivo by an inducible transcriptional repressor directed at the PAX3-FKHR oncogene. Cancer Res. 2000;60:5803–5814. [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Berger SL. An embarrassment of niches: The many covalent modifications of histones in transcriptional regulation. Oncogene. 2001;20:3007–3013. doi: 10.1038/sj.onc.1204324. [DOI] [PubMed] [Google Scholar]
- Bochar DA, Savard J, Wang W, Lafleur DW, Moore P, Cote J, Shiekhattar R. A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc Natl Acad Sci. 2000;97:1038–1043. doi: 10.1073/pnas.97.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs BA, Cheung P, Heard E, Spector DL, Chinault AC, Allis CD. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat Genet. 2002;30:73–76. doi: 10.1038/ng787. [DOI] [PubMed] [Google Scholar]
- Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes & Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capili AD, Schultz DC, Rauscher FJ, III, Borden KL. Solution structure of the PHD domain from the KAP-1 corepressor: Structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J. 2001;20:165–177. doi: 10.1093/emboj/20.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad BW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, Rauscher FJ., III KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes & Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- Harte PJ, Wu W, Carrasquillo MM, Matera AG. Assignment of a novel bifurcated SET domain gene, SETDB1, to human chromosome band 1q21 by in situ hybridization and radiation hybrids. Cytogenet Cell Genet. 1999;84:83–86. doi: 10.1159/000015220. [DOI] [PubMed] [Google Scholar]
- Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727–738. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Taverna SD, Zhang Y, Briggs SD, Li J, Eissenberg JC, Allis CD, Khorasanizadeh S. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 2001;20:5232–5241. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, Chodosh LA, Ishov AM, Tommerup N, Vissing H, Sekido Y, et al. BAP1: A novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Katsani KR, Arredondo JJ, Kal AJ, Verrijzer CP. A homeotic mutation in the trithorax SET domain impedes histone binding. Genes & Dev. 2001;15:2197–2202. doi: 10.1101/gad.201901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Lechner MS, Begg GE, Speicher DW, Rauscher FJ., III Molecular determinants for targeting heterochromatin protein 1-mediated gene silencing: Direct chromoshadow domain-KAP-1 corepressor interaction is essential. Mol Cell Biol. 2000;20:6449–6465. doi: 10.1128/mcb.20.17.6449-6465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken β-globin locus. Science. 2001;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- Mark C, Abrink M, Hellman L. Comparative analysis of KRAB zinc finger proteins in rodents and man: Evidence for several evolutionarily distinct subfamilies of KRAB zinc finger genes. DNA Cell Biol. 1999;18:381–396. doi: 10.1089/104454999315277. [DOI] [PubMed] [Google Scholar]
- Maul GG, Jensen DE, Ishov AM, Herlyn M, Rauscher FJ., III Nuclear redistribution of BRCA1 during viral infection. Cell Growth Differ. 1998;9:743–755. [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A. COMPASS: A complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, Reinberg D. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes & Dev. 2002;16:479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nislow C, Ray E, Pillus L. SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol Biol Cell. 1997;8:2421–2436. doi: 10.1091/mbc.8.12.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- Peng H, Begg GE, Harper SL, Friedman JR, Speicher DW, Rauscher FJ., III Biochemical analysis of the Kruppel-associated box (KRAB) transcriptional repression domain. J Biol Chem. 2000a;275:18000–18010. doi: 10.1074/jbc.M001499200. [DOI] [PubMed] [Google Scholar]
- Peng H, Begg GE, Schultz DC, Friedman JR, Jensen DE, Speicher DW, Rauscher FJ., III Reconstitution of the KRAB-KAP-1 repressor complex: A model system for defining the molecular anatomy of RING-B box–coiled-coil domain-mediated protein–protein interactions. J Mol Biol. 2000b;295:1139–1162. doi: 10.1006/jmbi.1999.3402. [DOI] [PubMed] [Google Scholar]
- Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. Loss of the suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan RF, Schultz DC, Ayyanathan K, Singh PB, Friedman JR, Fredericks WJ, Rauscher FJ., III KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: A potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol Cell Biol. 1999;19:4366–4378. doi: 10.1128/mcb.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Friedman JR, Rauscher FJ., III Targeting histone deacetylase complexes via KRAB-zinc finger proteins: The PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2α subunit of NuRD. Genes & Dev. 2001;15:428–443. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Ohba R, Cook RG, Allis CD. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc Natl Acad Sci. 1999;96:14967–14972. doi: 10.1073/pnas.96.26.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- Wade PA, Wolffe AP. ReCoGnizing methylated DNA. Nat Struct Biol. 2001;8:575–577. doi: 10.1038/89593. [DOI] [PubMed] [Google Scholar]