Abstract
Four sourdoughs (A to D) were produced under practical conditions by using a starter mixture of three commercially available sourdough starters and a baker's yeast constitutively containing various species of lactic acid bacteria (LAB). The sourdoughs were continuously propagated until the composition of the LAB flora remained stable. Two LAB-specific PCR-denaturing gradient gel electrophoresis (DGGE) systems were established and used to monitor the development of the microflora. Depending on the prevailing ecological conditions in the different sourdough fermentations, only a few Lactobacillus species were found to be competitive and became dominant. In sourdough A (traditional process with rye flour), Lactobacillus sanfranciscensis and a new species, L. mindensis, were detected. In rye flour sourdoughs B and C, which differed in the process temperature, exclusively L. crispatus and L. pontis became the predominant species in sourdough B and L. crispatus, L. panis, and L. frumenti became the predominant species in sourdough C. On the other hand, in sourdough D (corresponding to sourdough C but produced with rye bran), L. johnsonii and L. reuteri were found. The results of PCR-DGGE were consistent with those obtained by culturing, except for sourdough B, in which L. fermentum was also detected. Isolates of the species L. sanfranciscensis and L. fermentum were shown by randomly amplified polymorphic DNA-PCR analysis to originate from the commercial starters and the baker's yeast, respectively.
The production of sourdough bread can be traced back to ancient times (24). The products are characterized by their unique flavor, enhanced shelf life and nutritional value, and favorable technological properties (16, 25). Sourdough is an intermediate product and contains metabolically active microorganisms. The microbial ecology of the sourdough fermentation is determined by ecological factors described by Hammes and Gänzle (16). Endogenous factors are determined by the chemical and microbiological components of the dough, and exogenous factors are determined by temperature and atmosphere. In practice, strong effects are exerted by process parameters such as dough yield, amount and composition of the starter, number of propagation steps, and fermentation time. The impact of these parameters during continuous propagation of sourdough causes the selection of a characteristic microflora consisting of lactic acid bacteria (LAB) and usually yeasts. Microbiological studies have revealed that 43 species of LAB, mostly species of the genus Lactobacillus, and more than 23 species of yeasts, especially species of the genera Saccharomyces and Candida (6, 23), occur in this ecological niche. As shown for certain industrial sourdough processes (4), such microbial associations may endure for years, although the fermentation process runs under nonaseptic conditions.
Based on common principles used in artisanal and industrial processes, Böcker et al. (5) defined three types of sourdough. Type I sourdoughs are produced with traditional techniques and are characterized by continuous (daily) propagation to keep the microorganisms in an active state, as indicated by high metabolic activity, above all with regard to leavening, i.e., gas production. The process is performed at temperatures of <30°C, and examples of baked goods so obtained are San Francisco sourdough French bread, panettone, and three-stage sourdough rye bread. The industrialization of the baking process for rye bread led to the development of type II sourdoughs, which serve mainly as dough acidifiers. These sourdoughs are fermented for long periods (up to 5 days) at temperatures of >30°C, and high dough yields permit pumping of the dough. The microorganisms are commonly in the late stationary phase and therefore exhibit restricted metabolic activity only. Type III sourdoughs are dried doughs which are used as acidifier supplements and aroma carriers. Doughs of types II and III require the addition of baker's yeast for leavening. Yeast preparations usually contain LAB, which in predoughs used for the production of soda crackers contribute acidification and aroma development (13).
For investigation of the microbial populations of sourdoughs, traditional cultivation methods in combination with phenotypic (physiological and biochemical) and genotypic (randomly amplified polymorphic DNA [RAPD] and specific PCR) identification techniques have been used (5, 8, 9, 22, 27, 33). These studies, based on culturing techniques, are laborious and time-consuming. Moreover, such studies have focused commonly on the analysis of end products. Denaturing gradient gel electrophoresis (DGGE) of PCR-amplified fragments of the 16S rRNA gene has the potential to characterize the fermentation flora quickly and economically and is culture independent (21). Total bacterial DNA from the habitat of interest is extracted, and a variable region of the 16S rRNA gene is amplified by PCR. The resulting mixture of 16S ribosomal DNA (rDNA) fragments is subjected to polyacrylamide gel electrophoresis by using a denaturating gradient established with urea and formamide in order to separate the fragments and generate a genetic fingerprint of the community. Thus, PCR-DGGE is a suitable tool for investigating microbial community dynamics during whole-food fermentation processes, as demonstrated, for example, for the fermentation of maize dough pozol (3), malt whisky (2), and Italian sausages (7).
In this study, we used two LAB-specific PCR-DGGE systems to monitor changes in the bacterial population at the species level during four sourdough (type I and II) fermentations performed under different ecological conditions. The fermentations were inoculated with the same starter mixture and continuously propagated until a stable microflora had been established. Strains of the various LAB species were isolated by culturing, and their origins were traced back to the starter mixture by using an RAPD-PCR system.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains Lactobacillus acidophilus DSM 20079T, L. amylovorus DSM 20531T, L. brevis DSM 20054T, L. buchneri DSM 20057T, L. crispatus DSM 20584T, L. delbrueckii subsp. delbrueckii DSM 20074T, L. delbrueckii subsp. lactis DSM 20072T, L. farciminis DSM 20184T, L. fermentum DSM 20052T, L. fructivorans DSM 20203T, L. frumenti DSM 13145T, L. johnsonii DSM 20533T, L. panis DSM 6035T, L. paracasei DSM 5622T, L. plantarum DSM 20174T, L. pontis DSM 8475T, L. pontis LTH 1735, L. reuteri DSM 20016T, L. reuteri LTH 3569, L. sanfranciscensis DSM 20451T, L. sanfranciscensis LTH 1729, Pediococcus acidilactici DSM 20284T, P. pentosaceus DSM 20336T, Weissella confusa DSM 20196T, and W. viridescens DSM 20410T were used as reference strains. LAB were cultured in a modified atmosphere (2% O2, 10% CO2, 88% N2) at 30 or 37°C on MRS5 medium, containing, per liter, 10 g of tryptone, 5 g of meat extract, 5 g of yeast extract, 10 g of maltose, 5 g of fructose, 5 g of glucose, 5 g of C2H3NaO2 · 3H2O, 3 g of ammonium chloride, 2.6 g of K2HPO4 · 3H2O, 4 g of KH2PO4, 0.1 g of MgSO4 · 7H2O, 0.05 g of MnSO4 · 4H2O, 0.5 g of cysteine-HCl, 1 ml of Tween 80, and 1 ml of a vitamin mixture (pH 5.8). Sugars were autoclaved separately, and the vitamin mixture, containing cobalamin, folic acid, nicotinic acid amide, pantothenic acid, pyridoxal phosphate, and thiamine (0.2 g/liter each), was sterilized by filtration. For the isolation of LAB and the determination of viable cell counts, MRS5 agar containing 0.1 g of cycloheximide/liter was used, and the plates were incubated at 30°C for 48 h.
Sourdough fermentations and sampling.
Four fermentation batches (A to D) were designed as shown in Table 1. Doughs were prepared from water and rye flour or rye bran and provided different yields. Fermentations were started by adding a starter mixture (M) consisting of three commercial sourdoughs available for industrial use (type I, S1 and S2; type II, S3) and present in equal quantities and 1% baker's yeast (Y). Sourdoughs of type I (A) and type II (B to D) were obtained through continuous propagation by back-slopping of ripe dough for several days. At differing refreshment times, the ripe sourdough was used as an inoculum for the subsequent fermentation cycle. At each refreshment step, samples were taken from the ripe sourdough and investigated as follows. The pH and the total titratable acids (TTA) were determined with 10 g of sourdough suspended in 100 ml of distilled water. The TTA value is expressed as the amount (in milliliters) of 0.1 M NaOH needed to achieve a final pH of 8.5. For microbial counting, samples were serially diluted 1:10 with saline-tryptone diluent (containing, per liter, 8.5 g of NaCl and 1.0 g of tryptone [pH 6.0]) and plated on MRS5 agar. Sourdoughs were propagated until a stable microflora had been established, as indicated by the appearance of approximately the same numbers of different colony forms on MRS5 agar plates. Finally, for DNA extraction and PCR-DGGE analysis, 10-g samples were taken from the starter mixture (M), the starters (S1, S2, and S3), the baker's yeast (Y), and the sourdoughs (A to D) at each refreshment step and stored at −20°C.
TABLE 1.
Design of the sourdough fermentation batches
| Sourdough fermentation batch | Type | Temp (°C) | Stirring | Ingredientsa | Dough yieldb | Amt of starter mixture/inoculum for refreshing (%, wt/wt) | Refreshment time (h) |
|---|---|---|---|---|---|---|---|
| A | I | 25 | No | Rye flour (type 1150), tap water | 200 | 10 | 12 |
| B | II | 30 | No | Rye flour (type 1150), tap water | 200 | 37.5 | 24 |
| C | II | 40 | No | Rye flour (type 1150), tap water | 200 | 37.5 | 24 |
| D | II | 40 | Yes (200 rpm) | Rye bran, tap water | 367 | 10 | 48 |
Type 1150: ca. 1.15 g of ash content/kg of flour dry weight.
(Mass of dough/mass of flour) × 100.
At the end of fermentation, 36 to 43 colonies were picked randomly from an MRS5 agar plate containing 100 to 300 colonies. Additionally, from the starters (S1 to S3) and the baker's yeast (Y), 7 to 10 colonies were picked, with all different colony forms being taken into consideration. Species identification of the isolates was performed as follows. DNA was isolated from pure cultures as described below. DNAs were subjected to PCR-DGGE. Bands showing the same migration distances were considered to belong to the same species. DNA from one representative of each species was subjected to sequencing of the first 1,000 bp or the complete (L. mindensis) sequence of the 16S rDNA.
DNA extraction.
Total DNA was isolated from single colonies grown on MRS5 agar plates by resuspending the cells in 1 ml of sterile phosphate-buffered saline (containing, per liter, 8.0 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4, and 0.24 g of KH2PO4 [pH 8.3]) and applying a High Pure PCR template preparation kit (Roche Molecular Biochemicals, Mannheim, Germany). Lysozyme (20 mg/ml) and mutanolysin (1,000 U/ml) were used as lysing enzymes, and the mixture was incubated at 37°C for 60 min. For the extraction of total DNA from the starter, baker's yeast, and sourdough samples, 10 g was homogenized for 5 min in a stomacher bag containing 90 ml of saline-tryptone diluent. An aliquot (50 ml) was centrifuged at 4°C for 5 min at 200 × g (fermentation batches A to C) or for 5 min at 1,500 × g (fermentation batch D). Finally, to harvest the cells, the supernatant was centrifuged for 15 min at 5,000 × g, and the cell pellet was stored at −20°C. The frozen pellet was thawed on ice and washed three times with 1 ml of phosphate-buffered saline and once with 1 ml of water. The pellet was resuspended in 130 μl of lysis buffer (6.7% sucrose, 50 mM Tris HCl [pH 8.0], 1 mM EDTA, 20 mg of lysozyme per ml, 1,000 U of mutanolysin per ml, 100 μg of RNase per ml). After incubation for 1 h at 37°C, 30 μl of NaCl (5 M) and 25 μl of cetyltrimethylammonium bromide (10% CTAB in 0.7 M NaCl) were added, and the mixture was incubated for 10 min at 65°C. Afterward, 20 μl of proteinase K solution (15 mg/ml) and 10 μl of sodium dodecyl sulfate (20%) were added, and the mixture was incubated for 30 min at 60°C. Finally, 200 μl of phenol (65°C; pH 7) was added and mixed, and the mixture was incubated for 6 min at 65°C. After the mixture was cooled on ice, 220 μl of Tris HCl (10 mM; pH 8.0) was added, and the mixture was extracted twice with equal volumes of phenol-chloroform-isoamyl alcohol (25:24:1) and twice with chloroform. After ethanol precipitation, the DNA was dissolved in 100 μl of Tris HCl (10 mM; pH 8.0).
Primer design.
The PROBE-DESIGN tool of the software package ARB (http://www.biol.chemie.tu-muenchen.de/pub/ARB/documentation/arb.ps) was used for the construction of 16S rDNA-targeted primers. The site within the 16S rDNA which was used previously to design primer Lac1, specific for some LAB, including lactobacilli (35), was chosen as a target site for the primers. To improve the effect of mismatches to mitochondrial plant DNA, the target site was shifted one base to the 5′ end of the 16S rDNA, resulting in primer L1 (5′-CAGCAGTAGGGAATCTTCC-3′). The potential target sequences for the deduced primer were compared with all sequences in the ARB database by using the PROBE-MATCH tool. The melting behavior of PCR fragments obtained with primer L1 and universal primer HDA2 (34), as revealed by the software of the Poland Server (http://www.biophys.uni-duesseldorf.de/POLAND/poland.html), served as a basis for primer optimization. For PCR-DGGE, a 40-bp GC clamp described previously (35) was attached to primer L1, and the resulting primer was designated L1GC.
PCR amplification.
Amplification of 16S rDNA fragments for PCR-DGGE was carried out by using a Primus thermocycler (MWG-Biotech, Ebersberg, Germany) and primers L1GC and HDA2. The reaction mixture (50 μl) contained 25 pmol of each primer, 0.2 mM each deoxyribonucleotide triphosphate, reaction buffer (final concentrations, 10 mM Tris HCl [pH 8.3], 50 mM KCl, and 1.5 mM magnesium acetate), 2.5 U of Taq polymerase (Eppendorf, Hamburg, Germany), and 1 μl of DNA solution. The amplification program was 95°C for 2 min; 30 cycles of 94°C for 30 s, 66°C for 30 s, and 68°C for 1 min; and 68°C for 7 min. Additionally, 16S rDNA fragments for PCR-DGGE were generated by PCR with primers Lac1 and Lac2GC as described previously (35). For identification of pure cultures, the 16S rDNA was amplified by using primers 616V (5′-AGAGTTTGATYMTGGCTCA-3′) and 630R (5′-AAGGAGGTGATCCARCC-3′).
DGGE and excision of DNA fragments.
DGGE was performed as described previously (34) by using a gradient of 32.5 to 50% urea and formamide. Excision and purification of DNA fragments from DGGE gels were performed as described by ben Omar and Ampe (3).
Sequence analysis.
DNA sequences of PCR fragments obtained from purified DGGE bands and pure cultures were determined by the dideoxy chain termination method with an AutoRead sequencing kit (Amersham Pharmacia Biotech), a LI-COR system (MWG-Biotech), and IRD 800-labeled primers L1seq or HDA2seq and 616V, respectively. To determine the closest relatives of 16S rDNA sequences, a search of the GenBank DNA database was conducted by using the BLAST algorithm (1). A similarity of >98% to 16S rDNA sequences of type strains was used as the criterion for identification.
RAPD-PCR.
RAPD-PCR with primer M13V (MWG-Biotech) was performed as described by Müller et al. (20) but with the following modifications. The reaction mixture (50 μl) contained 100 pmol of primer M13V, 0.2 mM each deoxyribonucleotide triphosphate, 3.5 mM MgCl2, reaction buffer, 1.5 U of Taq polymerase, and 1 μl of DNA solution. PCR was carried out by using a GeneAmp 2400 thermocycler (Perkin-Elmer). The amplification program was 94°C for 45 s; 3 cycles of 94°C for 3 min, 40°C for 5 min, and 72°C for 5 min; and 32 cycles of 94°C for 1 min, 60°C for 2 min, and 72°C for 3 min. All PCR products (15 μl) were electrophoretically separated in a 1.5% agarose gel.
RESULTS
Development of specific PCR-DGGE systems.
To avoid the coamplification of mitochondrial plant DNA, the specific PCR primer L1 was constructed and combined with the universal primer HDA2 (34) for the amplification of 185 bp of the 16S rRNA genes of Lactobacillus spp., Weissella spp., Pediococcus spp., and Leuconostoc spp. In silico primer specificity analysis with L1 showed that the Lactobacillus species with relevance in sourdough fermentations are included. For DGGE analysis, a GC clamp of 40 bases was linked to primer L1, resulting in primer L1GC. The PCR-amplified 16S rDNA fragments of various lactobacilli containing this GC clamp exhibited a melting behavior suitable for DGGE. DGGE of the PCR amplicons from representative sourdough lactobacilli showed that the Lactobacillus species could be differentiated according to the migration distances of their respective 16S rDNA fragments, with the exception of L. crispatus, which comigrated with L. acidophilus (hereafter designated L. crispatus/L. acidophilus), and L. sanfranciscensis, which comigrated with L. pontis (hereafter designated L. sanfranciscensis/L. pontis). Differentiation of these species required sequence analysis of the excised DGGE bands. For rapid identification of DGGE bands, an identification ladder of DNA fragments of 13 Lactobacillus reference strains was used (Fig. 1 and 2, lanes RI).
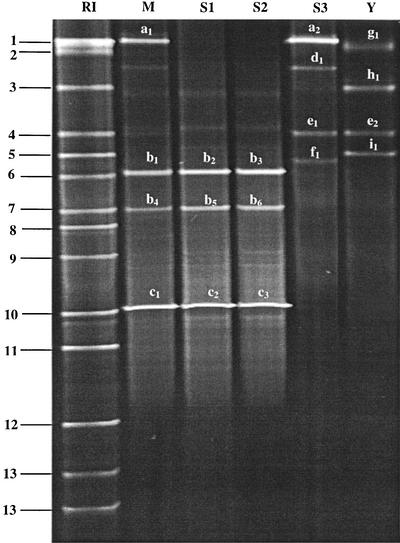
FIG. 1.
DGGE profiles of PCR products obtained with primer pair L1GC-HDA2 and DNA isolated from commercially available sourdough starters and baker's yeast. Lane RI, reference strains: 1, W. confusa; 2, L. johnsonii; 3, L. fermentum; 4, L. brevis; 5, L. crispatus/L. acidophilus; 6, P. pentosaceus; 7, L. farciminis; 8, L. panis; 9, P. acidilactici; 10, L. sanfranciscensis/L. pontis; 11, L. frumenti; 12, L. reuteri; and 13, L. paracasei. Lane M, starter mixture (S1, S2, S3, and Y). Lanes S1 to S3, sourdough starters. Lane Y, baker's yeast. Sequence characterization of the excised fragments indicated the presence of the following: a1 and a2, W. confusa; b1 to b6, L. mindensis; c1 to c3, L. sanfranciscensis; d1, W. viridescens; e1 and e2, L. brevis; f1, L. curvatus; g1, L. plantarum; h1, L. fermentum; and i1, L. crispatus.
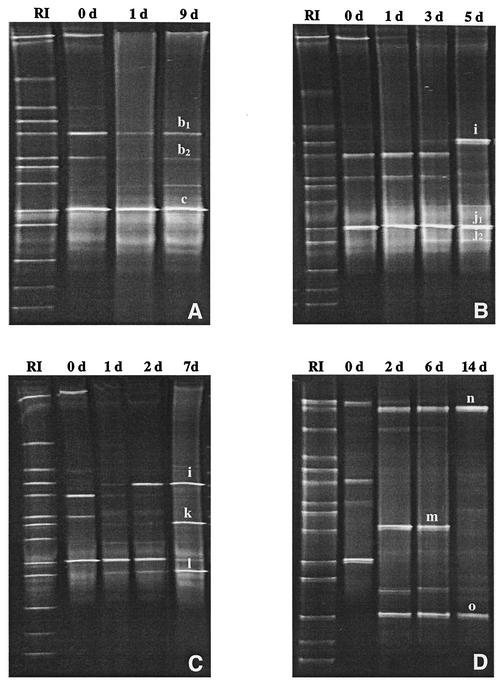
FIG. 2.
DGGE profiles of PCR-amplified 16S rDNA fragments obtained with primer pair L1GC-HDA2 and DNA isolated during sourdough fermentations A, B, C, and D at different fermentation days. (A) After 0 (0 d), 1 (1 d), and 9 (9 d) days. (B) After 0 (0 d), 1 (1 d), 3 (3 d), and 5 (5 d) days. (C) After 0 (0 d), 1 (1 d), 2 (2 d), and 7 (7 d) days. (D) After 0 (0 d), 2 (2 d), 6 (6 d), and 14 (14 d) days. The fragments were excised and sequenced at the end of the fermentation times. Based on sequence comparisons, the fragments were allotted to the following species: b1 and b2, L. mindensis; c, L. sanfranciscensis; i, L. crispatus; j1 and j2, L. pontis; k, L. panis; l, L. frumenti; m, P. acidilactici; n, L. johnsonii; and o, L. reuteri. Lanes RI, reference strains.
Additionally, primers Lac1 and Lac2GC were used to amplify a 340-bp fragment of the V3 region of the 16S rRNA gene of all reference strains. PCR-DGGE analysis of the resulting PCR products revealed migration distances which differed from those observed with primers L1GC and HAD2 (data not shown). Remarkably, the species L crispatus/L. acidophilus and L. sanfranciscensis/L. pontis could be differentiated and identified with primers Lac1 and Lac2GC by using a second identification ladder (Fig. 3, lane RII).
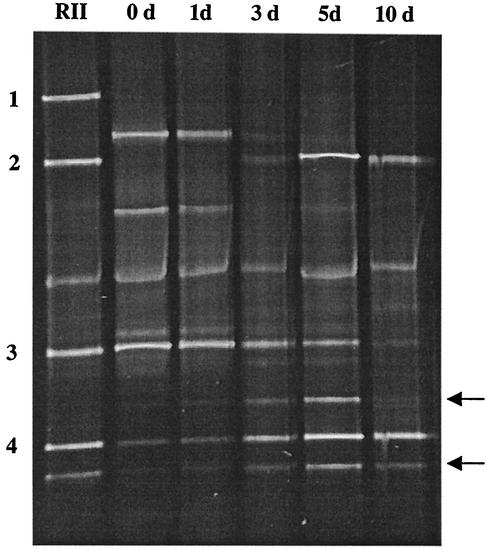
FIG. 3.
DGGE profiles of PCR-amplified 16S rDNA fragments obtained with primer pair Lac1-Lac2GC and DNA isolated from sourdough fermentation batch B after various fermentation times. Fermentation was carried out for 0 (0 d), 1 (1 d), 3 (3 d), 5 (5 d), and 10 (10 d) days. Lane RII, reference strains: 1, L. acidophilus; 2, L. crispatus; 3, L. sanfranciscensis; and 4, L. pontis. Nonspecific bands of L. pontis which could be observed by using pure DNA of the type culture and by using L. pontis strains isolated from dough are indicated by arrows.
Microbial counts, pH, and TTA.
In general, for all fermentations, cell counts for LAB of 108 to 109 CFU/g were obtained (Table 2). These cell counts were reached during the first few fermentation days, with the exception of fermentation batch C, which took 5 days to reach these numbers. Depending on the applied fermentation conditions, especially refreshment time and substrate, the pH and TTA values differed (Table 2). The lowest pH value (3.4 to 3.6) was reached in fermentation batch C, and the highest (3.8 to 3.9) was reached in fermentation batch A. In addition, the smallest amount of TTA (15.2 to 17.7) was reached in fermentation batch A, and the largest amount (69.1 to 79.3) was reached in fermentation batch D (made from rye bran). These data indicated the growth and metabolic activity of the LAB during the whole fermentation period.
TABLE 2.
Time and measured chemical as well as microbial characteristics of the sourdough fermentations
| Sourdough fermentation batch | Fermentation time (days) | pH rangea
|
TTA rangea
|
Cell count (CFU/g) | ||
|---|---|---|---|---|---|---|
| Initial | Final | Initial | Final | |||
| A | 9.5 | 5.5-5.6 | 3.8-3.9 | 4.3-6.7 | 15.2-17.7 | 3.1 × 108-1.8 × 109 |
| B | 10 | 4.1-4.4 | 3.6-3.8 | 7.6-12.4 | 20.3-26.4 | 4.2 × 108-2.0 × 109 |
| C | 13 | 4.1-4.4 | 3.4-3.6 | 7.6-13.5 | 23.6-29.9 | 6.9 × 107-3.2 × 109 |
| D | 16 | 6.2-6.3 | 3.5-3.8 | 12.3-20.5 | 69.1-79.3 | 5.5 × 108-2.9 × 109 |
Measured at the beginning and at the end of each fermentation cycle.
PCR-DGGE fingerprinting of LAB in the starter mixture and sourdough samples.
To characterize the LAB species composition of the starter mixture, PCR-DGGE with primers L1GC and HDA2 was performed with DNA extracted from the starter mixture (Fig. 1, lane M) as well as each commercial starter (Fig. 1, lanes S1 to S3) and the baker's yeast (Fig. 1, lane Y), which were used to prepare the starter mixture. For starters S1 and S2, identical DGGE profiles were obtained, and these clearly differed from that of starter S3; these results indicate that in commercial starters, different LAB species can dominate. Sequence analysis of 16S rDNA fragments from the DGGE gel of the starter mixture (Fig. 1, lane M) showed that these originated from the species L. sanfranciscensis and W. confusa as well as from the new species L. mindensis (10) (EMBL accession number AJ313530). For L. mindensis, two bands were obtained in the DGGE gel; one comigrated with the band obtained for L. farciminis (Fig. 1, lane M, band 7). Multiple fragments with different migration distances were also found for pure cultures of the species L. paracasei, L. pontis, and L. reuteri. There was, however, a major fragment (most dense in the DGGE gel) which indicated the identity of the isolate.
During the fermentation processes, the LAB microflora was monitored by PCR-DGGE with primers L1GC and HDA2, and the fluctuation of the populations is shown in Fig. 2. Changes in the DGGE profiles in all fermentations were observed within a few days, and unique profiles were obtained at the end of the propagation process. In fermentation batch A, L. mindensis and L. sanfranciscensis predominated after just 1 day of fermentation (Fig. 2A), and no further alteration in the DGGE profile was observed up to 9.5 days. After 5 days of fermentation, L. crispatus and L. pontis predominated in fermentation batch B (Fig. 2B); after 7 days of fermentation, L. crispatus, L. frumenti, and L. panis predominated in fermentation batch C (Fig. 2C); and after 14 days of fermentation, L. johnsonii and L. reuteri predominated in fermentation batch D (Fig. 2D). The respective DGGE profiles remained stable until the end of fermentation (Table 2).
As species L. crispatus/L. acidophilus and L. sanfranciscensis/L. pontis could not be differentiated by migration distances in PCR-DGGE with primers L1GC and HAD2, primers Lac1 and Lac2GC were used to monitor the microbial community dynamics in fermentation batch B (Fig. 3). On the basis of a migration distance comparison, L. sanfranciscensis (fragment 3) and L. pontis (fragment 4) could be detected as the dominant species at the start of fermentation (Fig. 3, lane 0 d). During the first 5 days of fermentation, L. sanfranciscensis was detectable and decreased in numbers over time, as indicated by faint bands. On the other hand, the apparent abundance of L. pontis increased, and this species dominated at the end of fermentation. In addition, L. crispatus (fragment 2) could be identified directly in the sourdough without sequencing, since in this system, the migration distance of the PCR amplicon differed from that of L. acidophilus (fragment 1).
Comparison of results of PCR-DGGE analyses of sourdough samples with those of bacteriological culturing.
The sourdough samples taken at the end of the fermentations were subjected to bacteriological culturing on MRS5 agar, and the isolates were subjected to species identification by 16S rDNA sequence analysis. These subcultured isolates represented the predominant strains comprising the population selected on MRS5 agar (18). As shown in Table 3, the species detected by PCR-DGGE were in agreement with those detected by culturing, except for L. fermentum in sourdough fermentation batch B, which was detected only by culturing.
TABLE 3.
LAB composition of sourdough fermentation batches A, B, C, and D, starters (S1, S2, and S3), and baker's yeast (Y), as detected by PCR-DGGE and bacteriological culturing
| Species | Resulta for the following sample (no. tested):
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Sourdough fermentation batch
|
Starter
|
Baker's yeast (9) | ||||||
| A (36) | B (36) | C (38) | D (43) | S1 (10) | S2 (9) | S3 (7) | ||
| L. mindensis | + (2) | + (4) | + (4) | |||||
| L. sanfranciscensis | + (34) | + (3) | + (2) | |||||
| L. crispatus | + (9) | + (2) | − (1) | + (0) | ||||
| L. fermentum | − (5) | − (2) | − (3) | − (3) | + (3) | |||
| L. pontis | + (22) | |||||||
| L. frumenti | + (17) | |||||||
| L. panis | + (19) | |||||||
| L. johnsonii | + (20) | |||||||
| L. reuteri | + (23) | |||||||
| Lactococcus lactis | − (4) | |||||||
| L. brevis | + (1) | + (1) | ||||||
| L. curvatus | + (1) | |||||||
| L. paracasei | − (1) | |||||||
| L. plantarum | + (1) | |||||||
| W. confusa | + (1) | |||||||
| W. viridescens | + (0) | |||||||
+, detected by PCR-DGGE and sequencing of the DNA fragment upon excision from the gel; −, not detected by PCR-DGGE; values in parentheses are numbers of colonies detected by culturing and identified by PCR-DGGE by migration distance comparison and/or sequencing of 16S rDNA.
Investigation of the origins of the predominant LAB strains.
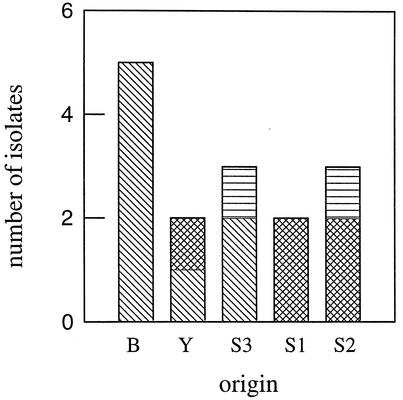
To trace back the origins of the predominant LAB strains, LAB were isolated from the starters (S1 to S3) and the baker's yeast (Y) and subjected primarily to species identification (Table 3). For further differentiation, strains of the species L. crispatus, L. fermentum, and L. sanfranciscensis were compared by RAPD-PCR with those in the fermentation batches. From the starters and the baker's yeast, all isolates were investigated, and from each fermentation batch (A to C), up to nine randomly selected isolates of each species were investigated. All isolates of L. sanfranciscensis obtained from fermentation batch A and starters S1 and S2 had the same RAPD type. For isolates of L. crispatus, different RAPD types occurred in fermentation batches B and C, and these differed from that in starter S1. For L. fermentum, three different RAPD types were obtained from the starters and the baker's yeast. As shown in Fig. 4 only one RAPD type, which could also be found in the baker's yeast and in starter S3, could compete in fermentation batch B.
FIG. 4.
L. fermentum isolates detected in sourdough fermentation batch B (B), baker's yeast (Y), and starters (S1 to S3). Different fills in the bars indicate that different RAPD types were detected.
In addition, DNA was extracted from L. pontis and L. reuteri isolates of fermentation batches B and D, respectively, and subjected to RAPD-PCR. The patterns were compared with these obtained from strains L. pontis LTH 1735 and L. reuteri LTH 3569 (5), which were isolated 9 years ago from commercial sourdough starters (S1 and S2) from the same sources. All isolates of L. pontis and half of the isolates of L. reuteri had the same RAPD type as LTH 1735 and LTH 3569, respectively. Moreover, L. sanfranciscensis LTH 1729 (4), which was isolated 12 years ago from starter S1, had the same RAPD type as the L. sanfranciscensis isolates of fermentation batch A.
DISCUSSION
Numerous studies of sourdoughs by bacteriological culturing showed that species of the genus Lactobacillus belonged to the dominant bacterial flora in the end products of the fermentation process. These sourdoughs originated mainly from industrial processes and had been continuously propagated over a long time, sometimes years (5, 19, 28, 32). As the ecological conditions remained unchanged, a stable microflora without changes in the species composition could be detected. Using LAB-specific PCR-DGGE systems, we have monitored for the first time the development of a Lactobacillus population which is specific for sourdough types I and II. Of several LAB species contained in the starter mixture, only a few Lactobacillus species competed under the prevailing ecological conditions and became dominant, but they did so within a remarkably short time. These findings are consistent with the rule that growth rate and yield of microorganisms are governed by a multitude of ecological factors. In sourdoughs, these factors are temperature, ionic strength, dough yield, and microbial products, such as lactate, acetate, CO2, and ethanol, as well as factors resulting from substrates present in the cereal fraction and from enzymatic reactions (6, 15).
For the type I sourdough (batch A), we observed that L. sanfranciscensis dominated during the whole fermentation process. This finding is consistent with the results of investigations of sourdoughs taken from industrial processes (5, 8, 14, 17). Remarkably, all isolates, including L. sanfranciscensis LTH 1729, which was isolated from a commercial starter (S1) from the same source 12 years ago (4), had the same RAPD type. Thus, strains of that type are highly competitive under the prevailing ecological conditions and may persist for decades in continuously propagated type I sourdoughs. Interestingly, we confirmed by analysis of the 16S rDNA sequence the finding of the new species L. mindensis by PCR-DGGE and bacteriological culturing. This organism has already been isolated from a commercial sourdough starter (10).
Continuous propagation of type II sourdoughs (batches B to D) resulted in the enrichment of L. frumenti, L. fermentum, L. johnsonii, L. panis, L. pontis, and L. reuteri. All of these species had been described as belonging to the predominant lactobacilli of type II sourdoughs (16, 19). RAPD analysis revealed that isolates of L. pontis and L. reuteri from fermentation batches B and D were identical to those obtained previously from commercial type II sourdough starters and thus have remained dominant for a long time during continuous propagation, supporting their important role in type II sourdough fermentations. A comparison of the population dynamics of fermentation batches B and C (Table 3), which were set up under identical ecological conditions except for temperature, revealed the selection of the species L. frumenti and L. panis in batch C (40°C). These species have been described as being more adapted to higher temperatures (19, 36). Thus, the process parameter temperature is an important ecological factor strongly affecting the competitiveness of lactobacilli in sourdough fermentations. This result differs from the conclusion of Müller et al. (20) that the qualitative composition of the Lactobacillus flora is not affected by temperature. However, in their studies, the type II sourdough fermentation was run for 5 days only without any refreshment, and a type II sourdough was used as a starter. Remarkably, we found that L. crispatus dominated in fermentation batches B and C, although L. crispatus has not been considered important in the ecology of sourdoughs until now (5). As shown by RAPD analysis, the dominant strains in fermentation batches B (30°C) and C (40°C) differed, suggesting that within that species, adaptations to different temperatures exist. As their RAPD types also differed from that of the strain isolated from starter S1, they must have been present either in the starter mixture in small numbers, thus escaping detection by PCR-DGGE and culturing, or in the rye flour.
Bacteriological culturing of fermentation samples and especially of the starters resulted in a more complex flora than did DGGE analysis. With regard to the starters, it should be considered that DGGE detects only the predominant 90 to 99% of the bacteria present in an ecosystem (21). As the isolates were picked based on colony forms from agar plates containing up to 300 colonies, isolates were included which represented <1% of the total LAB flora. With regard to the fermentations, L. fermentum was not detected by PCR-DGGE in batch B, although random picking of 36 colonies indicated that this species was one of the predominant members of the flora. These results are consistent with the findings of Ercolini et al. (11), who found that L. fermentum was not detectable in whey cultures of water buffalo mozzarella by PCR-DGGE but was detectable by culturing. Such a bias may be caused by differences in the efficiency of DNA extraction (37) or by preferred amplification of certain templates in PCR (29).
Characterization of the microflora of sourdough fermentations by bacteriological culturing is logistically daunting, because large amounts of colonies must be picked, purified, and identified to the species level by phenotypic or genotypic identification methods. PCR-DGGE has the advantage of allowing study of the species diversity of predominant members of the ecosystem, which can be achieved simply and economically by a single PCR. Sequencing of bands is laborious, and inaccurate identification may occur due to the use of short sequences in combination with sequencing failures. We consider it easier to identify bacteria by comparison of the PCR amplicon migration distances in DGGE gels with those of reference strains, i.e., by using an identification ladder (34). The drawback that even different sequences give rise to bands with similar distances in DGGE gels (2, 7, 21, 34) could be compensated for by using a second primer pair (Lac1 and Lac2GC). As the identification ladders used in this study comprised all important LAB species of sourdough fermentations, all bands of the genetic fingerprint of the sourdough populations could be allotted to species by combined use of the two PCR-DGGE systems without the need for further sequence analysis of PCR fragments. Provided that no PCR bias exists, a semiquantitative prediction of the relative abundance of a species within a population is possible due to the intensity of a PCR fragment in the DGGE gel (31). However, a quantitative estimation of counts requires the use of more complex approaches, e.g., a combination of competitive PCR with DGGE or temperature gradient gel electrophoresis, as shown, for example, by Felske et al. (12).
Species of sourdough lactobacilli exhibit unique technological properties related to the flavor, texture, staling, and shelf life of sourdough bread (16). For example, strains of the species L. sanfranciscensis and L. pontis were shown to improve the taste and flavor of bread (26, 30). Our PCR-DGGE systems are useful tools for the design of sourdough fermentation processes to ensure the development of a desired microflora dominated, for example, by L. sanfranciscensis and L. pontis. Stable composition and activity of the sourdough microflora are important for achieving a constant quality of sourdough bread. Undesirable changes in the bacterial population caused by fluctuating qualities of ingredients or false fermentation conditions can be rapidly detected by using PCR-DGGE and thus adjusted by technological measures.
Acknowledgments
We thank M. Kranz for excellent technical assistance during sequencing.
This study was supported by a grant from LGFG, Baden-Württemberg, Germany.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Beek, S., and F. G. Priest. 2001. Evolution of the lactic acid bacterial community during malt whisky fermentation: a polyphasic study. Appl. Environ. Microbiol. 68:297-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.ben Omar, N., and F. Ampe. 2000. Microbial community dynamics during production of the Mexican fermented maize dough pozol. Appl. Environ. Microbiol. 66:3664-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Böcker, G., R. F. Vogel, and W. P. Hammes. 1990. Lactobacillus sanfrancisco als stabiles Element in einem Reinzucht-Sauerteig-Präparat. Getreide Mehl Brot 44:269-274. [Google Scholar]
- 5.Böcker, G., P. Stolz, and W. P. Hammes. 1995. Neue Erkenntnisse zum Ökosystem Sauerteig und zur Physiologie der sauerteigtypischen Stämme Lactobacillus sanfrancisco und Lactobacillus pontis. Getreide Mehl Brot 49:370-374. [Google Scholar]
- 6.Brandt, M. J. 2001. Mikrobiologische Wechselwirkungen von technologischer Bedeutung. Ph.D. thesis. University of Hohenheim, Honenheim, Germany.
- 7.Cocolin, L., M. Manzano, C. Cantoni, and G. Comi. 2001. Denaturing gradient gel electrophoresis analysis of the 16S rRNA gene V1 region to monitor dynamic changes in the bacterial population during fermentation of Italian sausages. Appl. Environ. Microbiol. 67:5113-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corsetti, A., P. Lavermicocca, M. Morea, F. Baruzzi, N. Tosti, and M. Gobbetti. 2001. Phenotypic and molecular identification and clustering of lactic acid bacteria and yeasts from wheat (species Triticum durum and Triticum aestivum) sourdoughs of southern Italy. Int. J. Food Microbiol. 64:95-104. [DOI] [PubMed] [Google Scholar]
- 9.Creemers-Molenaar, T., M. J. R. Nout, and T. M. G. Bonants-Van Laarhoven. 1985. Microbiological aspects of wheat sour-dough. Antonie Leeuwenhoek 51:607-608. [Google Scholar]
- 10.Ehrmann, M. A., M. R. A. Müller, and R. F. Vogel. 2002. Molecular analysis of sourdough reveals Lactobacillus mindensis sp. nov. Int. J. Syst. Evol. Microbiol., in press. [DOI] [PubMed]
- 11.Ercolini, D., G. Moschetti, G. Blaiotta, and S. Coppola. 2001. The potential of a polyphasic PCR-DGGE approach in evaluating microbial diversity of natural whey cultures for water-buffalo mozzarella cheese production: bias of culture-dependent and culture-independent analyses. Syst. Appl. Microbiol. 24:610-617. [DOI] [PubMed] [Google Scholar]
- 12.Felske, A., A. D. L. Akkermans, and W. M. de Vos. 1998. Quantification of 16S rRNAs in complex bacterial communities by multiple competitive reverse transcription-PCR in temperature gradient gel electrophoresis fingerprints. Appl. Environ. Microbiol. 64:4581-4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fields, M. L., R. C. Hoseney, and E. Varriano-Marston. 1982. Microbiology of cracker sponge fermentation. Cereal Chem. 59:23-26. [Google Scholar]
- 14.Foschino, R., C. Arrigoni, C. Picozzi, D. Mora, and A. Galli. 2001. Phenotypic and genotypic aspects of Lactobacillus sanfranciscensis strains isolated from sourdoughs in Italy. Food Microbiol. 18:277-285. [Google Scholar]
- 15.Gänzle, M. G., M. Ehmann, and W. P. Hammes. 1998. Modeling of growth of Lactobacillus sanfranciscensis and Candida milleri in response to process parameters of the sourdough fermentation. Appl. Environ. Microbiol. 64:2616-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammes, W. P., and M. G. Gänzle. 1997. Sourdough breads and related products, p. 199-216. In B. J. B. Wood (ed.), Microbiology of fermented foods. Chapman & Hall, Ltd., London, England.
- 17.Kline, L., and T. F. Sugihara. 1971. Microorganisms of the San Fransisco sour dough bread process. II. Isolation and characterization of undescribed bacterial species responsible for the souring activity. Appl. Microbiol. 21:459-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCartney, A. L., W. Wang, and G. W. Tannock. 1996. Molecular analysis of the composition of the bifidobacterial and Lactobacillus microflora of humans. Appl. Environ. Microbiol. 62:4608-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Müller, M. R. A., M. A. Ehrmann, and R. F. Vogel. 2000. Lactobacillus frumenti sp. nov., a new lactic acid bacterium isolated from rye-bran fermentations with a long fermentation period. Int. J. Syst. E vol. Microbiol. 50:2127-2133. [DOI] [PubMed] [Google Scholar]
- 20.Müller, M. R. A., G. Wolfrum, P. Stolz, M. A. Ehrmann, and R. F. Vogel. 2001. Monitoring the growth of Lactobacillus species during a rye flour fermentation. Food Microbiol. 18:217-227.
- 21.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 22.Okada, S., M. Ishikawa, I. Yoshida, T. Uchimura, N. Ohara, and M. Kozaki. 1992. Identification and characteristics of lactic acid bacteria isolated from sour dough sponges. Biosci. Biotechnol. Biochem. 56:572-575. [DOI] [PubMed] [Google Scholar]
- 23.Ottogalli, G., A. Galli, and R. Foschino. 1996. Italian bakery products obtained with sour dough: characterization of the typical microflora. Adv. Food Sci. 18:131-144. [Google Scholar]
- 24.Rothe, M., R. Schneeweiss, and R. Ehrlich. 1973. Zur historischen Entwicklung von Getreideverarbeitung und Getreideverzehr. Ernährungsforschung 18:249-283. [Google Scholar]
- 25.Salovaara, H. 1998. Lactic acid bacteria in cereal products, p. 115-138. In S. Salminen and A. Von Wright (ed.), Lactic acid bacteria—technology effects, 2nd ed. Marcel Dekker, Inc., New York, N.Y.
- 26.Spicher, G., R. Schröder, and H. Stephan. 1980. Die Mikroflora des Sauerteiges. X. Mitteilung. Die backtechnische Wirkung in der “Reinzuchtsauern” auftretenden Milchsäurebakterien (Genus Lactobacillus Beijerinck). Z. Lebensm. Unters. Forsch. 171:119-124. [DOI] [PubMed] [Google Scholar]
- 27.Spicher, G. 1984. Die Mikroflora des Sauerteiges. XVII. Mitteilung. Weitere Untersuchungen über die Zusammensetzung und die Variabilität der Mikroflora handelsüblicher Sauerteig-Starter. Z. Lebensm. Unters. Forsch. 178:106-109. [Google Scholar]
- 28.Strohmar, W., and H. Diekmann. 1992. Die Mikroflora eines Langzeit-Sauerteiges. Z. Lebensm. Unters. Forsch. 194:536-540. [Google Scholar]
- 29.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thiele, C., M. G. Gänzle, and R. F. Vogel. 2002. Contribution of sourdough lactobacilli, yeast, and cereal enzymes to the generation of amino acids in dough relevant for bread flavor. Cereal Chem. 79:45-51. [Google Scholar]
- 31.Vaughan, E. E., M. C. de Vries, E. G. Zoetendal, K. Ben-Amor, A. D. L. Akkermans, and W. M. de Vos. 2002. The intestinal LABs. Antonie Leeuwenhoek 82:341-352. [PubMed] [Google Scholar]
- 32.Vogel, R. F., G. Böcker, P. Stolz, M. A. Ehrmann, D. Fanta, W. Ludwig, B. Pot, K. Kersters, K. H. Schleifer, and W. P. Hammes. 1994. Identification of lactobacilli from sourdough and description of Lactobacillus pontis sp. nov. Int. J. Syst. Bacteriol. 44:223-229. [DOI] [PubMed] [Google Scholar]
- 33.Vogel, R. F., M. Müller, P. Stolz, and M. Ehrmann. 1996. Ecology in sourdoughs produced by traditional and modern technologies. Adv. Food Sci. 18:152-159. [Google Scholar]
- 34.Walter, J., G. W. Tannock, A. Tilsala-Timisjarvi, S. Rodtong, D. M. Loach, K. Munro, and T. Alatossava. 2000. Detection and identification of gastrointestinal Lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl. Environ. Microbiol. 66:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walter, J., C. Hertel, G. W. Tannock, C. M. Lis, K. Munro, and W. P. Hammes. 2001. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:2578-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiese, B. G., W. Strohmar, F. A. Rainey, and V. Diekmann. 1996. Lactobacillus panis sp. nov., from sourdough with a long fermentation period. Int. J. Syst. Bacteriol. 46:449-453. [DOI] [PubMed] [Google Scholar]
- 37.Zoetendal, E. G., K. Ben-Amor, A. D. Akkermans, T. Abee, and W. M. de Vos. 2001. DNA isolation protocols affect the detection limit of PCR approaches of bacteria in samples from the human gastrointestinal tract. Syst. Appl. Microbiol. 24:405-410. [DOI] [PubMed] [Google Scholar]