Abstract
In response to a bioterrorism attack in the Washington, D.C., area in October 2001, a mobile laboratory (ML) was set up in the city to conduct rapid molecular tests on environmental samples for the presence of Bacillus anthracis spores and to route samples for further culture analysis. The ML contained class I laminar-flow hoods, a portable autoclave, two portable real-time PCR devices (Ruggedized Advanced Pathogen Identification Device [RAPID]), and miscellaneous supplies and equipment to process samples. Envelopes and swab and air samples collected from 30 locations in the metropolitan area once every three days were subjected to visual examination and DNA extraction, followed by real-time PCR using freeze-dried, fluorescent-probe-based reagents. Surface swabs and air samples were also cultured for B. anthracis at the National Veterinary Service Laboratory (NVSL) in Ames, Iowa. From 24 October 2001 to 15 September 2002, 2,092 pieces of mail were examined, 405 real-time PCR assays were performed (comprising 4,639 samples), and at the NVSL 6,275 samples were subjected to over 18,000 platings. None of the PCR assays on DNA extracted from swab and air samples were positive, but viable spores were cultured from surface swabs taken from six locations in the metropolitan area in October, November, and December 2001 and February, March, and May 2002. DNA extracted from these suspected B. anthracis colonies was positive by real-time and conventional PCRs for the lethal factor, pXO1, and for capA and vrr genes; sequence analysis of the latter amplicons indicated >99% homology with the Ames, vollum, B6273-93, C93022281, and W-21 strains of B. anthracis, suggesting they arose from cross-contamination during the attack through the mail. The RAPID-based PCR analysis provided fast confirmation of suspect colonies from an overnight incubation on agar plates.
On 27 September 2001, a 63 year-old male working as a photo editor at the offices of the tabloid newspaper The Sun in Boca Raton, Fla., fell ill with flu-like symptoms. On 2 October, he was admitted to the hospital, and on 4 October, he was diagnosed with inhalation anthrax; he died on 5 October. Other cases of cutaneous anthrax were reported among workers at the National Broadcasting Company, American Broadcasting Company, Columbia Broadcasting Company, and New York Post offices in New York City, N.Y. Both cutaneous and inhalation anthrax cases were reported among workers at a post office in Trenton, N.J. On October 15, in Washington, D.C., an aide to U.S. Senator Tom Daschle opened a letter containing anthrax spores, which led to the closure of 12 Senate offices and the testing of hundreds of exposed individuals. The next day, workers at the Brentwood post office in Washington, D.C., fell ill; on October 21 one worker died of inhalation anthrax, and this was followed by the death of another worker the next day. Testing of a number of mail rooms located in the Washington, D.C., area revealed viable spores of Bacillus anthracis. It became apparent that mail processing equipment in the Brentwood facility had caused the deposition of spores on a large number of envelopes, which in turn had been delivered to satellite mail rooms throughout the Washington, D.C., area (3).
In response to this situation and to evaluate contamination of U.S. Department of Agriculture (USDA) buildings in the Washington, D.C., area, the USDA established a mobile laboratory (ML) for the detection of B. anthracis spores from environmental samples. Located in the Southeast Federal Facility in Washington, D.C., the laboratory received swab and air samples collected on a daily basis from mail rooms located in as many as 30 buildings throughout the Washington, D.C., area, including the Forest Service Yates and Rosslyn Buildings, the National Arboretum, and the Veterans Administration Building. While the laboratory was utilized primarily as a staging area for examining and sending samples for culture to the Animal and Plant Health Inspection Service (APHIS) National Veterinary Services Laboratories (NVSL) in Ames, Iowa, all samples were tested by real-time PCR analysis prior to shipment to Ames, and initially overnight blood agar cultures of some samples allowed selection of suspect colonies and rapid screening by PCR. (Subsequently, because of safety concerns as well as the large volume of samples requiring processing, overnight culturing was halted in the trailer and only PCR assays were performed.) The rapid deployment of the trailer near the site of contamination, along with the real-time PCR machines, allowed for an evaluation of the potential usefulness of such an approach in emergency response to bioterrorism events.
MATERIALS AND METHODS
ML.
The ML was housed in a 30-ft trailer, located in a warehouse in the Southeast Federal Facility (the “Navy Yard”) in Washington, D.C., from 24 October 2001 to 9 March 2002, after which time it was relocated to the campus of the Beltsville Agricultural Research Center in Beltsville, Md., and operated until late September 2002. The trailer was provided with water and electrical lines. Two class I laminar-flow hoods were placed at one end of the trailer and used for sample processing steps such as manipulation of swabs and air samples, examination of suspicious envelopes and other items, and nucleic acid extraction. The height of the trailer was such that class II hoods could not be reasonably accommodated. The class I hoods afford protection to the operator only (not the sample). On bench space at the other end of the trailer, two RAPID (Ruggedized Advanced Pathogen Identification Device; Idaho Technology [IT], Inc., Salt Lake City, Utah) instruments and associated laptop computers were stationed, as well as centrifuges, heat blocks, and other laboratory equipment. The RAPID is a field-deployable version of the LightCycler (Roche, Indianapolis, Ind.) instrument, allowing real-time monitoring of fluorescence emissions associated with the use of fluorogenic probes in the PCR. A small autoclave was also installed in the trailer, as well as telephone and Internet access lines. Two incubators located under the benches were used as a refrigerator (4°C) and a growth chamber (37°C). The capacity of the ML was four people.
Sample handling and processing.
During deployment of the trailer 30 sites in the Washington, D.C., area, representing office buildings whose mail rooms received mail from the Brentwood distribution facility, were sampled once every three days by one of two rotating work teams that took air and surface swab samples. For sites that tested positive, more extensive daily sampling would continue until decontamination was carried out, after which sampling of the site continued on a daily basis until subsequent samples tested negative. For those sites undergoing the air sampling procedure, the teams placed a SpinCon (Lares/Camber, Arlington, Va.) air sampling device in the room, preferably near an area of air disturbance (such as a heating, ventilating, air-conditioning vent), and sampled the air for 15 to 30 min, depending on the size of the room, with a sampling rate of 450 liters of air/min. In addition to air sampling the team used pairs of cotton or Dacron-tipped swab sticks to sample mail bins, envelope slots, countertops, computer keyboards, and other office furniture. Air samples (in 10 ml of surfactant, consisting of 0.3% Triton X-100 in 1× phosphate-buffered saline [pH 7.4]), into which airborne particles are deposited during the operation of the SpinCon device, and swabs were placed in plastic bags for transport back to the ML. Envelopes and other suspect materials from various sites, delivered to the ML by Federal Protective Services and other law enforcement agency personnel, also were transported in plastic bags. Upon receipt at the ML, the outsides of all bags were sprayed with a 1% sodium hypochlorite solution prior to being brought into the ML, where they were examined in class I hoods by personnel wearing laboratory coats and surgical masks. If the envelopes, clothing, containers, etc. contained powder, the powder was added to water to obtain an extractable suspension or solution, and the solution was subjected to DNA extraction as described below.
Surface swabs were processed for DNA extraction as follows. Swabs were grouped by location, and up to 15 were placed into 15- or 50-ml conical centrifuge tubes containing 3 to 5 ml of sterile water. The swabs were vigorously agitated to dislodge particulate material into the water, after which 1 ml of this “swab washing” was transferred to a 2-ml microcentrifuge tube and centrifuged at 16,168 × g for 2 min to pellet the contents. All but approximately 100 μl of supernatant was removed, and the pellet was resuspended and transferred to the “bead” tube (containing 250 μl of a liquid suspension of 0.1-mm-diameter zirconia-silica beads) of the 1,2,3 RAPID DNA extraction kit (IT). Samples were agitated for 5 min on a Vortex Genie II (Fisher Scientific, Pittsburgh, Pa.), after which 350 μl of buffer 1 was added to the bead tube. As much of the liquid in the bead tube as possible (approximately 500 μl) was then transferred to a 1,2,3 kit spin column and centrifuged at 16,168 × g for 1.5 min. Buffer 2 (550 μl) was added to the column, followed by consecutive centrifugation steps of 1.5 and 3 min at 16,168 × g to ensure complete removal of buffer 2 from the filter. In the final step, DNA was eluted from the column in 100 μl of buffer 3 by centrifugation at 16,168 × g for 1.5 min and stored at 4°C until used. For long-term storage, samples were transferred to −20°C freezers located in the investigators' laboratories in Beltsville, Md.
The initial culture protocol involved heating swab washings at 65°C for 15 min, which was followed by streaking on sheep red blood cell (SRBC) (Remel, Lenexa, Kans.) agar plates and incubation overnight at 37°C, with plates checked for suspicious B. anthracis colony formation the next morning. After the first week of operation, however, the number of samples and safety considerations had increased to the point where it was decided to have all the culture work performed in the biosafety level-3 facilities of the NVSL in Ames, Iowa. Subsequently, starting on 1 November 2001, samples were shipped with ice packs to the NVSL in appropriate biosafety containers using a commercial overnight delivery service, and upon receipt, swabs were streaked onto HiAB (heart infusion agar with 5% bovine red blood cells) plates and PLET (polymyxin-lysozyme-EDTA-thallous acetate) agar, as well as inoculated into PLET broth, for overnight incubation. The composition of the PLET agar was as follows: heart infusion agar (40%; Difco, Becton Dickinson, Sparks, Md.), thallium acetate solution (Fisher, Fair Lawn, N.J.), polymyxin B sulfate (Bedford Laboratories/Pfizer, Bedford, Ohio), lysozyme (Difco, Detroit, Mich.), and EDTA (Fisher) (for PLET broth, the concentration of heart infusion agar was reduced to 25%). If colonies with the appearance of B. anthracis were observed on the plates the next morning, they were transferred to a fresh HiAB plate, a sodium bicarbonate agar plate, and a tube of motility medium and incubated at 37°C overnight. If the suspect colony was nonhemolytic, nonmotile, and mucoid and bicarbonate positive, confirmatory tests were performed, including tests for morphology of bacterial cells under light microscopy and Gram staining, penicillin sensitivity, and gamma phage susceptibility (using 1,000 PFU of phage applied to the bacterial lawn on the plate in a 10-μl volume, kindly supplied by Melissa Libal, Texas A&M University, College Station, and maintained at the NVSL on B. anthracis Sterne strain). In late November 2001 the NVSL acquired a RAPID instrument and began conducting real-time PCR assays on suspect colony DNA.
Air samples were processed as follows: following agitation to resuspend any particulates, 1 ml of surfactant (of the total collection volume of 10 ml) was transferred to a 2-ml microcentrifuge tube and centrifuged at 16,168 × g for 2 min. All but 100 μl of supernatant was discarded, and the pellet was resuspended and transferred to a 1,2,3 kit bead tube and subjected to DNA extraction as described above. For culture, 1 ml of the resuspended surfactant was drawn into a 1-ml borosilicate glass pipette, and 1 drop (approximately 20 μl) was deposited onto an HiAB agar plate and another drop was deposited onto a PLET agar plate. The remaining 960 μl of air sample was dispensed into a tube of PLET broth and incubated overnight at 37 ± 2°C (mean ± standard deviation).
For extraction of DNA from bacterial colonies, sterile plastic loops were used to remove a portion of the colony and placed in the 1,2,3 kit bead tube, and gentle agitation was used to dislodge bacteria from the loop. The bacterium-bead suspension was then subjected to DNA extraction as described above.
Real-time PCR and sequencing.
Initially, 2-, 5-, and 10-μl aliquots of DNA extracted from swab washings and air samples were spiked with an internal positive control (IPC) (consisting of a 3.2-kb plasmid containing a 350-bp insert of a Cryptosporidium parvum 15-kDa gene segment, routinely used in our laboratories to monitor extracted DNA for the absence of PCR inhibitors), and tested by PCR using primers specific for the IPC. Results indicated that IPC PCR was inhibited when more than 2 μl of DNA extracted from “dirty” samples (e.g., swab washing that displayed a noticeable turbidity) was used as the template; consequently, 2 μl was the volume of extracted DNA used for real-time PCR (below). The DNA yield from swab and air samples was too small to be accurately measured spectrophotometrically, even using a 50-μl cuvette. For agar plate colonies of B. anthracis, the yield of DNA via the IT 1,2,3 kit was approximately 27 ng/μl.
For real-time PCR, freeze-dried reagents containing two fluorescence resonance energy transfer (FRET) hybridization probes for the B. anthracis lethal factor (LF) gene (obtained from IT) were reconstituted in 40 μl of sterile water; 18 μl was placed in the RAPID capillary, along with 2 μl of template DNA. LF kit positive and negative control reactions were included with every PCR run. Reaction conditions on the RAPID were as follows: 1 min at 95°C followed by 45 cycles of 95°C for 0 s and 60°C for 20 s. Fluorescence from the LF FRET probes (labeled with LightCycler red dye 640) was monitored via real-time graph. The RAPID software's Batch Test program was used to automatically identify positive reactions at the completion of the run; the negative controls were given a fluorescence score of 20, and positive samples were those with fluorescence scores (corrected for background) exceeding those of the negative control (for example, the LF freeze-dried reagent kit positive controls routinely gave fluorescence scores of 185 to 200). In addition to the freeze-dried LF reagents, freeze-dried reagents containing a TaqMan probe (Applied Biosystems, Foster City, Calif.) for the pXO1 locus were also used (Cepheid, Sunnyvale, Calif.) on the Smart Cycler real-time PCR instrument, with the same thermal cycling parameters, reaction volume (20 μl), and quantity of template (2 μl) used on the RAPID instrument.
Samples positive by RAPID PCR were also analyzed by capA and vrr gene PCRs using the primers and protocols of Jackson et al. (6). These reactions were performed in conventional 0.2-ml PCR tubes with 1 U of Taq polymerase, 1.5 mM MgCl2, 5 μl of 10× PCR buffer, 50 pmol of each primer, and 10 mM concentrations of deoxynucleoside triphosphates (Invitrogen, Gaithersburg, Md.), with 2 to 5 μl of DNA as the template, and analyzed via 1% agarose gels electrophoresed for 90 min at 50 V and ethidium bromide staining. PCR amplicons were sequenced using dye-terminator chemistry on the ABI model 3100 automated fluorescence sequencing instrument (Applied Biosystems) and analyzed using NCBI BLAST and MacVector (Oxford Molecular, Madison, Wis.). The DNASTAR (DNASTAR, Inc., Madison, Wis.) software's ClustalW and Sequence Distance programs were used to analyze the data. Reference sequences from GenBank included B. anthracis strain B6273/93 (accession no. U63965), strain C93022281 (accession no. U639 67), strain W-21 (accession no. U63964), strain vollum (accession no. U63968), and strain Ames (accession no. U63966.1) (5).
Spiked swab and air samples.
To determine the sensitivity of assays conducted on swab washings and aliquots of air samples, standards containing known quantities of B. anthracis Sterne spores in 10 ml of sterile water were made. Spores from a provisional stock solution (106 spores/ml) were counted using a Neubauer A-2901 hemacytometer (Clay-Adams, New York, N.Y.) and phase-contrast microscopy at a magnification of ×400, with two determinations (two grids) per 0.1 μl examined (approximately 100 spores per grid). Subsequent serial dilutions used for spiking experiments were made based on those counts. To determine the CFU present in each sample, aliquots (200 μl or 1 ml) of standards (the larger volume was used with the dilutions containing the smaller quantity of spores per unit volume) were subjected to a 15-min heating at 65°C and cultured overnight on SRBC plates. All colonies were counted using a Q-Count instrument (Spiral Biotech, Norwood, Mass.); subsequently, those with an appearance suggestive of B. anthracis were counted by direct observation, to arrive at a determination of the number of B. anthracis colonies per total number of colonies per plate.
Moistened Dacron-tipped swabs were used to sample the interior of a standard U.S. Postal Service white plastic mail bin (43 by 28 by 30 cm). Quantities of 1,000, 100, 10, and 1 spores, in 2.5 μl, were then pipetted directly onto the swabs, with three swabs per quantity. Extraction control swabs received sterile water. The swabs were then subjected to DNA extraction using the 1,2,3 kit as described above, and 2 μl of eluted DNA was used as the template on the RAPID unit. Another set of spiked swabs was subjected to culture; here, individual swabs were washed in 200 μl of sterile water, which was then heated at 65°C for 15 min and plated overnight on SRBC plates.
For air samples, 1-ml aliquots of surfactant used in the SpinCon devices were spiked with 10,000, 5,000, 1,000, 100, 10, and 1 spores; agitated for 15 s; and then processed for DNA extraction as described above, with 2 μl of eluted DNA used as the template. Extraction controls received sterile water. A second set of spiked surfactants was spread onto SRBC plates and cultured overnight.
To confirm that positive RAPID PCR results using spiked samples were not benefiting from the presence of extracellular DNA (carried over from vegetative cells when the spores were first prepared), 1 ml of a 106-spore suspension was centrifuged at 16,168 × g for 5 min to pellet the spores, and three 100-μl aliquots of supernatant were removed and subjected to DNA extraction using the IT 1,2,3. RAPID DNA kit. RAPID PCR was performed on 5 μl of the resultant DNA.
RESULTS
Culture and analysis of B. anthracis from samples.
Between 24 October 2001 and 15 September 2002, over 3,000 swab samples, 300 air samples, and 2,092 pieces of mail and other objects (Tupperware containers, jackets, and handbags, etc.) had been processed at the ML. Some of the letters submitted for examination displayed idiosyncratic behavior on the part of the writer. Determining which of the many envelopes and letter showed evidence of threatening intent on the part of the writer (and consequently required oversight by law enforcement personnel during the examination and testing process in order to preserve custody of evidence that later may have been used for prosecutorial purposes) was usually straightforward. Some of these “genuinely” threatening letters contained powders but no B. anthracis spores were detected. None of the results of real-time PCR assays performed on DNA extracted from swab washings, air samples, or envelope contents (a total of 4,639 reactions as of 15 September 2002) were positive.
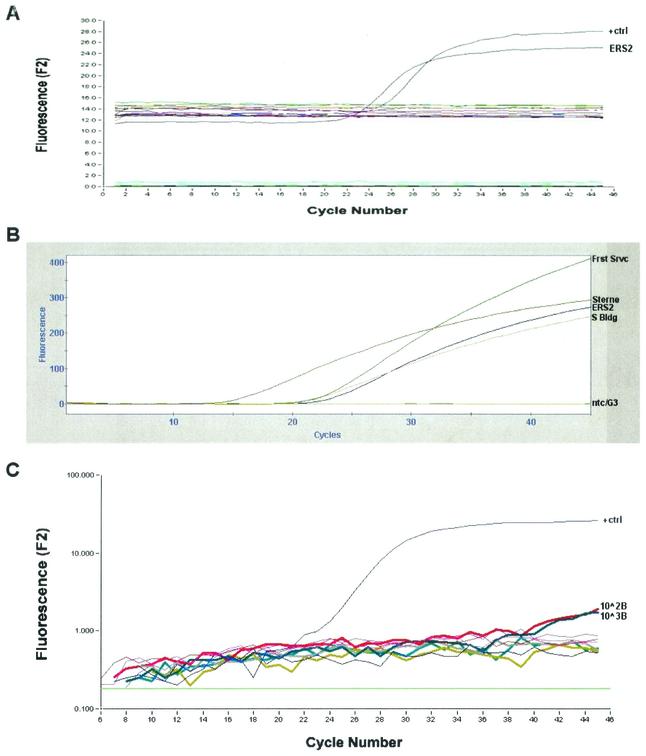
As of 15 September 2002, over 18,000 plates, representing over 6,200 samples, have been used for culture of B. anthracis at the NVSL in Ames. A variety of colony morphologies were observed on the plates inoculated with swab and air samples, some of which were hemolytic Bacillus spp., as well as other genera of bacteria and fungi. B. anthracis was recovered in culture from swabs taken at the Economic Research Service (ERS-2) mail room in Washington, D.C. (October 2001); the Forest Service mail room in Arlington, Va. (November 2001); the USDA South Building mail room in Washington, D.C. (November 2001); the Office of Personnel Management mail room in Washington, D.C. (December 2001); the Veterans Administration mail room (February 2002); the Small Business Administration mail room (March 2002); and the Office of Personnel Management mail room (May 2002). DNA extracted from six of these seven colonies (the exception being the Office of Personnel Management sample from May 2002) was available for analysis, and all six samples subsequently tested positive when assayed by real-time PCR for the LF gene on the RAPID instruments; an example of one such reaction, for the ERS-2 sample, is shown in Fig. 1A. The DNA extracted from three isolates also generated positive real-time PCR signals when assayed using a different freeze-dried reagent, for pXO1-associated DNA, on the Cepheid Smart Cycler (Fig. 1B).
FIG. 1.
(A) Results of real-time B. anthracis LF PCR performed with the RAPID instrument using DNA extracted from a bacterial colony with morphology suggestive of B. anthracis, cultured from a swab sample taken from the Economic Research Service (ERS2) mail room in Washington, D.C., October 2001. Relative fluorescence is plotted on the y axis, and cycle number is plotted on the x axis. The plot for the positive control is indicated; the horizontal plots represent negative controls and non-B. anthracis samples. (B) Results of real-time PCR for the pXO1 locus performed on the Smart Cycler on DNA extracted from suspected B. anthracis colonies cultured from samples taken at the Economic Research Service (ERS2), the Forest Service (Frst Srvc) (November 2001), and the USDA South Building (S Bldg) (Washington, D.C., November 2001). The Sterne strain positive control (Sterne), no template control (ntc), and extraction control (G3) are indicated. (C) Results of real-time B. anthracis LF PCR performed with the RAPID instrument using DNA extracted from swabs spiked with 1,000 and 100 spores of B. anthracis Sterne. Plots for the positive control (+ctrl) and the 1,000- and 100-spore-spiked samples are indicated; horizontal plots represent negative samples (these include other replicates spiked with 1,000, 100, 10, and 1 spores, and extraction and no template controls).
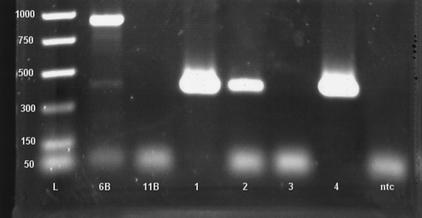
When samples that were positive and negative by RAPID PCR were also assayed by capA conventional PCR, nonspecific amplicons were occasionally observed on agarose gels of the PCR products. For example, a swab DNA sample (sample 6B) giving a faint positive signal for the LF PCR generated a strong band at approximately 900 bp, while colony DNAs positive by LF PCR on the RAPID generated bands of the expected size (i.e., 397 bp) (Fig. 2). Sequence analysis of approximately 800 bp of the amplicon from sample 6B indicated no homologies with GenBank depositions for B. anthracis. Assay of other, non-B. anthracis colonies with this primer set also generated amplicons of different sizes (data not shown). Interestingly, use of the same volume of template that generated strong positives with the capA primers was unsuccessful for the vrr primers for the Forest Service and South Building colony DNAs; a nested PCR was necessary to generate a positive result for vrr. However, the vrr PCR generated amplicons of the predicted size range (377 to 425 bp) from six B. anthracis colony DNAs; sequence analysis of these amplicons indicated >99% homology with the Ames, B6273/93, C93022281, vollum, and W-21l strains of B. anthracis (data not shown).
FIG. 2.
Results of capA gene PCR (6) performed on DNA extracted from swabs and bacterial colonies suspected of harboring or being B. anthracis. Lane L, DNA ladder with rung sizes indicated; lanes 6B and 11B, DNAs extracted from swab samples; lane 1, DNA extracted from a colony cultured from the USDA South Building mail room, Washington, D.C., November, 2001; lane 2, DNA extracted from a colony cultured from the Forest Service mail room, November 2001; lane 3, DNA extraction control; lane 4, B. anthracis Pasteur strain positive control; lane ntc, no template control.
As of September 2002, 405 runs, comprising 4,639 samples, had been performed on the RAPID instruments in the ML; with 25 runs having to be repeated because the positive control either failed to amplify or gave a fluorescence signal below that of the no-template control, this gives a run failure rate of 6%. Out of the 405 runs, fewer than 20 gave results in which one or more samples displayed a fluorescence reading that was weakly positive and that could possibly be considered a false positive. When these samples were subsequently reanalyzed using the RAPID's melt curve program, they were all negative.
Spiked swab and air samples.
When assayed by real-time PCR on the RAPID instrument, one each of the three replicates for the 1,000- and 100-spore-spiked swabs was weakly positive (Fig. 1C). A faint positive signal was observed for one of the three swabs spiked with 10 spores, but the sample was considered negative by the RAPID software's automatic sample calling software. The positive results were associated with the cleaner swabs, for example, if after the swab's placement in the 200 μl of bead lysis buffer used in the first step of the 1,2,3 kit extraction protocol the buffer was clear, as opposed to brown in color. When washes from the 10-spore-spiked swabs were examined, 10, 2, and 7 B. anthracis colonies were observed per total number of colonies (110, 64, and 109, respectively). More numerous B. anthracis colonies were detected on the plates receiving washes from the swabs spiked with 100 spores (98 colonies of 465, 50 colonies of 640, and 87 colonies of 495). B. anthracis colonies on plates receiving washes from swabs spiked with 1,000 spores had in excess of 400 colonies, while plates receiving washes from swabs spiked with only one spore had no B. anthracis colonies, as did the extraction control plate. When seven putative colonies of B. anthracis and one non-B. anthracis colony were picked from the 10-spore-spiked plates and subjected to DNA extraction and analyzed by PCR using the RAPID instrument, all seven of the B. anthracis colonies were positive and the non-B. anthracis colony was negative, confirming our designation of the colonies as genuine B. anthracis.
When 1-ml aliquots of the surfactant used in the SpinCon device were spiked with log dilutions of spores, subjected to DNA extraction, and assayed by real-time PCR, results for four of six replicates using a spiking dose of 10,000 spores were positive. Four of four replicates with a 5,000-spore spike were positive. None of the replicates using 1,000, 100, or 10 spores were positive. However, when samples of 1 ml of spiked surfactant were plated on SRBC plates, positive colonies were observed for all dilutions. For example, the 100-spore-spiked samples had 146, 115, and 153 colonies; the 10-spore-spiked plates had 12, 14, and 30 colonies; and the samples spiked with 1 spore had 2, 0, and 2 colonies. The extraction control plate had one, non-B. anthracis colony, and plates receiving surfactant spiked with 1,000 spores had in excess of 400 colonies. When 1,000 spores were incubated in 1 ml of the surfactant solution for 5 days at ambient temperature (the usual time interval between air sample collection in the Washington, D.C., area and culture at the NVSL in Ames), spores were recovered on plates at approximately 40% efficiency.
RAPID PCR performed on DNA extracted three 100-μl aliquots of supernatant from a pelleted 106-spore/ml suspension was negative, indicating that positive PCR results were not being inflated due to the presence of appreciable quantities of extracellular B. anthracis DNA (presumably deposited by vegetative cells during the sporulation process) in the spiking dose.
Based on the results of the spiking experiments, what is the theoretical detection limit of the RAPID PCR for spores in air samples? We will assume that the average room size sampled by one SpinCon device is 13.5 m3 and that the particulates in this volume of air are deposited into the 10 ml of surfactant with almost 100% efficiency. Therefore, one ml of surfactant contains particulates from 1.35 m3 of air; if our PCR detection limit is 5,000 spores per 1 ml, then this would translate into a detection limit of 3,700 spores for 1 m3.
DISCUSSION
The ML was implemented to provide testing of workplace (particularly mail room) environments for the presence of B. anthracis spores during a bioterrorism event in the Washington, D.C., area in October of 2001. While originally envisaged as monitoring USDA-associated properties, the sample locations were soon expanded to include other government and private facilities, in part because the large number of samples delivered to the Centers for Disease Control and Prevention, U.S. Army Medical Research Institute for Infectious Diseases, and other permanent laboratories had created a testing backlog. The ML's proximity to many of the federal buildings receiving mail from the Brentwood post office allowed for prompt examination of suspicious envelopes and other items provided by Federal Protective Services, Capitol Police, Secret Service, and Federal Bureau of Investigation personnel, as well as rapid analysis of swab and air samples taken from a number of mail rooms in the Washington, D.C., area. Using a combination of culture followed by real-time PCR, it was possible to have results within 24 h of sample submission. Eschewing culture and using DNA extraction and real-time PCR, it was possible to have results from up to 40 samples in 5 h, provided, of course, that there were sufficient organisms present in the sample. While considerably more expensive than conventional PCR reagents (>$10.00 per reaction), the freeze-dried B. anthracis LF PCR kits were easy to work with, reduced the likelihood of PCR contamination within the limited bench space of the ML, and enabled personnel without molecular biology experience to readily conduct real-time PCR assays with only several hours of training. We were also interested in learning if reagent kits and real-time PCR platforms developed by other vendors could be of use in our investigations; consequently, assays were conducted with Cepheid's freeze-dried reagents and the Smart Cycler real-time PCR instrument on DNA extracted with the IT 1,2,3 RAPID DNA extraction kit. These assays were successful (Fig. 1B), indicating that alternative reagent kits and associated platforms may be available for use by personnel investigating a threat situation.
We operated the ML under the assumption that molecular technique-based detection of spores would be successful only with those environmental samples harboring very large (>1,000) numbers of spores, given the likelihood of a swab successfully picking up spores from a sample surface, adequate extraction of spore DNA from a diluted swab washing, and the use of only 2 μl (2%) of DNA as template in the RAPID instrument. The small reaction volume (20 μl) of the real-time PCR assays may exacerbate the effects of inhibitory substances in the template DNA, further hampering the sensitivity of any PCR-based analysis. In an attempt to determine the detection level of our assays, we used standards of B. anthracis Sterne spores to spike swab and SpinCon surfactant samples. For spiked swabs, only one replicate each of the 1,000- and 100-spore-spiked swabs allowed amplification, and this appeared to be related to the dirt and dust content of the swabs. For the SpinCon air sampling device surfactant, positive real-time PCR results were obtained only with the 5,000-spore/ml spiking dose. However, better sensitivity was obtained using culture, with as little as 10 spores spiked into the swabs resulting in positive colonies, and 1 spore spiked into the surfactant generating colonies. Based on these results, we feel it prudent to operate under the assumption that the real-time PCR assays conducted on the swab sample-derived DNAs would at best detect only large (>1,000) numbers of spores in the 100 μl of pelleted particulates that was subjected to DNA extraction. We also conclude that, in our experience, culture followed by next-day RAPID identification of suspect colonies is the best method for detection of spores in air samples.
Previous reports on the sensitivity of the real-time PCR for B. anthracis detection utilized DNA obtained from vegetative cells or large numbers of spores as the template or introduced purified spores into reactions. For example, Qi et al. (8) used a LightCycler instrument and FRET probes to the rpoB gene to selectively identify B. anthracis vegetative cell-derived DNA from that of other Bacillus spp., with a detection limit of 1 pg. Using the 7700 model sequence detector instrument and TaqMan probes to the capA and lef genes, Dang et al. (4) obtained satisfactory real-time signals when 106 spores were used as the template. In other experiments, Makino et al. (7) passed 100 liters of air through a 0.45-μm-pore-size membrane filter and suspended the filter in 1 ml of phosphate-buffered saline, to which various numbers of B. anthracis Pasteur strain spores were added. Ten-microliter aliquots were then subjected to DNA extraction and analyzed on a LightCycler using SYBR Green dye and primers to the cap gene; the detection limit was one spore. Obviously our assays, performed on actual environmental samples, do not approach this level of sensitivity; we were able to amplify swab DNA samples from only one of three replicates spiked with 1,000 and 100 spores. Presumably, more rigorous extraction protocols (involving phenol-chloroform-isoamyl alcohol) could yield purer DNA from the swab and air samples and thus improve the detection limit; however, working with these hazardous reagents is problematic in the ML. We did observe that swabs and air samples spiked with as few as 10 and 1 spores, respectively, yielded viable colonies on plates; this finding indicates that in our hands, culture methods are superior to molecular methods when attempting to detect spores in environmental samples.
B. anthracis was cultured from swabs taken at seven different locations, but the result of a real-time PCR conducted on DNA extracted from these swabs was negative. A similar lack of congruence between culture and molecular assays was observed by other investigators (2), who found that of 107 samples taken from the Brentwood post office, 8 (0.07%) were positive by PCR and 6 (0.05%) were positive by culture; two of the PCR-positive samples were also positive by culture. However, reference 2 does not provide information on the sampling methods, DNA extraction techniques, or the PCR assay used, so it is difficult to draw conclusions as to the comparative efficiency of their PCR and culture protocols and ours. An updated account of the Brentwood facility testing indicated that 8 of 114 swab samples (7%) were positive by culture, while 27 of 39 (69%) vacuum dust samples were positive (3). None of 12 air samples obtained from the Brentwood facility were positive (3). B. anthracis concentrations in vacuum dust samples showed great variability, ranging from 3 CFU/g to 9.7 × 10 6 CFU/g; it is unclear how these calculations were made, and we have not attempted to apply them to our culture results.
In addition to using freeze-dried real-time PCR reagents, we analyzed samples by published PCR protocols for the capA and vrr genes; the latter has previously been used for differentiation of strains of B. anthracis (1, 5, 6). In our hands, spurious amplicons were generated from non-B. anthracis samples by using the capA primers, while the variable repeat region (vrr) locus PCR did not give such results; consequently, we used this locus to obtain DNA sequence information that indicated our isolates had >99% homology with B. anthracis sequences deposited in GenBank. Given that other isolates taken from other locations in the Washington, D.C., area were typed as the Ames strain, we feel it reasonable to conclude that our isolates are representative of the original stock of B. anthracis used to contaminate the letter(s) which served as the vehicle(s) for the dissemination of this agent.
Keeping in mind that the ML was a rapid and previously untested response to an emergency situation, we have identified areas for improvement. First, provision of a real-time PCR internal or endogenous control, preferably in a multiplex format with existing B. anthracis probes and primers, would allow for accurate identification of false-negative PCR assays. While the ability of the RAPID instrument to simultaneously monitor up to three different fluorescent dyes (6-carboxyfluorescein, LightCycler red 640, and LightCycler red 705) makes such an approach feasible, synthesizing a multiplex PCR freeze-dried reagent has not been accomplished to date and may be considerably more expensive than using the existing one-target reagents (which are approximately $10 to 12 per assay). Therefore, ML personnel may be required to mix and apply such internal controls to the freeze-dried reagents. Second, our experiments using spiked swab and air samples indicate that PCR inhibitors may not be adequately removed with our DNA extraction protocol, particularly in the case of the dirtier swabs. An extraction method that can more effectively remove the inhibitory substance(s), while offering the relative ease, convenience, and use of nonhazardous reagents of the 1,2,3 kit, would be welcome. Third, in light of the fact that Jackson et al. (5) successfully used nested primers to improve the yield of B. anthracis vrr region amplicons from DNA extracted from formalin-preserved tissue specimens, use of a nested PCR with the RAPID instrument may improve sensitivity for environmental samples as well. Because the glass capillaries used in the RAPID system are not very amenable to postamplification handling, and given the space limitations within a ML or small biosafety level-3 facility, a single-tube nested reaction would be necessary.
In conclusion, the ML provided law enforcement personnel with a conveniently located and readily accessible facility for examination of potentially infectious envelopes in the midst of a fatal bioterrorism event in the Washington, D.C., metropolitan area. It also provided rapid testing of environmental samples for the presence of B. anthracis spores—albeit within the limitations of assay sensitivity—and reassured employees that government agencies were actively involved in efforts to safeguard their health and well-being. Finally, the combined efforts of the NVSL culture work and the accompanying molecular analyses indicated that viable spores were present at several locations receiving mail from the Brentwood facility. This information was used by USDA authorities to arrange for select decontamination of the affected sites. As of 15 September 2002, no anthrax cases have been reported by workers at these facilities.
Acknowledgments
We thank Christina Hohn, Sebastian Botero, Becky Garbarino, Amy Barringer, Dave Prevar, Mike Kiley, and Bob Gililland (USDA-ARS); Elliot Grollman (FPS); and David House, Jessica White, Pat Minix, Steve Medaglia, Terrie McConnell, and Matt Roberts (Lares/CRE/Camber) for providing technical and administrative assistance. Catherine Senselau, University of Maryland at College Park, kindly provided the B. anthracis Sterne strain vegetative cells.
REFERENCES
- 1.Anderson, G. L., J. M. Simchock, and K. H. Wilson. 1996. Identification of a region of genetic variability among Bacillus anthracis strains and related species. J. Bacteriol. 178:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. 2001. Use of onsite technologies for rapidly assessing environmental Bacillus anthracis contamination on surfaces in buildings. Morb. Mortal. Wkly. Rep. 50:1087. [PubMed]
- 3.Anonymous. 2001. Evaluation of Bacillus anthracis contamination inside the Brentwood Mail Processing and Distribution Center, District of Columbia, October 2001. Morb. Mortal. Wkly. Rep. 50:1129-1133. [Google Scholar]
- 4.Dang, J. L., K. Heroux, J. Kearney, A. Arasteh, M. Gostomski, and P. E. Emanuel. 2001. Bacillus spore inactivation methods affect detection assays. Appl. Environ. Microbiol. 67:3665-3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson, P. J., E. A. Walthers, A. S. Kalif, K. L. Richmond, D. M. Adair, K. K. Hill, C. R. Kuske, G. L. Anderson, K. H. Wilson, M. E. Hugh-Jones, and P. Keim. 1997. Characterization of the variable-number tandem repeats in vrrA from different Bacillus anthracis isolates. Appl. Environ. Microbiol. 63:1400-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson, P. J., M. E. Hugh-Jones, D. M. Adair, G. Green, K. K. Hill, C. R. Kuske, L. M. Grinberg, F. A. Abramova, and P. Keim. 1998. PCR analysis of tissue samples from the 1979 Sverdlovsk anthrax victims: the presence of multiple Bacillus anthracis strains in different victims. Proc. Natl. Acad. Sci. USA 95:1224-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makino, S.-I., H. I. Cheun, M. Watarai, I. Uchida, and K. Takeshi. 2001. Detection of anthrax spores from the air by real-time PCR. Lett. Appl. Microbiol. 33:237-240. [DOI] [PubMed] [Google Scholar]
- 8.Qi, Y., G. Patra, X. Liang, L. E. Williams, S. Rose, R. J. Redkar, and V. G. Del Vecchio. 2001. Utilization of the rpoB gene as a specific chromosomal marker for real-time PCR detection of Bacillus anthracis. Appl. Environ. Microbiol. 67:3720-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]