Abstract
In a longitudinal study in a Finnish cattle finishing unit we investigated excretion and sources of Escherichia coli O157 in bulls from postweaning until slaughter. Three groups of 31 to 42 calves were sampled in a calf transporter before they entered the farm and four to seven times at approximately monthly intervals at the farm. All calves sampled in the livestock transporter were negative for E. coli O157 on arrival, whereas positive animals were detected 1 day later. During the fattening period the E. coli O157 infection rate varied between 0 and 38.5%. The animals were also found to be shedding during the cold months. E. coli O157 was isolated from samples taken from water cups, floors, and feed passages. E. coli O157 was detected in 9.7 to 38.9% of the fecal samples taken at slaughter, while only two rumen samples and one carcass surface sample were found to be positive. E. coli O157 was isolated from barn surface samples more often when the enrichment time was 6 h than when the enrichment time was 24 h (P < 0.0001). Fecal samples taken at the abattoir had lower counts (≤0.4 MPN/g) than fecal samples at the farm (P < 0.05). E. coli O157 was isolated more often from 10-g fecal samples than from 1-g fecal samples (P < 0.0001). Most farm isolates belonged to one pulsed-field gel electrophoresis (PFGE) genotype (79.6%), and the rest belonged to closely related PFGE genotypes. In conclusion, this study indicated that the finishing unit rather than introduction of new cattle was the source of E. coli O157 at the farm and that E. coli O157 seemed to persist well on barn surfaces.
Infection with shigatoxigenic Escherichia coli O157:H7 is an important cause of serious illness in humans. The pathogenicity of shigatoxigenic E. coli is mainly mediated by genes coding for Shiga toxins (stx1 and stx2) and attaching and effacing mechanisms (eae) (29). Cattle are considered a major reservoir of this organism (8, 33). Humans acquire the infection through ingestion of contaminated food or drinking water, via direct or indirect cattle contact, or through person-to-person transmission. In a recent Scottish case control study, contact and likely contact with animal feces were strong risk factors for E. coli O157 infection (31). Surveys of cattle at slaughter have given point prevalence values ranging from 1.3 to 28% (4, 6-8, 14, 24, 35, 40). In a 1997 Finnish survey, a low prevalence, 1.3%, was found among cattle at slaughter (28). Elsewhere, surveys performed at the farm level have shown herd prevalence values ranging from 7.1% (15) to 28% (41). Prevalence in cattle may be higher during the summer months and early autumn than in the winter (7). Weaned calves and heifers reportedly shed the agent more often than adult cattle (19) and more often than calves that are less than 8 weeks old (18). In several longitudinal studies performed in the United States it has been noted that E. coli O157:H7 is a ubiquitous organism present on most cattle farms (21).
Cattle finishing units receive animals from several dairy farms and raise them until slaughter. Therefore, they might be a continuous contamination source of E. coli O157 for meat intended for human consumption. The herd in this study was identified in a survey of slaughter cattle (28). Mainly indistinguishable pulsed-field gel electrophoresis (PFGE) genotypes of E. coli O157 were repeatedly isolated from fecal samples taken from the bulls at the cattle finishing unit, raising the question of the source of infection. This study was conducted at the farm to investigate shedding of E. coli O157 from fed cattle from the postweaning period to slaughter in order to explore possible seasonal variation in shedding and to discover if introducing new animals posed a risk of transmission of E. coli O157.
MATERIALS AND METHODS
Description of the farm.
The farm studied was situated in southern Finland, and the herd comprised about 300 cattle. Approximately 3-month-old uncastrated bull calves arrived on the farm after weaning, mainly in lots from dairy farms. The pens in the calf department were routinely cleaned with a high-pressure cleaner the day before arrival of a new lot. The bull calves in this study were delivered by the slaughterhouse organization and first housed in a separate calf department containing pens for four animals before they were moved later to bigger pens for seven or eight animals. When the calves arrived, there were always some calves from the previous group still in the calf department. There was no empty period between the groups. The animals were housed indoors throughout the year and were slaughtered when they were 14 to 17 months old. The pens had a slatted concrete floor, and manure and urine were removed by mechanical scrapes. Slurry was stored in tanks outdoors, treated by aeration, and applied to fields with a slurry injector in the spring. The animals received concentrated feed twice daily, and the maximum portion was 2.8 kg. The concentrated feed comprised barley (2 kg), turnip rape (0.6 kg), and dried mash (0.2 kg). Calcium, minerals, and salt were the only feed additives used. The animals were treated with antimicrobial agents only if there was a veterinary medical indication. No other medications were given. Predried grass silage (1,000 kg) was given to the herd at each feeding, along with concentrated feed fresh mash (1,000 kg).
Sampling. (i) Farm.
Individual rectal samples of fecal material were taken with plastic gloves from new bull calves entering the farm in May 1999 (group 1, n = 40), July 1999 (group 2, n = 21), September 1999 (group 3, n = 31), and December 1999 (group 4, n = 42). Samples from calves in groups 1, 3, and 4 were obtained in a livestock transporter before the calves entered the barn, and samples from calves in group 2 were obtained in the barn 1 day after arrival. Individual rectal samples were taken from the bulls in group 1 from May to November 1999, from the bulls in group 3 from September 1999 to March 2000, and from the bulls in group 4 from December 1999 to May 2000 (Fig. 1). Samples were obtained from the bulls in group 2 on arrival at the farm. Each time, surface samples were also taken from the feed passages, water bowls, water nipples, and floors of pens by using three sterile gauze pads (7.5 by 7.5 cm) that were wetted with sterile buffered peptone water. Before the calves entered the farm, surface samples were taken from the calf department in July, September, and December. Samples were also taken from aerated slurry (n = 3) and nonaerated slurry (n = 6) in May and from mash (n = 1) and hay (n = 1) in August.
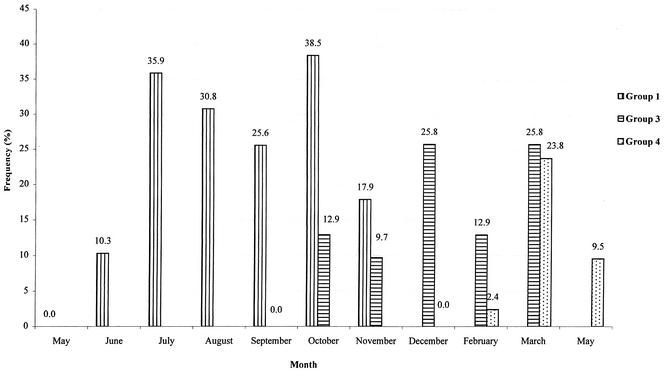
FIG. 1.
Fecal excretion of E. coli O157 in bulls belonging to groups 1, 3, and 4 during farm sampling. A 1-g sample size was used.
(ii) Abattoir.
At the abattoir, samples from feces, rumen contents, and carcass surfaces were taken after stunning, dehiding, and evisceration. Carcass surfaces were swabbed by using wetted gauze pads similar to those used for taking surface samples in the barn. The sampling area included the brisket, the inner and outer sides of the thigh, and the pelvic cavity. Group 1 animals were slaughtered at three different abattoirs between March and June 2000, group 2 animals were slaughtered at two abattoirs between June and December 2000, and group 3 animals were slaughtered at one abattoir between August 2000 and January 2001. Samples were not obtained from group 4 animals at slaughter.
Samples.
All the samples were transported immediately to the National Veterinary and Food Research Institute, Helsinki, Finland, where the analysis began on the same day. The fecal samples taken at the farm were analyzed by using a sample size of 1 g, while the fecal samples taken at the abattoir were analyzed by using sample sizes of 1 and 10 g. The rumen contents were analyzed by using a sample size of 25 g.
Isolation and confirmation.
Isolation was performed by using the ISO 16654 method (25), with some modifications, as described previously (28). Briefly, an enrichment time of 6 h was used for fecal samples, and enrichment times of 6 and 24 h were used for the other samples. After enrichment in modified tryptone soya broth (MTSB), immunomagnetic separation (IMS) was performed; after this, 50 μl was cultured on sorbitol MacConkey agar with and without cefixime and tellurite. Serological testing of the O157 and H7 antigens was performed by using a slide agglutination test (Denka Seiken, Tokyo, Japan). To eliminate the possibility of a false-positive O157 agglutination reaction, every isolate showing positive agglutination was suspended in saline and boiled at 100°C for 1 h. The agglutination test was then repeated. The enterohemolytic phenotype was detected as described previously by using washed sheep blood cells (3). A blood agar with unwashed sheep blood was used in a comparison plate.
MPN method.
Quantitative determination by the most-probable-number (MPN) method was performed for 14 fecal samples that yielded E. coli O157 after enrichment in 1-g samples from the abattoir and for 24 randomly chosen fecal samples taken at the farm in July, September, and October. One gram of fecal material was added to each of five tubes containing 9 ml of MTSB. Each sample tube was diluted 1:10−6; then 1-ml portions from each dilution were added to five tubes with MTSB, and the analysis was continued as described above for fecal samples. The MPN was determined by determining the sum of the positive and negative tubes by a previously described protocol (34).
Genotyping.
Genotyping of the isolates was performed by using PFGE with restriction endonuclease XbaI (28). If possible, up to five colonies per environmental isolate were analyzed by PFGE. The coding system used for genotypes was the system described previously (28). The presence of the virulence genes stx1, stx2, and eae was studied by using a previously described PCR procedure (28).
Statistical analyses.
Enrichment times of 6 and 24 h were compared, as were the 1- and 10-g sample sizes, by using McNemar's test. The quantitative counts of E. coli O157 for the fecal samples originating at the abattoir or farm were compared. The analyses were performed with the SAS statistical program (version 8.0; SAS Institute, Cary, N.C.).
RESULTS
Background information.
The mean ages of the bull calves on arrival were as follows: group 1, 3.4 months (standard deviation, 1.3 months); group 2, 2.9 months (standard deviation, 0.93 month); group 3, 3.1 months (standard deviation, 0.77 month); and group 4, 3.3 months (standard deviation, 0.56 month). The bulls (n = 134) in the four groups originated from 67 farms; the group 1 bulls originated from 27 farms, the group 2 bulls originated from 11 farms, the group 3 bulls originated from 13 farms, and the group 4 bulls originated from 18 farms. All animals were clinically healthy on arrival, although one bull in group 1 died 13 days later. The mean ages at slaughter were as follows: group 1, 16.1 months (standard deviation, 1.25 months); group 2, 15.4 months (standard deviation, 1.37 months); and group 3, 16.0 months (standard deviation, 1.29 months).
Shedding of E. coli O157 on the farm.
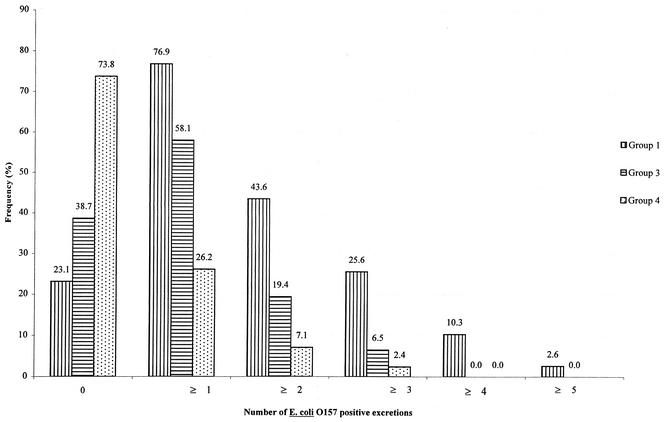
Of the 688 fecal samples taken at the farm, 110 (16%) were positive for E. coli O157. All animals in groups 1, 3, and 4 sampled in the livestock transporter were negative for E. coli O157 on arrival at the farm, whereas six animals (28.6%) in group 2 sampled 1 day after arrival were positive. During the fattening period the E. coli O157 infection rates in groups 1, 3, and 4 varied from 10.3 to 38.5%, from 9.68 to 25.81%, and from 0 to 23.81%, respectively (Fig. 1). No seasonal variation in the shedding of E. coli O157 was found in any of the groups studied. At the times of the peak shedding rates, the mean ages of the bulls were 8.24 months (standard deviation, 1.18 months) for group 1, 6.01 months (standard deviation, 0.77 month) and 9.17 months (standard deviation, 0.77 month) for group 3, and 6.86 months (standard deviation, 0.56 month) for group 4. At seven, six, and four sampling times for the bulls in groups 1, 3, and 4, respectively, 76.9, 58.1, and 26.2% of the animals, respectively, were shedding E. coli O157 at least once (Fig. 2).
FIG. 2.
Fecal excretion of E. coli O157 in bulls belonging to groups 1, 3, and 4 during farm sampling. Group 1 was sampled seven times, group 3 was sampled six times, and group 4 was sampled four times.
Surface samples at the farm.
Of the 203 barn surface samples taken at the farm, 49 (24.6%) yielded E. coli O157. Between 9 and 21 surface samples were taken during each session. E. coli O157 was isolated from samples taken from water cups (18 of 74 samples; 24.3%), floors (8 of 10 samples; 80%), and feed passages (24 of 65 samples; 36.9%). No E. coli O157 was isolated from the 51 water nipple samples. In December, 13 of 20 samples were positive, and the percentage of positive barn surface samples varied from 0 to 55% for the different sampling times. E. coli O157 was isolated from surface samples more often when an enrichment time of 6 h was used than when an enrichment time of 24 h was used (P < 0.0001).
Positive animals at slaughter.
At the abattoir, E. coli O157 was isolated more often from the 10-g fecal samples than from the 1-g fecal samples (P < 0.0001). Samples were taken from 38 bulls in group 1, from 18 bulls in group 2, and from all 31 animals in group 3 (Table 1). E. coli O157-positive samples were obtained most often from animals in group 2 (38.9%) and least often from animals in group 3 (9.7%). E. coli O157 was isolated from 9 of 11 samples taken in August and from 10 of 41 samples taken in June, whereas only 1 of the 27 samples taken from October to January was positive. Two rumen samples were positive, one in May and the other in August. Both animals with positive rumen samples also had positive fecal samples. Only one carcass surface sample was positive (group 2, August).
TABLE 1.
E. coli O157 in fecal samples taken at the abattoir from bulls belonging to groups 1, 2, and 3
| Month | No. of samples | Groupa | No. positive for E. coli O157 with:
|
PFGE geno- type | Positive for E. coli 0157
|
||
|---|---|---|---|---|---|---|---|
| 1-g fecal sample | 10-g fecal sample | Total no. per mo | % per mo | ||||
| May | 8 | 1 | 1 | 1 | 1.62 | 1 | 12.5 |
| June | 30 | 1 | 4 | 10 | 1.1b | 10 | 24.4 |
| June | 11 | 2 | 0 | 0 | |||
| August | 7 | 2 | 6 | 7 | 1.1 | 7 | 63.6 |
| August | 4 | 3 | 0 | 2 | 1.1 | ||
| October | 23 | 3 | 1 | 1 | 1.1 | 1 | 4.3 |
| November | 1 | 3 | 0 | 0 | 0 | 0.0 | |
| January | 3 | 3 | 0 | 0 | 0 | 0.0 | |
| Total | 38 | 12 (31.6)c | 21 (55.3) | ||||
One group 1 animal and three group 2 animals were not sampled.
A rumen isolate from one bull was a PFGE genotype 1.63 isolate.
The numbers in parentheses are the percentages of samples positive for E. coli O157.
MPN analysis.
For five abattoir and two farm samples, the counts were below the detection limit (<0.2 MPN/g), while the highest count exceeded 1.6 × 105 MPN/g (Table 2). Low counts (≤0.4 MPN/g) were encountered more often in the samples from the abattoir than in the samples from the farm (odds ratio, 7.00; 95% confidence interval, 1.13 to 50.76; P = 0.02). The samples were stored refrigerated for 8 to 157 days (mean, 45.6 days; standard deviation, 46.3 days). The levels in 19% of the samples (7 of 37 samples) were below the detection limit even when the samples were positive as determined by the qualitative analysis.
TABLE 2.
Counts of E. coli O157 in fecal samples from bulls at the farm and abattoir as determined by the MPN method
| Bull no. | MPN/g | Group | Mo of isolation | Sampling site |
|---|---|---|---|---|
| 2117 | 7,900 | 1 | June | Farm |
| 2117 | 22 | 1 | July | Farm |
| 2735 | >1,600 | 1 | June | Farm |
| 7511 | 33 | 1 | June | Farm |
| 2735 | 1,100 | 1 | July | Farm |
| 2735 | 2.3 | 1 | June | Abattoir |
| 782 | >160,000 | 1 | September | Farm |
| 782 | 1,300 | 1 | October | Farm |
| 8837 | 0.4 | 1 | September | Farm |
| 1412 | <0.2 | 1 | June | Farm |
| 1412 | 26 | 1 | October | Farm |
| 804 | 11,000 | 1 | October | Farm |
| 6247 | 17 | 1 | October | Farm |
| 4188 | <0.2 | 1 | June | Abattoir |
| 6981 | 22 | 1 | October | Farm |
| 6981 | 13,000 | 1 | June | Abattoir |
| 3072 | 1,700 | 1 | October | Farm |
| 4261 | 4.9 | 1 | May | Abattoir |
| 9967 | 1.4 | 1 | September | Farm |
| 6248 | 240 | 1 | September | Farm |
| 8837 | 1,300 | 1 | October | Farm |
| 2259 | <0.2 | 1 | June | Abattoir |
| 3533 | 22,000 | 1 | June | Abattoir |
| 7969 | <0.2 | 2 | July | Farm |
| 7970 | 9 | 2 | July | Farm |
| 5611 | 54 | 2 | July | Farm |
| 1004 | 140 | 2 | July | Farm |
| 4265 | 900 | 2 | July | Farm |
| 7373 | 1,100 | 2 | July | Farm |
| 3197 | <0.2 | 2 | August | Abattoir |
| 4014 | <0.2 | 2 | August | Abattoir |
| 2264 | 0.4 | 2 | August | Abattoir |
| 4265 | 0.4 | 2 | August | Abattoir |
| 5612 | 1.3 | 2 | August | Abattoir |
| 6898 | 11 | 2 | August | Abattoir |
| 8947 | <0.2 | 3 | October | Abattoir |
| 7965 | 17 | 3 | October | Farm |
| 7061 | 13 | 4 | October | Abattoir |
Characterization of the isolates.
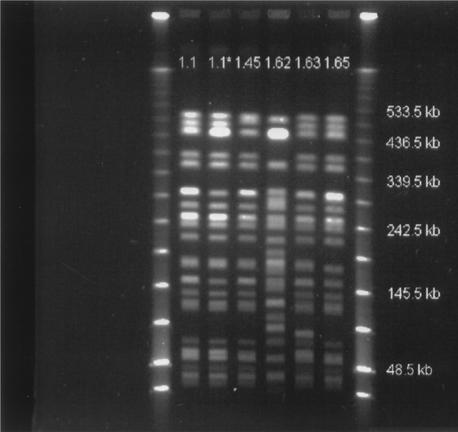
Of the 113 fecal isolates from the farm, 90 (79.6%) belonged to PFGE genotype 1.1, 17 (14.9%) belonged to PFGE genotype 1.1*, 5 (4.4%) belonged to PFGE genotype 1.45, and 1 (0.88%) belonged to PFGE genotype 1.65 (Table 3 and Fig. 3). All of the PFGE genotypes of the farm fecal isolates were closely related (Fig. 3). A PFGE genotype 1.62 isolate that clearly differed from the other genotypes was isolated from the rumen contents and fecal sample of the same bull. A PFGE genotype 1.63 isolate was detected in a sample from the rumen contents of a bull whose fecal isolate was a genotype 1.1 isolate. Bulls that excreted E. coli O157 more than once tended to have isolates with indistinguishable PFGE genotypes from repeated samplings, although a closely related genotype was detected at least once for 8 of 30 bulls with more than one positive isolate (Table 4). The farm surface isolates were PFGE genotype 1.1, 1.1*, and 1.45 isolates. A mean of 3.02 colonies (standard deviation, 1.09 colonies) was analyzed from each positive farm surface sample. In six of 49 cases (12.2%), two different PFGE genotypes were identified from the farm surface sample. All isolates had stx2 and eae genes and the enterohemolytic phenotype. Two fecal isolates from samples taken in February yielded Shiga toxin 1 and 2 genes; the PFGE genotype of one isolate was PFGE genotype 1.1, and the PFGE genotype of the other was PFGE genotype 1.45. Both animals yielded only stx2 in either previous or later samples. Of the 134 fecal isolates, 6 (4.5%) were nonmotile.
TABLE 3.
Distribution of PFGE genotypes of E. coli O157 isolates (n = 194) taken from bulls in groups 1 to 4 and from barn surfaces
| PFGE type | No. of isolates
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Group 1
|
Group 2
|
Group 3
|
Group 4
|
Barn surfaces | Total | |||||
| Farm | Abattoir | Farm | Abattoir | Farm | Abattoir | Farm | Abattoir | |||
| 1.1 | 45 | 10 | 6 | 9 | 27 | 3 | 12 | 2a | 34 | 146 |
| 1.1* | 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 22 |
| 1.45 | 2 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 17 | 22 |
| 1.62 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| 1.63 | 0 | 1b | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 1.65 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Total | 65 | 13 | 6 | 9 | 27 | 3 | 15 | 0 | 56 | 194 |
One rumen isolate and one fecal isolate from one bull.
One rumen isolate.
FIG. 3.
PFGE genotypes of E. coli O157 isolates obtained from bulls at the farm and abattoir and from barn surfaces. Lanes 1 and 8 from left, low-range PFGE marker 350 (New England Biolabs, Beverly, Mass.); lane 2, strain 363; lane 3, strain 376; lane 4, strain 422; lane 5, strain 531; lane 6, strain 556; lane 7, strain 461.
TABLE 4.
PFGE and Shiga toxin genotypes, expression of flagella, and sampling months for E. coli O157 isolates from bulls with two to five excretions of E. coli 0157
| Bull no. | Group | First excretion
|
Second excretion
|
Third excretion
|
Fourth excretion
|
Fifth excretion
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mo | Flagella | stx gene | PFGE genotype | Mo | Flagella | stx gene | PFGE genotype | Mo | Flagella | stx gene | PFGE genotype | Mo | Flagella | stx gene | PFGE genotype | Mo | Flagella | stx gene | PFGE genotype | ||
| 2117 | 1 | June | NMa | 2 | 1.1* | August | NM | 2 | 1.1* | ||||||||||||
| 2735 | 1 | June | NM | 2 | 1.1* | July | H7 | 2 | 1.1 | June | H7 | 2 | 1.1 | ||||||||
| 7511 | 1 | June | H7 | 2 | 1.1 | July | H7 | 2 | 1.1 | August | H7 | 2 | 1.1 | ||||||||
| 782 | 1 | July | H7 | 2 | 1.1* | September | H7 | 2 | 1.1 | October | H7 | 2 | 1.1* | ||||||||
| 8837 | 1 | July | H7 | 2 | 1.1* | August | H7 | 2 | 1.1* | September | H7 | 2 | 1.1* | October | H7 | 2 | 1.1* | ||||
| 1412 | 1 | June | NDb | ND | ND | October | H7 | 2 | 1.1 | ||||||||||||
| 3071 | 1 | October | H7 | 2 | 1.1 | November | H7 | 2 | 1.65 | ||||||||||||
| 2116 | 1 | September | H7 | 2 | 1.1 | October | H7 | 2 | 1.1 | June | H7 | 2 | 1.1 | ||||||||
| 804 | 1 | July | H7 | 2 | 1.1 | September | H7 | 2 | 1.45 | October | H7 | 2 | 1.1 | November | H7 | 2 | 1.1 | ||||
| 8197 | 1 | July | H7 | 2 | 1.1 | October | H7 | 2 | 1.1 | ||||||||||||
| 2257 | 1 | July | H7 | 2 | 1.1 | August | H7 | 2 | 1.1 | October | H7 | 2 | 1.1 | ||||||||
| 6247 | 1 | August | H7 | 2 | 1.1 | September | H7 | 2 | 1.45 | October | H7 | 2 | 1.1 | ||||||||
| 7510 | 1 | July | H7 | 2 | 1.1 | August | H7 | 2 | 1.1 | September | H7 | 2 | 1.1 | November | H7 | 2 | 1.1 | ||||
| 4188 | 1 | August | H7 | 2 | 1.1 | October | H7 | 2 | 1.1 | June | H7 | 2 | 1.1 | ||||||||
| 6981 | 1 | July | NM | 2 | 1.1* | August | H7 | 2 | 1.1 | September | H7 | 2 | 1.1 | October | H7 | 2 | 1.1 | June | H7 | 2 | 1.1 |
| 2253 | 1 | August | H7 | 2 | 1.1 | October | H7 | 2 | 1.1 | November | H7 | 2 | 1.1 | ||||||||
| 3072 | 1 | July | NM | 2 | 1.1* | August | H7 | 2 | 1.1 | October | H7 | 2 | 1.1 | ||||||||
| 4260 | 1 | August | H7 | 2 | 1.1 | November | H7 | 2 | 1.1 | June | H7 | 2 | 1.1 | ||||||||
| 4261 | 1 | October | H7 | 2 | 1.1 | May | NM | 2 | 1.62 | ||||||||||||
| 683 | 3 | December | H7 | 2 | 1.1 | August | H7 | 2 | 1.1 | ||||||||||||
| 685 | 3 | December | H7 | 2 | 1.1 | August | H7 | 2 | 1.1 | ||||||||||||
| 9572 | 3 | December | H7 | 2 | 1.1 | February | H7 | 2 | 1.1 | March | H7 | 2 | 1.1 | ||||||||
| 8947 | 3 | February | H7 | 2 | 1.1 | March | H7 | 2 | 1.1 | October | H7 | 2 | 1.1 | ||||||||
| 7965 | 3 | October | H7 | 2 | 1.1 | November | H7 | 2 | 1.1 | ||||||||||||
| 8945 | 3 | November | H7 | 2 | 1.1 | December | H7 | 2 | 1.1 | February | H7 | 1.2 | 1.1 | ||||||||
| 2087 | 3 | October | H7 | 2 | 1.1 | November | H7 | 2 | 1.1 | ||||||||||||
| 8944 | 3 | February | H7 | 2 | 1.1 | March | H7 | 2 | 1.1 | ||||||||||||
| 7061 | 4 | February | H7 | 1.2 | 1.45 | March | H7 | 2 | 1.45 | May | H7 | 2 | 1.45 | October | H7 | 2 | 1.1 | ||||
| 7971 | 4 | March | H7 | 2 | 1.1 | May | H7 | 2 | 1.1 | ||||||||||||
| 7970 | 4 | March | H7 | 2 | 1.1 | May | H7 | 2 | 1.1 | ||||||||||||
NM, nonmotile.
ND, not determined.
DISCUSSION
Our investigations revealed that the calves most probably acquired E. coli O157 infections at the farm; the organism could not be isolated from any of the 113 animals sampled in the livestock transporter before arrival, whereas 6 of the 31 calves in group 2 sampled 1 day after arrival were shedding E. coli 0157. It should be noted, however, that the transporter surfaces were not sampled, nor were the group 2 animals. Moreover, a majority of the isolates obtained from calves during the fattening period belonged to PFGE genotype 1.1, and the rest belonged to closely related genotypes which were all also found in surface samples taken at the farm before the animals arrived. This is further evidence that the source of infection was at the farm. In contrast to our findings, 3.8% of the calves entering an Italian heifer raising operation (13 of 341 calves) were positive for E. coli O157 on arrival (11), and 6.9% of calves in a United States range beef herd were shedding E. coli O157 at weaning (26). Most bull calves entering cattle finishing units in Finland arrive from dairy herds that are rather closed, with only a few new animals entering the farm, which might explain why the calves were not shedding on arrival. In a cohort study on dairy farms, the animals first tested positive when they were moved from individual housing to group housing (38).
The swab samples taken from the barn surfaces before introduction of new animals showed that the washing procedures had failed to remove E. coli O157. The pens were emptied and washed by using a high-pressure cleaner, but they were not disinfected the day before arrival of the new calves. Thus, the washing procedure and especially the empty period of approximately 1 day were not sufficient to destroy E. coli O157; the floor was still visibly wet when the new calves arrived. In addition, the calf department was not completely vacated between the different calf lots as in each case some of the animals from the previous lot remained, even though calves from different lots were not mixed in the same pens.
A total of 59 (52.2%) of all the bulls studied shed E. coli 0157 at least once during the fattening period. Other longitudinal studies have also found high percentages of animals excreting this pathogen; for example, a Czech study found that 72 (19.7%) of 365 cattle from which individual fecal samples were obtained were positive for E. coli O157 (10), and an Italian study found that 138 (10.7%) of 1,293 cattle from which individual fecal samples were obtained were positive for E. coli O157 (11). In studies in which pat samples have been used, much lower percentages of positive samples have been found, ranging from 0.26 to 3.6% (16, 19, 22, 23, 39). Pat sampling might underestimate the true herd prevalence because negative samples dilute the numbers of E. coli O157, and the survival of E. coli O157 may also decrease in pat samples.
Comparison of results from different prevalence studies is also complicated by the variety of microbiological methods used. In the earliest studies, direct culture without enrichment on a selective medium was used (19), and in some studies IMS was not used (39). Sample sizes have also varied in different studies, from cotton swabs to 1- or 10-g samples. By use of 10-g samples, enrichment and IMS before selective culture have been shown to increase the sensitivity of the method severalfold (2, 9, 13, 37, 41). Our findings also revealed that a sample size of 10 g gave significantly more positives than a sample size of 1 g.
High percentages of the bulls (26 to 77% in groups 1, 3, and 4) shed at least once. The barn surfaces were continuously contaminated, and spreading of the infection was possible. Horizontal transmission has been demonstrated experimentally (5). Although the other groups were not sampled as many times as group 1, it is likely that a higher percentage of shedders would also have been noted in them with more sampling. Thus, the status of shedding in an individual animal cannot be estimated from just one sampling. Persistent shedders were not common. Four of 113 animals (3.53%) were shedding E. coli O157 at four sampling times on the farm, and 13 (11.5%) of these animals were shedding E. coli O157 at three sampling times. This might reflect continuous exposure. Shedding in dairy herds has been found to have a duration of less than 1 month (2).
We found that the animals were also shedders in the colder months. The outside temperature during the sampling in October was 3°C, and in November the outside temperature was 0°C. The shedding increased in the calves during the study period; it was lowest 1 month after arrival and highest when the bulls were 6 to 9 months old. The animals were not on pasture but were fed grass during the summer months (June to August). In a longitudinal study of Wisconsin dairy herds, seasonal shedding was not observed (38), whereas in a British longitudinal study no excretion was detected in a dairy herd between November and May (32). Higher prevalence values were obtained for other herds tested during the summer months than for the herds tested during the spring (17), as well as in a heifer raising operation in Italy (11).
In our study the shedding rate at slaughter seemed to be higher during the warmer months. There were no significant differences in slaughter ages among the groups. A high percentage of the animals were shedding at slaughter: 31.6% as determined with the 1-g samples and 55.3% as determined with the 10-g samples. Most shedders were detected in August. The reason for seasonality in shedding at slaughter remains to be determined. A higher point prevalence in summer was also noted in a Finnish survey of slaughter cattle (28).
The farm surface samples, especially those from water cups and feed passages, were positive. In an experimental study, E. coli O157 was isolated from tonsils (12). E. coli O157 has been isolated from drinking water used for cattle (20, 38, 39), and water has been suggested to be a vehicle for transmission on farms. The bacterium can survive in water for weeks (30). In our study the drinking water of bulls and farmers came from the same source, although the farm family had not experienced episodes of diarrhea. Even though we did not analyze the water for the presence of E. coli O157, it is unlikely that water was the source.
Carcass surfaces were seldom found to be contaminated; only one positive carcass swab sample was detected. The animals were very clean on visible antemortem inspection, and their hides were dry and clean at the farm. In a United States study much higher percentages of carcasses were positive for E. coli O157, and they seemed to be contaminated by neighboring carcasses (14). Differences in slaughter hygiene and farm management, as well as in sampling sites and techniques, may explain such disparities. The animals in the present study were housed indoors and not washed before slaughter; carcasses are minimally washed and not sprayed at Finnish abattoirs. These procedures might decrease cross-contamination between carcasses and survival of the bacterium in the farm environment.
The MPN in fecal samples taken at abattoirs were lower than the MPN in samples taken on the farm. In an experimental study, calves shed higher numbers of E. coli O157 (expressed as CFU per gram) than adult animals shed (12), which could explain why the bulls in our study shed lower numbers at the abattoir than at the farm. The MPN method is the only possible method for quantitative analysis when enrichment and IMS are used. The MPN method used in this study detected low counts (>0.2 MPN/g). In previous studies counts have been detected by direct plating of diluted samples. Direct plating detects samples with concentrations of >102 cells/g, so it would have detected organisms in only 13 (30%) of 37 samples in our study. Differences in methods mean that counts obtained in other studies cannot be directly compared to our results. In another study in which direct plating was used, the numbers of E. coli O157 in the feces of calves ranged from 103 to 105 CFU/g, and 15 (48%) of 31 fecal samples were positive with enrichment only; the population sizes were <102 CFU/g (41). Our MPN counts were possibly lower than the true values because the samples were stored refrigerated for 8 to 157 days (mean, 45.6 days; standard deviation, 46.3 days) and only counts were analyzed when the qualitative results were obtained. This might explain why the levels for 7 (19%) of 37 samples were below the detection limit even when the samples were positive in the qualitative analysis.
An enrichment time of 6 h was better than an enrichment time of 24 h for analyzing the environmental samples. However, when samples with stressed cells are analyzed, the longer enrichment time is superior. In highly contaminated samples, the shorter enrichment time results in more positives because the bacterium grows rapidly and contaminants are suppressed.
Most isolates in our study were members of a single PFGE genotype or a closely related PFGE genotype. PFGE genotype 1.1 has been one of the predominant genotypes in Finnish cattle (27, 28) and in humans too (36). In other studies one genotype (38) or very similar genotypes (15) were found. However, in a longitudinal study of a dairy herd, different PFGE types were present simultaneously (2). This might suggest that the sources on farms can be either single or multiple. In our study only one colony from each positive fecal sample was analyzed because during previous samplings at this farm a mean of 4.5 colonies from 44 positive fecal samples were analyzed and the PFGE genotypes were similar. In 12.2% of the cases at least one parallel colony from the farm surfaces had a different PFGE genotype, but on most occasions the genotypes of the parallel colonies were indistinguishable. Multiple colonies analyzed from single fecal enrichments have usually been shown to have a single PFGE genotype (2). However, other closely related or completely different genotypes could be isolated from parallel enrichments of the same sample, as more than one genotype, even though closely related, was isolated from repeated samples from some bulls in our study. One PFGE genotype (PFGE genotype 1.62) isolated from feces and rumen contents of a bull at slaughter differed considerably from the other genotypes and was never isolated from any of the sampling sites in the present study. The strain with this genotype could have been acquired during the transfer or from the abattoir.
In conclusion, this study showed that the finishing unit and not introduction of new cattle seemed to be the source of E. coli O157 at the farm studied, although the possibility that transporter surfaces were the infection site could not be ruled out. E. coli O157 seemed to persist well on barn surfaces. In order to decrease the numbers of E. coli O157 at the farm, proper cleaning and disinfection and a longer empty period between lots would be needed. The surfaces should be dry before introduction of new animals, and the production system should preferably be an all in-all out system. Seasonal variation in the shedding of E. coli O157 was detected in bulls at slaughter, but there was no absence of shedding at the farm during the cold months. Older animals had lower counts of E. coli O157 than young animals. The counts shed by bulls varied from very low (less than 10 CFU/g) to 105 CFU/g, which emphasizes the importance of using sensitive detection methods.
Acknowledgments
We thank the farmer for cooperation and for help with taking samples and the laboratory technicians for their expert technical assistance.
We thank the Farmos Research Foundation for financial support.
REFERENCES
- 1.Bender, J. B., C. W. Hedberg, J. M. Besser, D. J. Boxrud, K. L. MacDonald, and M. T. Osterholm. 1997. Surveillance for Escherichia coli O157:H7 infections in Minnesota by molecular subtyping. N. Engl. J. Med. 337:388-394. [DOI] [PubMed] [Google Scholar]
- 2.Besser, T. E., D. D. Hancock, L. C. Pritchett, E. M. McRae, D. H. Rice, and P. I. Tarr. 1997. Duration of detection of fecal excretion of Escherichia coli O157:H7 in cattle. J. Infect. Dis. 175:726-729. [DOI] [PubMed] [Google Scholar]
- 3.Beutin, L., M. A. Montenegro, I. Ørskov, F. Ørskov, J. Prada, S. Zimmermann, and R. Stephan. 1989. Close association of verotoxin (Shiga-like toxin) production with enterohemolysin production in strains of Escherichia coli. J. Clin. Microbiol. 27:2559-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonardi, S., E. Maggi, A. Bottarelli, M. L. Pacciarini, A. Ansuini, G. Vellini, S. Morabito, and A. Caprioli. 1999. Isolation of verocytotoxin-producing Escherichia coli O157:H7 from cattle at slaughter in Italy. Vet. Microbiol. 67:203-211. [DOI] [PubMed] [Google Scholar]
- 5.Brown, C. A., B. G. Harmon, T. Zhao, and M. P. Doyle. 1997. Experimental Escherichia coli O157:H7 carriage in calves. Appl. Environ. Microbiol. 63:27-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman, P. A., A. T. Cerdán Malo, M. Ellin, R. Ashton, and M. A. Harkin. 2001. Escherichia coli O157 in cattle and sheep at slaughter, on beef and lamb carcasses and in raw beef and lamb products in South Yorkshire, UK. Int. J. Food Microbiol. 64:139-150. [DOI] [PubMed] [Google Scholar]
- 7.Chapman, P. A., C. A. Siddons, A. T. Cerdan Malo, and M. A. Harkin. 1997. A 1-year study of Escherichia coli O157 in cattle, sheep, pigs and poultry. Epidemiol. Infect. 119:245-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman, P. A., C. A. Siddons, D. J. Wright, P. Norman, J. Fox, and E. Crick. 1993. Cattle as a possible source of verocytotoxin-producing Escherichia coli O157 infections in man. Epidemiol. Infect. 111:439-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman, P. A., D. J. Wright, and C. A. Siddons. 1994. A comparison of immunomagnetic separation and direct culture for the isolation of verocytotoxin-producing Escherichia coli O157 from bovine faeces. J. Med. Microbiol. 40:424-427. [DOI] [PubMed] [Google Scholar]
- 10.Čízěk, A., P. Alexa, I. Literák, J. Hamřik, P. Novák, and J. Smola. 1999. Shiga toxin-producing Escherichia coli O157 in feedlot cattle and Norwegian rats from a large-scale farm. Lett. Appl. Microbiol. 28:435-439. [DOI] [PubMed] [Google Scholar]
- 11.Conedera, G., P. A. Chapman, S. Marangon, E. Tisato, P. Dalvit, and A. Zuin. 2001. A field survey of Escherichia coli O157 ecology on a cattle farm in Italy. Int. J. Food Microbiol. 66:85-93. [DOI] [PubMed] [Google Scholar]
- 12.Cray, W. C., and H. W. Moon. 1995. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Appl. Environ. Microbiol. 61:1586-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Boer, E., and A. E. Heuvelink. 2000. Methods for the detection and isolation of Shiga toxin-producing Escherichia coli. Symp. Ser. Soc. Appl. Microbiol. 29:133S-143S. [DOI] [PubMed]
- 14.Elder, R. O., J. E. Keen, G. R. Siragusa, G. A. Barkocy-Gallagher, M. Koohmaraie, and W. W. Laegreid. 2000. Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc. Natl. Acad. Sci. USA 97:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faith, N. G., J. A. Shere, R. Brosch, K. W. Arnold, S. E. Ansay, M.-S. Lee, J. B. Luchansky, and C. W. Kaspar. 1996. Prevalence and clonal nature of Escherichia coli O157:H7 on dairy farms in Wisconsin. Appl. Environ. Microbiol. 62:1519-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galland, J. C., D. R. Hyatt, S. S. Crupper, and D. W. Acheson. 2001. Prevalence, antibiotic susceptibility, and diversity of Escherichia coli O157:H7 isolates from a longitudinal study of beef cattle feedlots. Appl. Environ. Microbiol. 67:1619-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garber, L., S. Wells, L. Schroeder-Tucker, and K. Ferris. 1999. Factors associated with fecal shedding of verotoxin-producing Escherichia coli O157 on dairy farms. J. Food Prot. 62:307-312. [DOI] [PubMed] [Google Scholar]
- 18.Garber, L. P., S. J. Wells, D. D. Hancock, M. P. Doyle, J. Tuttle, J. A. Shere, and T. Zhao. 1995. Risk factors for fecal shedding of Escherichia coli O157:H7 in dairy calves. J. Am. Vet. Med. Assoc. 207:46-49. [PubMed] [Google Scholar]
- 19.Hancock, D. D., T. E. Besser, M. L. Kinsell, P. I. Tarr, D. H. Rice, and M. G. Paros. 1994. The prevalence of Escherichia coli O157:H7 in dairy and beef cattle in Washington State. Epidemiol. Infect. 113:199-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hancock, D. D., T. E. Besser, D. H. Rice, E. D. Ebel, D. E. Herriott, and L. V. Carpenter. 1998. Multiple sources of Escherichia coli O157 in feedlots and dairy farms in the Northwestern USA. Prev. Vet. Med. 35:11-19. [DOI] [PubMed] [Google Scholar]
- 21.Hancock, D. D., T. E. Besser, D. H. Rice, D. E. Herriott, and P. I. Tarr. 1997. A longitudinal study of Escherichia coli O157 in fourteen cattle herds. Epidemiol. Infect. 118:193-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hancock, D. D., D. H. Rice, L. E. Thomas, D. A. Dargatz, and T. E. Besser. 1997. Epidemiology of Escherichia coli O157 in feedlot cattle. J. Food Prot. 60:462-465. [DOI] [PubMed] [Google Scholar]
- 23.Herriott, D. E., D. D. Hancock, E. D. Ebel, L. V. Carpenter, D. H. Rice, and T. E. Besser. 1998. Association of herd management factors with colonization of dairy cattle by shiga toxin-positive Escherichia coli O157. J. Food Prot. 61:802-807. [DOI] [PubMed] [Google Scholar]
- 24.Heuvelink, A. E., F. L. A. M. van den Biggelaar, E. de Boer, R. G. Herbes, W. J. G. Melchers, J. H. J. Huis in’t Veld, and L. A. H. Monnens. 1998. Isolation and characterization of verocytotoxin-producing Escherichia coli O157 strains from Dutch cattle and sheep. J. Clin. Microbiol. 36:878-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.International Organization for Standardization. 2001. Microbiology of food and animal feeding stuffs—horizontal method for the detection of Escherichia coli O157.16654:2001. International Organization for Standardization, Geneva, Switzerland. [DOI] [PubMed]
- 26.Laegreid, W. W., R. O. Elder, and J. E. Keen. 1999. Prevalence of Escherichia coli O157:H7 in range beef calves at weaning. Epidemiol. Infect. 123:291-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lahti, E., M. Eklund, P. Ruutu, A. Siitonen, L. Rantala, P. Nuorti, and T. Honkanen-Buzalski. 2002. Use of phenotyping and genotyping to verify transmission of Escherichia coli O157:H7 from dairy farms. Eur. J. Clin. Microbiol. Infect. Dis. 21:189-195. [DOI] [PubMed] [Google Scholar]
- 28.Lahti, E., M. Keskimäki, L. Rantala, P. Hyvönen, A. Siitonen, and T. Honkanen-Buzalski. 2001. Occurrence of Escherichia coli O157 in Finnish cattle. Vet. Microbiol. 79:239-251. [DOI] [PubMed] [Google Scholar]
- 29.Law, D. 2000. Virulence factors of Escherichia coli O157 and other Shiga toxin-producing E. coli. J. Appl. Microbiol. 88:729-745. [DOI] [PubMed] [Google Scholar]
- 30.LeJeune, J. T., T. E. Besser, and D. D. Hancock. 2001. Cattle water troughs as reservoirs of Escherichia coli O157. Appl. Environ. Microbiol. 67:3053-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Locking, M. E., S. J. O'Brien, W. J. Reilly, E. M. Wright, D. M. Campbell, J. E. Coia, L. M. Browning, and C. N. Ramsay. 2001. Risk factors for sporadic cases of Escherichia coli O157 infection: the importance of contact with animal excreta. Epidemiol. Infect. 127:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mechie, S. C., P. A. Chapman, and C. A. Siddons. 1997. A fifteen month study of Escherichia coli O157:H7 in a dairy herd. Epidemiol. Infect. 118:17-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ørskov, F., I. Ørskov, and J. A. Villar. 1987. Cattle as reservoir of verotoxin-producing Escherichia coli O157:H7. Lancet i:276. [DOI] [PubMed]
- 34.Peeler, J. T., G. A. Houghtby, and A. P. Rainosek. 1992. The most probable number technique, p. 105-119. In C. Vanderzant and D. F. Splittstoesser (ed.), Compendium of methods for the microbiological examination of foods, 3rd ed. American Public Health Association, Washington, D.C.
- 35.Rice, D. H., E. D. Ebel, D. D. Hancock, T. E. Besser, D. E. Herriott, and L. V. Carpenter. 1997. Escherichia coli O157 in cull dairy cows on farm and slaughter. J. Food Prot. 60:1386-1387. [DOI] [PubMed] [Google Scholar]
- 36.Saari, M., T. Cheasty, K. Leino, and A. Siitonen. 2001. Phage types and genotypes of Shiga toxin-producing Escherichia coli O157 in Finland. J. Clin. Microbiol. 39:1140-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanderson, M. W., J. M. Gay, D. D. Hancock, C. C. Gay, L. K. Fox, and T. E. Besser. 1995. Sensitivity of bacteriologic culture for detection of Escherichia coli O157:H7 in bovine feces. J. Clin. Microbiol. 33:2616-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shere, J. A., K. J. Bartlett, and C. W. Kaspar. 1998. Longitudinal study of Escherichia coli O157:H7 dissemination on four dairy farms in Wisconsin. Appl. Environ. Microbiol. 64:1390-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Donkersgoed, J., J. Berg, A. Potter, D. Hancock, T. Besser, D. Rice, J. LeJeune, and S. Klashinsky. 2001. Environmental sources and transmission of Escherichia coli O157 in feedlot cattle. Can. Vet. J. 42:714-720. [PMC free article] [PubMed] [Google Scholar]
- 40.van Donkersgoed, J., T. Graham, and V. Gannon. 1999. The prevalence of verotoxins, Escherichia coli O157:H7, and Salmonella in the feces and rumen of cattle at processing. Can. Vet. J. 40:332-338. [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao, T., M. P. Doyle, J. Shere, and L. Garber. 1995. Prevalence of enterohemorrhagic Escherichia coli O157:H7 in a survey of dairy herds. Appl. Environ. Microbiol. 61:1290-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]