Abstract
A novel phosphodiesterase (PdeA) was purified from Delftia acidovorans, the gene encoding the enzyme was cloned and expressed in Escherichia coli, and the recombinant enzyme was purified to apparent homogeneity and characterized. PdeA is an 85-kDa trimer that exhibits maximal activity at 65°C and pH 10 even though it was isolated from a mesophilic bacterium. Although PdeA exhibited both mono- and diesterase activity, it was most active on the phosphodiester bis(p-nitrophenyl)phosphate with a Km of 2.9 ± 0.1 mM and a kcat of 879 ± 73 min−1. The enzyme showed sequence similarity to cyclic AMP (cAMP) phosphodiesterase and cyclic nucleotide phosphodiesterases and exhibited activity on cAMP in vivo when the gene was expressed in E. coli. The IS1071 transposon insertion sequence was found downstream of pdeA.
Biological systems use organophosphates as energy carriers, signaling molecules, and major components of the cell (DNA, RNA, and phospholipids). As such, organisms contain a number of organophosphate esterases to regulate the levels of these compounds. Due to the low solubility of inorganic phosphate in the environment, organisms also produce organophosphate esterases to liberate inorganic phosphate for use in growth. The most studied organophosphate esterase is the monoesterase alkaline phosphatase (16). Recently, there has been significant interest in phophotriesterases because of their ability to detoxify the organophosphate triester nerve agents and pesticides (6, 18). The phosphodiesterases remain the least studied of the phosphoesterases.
Phosphodiesterases from mammalian sources have attracted some attention due to their importance in regulation (19). These phosphodiesterases are high-affinity enzymes, whereas phosphodiesterases from bacterial sources show much lower affinities (12). Bacterial phosphodiesterases have been purified from a variety of sources including Escherichia coli (10), Haemophilus influenzae (14), and Burkholderia caryophylli PG2982 (7). The phosphodiesterases from E. coli and H. influenzae are similar in sequence, and both moderate intracellular cyclic AMP (cAMP) levels. A phosphodiesterase with no sequence similarity to the enzymes of E. coli and H. influenzae that was isolated from B. caryophylli PG2982 initially for its monoesterase activity was later found to have diesterase activity (7). This enzyme could not be clearly ascribed a function but was thought to play a role in a xenobiotic degradation pathway because it degraded glycerol glyphosate.
Delftia acidovorans (formerly Comamonas acidovorans), originally isolated from a waste treatment facility, can utilize diethylphosphate (DEP) and diethylthiophosphate as sole sources of phosphorus (5). To date, the phosphodiesterase responsible for this activity has not been isolated or cloned. Since many organophosphate xenobiotics such as pesticides and chemical warfare agents contain ester bonds, this phosphodiesterase could be applied to biodegradation of this class of pollutants. Other enzymes isolated from D. acidovorans have been implicated in xenobiotic degradation pathways for compounds such as 3-chloroanaline, aniline, and nitrobenzene (3, 9, 11).
In the present study, we isolated the phosphodiesterase from D. acidovorans, purified it to apparent homogeneity, and studied its kinetic properties. The purified enzyme was shown to act on a broad range of organophosphate substrates. The optimal pH and temperature for PdeA were also determined.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains used were E. coli DH10B (F− mcrA Δ[mrr-hsdRMS-mcrBC] 80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araΔ139 Δ[ara leu]7697 galU galK l− rpsL nupG), purchased as electrocompetent cells from Invitrogen (Carlsbad, Calif.), E. coli BL21(DE3) (F− ompT hsdSB [rB− mB−] gal dcm [DE3]), purchased as chemically competent cells from Novagen (Madison, Wis.), and D. acidovorans from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ 1621). The plasmid pBluescript SK(−) (Stratagene) was used for generating a gene library and subcloning the pdeA gene, and pTYB1 (New England Biolabs) was used for production of the enzyme in E. coli. E. coli was grown in either Luria broth (LB) or Terrific broth with appropriate antibiotics. D. acidovorans was grown in 3-morpholinopropanesulfonic acid (MOPS)-buffered minimal medium (17).
Isolation of gene encoding phosphodiesterase activity.
Genomic DNA from D. acidovorans was purified with a Qiagen genomic DNA extraction kit, digested with Asp718, BamHI, BbrPI, ClaI, DraIII, EcoRI, EcoRV, HindIII, MunI, PstI, PvuII, SacI, SalI, XbaI, XhoI, EcoRI and SalI, or EcoRI and ClaI, and electrophoresed on a 0.7% agarose gel. The DNA was blotted onto nylon membranes (Turboblotter system; Schleicher and Schuell, Keene, N.H.) and probed with the 27-bp degenerate probe labeled with [γ-32P]ATP in hybridization solution (5× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA {pH 7.7}], 10× Denhardt's solution, 0.5% sodium dodecyl sulfate [SDS], and 1 mg of herring sperm DNA/ml). Hybridization was carried out at 40°C. The posthybridization washes contained 1% SDS and 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate). The membrane was washed for 2 h at 40°C after hybridization, followed by 10-min washes at 45, 50, or 55°C. The radioactive signal was captured on X-ray film.
To isolate the pdeA gene, genomic DNA was digested with ClaI and separated on an agarose gel. The 7-kb region of digested DNA was gel extracted and ligated into pBluescript cut with the same enzyme. The partial library was transformed into E. coli DH10B, and the transformed E. coli was plated onto LB agar containing 100 μg of ampicillin/ml and 32 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)/ml. White colonies were plated in a grid onto fresh LB agar plates, grown overnight, and replica plated onto a nylon membrane (Nytran N; Schleicher and Schuell). The cells were lysed by soaking the filter in a solution containing 0.5 N NaOH and 1.5 M NaCl and then neutralized with 1 M Tris-HCl (pH 8). The DNA was cross-linked to the membrane by baking at 80°C for 1 h. Cell debris was removed by soaking the filter in 200 μg of proteinase K/ml dissolved in 1× TBS (137 mM NaCl, 25 mM Tris base, and 2.7 mM KCl adjusted to pH 7.4). The membranes were hybridized using essentially the same procedures outlined for Southern blots. The positive clone pSKT1 was sent for nucleotide sequencing by Genemed Inc. (South San Francisco, Calif.) and the DNA Sequencing Facility at the University of California at Berkeley.
Phosphodiesterase activity assay.
Phosphodiesterase (PdeA) activity from D. acidovorans was determined by measuring the rate of hydrolysis of bis(p-nitrophenyl)phosphate (BNP) by the procedure of Bessey et al. (2). The hydrolysis of 1 mol of BNP yielded 1 mol of p-nitrophenol (PNP). PNP is yellow at basic pH, and the absorbance at a wavelength of 402 nm (A402) was measured with a Beckman DU640 spectrophotometer. BNP (13.2 mM, 100 μl) and alkaline buffer (2-amino-2-methyl-1-propanol; pH 10.3, 1.5 M, 100 μl) were added to a 1.6-ml microcentrifuge tube and incubated at 30°C. The protein sample (5 to 20 μl) was added to the buffered BNP mixture and reacted for 20 to 60 min at 30°C, at which time the reaction was terminated by addition of 1 ml of 0.05 N HCl. To determine background absorbance, 20 μl of concentrated HCl was added to the mixture to remove the yellow color and the A402 was measured. The difference between the two A402 readings was compared to a standard curve to determine the concentration of the PNP that evolved. This end point assay was found to be linear over a 90-min time period, thus eliminating the need for a continuous assay. One unit of activity is defined as the number of moles of substrate utilized per minute at 30°C. For determining kinetic parameters, the range of substrates used was 0.1 to 5 times the Km and the data were fitted directly to the Michaelis-Menten equation by a least-squares method.
Purification of native phosphodiesterase (PdeA).
D. acidovorans was grown in a 15-liter fermentor at 30°C in MOPS medium with diethylthiophosphate as the sole phosphorus source and 4-hydroxybenzoic acid as the carbon source. At an optical density at 600 nm (OD600) of 0.9, the cells were removed from the fermentor, centrifuged at 17,600 × g, suspended in 25 ml of 50 mM Tris-HCl (pH 8.0) containing 10% sucrose, and stored at −80°C prior to lysis. The cells were thawed on ice, and DNase I was added to a final concentration of 100 μg/ml. A French press was used to lyse the cells at 14,000 lb/in2. The resulting sample was centrifuged at 31,000 × g for 1 h, and the supernatant (crude cell extract) was saved.
Ion-exchange chromatography was used to purify the native protein. The crude cell extract was dialyzed overnight in 50 mM Tris-HCl (pH 7.7). Approximately 400 mg of total protein was dialyzed against 12 liters of buffer at 4°C with the Pierce Slide-A-Lyzer dialysis assembly (cutoff at a molecular weight of 10,000). The dialyzed protein was loaded onto a 20-ml column containing DEAE-650 M anion-exchange medium (Amersham Pharmacia, Piscataway, N.J.) equilibrated in 50 mM Tris-HCl (pH 7.5). A step-wise gradient from 20 mM to 2 M NaCl was used to elute the protein. To analyze for phosphodiesterase activity, the protein fractions from the column were separated on a NuPAGE 4 to 12% Tris-glycine (Invitrogen) polyacrylamide native gel with a Tris-glycine running buffer. The gel was run at 125 mV (8 to 20 mA) for 2 h 20 min, followed by an incubation in the alkaline buffer-BNP used in the PdeA assay for 20 min. The appearance of a yellow band indicated the location of a protein with PdeA activity. This band was then cut from the native gel and separated by denaturing SDS-polyacrylamide gel electrophoresis (PAGE) (NuPAGE) at 125 mV (8 to 20 mA) for 2 h 20 min to further purify the protein. The gel was electroblotted onto a polyvinylidene difluoride membrane at 135 V for 45 min, and the band on the membrane was sequenced at its N terminus by using Edman degradation (Molecular Structure Facility at the University of California at Davis). From theN-terminal sequence (Met-His-Lys-Phe-Ile-His-Ile-Thr-Asp-?-His-Leu-Val-Glu-Gln-Gly-Arg [note that residue 10 was unknown from protein sequencing but was determined to be Ile from subsequent DNA sequencing]) and a codon usage table for D. acidovorans (Codon usage table for D. acidovorans, K. D. R. Institute [http://www.kazusa.or.jp/codon/]), a degenerate 27-base oligonucleotide encoding the first nine amino acids (ATG CAC AAG TTC/T ATC CAC ATC ACC/G GAC/T) was designed for Southern blotting analysis.
Cloning and overexpression of PdeA.
With pSKT1 plasmid DNA as a template for the PCR, an NdeI site was added directly upstream of the open reading frame (ORF), the stop codon of pdeA was removed, and a SapI site was added directly downstream. The gene was then cloned into the NdeI and SapI restriction sites of the pTYB1 vector to add a C-terminal intein tag to PdeA.
E. coli BL21(DE3) harboring the pTYB1-pdeA plasmid was used to overexpress pdeA prior to purification of PdeA. Cultures were grown at 37°C in 2-liter flasks containing 400 ml of Terrific broth supplemented with 4 g of glucose and 20 mg of carbenicillin to an OD600 of 0.6 to 0.8. At this time, the culture was cooled to 16°C and IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 0.3 mM. The cultures were grown for an additional 16 to 20 h at 16°C and then removed from the shake flasks and centrifuged at 17,600 × g. The resulting pellet was stored at −80°C until lysis. Prior to lysis, 2 g of cell paste was suspended in 9 ml of 20 mM Tris-HCl (pH 8.0)-10 mM MgCl2-500 mM NaCl (buffer A). The mixture was immediately sonicated by using four 20-s pulses while kept on ice. The cell debris was separated from the protein supernatant (crude cell extract) by centrifugation at 17,530 × g and 4°C for 45 min.
Purification of recombinant phosphodiesterase.
The chitin column purification of the recombinant protein was carried out at 4°C by gravity flow. Approximately 7.5 ml of cell supernatant was loaded onto a 5-ml chitin column (New England Biolabs, Beverly, Mass.) previously equilibrated with buffer A. After protein loading and extensive washing with 25 column volumes, the column was flushed with 50 mM dithiothreitol (DTT) in buffer A and left overnight to allow cleavage of the intein tag from PdeA. After 18 h of cleavage, the PdeA was eluted from the column with buffer A in seven 2-ml fractions. Fractions I and II from the chitin column were loaded onto a Sephacryl S200 (Amersham Pharmacia) size exclusion column and eluted by using buffer A with 1 mM DTT. The column was calibrated with gel filtration standards from Bio-Rad. The fractions containing PdeA activity were pooled and used for all characterization studies. The purity of the enzyme was evaluated with 4 to 12% Tris-glycine SDS-PAGE gels by using a benchmark protein ladder (Invitrogen) as a molecular weight standard.
Protein concentration was determined by the method of Bradford (4) with a Bio-Rad protein dye staining solution.
β-Galactosidase assay.
Strains were grown in LB medium to an OD600 of 0.07 to 0.1, at which time IPTG, to a final concentration of 1 mM, was added to the culture. The cultures were grown until the OD600 reached 0.35 to 0.45. The β-galactosidase activity was measured according to the method of Miller (15). The experiment was repeated three times, with each experiment completed in triplicate.
Nucleotide sequence accession number.
The nucleotide sequence reported in this study has been submitted to GenBank under accession no. AF548455.
RESULTS
Identification of PdeA activity in and cloning of pdeA gene from D. acidovorans.
PdeA activity was monitored through purification by using BNP in a colorimetric assay. The first step in the purification of the native enzyme from D. acidovorans used a DEAE column. The specific activity increased 5.7-fold, with a yield of 37%. Because the use of several cation exchange and size exclusion columns resulted in a significant loss of activity without purification (data not shown), the protein was purified electrophoretically by using a native gel followed by a denaturing gel. The N-terminal sequence of the band with PdeA activity was determined.
A degenerate 27-bp oligonucleotide was constructed from the N-terminal sequence and used in Southern blots to probe digested D. acidovorans genomic DNA. Based on these Southern blots, a restriction map was constructed. A partial ClaI genomic library was constructed by choosing fragments likely to contain the entire pdeA gene (∼7 kb) and probed with the 27-bp oligonucleotide. Three hundred colonies were screened, resulting in three positive clones. The plasmid DNA of the positive clones was also isolated, probed with the labeled oligonucleotide, and confirmed as being that of positive clones.
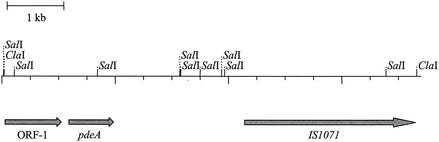
When one ClaI-positive library clone, pSKT1, was sequenced, the sequence encoding the N terminus of PdeA was found at the beginning of a 792-bp ORF (pdeA) (Fig. 1). A search for sequence similarity using the BLAST network service on the National Center for Biotechnology Information website (1) yielded moderate-to-high identity (44 to 58%) to cyclic nucleotide phosphodiesterases from Mesorhizobium loti and Streptomyces avermitilis and a cyclic 3′,5′-AMP phosphodiesterase from Salmonella enterica serovar Typhimurium LT2. On the basis of this putative ORF, the deduced size of the protein subunit was 29,968 Da. We also investigated the sequences surrounding pdeA. Directly upstream was an ORF that showed similarity to that encoding a sugar binding protein of an ABC transporter and downstream was the transposon insertion sequence IS1071.
FIG. 1.
Physical map of the 7.3-kb genomic DNA fragment cloned from D. acidovorans.
Characterization of recombinant phosphodiesterase.
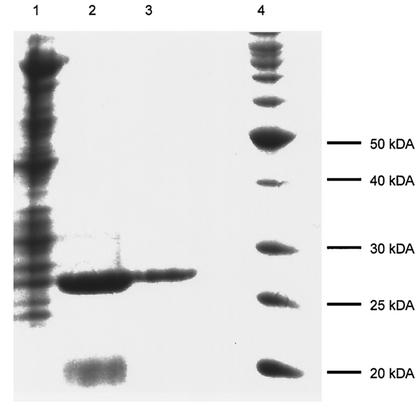
The gene encoding PdeA was overexpressed in E. coli BL21(DE3), and then PdeA was purified to homogeneity by affinity chromatography followed by size exclusion chromatography. The enzyme had an apparent native molecular weight of 85,000 as determined by chromatography on a Sephacryl S200 size exclusion column (data not shown). An SDS-PAGE gel showed a single peptide chain of approximately 30,000 Da, indicating that the protein was a homotrimer (Fig. 2). The specific activity increased 12-fold, with a yield of 36%.
FIG. 2.
SDS-PAGE analysis of samples from PdeA purification. Lane 1, total-cell lysate; lane 2, chitin column fraction; lane 3, Sephacryl S200 fraction; lane 4, molecular mass markers. The gel was stained with Coomassie brilliant blue.
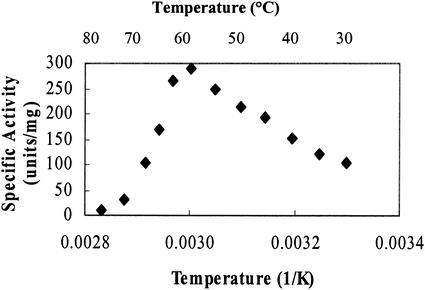
We found that the enzyme was active at higher-than-expected temperatures. Although the enzyme was isolated from a mesophilic soil bacterium, the activity appeared to be highest at 65°C (Fig. 3). The activity of the enzyme increased 64% from 30 to 65°C and then rapidly fell off at higher temperatures. The temperature stability of the enzyme was also investigated. As expected, the enzyme was less stable at 60°C than at 30 and 45°C. The half-lives at 30, 45, and 60°C were 133, 86, and 26 min, respectively.
FIG. 3.
Effect of temperature on PdeA activity. Two experiments were performed, each in duplicate. The data from one experiment are shown. The standard deviations among the four sets of data were less than 7%.
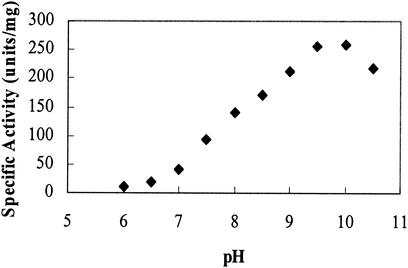
The enzyme activity also depended on the pH of the assay mixture and the buffer. The enzyme was active at more-alkaline pH, with the maximal activity occurring at approximately 10 (Fig. 4). An assay performed in 100 mM Tricine at pH 8.0 showed approximately 30% less activity than an assay performed in 100 mM Tris at the same pH. A buffer system of constant ionic strength was utilized to eliminate the variability of ionic strength among buffers (8).
FIG. 4.
Effect of pH on PdeA activity. A three-buffer system of constant ionic strength was utilized (see Materials and Methods). The experiment was performed in triplicate, with the standard deviations less than 5%.
We found that the activity of PdeA also depended on the presence of metals. During the initial purification from D. acidovorans, addition of 10 mM EDTA in the dialysis and elution buffers was found to reduce activity by 50 to 80% (data not shown). All subsequent purifications did not use EDTA. When the purified recombinant enzyme was subjected to 200 mM EDTA, activity decreased 80%. Addition of 5 mM Mg2+ (in the form of MgCl2) was able to stimulate activity by 105%.
We investigated the kinetic properties of PdeA by studying the catalytic efficiency of the enzyme on several substrates. The enzyme had a Km for BNP of 2.9 ± 0.1 mM and a kcat on BNP of 879 ± 73 min−1 (Table 1). The kinetics of the enzyme showed an excellent fit to the Michaelis-Menten equation (data not shown). PdeA was also tested on a variety of mono-, di-, and triesterase substrates. The enzyme demonstrated monoesterase activity on p-nitrophenyl phosphonate but not on p-nitrophenyl phosphate. No activity was found by using tris(p-nitrophenyl)phosphate as a substrate, indicating that PdeA did not possess triesterase activity.
TABLE 1.
Kinetics of PdeA on various substrates
| Substrate | Km (mM) | kcat (min−1) | kcat/Km (min−1 M−1) |
|---|---|---|---|
| BNP | 2.9 ± 0.1 | 879 ± 73 | 3.0 × 105 |
| p-Nitrophenyl phosphonate | 5.0 ± 0.2 | 171 ± 22 | 3.4 × 104 |
Because of pdeA's homology to the gene encoding the cAMP phosphodiesterase of E. coli (CpdA), the activity of PdeA on cAMP was tested in an in vivo assay determining the ability of the enzyme to hydrolyze cAMP in vivo and to inhibit the expression of lacZ in the absence of catabolite repression (10). The β-galactosidase activity in E. coli BL21(DE3) decreased from 136 ± 26 to 20 ± 8 β-galactosidase units when pdeA was expressed, a drop of nearly 85%.
DISCUSSION
The gene for a phosphodiesterase from D. acidovorans was cloned from genomic DNA and expressed in E. coli BL21(DE3), and its product was purified to apparent homogeneity and characterized. The sequence of PdeA has similarity to other bacterial phosphodiesterases, e.g., the 3′,5′ cyclic nucleotide phosphodiesterase from M. loti, indicating that it may function in D. acidovorans as a cyclic nucleotide phosphodiesterase.
The catalytic efficiency of PdeA (3.0 × 105 min− M−1) on BNP corresponds to kinetic parameters found for other bacterial phosphodiesterases (7, 10). With the ability to cleave both BNP and p-nitrophenyl phosphonate, this enzyme demonstrates both di- and monoesterase activities. Since PdeA was not able to cleave p-nitrophenyl phosphate, the monoesterase activity appears to be limited to monoesters of phosphonates and to not include monoesters of phosphates. The higher catalytic efficiency for BNP suggests that the enzyme may function primarily as a phosphodiesterase as opposed to a phosphomonoesterase. Interestingly, even though PdeA behaved similarly with respect to substrate range to a phosphodiesterase from B. caryophylli PG2982, there is no significant DNA sequence identity between the genes encoding the two enzymes.
The dependence of other phosphodiesterases on metals such as Mg2+ is well documented (7, 12, 13, 21). Partial recovery of activity upon addition of Mg2+ to PdeA treated with EDTA indicated that the removal of metal does not irreversibly denature the protein. The phosphodiesterase from B. caryophylli PG2982 was also found to depend on metals, and it was thought that the divalent cation was required to stabilize negative charges on the transition state intermediate (7).
We tested the ability of the PdeA to cleave cAMP and thus reduce expression of lacZ. The results showed an 85% decrease in β-galactosidase activity in E. coli BL21(DE3) with the expression of pdeA. The phosphodiesterase from E. coli (CpdA) exhibited very similar characteristics, with an 80% decrease in β-galactosidase activity in W3110 upon the expression of cpdA (10), suggesting that the D. acidovorans enzyme may regulate intracellular cAMP levels. A search for amino acid sequence similarity between pdeA and cpdA using the BLAST program on the National Center for Biotechnology Information website showed only 28% identity between the two sequences.
PdeA allows D. acidovorans to use DEP as a sole source of phosphorus. The bacterium will use DEP only upon phosphate starvation and loses this ability if repeatedly grown on rich media (data not shown). The loss of activity on rich media indicates that the enzyme may not be essential to the cell, and, therefore, PdeA may not function primarily as a cAMP phosphodiesterase.
The 7.3-kb genomic DNA fragment from which pdeA was derived contained two other interesting regions. ORF-1 showed 24 to 40% similarity to ORFs encoding sugar binding proteins of Brucella melitensis, M. loti, Agrobacterium tumefaciens, and Sinorhizobium meliloti. Downstream of pdeA was the transposon insertion sequence IS1071, identified in D. acidovorans strain B (20), Pseudomonas sp. strain ADP (GenBank accession no. AAK50274), Burkholderia cepacia (GenBank accession no. AAK81686), and Pseudomonas putida UCC22 (GenBank accession no. BAA12804.1). It has been suggested that this transposon is involved in the spread of catabolic genes among environmental bacterial species due to IS1071's prevalence in bacterial strains that metabolize xenobiotics such as aniline, carboxydiphenyl ethers, and 2-4-dichlorophenoxyacetate (20). The proximity of pdeA, a gene that may have applications in degradation of organophosphate xenobiotics, to this transposon suggests that pdeA also may encode a protein with a xenobiotic catabolic function.
The ability to use DEP as a phosphate source could have applications in the degradation of organophosphate nerve agents and pesticides. The ability of PdeA to act as a phosphomonoesterase in addition to a phosphodiesterase gives it a broad substrate range for degradation. The broad pH range for the enzyme also gives flexibility for the environment in which the enzyme can function.
Acknowledgments
This research was supported by grants from the National Science Foundation (BES-9814088) and the Office of Naval Research (N00014-99-1-0182) and by graduate fellowships to S.K.T. from the University of California at Berkeley and the University of California Toxic Substances and Research Teaching Program.
We thank Robert Frankenberg, Vincent J. J. Martin, and Jack D. Newman for useful discussions.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bessey, O. A., O. H. Lowry, and M. J. Brock. 1946. A method for the rapid determination of alkaline phosphatase with five cubic millimetres of serum. J. Biol. Chem. 164:321-329. [PubMed] [Google Scholar]
- 3.Boon, N., J. Goris, P. De Vos, W. Verstraete, and E. M. Top. 2001. Genetic diversity among 3-chloroaniline- and aniline-degrading strains of the Comamonadaceae. Appl. Environ. Microbiol. 67:1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Cook, A. M., C. G. Daughton, and M. Alexander. 1980. Desulfurization of dialkyl thiophosphoric acids by a Pseudomonas. Appl. Environ. Microbiol. 39:463-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Frank, J. J. 1991. Organophosphorus cholinesterase inhibitors: dexotification by microbial enzymes. Plenum Press, New York. N.Y.
- 7.Dotson, S. B., C. E. Smith, C. S. Ling, G. F. Barry, and G. M. Kishore. 1996. Identification, characterization, and cloning of a phosphonate monoester hydrolase from Burkholderia caryophilli PG2982. J. Biol. Chem. 271:25754-25761. [DOI] [PubMed] [Google Scholar]
- 8.Ellis, K. J., and J. F. Morrison. 1982. Buffers of constant ionic strength for studying pH-dependent processes. Methods Enzymol. 87:405-426. [DOI] [PubMed] [Google Scholar]
- 9.Hoffmann, D., S. Kleinsteuber, R. H. Mueller, and W. Babel. 2001. Development and application of PCR primers for the detection of the tfd genes in Delftia acidovorans P4a involved in the degradation of 2,4-D. Acta Biotechnol. 21:321-331. [Google Scholar]
- 10.Imamura, R., K. Yamanaka, T. Ogura, S. Hiraga, N. Fujita, A. Ishihama, and H. Niki. 1996. Identification of the cpdA gene encoding cyclic 3′,5′-adenosine monophosphate phosphodiesterase in Escherichia coli. J. Biol. Chem. 271:25423-25429. [DOI] [PubMed] [Google Scholar]
- 11.Lessner, D. J., G. R. Johnson, R. E. Parales, J. C. Spain, and D. T. Gibson. 2002. Molecular characterization and substrate specificity of nitrobenzene dioxygenase from Comamonas sp. strain JS765. Appl. Environ. Microbiol. 68:634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim, S. T., A. K. Krishnan, and K. K. Ong. 1989. Purification and characterization of the second form of cyclic 3′,5′-nucleotide phosphodiesterase from Rhizobium fredii. Biochim. Biophys. Acta 991:353-358. [Google Scholar]
- 13.Lim, S. T., U. M. Palanisamy, and K. K. Ong. 1986. Purification and properties of 3′,5′-cyclic nucleotide phosphodiesterase from Rhizobium fredii MAR-1. Arch. Microbiol. 146:142-146. [Google Scholar]
- 14.Macfadyen, L. P., C. Ma, and R. J. Redfield. 1998. A 3′,5′ cyclic AMP (cAMP) phosphodiesterase modulates cAMP levels and optimizes competence in Haemophilus influenzae Rd. J. Bacteriol. 180:4401-4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Neidhardt, F. C., R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.). 1996. Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 17.Neidhardt, F. C., P. L. Bloch, and D. F. Smit. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:737-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serdar, C. M., and D. T. Gibson. 1985. Enzymatic hydrolysis of organophosphates: cloning and expression of a parathion hydrolase from Psuedomonas diminuta. Bio/Technology 3:567-571. [Google Scholar]
- 19.Soderling, S. H., S. J. Bayuga, and J. A. Beavo. 1998. Cloning and characterization of a cAMP-specific cyclic nucleotide phosphodiesterase. Proc. Natl. Acad. Sci. USA 95:8991-8996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sota, M., M. Endo, K. Nitta, H. Kawasaki, and M. Tsuda. 2002. Characterization of a class II defective transposon carrying two haloacetate dehalogenase genes from Delftia acidovorans plasmid pUOh1. Appl. Environ. Microbiol. 68:2307-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogel, A., O. Schilling, M. Niecke, J. Bettmer, and W. Meyer-Klaucke. 2002. ElaC encodes a novel binuclear zinc phosphodiesterase. J. Biol. Chem. 277:29078-29085. [DOI] [PubMed] [Google Scholar]