Abstract
Phytochelatins (PCs) are metal-binding cysteine-rich peptides, enzymatically synthesized in plants and yeasts from glutathione in response to heavy metal stress by PC synthase (EC 2.3.2.15). In an attempt to increase the ability of bacterial cells to accumulate heavy metals, the Arabidopsis thaliana gene encoding PC synthase (AtPCS) was expressed in Escherichia coli. A marked accumulation of PCs was observed in vivo together with a decrease in the glutathione cellular content. When bacterial cells expressing AtPCS were placed in the presence of heavy metals such as cadmium or the metalloid arsenic, cellular metal contents were increased 20- and 50-fold, respectively. We discuss the possibility of using genes of the PC biosynthetic pathway to design bacterial strains or higher plants with increased abilities to accumulate toxic metals, and also arsenic, for use in bioremediation and/or phytoremediation processes.
Industrial and agricultural activities have led to substantial release of toxic heavy metals in the environment, which can constitute a major hazard for ecosystems and human health (16). Bioremediation, using bacteria or plants, is often regarded as a relatively inexpensive and efficient way of cleaning up wastes, sediments, or soils contaminated with toxic heavy metals (14). A promising way of improving bioremediation processes is to genetically engineer bacterial strains to confer increased abilities to accumulate toxic heavy metals. Attempts to enhance the metal content of bacterial cells have been made by overexpressing metal-binding peptides or proteins such as poly-histidines (23) or metallothioneins (14, 24).
When exposed to toxic heavy metals, plant cells and yeasts accumulate phytochelatins (PCs), which are n repeats (n = 2 to 5) of the γ-Glu-Cys dipeptide terminated by a Gly residue. PCs are enzymatically synthesized from glutathione (GSH) by PC synthase (EC 2.3.2.15) (7) and bind heavy metals such as cadmium, silver, copper, or arsenite by forming complexes (13, 21). PC synthase genes were initially cloned from wheat, Arabidopsis thaliana and Schizosaccharomyces pombe (4, 9, 27) and recently a gene encoding a functional PC synthase was found in the nematode Caenorhabditis elegans (5, 26). PCs are clearly involved in heavy metal detoxification. Indeed, plant and yeast mutants deficient in PC synthase are hypersensitive to cadmium (9), and expression of PC synthase genes in yeast confers augmented cadmium tolerance (4, 26, 27).
In an effort to increase the heavy metal content of bacterial cells, we expressed the A. thaliana PC synthase gene (AtPCS) in Escherichia coli. We show that expression of AtPCS allows PC synthesis resulting in a large increase in intracellular metal content when bacterial cells are grown on metal-enriched media. These results pave the way for using enzymes of the PC biosynthetic pathway to enhance the metal content of bacteria and plants and optimize bioremediation and/or phytoremediation processes of toxic metals.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Plasmid constructions and gene expression were performed in the E. coli strains DH10β and M15REP4 (Qiagen), respectively. Bacterial cells were grown in liquid Luria-Bertani (LB) medium supplemented with appropriate antibiotics. For expression of AtPCS, bacteria were grown at 37°C in liquid LB medium supplemented with carbenicillin and kanamycin (50 μg. ml−1 each). Expression was induced by addition of 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) when the optical density at 600 nm reached 0.25 and cultures were further incubated for 3 h unless otherwise indicated.
AtPCS cloning and plasmid constructs.
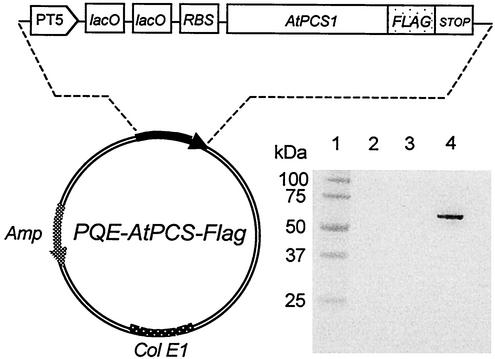
Total RNAs from A. thaliana ecotype Columbia (leaves) were extracted with an RNeasy plant mini kit (Qiagen) following the instructions provided by the manufacturer. The first strand cDNA was synthesized from total RNAs (1 μg) by reverse transcription (Enhanced Avian RT-PCR kit; Sigma) with an AtPCS specific primer (L-AtPCS: 5′-ATAGGCAGGAGCAGCGAGATC-3′) for 10 min at room temperature. The double-strand cDNA was obtained by PCR with the specific primers L-AtPCS and U-AtPCS (5′-CCATGGCTATGGCGAGTTTATATCGGC-3′) for 25 cycles (annealing temperature of 55°C, elongation time of 1 min 30 s). The reverse transcription-PCR product of 1,457 bp was sequenced and cloned into plasmid pBluescript (Stratagene) linearized with SmaI to obtain pBluescript-AtPCS. Because of differences in the nucleotide sequence among the AtPCS sequences published in the data banks, the sequence we obtained was submitted to GenBank (accession no. AF461180). To allow an immunodetection of overexpressed AtPCS, a short peptide, Flag (DIDYKDDDDK), was fused at the C-terminal part of AtPCS. This Flag epitope was made by hybridizing the primers L-Flag (5′-TTATTACTTATCGTCGTCATCCTTGTAATCGATATC-3′) and U-Flag (5′-GATATCGATTACAAGGATGACGACGATAAGTAATAA-3′) for 5 min at 95°C and 10 min at room temperature (the two stop codons in the Flag primer are underlined). Then, it was cloned into pBluescript-AtPCS linearized by EcoRV to obtain the plasmid pBluescript-AtPCS-Flag. Two primers (U-PCS-BamHI, 5′-GAGAGGATCCATGGCTATGGCGAGTTTATATCGGC-3′; L-Flag-BamHI, 5′-GAGAGGATCCGAGCTCGGGCTTATTACTTATCGTCGTC-3′) were de-signed to amplify by PCR the AtPCS-Flag sequence flanked with BamHI restriction sites at both ends (annealing temperature of 57°C, elongation time of 2 min). The DNA fragment containing the AtPCS-Flag gene was isolated after digestion with BamHI, sequenced and ligated into the unique BamHI site of plasmid PQE60 (Qiagen) in the orientation that places AtPCS-Flag under the control of the strong phage T5 promoter to obtain plasmid PQE-AtPCS-Flag (Fig. 1A).
FIG. 1.
Schematic representation of the AtPCS overexpression plasmid and expression of AtPCS in E. coli. Upper panel abbreviations: PT5, T5 phage promoter; lacO, lac operator; RBS, ribosomal binding site; AtPCS, A. thaliana PC synthase coding sequence from nucleotides 1 to 1476; FLAG, Flag epitope coding sequence of 24 bp; STOP, two-codon stop; Amp, β-lactamase coding sequence; colE1, origin of replication. The lower panel shows soluble proteins that were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (13% polyacrylamide), transferred to a nitrocellulose membrane, and probed with a monoclonal anti-Flag antibody. Lane 1, molecular mass markers; lane 2, E. coli extracts cells containing the vector without PCS as a control; lane 3 and 4, extracts of E. coli cells containing the overexpression plasmid, grown in the absence (lane 3) or in the presence (lane 4) of IPTG.
Protein techniques.
Five milliliters of culture cells were pelleted, resuspended in 300 μl of a 0.1 M Tris-Cl buffer (pH 8.0) containing 0.1 M KCl-0.1 mg · ml−1 fresh lysozyme, and incubated 1 h at 4°C. After incubation (30 min at 37°C) in the presence of DNase I (10 μg) and deoxycholic acid sodium salt (2 mg) and centrifugation at 10,000 × g for 10 min at 4°C (Centromix 1236 V, rotor 20RT), soluble proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 13% polyacrylamide gel (10) and stained with Coomassie blue or transferred to a nitrocellulose membrane. Flag-tagged AtPCS was detected with a Flag monoclonal antibody (Sigma). Protein concentration was determined according to Laemmli (12).
PC analysis.
Cells were harvested and rinsed with LB medium and stored at −80°C until PC analysis. Nonprotein thiols were extracted by homogenization of 5 to 7 mg of frozen bacteria in 0.5 to 0.7 ml of extraction buffer (6.3 mM diethylenetriamine pentaacetic acid [DTPA]-0.1% [vol/vol] trifluoroacetic acid) with a pestle and disruption of cells by sonication. Thirty microliters of 100 μM N-acetyl-l-cysteine was added as an internal standard. The homogenate was centrifuged at 10,000 × g for 15 min at 4°C (Centromix 1236 V, Rotor 20RT) and the supernatant was filtered (0.22 μm). The derivatization procedure was modified from Rijstenbil and Wijnholds (20). Filtered extracts (125 μl) were mixed with 225 μl of reaction buffer [0.2 M 4-(2-hydroxy-ethyl)-piperazine-1-propane-sulfonic acid pH 8.2 containing 6.3 mM DTPA] and 5 μl of 25 mM monobromobimane dissolved in acetonitrile. Following 15 min of incubation in the dark at room temperature, the reaction was stopped by adding 150 μl of 1 M methane sulfonic acid. The samples were stored at 4°C in the dark until high-performance liquid chromatography (HPLC) analysis. The bimane derivatives were separated on a reversed-phase Nova-Pak C18 analytical column (pore size, 60 Å; particle size, 4 μm; dimensions, 3.9 by 300 mm; Waters catalog no. 11695) using two eluents (0.1% [vol/vol] trifluoroacetic acid in water and acetonitrile) at a flow rate of 1 ml · min−1. Fluorescence was monitored by a Waters 464 detector (λexcitation = 380 nm; λemission = 470 nm). Calibration curves of glutathione were used in all measurements. Cysteine, GSH, γ-glutamylcysteine (γ-EC) (both from Sigma) and three PCs (n = 2, 3, and 4) chemically synthesized were used as standards.
Quantitative determination of metal content.
After growth for 3 h in the presence of IPTG and metal (ZnCl2, PbCl2, AsO4, CuCl2, CdCl2, HgCl2, or AgNO3), cells were harvested, rinsed three times using LB medium and dried at 55°C for 24 h. Following addition of 5 ml HNO3 (70%), mineralization was carried out in a microwave oven (Mars X; CEM Corp., Matthews, N.C.). Metal content was determined using an ICP-OES device (Varian); standard solutions were supplied by Merck.
RESULTS
Expression of AtPCS in E. coli and PC production.
The AtPCS was fused to a C-terminal Flag epitope to allow easy detection of the recombinant protein using an anti-Flag antibody (Fig. 1). It was previously reported that such a fusion leads to a functional enzyme able to catalyze in vitro Cd-dependent PC synthesis from GSH (27, 28). The engineered AtPCS-Flag sequence was placed under the control of T5 promoter/lac operator/repressor system that ensures high production of recombinant proteins. After induction with IPTG, a single band (56.3 kDa) of the expected size was recognized by a monoclonal antibody directed against the Flag sequence (Fig. 1).
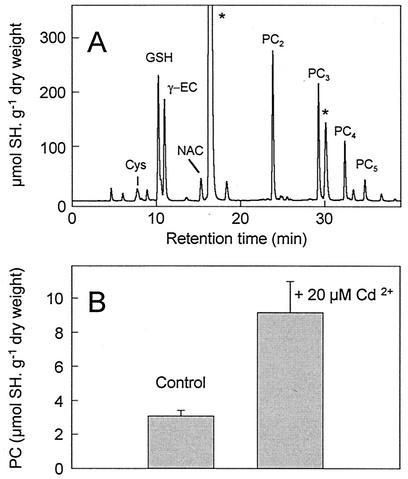
As a consequence of AtPCS expression, different PC forms (PC2, PC3, PC4, and to a lesser extent PC5, corresponding to repeats of γ-Glu-Cys dipeptide) were abundantly detected by HPLC analysis of nonprotein thiols (Fig. 2A). Interestingly, addition of 20 μM cadmium increased the total amount of PCs by about threefold (Fig. 2B). This enhancement, which was also observed upon addition of other metal ions such as Ag+, Hg2+, Zn2+, or Cu2+ (Table 1), is likely due to the metal activation of the enzyme (9, 28).
FIG. 2.
HPLC analysis of nonprotein thiols in E. coli cells expressing AtPCS. (A) Chromatogram of nonprotein thiols extracted from E. coli cells expressing AtPCS; different peaks are cysteine (Cys), GSH, N-acetyl-l-cysteine (NAC) used as an internal standard, γ-glutamylcysteine (γ-EC), PCs (PCn) and reagent (*). (B) Total PC content measured in E. coli cells expressing AtPCS in the absence (control) or presence of 20 μM Cd. Thiol concentrations are given in micromoles of GSH equivalent per gram (dry weight) of cells. Values are means of three independent experiments. Error bars, standard errors.
TABLE 1.
Metal and PC accumulation by bacterial cells expressing AtPCS
| Metal | Final concn (μM) | Mean total PC contenta ± SE (μmol of GSH · g−1) | Mean cell metal contenta ± SE (μmol · g−1)
|
|
|---|---|---|---|---|
| Controld | AtPCS | |||
| Cd(II) | 20 | 9.17 ± 1.81 | 0.37 ± 0.04 | 7.22 ± 1.78 |
| Zn(II) | 20 | 4.16 ± 0.037 | 0.78 ± 0.03 | 0.99 ± 0.11 |
| Pb(II) | 20 | 2.65 ± 0.3 | 1.83 ± 0.62 | 1.46 ± 0.3 |
| As(V) | 20 | —b | 0.03 ± 0.02 | 1.47 ± 0.25 |
| Cu(II) | 20 | 4.05 ± 0.37 | 0.44 ± 0.05 | 2.74 ± 0.19 |
| Hg(II) | 5 | 4.85 ± 0.56 | 1.4 ± 0.26 | 0.83 ± 0.08 |
| Ag(I) | 2.5 | 5.16 ± 0.45 | NDc | ND |
Based on dry weight and three independent experiments.
—, not determined.
ND, not detectable.
E. coli cells containing the vector without PCS were used as a control.
Metal accumulation.
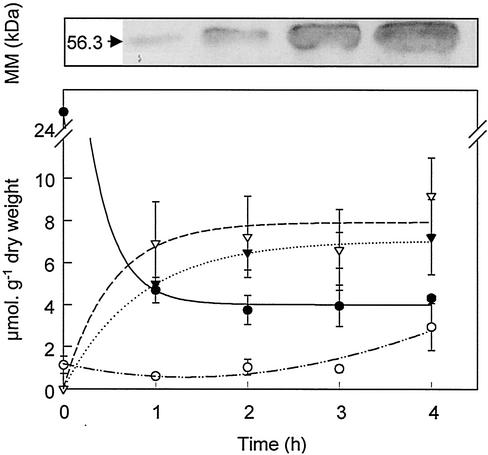
Upon induction by IPTG and addition of Cd to the culture medium, a relatively good correlation was observed between PC and intracellular Cd contents (Fig. 3). Both curves reached a plateau after 1 to 2 h, while the amount of AtPCS continuously increased during the 4 h of induction, likely indicating a metabolic limitation in PC synthesis. Concurrently, a severe drop in the GSH cellular content was observed, while the γ-glutamylcysteine (γ-EC) levels remained mostly unaffected, at least during the first 3 h (Fig. 3). We conclude from this experiment that the GSH supply most likely limits PC synthesis. This limitation might be overcome by overexpressing enzymes of the GSH biosynthetic pathway such as GSH synthase or γ-EC synthase.
FIG. 3.
Time course of AtPCS expression, PC synthesis, and Cd accumulation in E. coli cells expressing AtPCS after treatment with 20 μM Cd. The upper panel shows production of AtPCS followed by Western blotting with a monoclonal anti-Flag antibody. MM,: molecular mass. The lower panel shows PC synthesis and Cd accumulation. Cd concentrations were given in micromoles per gram (dry weight) of cells (▾). Thiol concentrations are given in micromoles of GSH equivalent per gram (dry weight) of cells (▿, PCs; •, GSH; ○, γ-EC). Values are means of three independent experiments. Error bars, standard errors.
Growing bacteria expressing AtPCS on Cd-enriched medium led to a 20-fold increase in the intracellular Cd content. Interestingly, marked increases in the Cu or As intracellular metal contents (respectively, 6- and 50-fold) were also observed when bacteria were grown on Cu- or As-enriched media, respectively. In contrast, despite the fact that addition of Ag, Hg, or Zn increased PC levels either in vivo (Table 1) or in vitro (data not shown; see also reference 9), no significant intracellular accumulation of these metals could be detected when bacteria were grown on metal-enriched media.
DISCUSSION
Different attempts have been made in the past to increase the metal content of bacterial cells by genetic engineering. Expression of metallothioneins in bacteria has encountered some difficulties, likely due to the instability of this cystein-rich protein (14). Relatively good expression was, however, obtained when metallothionein or its α subdomain were fused to a stabilizing protein such as the maltose-binding protein, resulting in a significant increase in cadmium accumulation (11, 19). Also, fusion of metallothioneins to proteins of the outer membrane significantly increased cadmium accumulation by bacterial cells (24). Fusion of poly-histidines, or other artificial peptides able to chelate divalent metal ions like Ni2+ or Zn2+ and Cd2+, to highly expressed proteins has also been found to significantly enhance metal accumulation by bacteria (15, 23). Recently, the peptide (Glu-Cys)n-Gly, in which Glu and Cys are linked by an α-carboxyamide bond (and not by a γ-carboxyamide bond, as seen in PCs), was successfully expressed as a fusion protein in E. coli, resulting in 15- or 20-fold increases in cadmium (1) or mercury (2) cellular contents, respectively. In contrast to ribosomally synthesized fusion peptides or to metallothioneins, PCs are synthesized enzymatically, presenting certain advantages for biotechnological applications. First, although the degradation of PCs has not been studied so far, PCs, unlike metallothioneins appear to be relatively stable compounds not subject to extensive proteolysis. Second, when compared to metallothioneins, PCs appear to have a different selectivity for metals, efficiently binding Cd and Cu, but also the metalloid As. Another interesting feature of PC synthesis is that it significantly increases when bacteria are treated by Cd, or by other heavy metals (Cu, Ag, or Zn), likely due to the activation of PC synthase by metals (Fig. 2; Table 1) (6, 9, 28). Also, PC synthesis might be optimized by overexpressing enzymes of its biosynthetic pathway, for instance, enzymes involved in glutathione or cysteine synthesis, the glutathione supply most likely limiting PC synthesis (Fig. 3).
In contrast to the cases of Cd, Cu, and As, no significant accumulation of Ag, Hg, and Zn was observed when bacteria were grown on media supplemented with these metals. Interestingly, Ag, Hg, or Zn were able to increase PC levels either in vivo (Table 1) or in vitro (data not shown), likely indicating, as already reported (9), that PC synthase can be activated by these metals. The absence of accumulation might be explained by the fact that these metals are either not efficiently bound by PCs or not available in sufficient amounts within bacterial cells, due for instance to inefficient transport or high affinity chelation by metallo-chaperones.
Surprisingly, although expression of AtPCS led to a significant increase in intracellular cadmium concentrations, we did not observe any increase in cadmium tolerance (data not shown). This contrasts with previous reports in yeasts. In S. cerevisiae, expression of AtPCS induced an increased cadmium tolerance in two different mutant strains, a mutant disrupted in an ABC transport protein responsible for vacuolar sequestration of Cd(GSH)2 complexes and a Cu2+-hypersensitive mutant deficient in metallothionein (27). In S. pombe, Cd is complexed to PCs and subsequently transported to the vacuole by HMT1, another ABC transport protein (17, 18). These contrasting results may be due to the fact that the bacterial strain used in this study is relatively tolerant to high levels of Cd (up to 200 to 400 μM). This might also reflect the fact that metal tolerance in yeasts results not only from the binding of metals by PCs, but may also require transport of PC-heavy metal complexes to the vacuole.
The case of arsenic appears particularly interesting. In higher plants, the existence of complexes between PCs and arsenite in vivo and in vitro has recently been observed (21). Bacteria generally accumulate very low arsenic levels because of a highly efficient export system, based on the reduction of arsenate to arsenite and export by a specific efflux pump (22). This explains why, despite the high toxicity of arsenic, most bacteria are naturally tolerant to high concentrations of this metalloid. In our hands, no effect of arsenic on bacterial growth was observed at concentration up to 500 μM in the medium. In bacteria cells expressing AtPCS, arsenic accumulation is likely due to the production of PCs which compete with the arsenic efflux transporter and sequester arsenic ions (likely arsenite [21]) in a nontoxic form. This, to our knowledge, is the first report of increased intracellular arsenic levels in a living organism based on genetic engineering. These results open the way to applications in arsenic bioremediation, since this metalloid is considered a major hazard for human health, responsible for large-scale contamination of groundwater in Bangladesh (3), for example.
In conclusion, overexpression of PC synthase in bacterial strains appears to be a promising way to improve the heavy metal (such as Cd) or metalloid (such as As) content of organisms for use in bioremediation processes. Such bioprocesses can be based on the use of bacterial strains adapted either to the formation of biofilms (8) or to the growth in bioreactors to clean up liquid wastes. Other applications may include the development of biofilters by expressing PC synthase in soil bacteria, in which PCs may scavenge heavy metals from polluted soils and decrease their toxic effects on plant growth, as recently shown in tobacco plants growing in the presence of Ralstonia eutropha cells expressing mouse metallothionein (25). Finally, overexpression of PC synthase in plants might optimize their metal-binding abilities and increase their potential use in phytoremediation processes.
Acknowledgments
We thank Claudio Vita (CEA Saclay, Direction des Sciences du Vivant) for synthesizing PC standards and André Verméglio (CEA Cadarache, Direction des Sciences du Vivant) and Michael DuBow (Institut de Génétique et de Microbiologie, Université Paris Sud, Orsay, France) for helpful discussions.
This work was supported by the Toxicologie Nucléaire program of the CEA.
REFERENCES
- 1.Bae, W., W. Chen, A. Mulchandani, and R. K. Mehra. 2000. Enhanced bioaccumulation of heavy metals by bacterial cells displaying synthetic phytochelatins. Biotechnol. Bioeng. 70:518-524. [DOI] [PubMed] [Google Scholar]
- 2.Bae, W., R. K. Mehra, A. Mulchandani, and W. Chen. 2001. Genetic engineering of Escherichia coli for enhanced uptake and bioaccumulation of mercury. Appl. Environ. Microbiol. 67:5335-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chowdhury, T. R., G. K. Basu, B. K. Mandal, B. K. Biswas, G. Samanta, U. K. Chowdhury, C. R. Chanda, D. Lodh, S. L. Roy, K. C. Saha, S. Roy, S. Kabir, Q. Quamruzzaman, and D. Chakraborti. 1999. Arsenic poisoning in the Ganges delta. Nature 401:545-546. [DOI] [PubMed] [Google Scholar]
- 4.Clemens, S., E. J. Kim, D. Neumann, and J. I. Schroeder. 1999. Tolerance to toxic metals by a gene family of phytochelatin synthases from plants and yeast. EMBO J. 18:3325-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clemens, S., J. I. Schroeder, and T. Degenkolb. 2001. Caenorhabditis elegans expresses a functional phytochelatin synthase. Eur. J. Biochem. 268:3640-3643. [DOI] [PubMed] [Google Scholar]
- 6.Cobbett, C. S. 2000. Phytochelatin biosynthesis and function in heavy-metal detoxification. Curr. Opin. Plant Biol. 3:211-216. [PubMed] [Google Scholar]
- 7.Cobbett, C. S. 2000. Phytochelatins and their roles in heavy metal detoxification. Plant Physiol. 123:825-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deziel, E., Y. Comeau, and R. Villemur. 2001. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J. Bacteriol. 183:1195-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ha, S. B., A. P. Smith, R. Howden, W. M. Dietrich, S. Bugg, M. J. O'Connell, P. B. Goldsbrough, and C. S. Cobbett. 1999. Phytochelatin synthase genes from arabidopsis and the yeast Schizosaccharomyces pombe. Plant Cell 11:1153-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 11.Li, Y., W. Cockburn, J. Kilpatrick, and G. C. Whitelam. 2000. Cytoplasmic expression of a soluble synthetic mammalian metallothionein-alpha domain in Escherichia coli. Enhanced tolerance and accumulation of cadmium. Mol. Bio/Technology 16:211-219. [DOI] [PubMed] [Google Scholar]
- 12.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 13.Maitani, T., H. Kubota, K. Sato, and T. Yamada. 1996. The composition of metals bound to class III metallothionein (phytochelatin and its desglycyl peptide) induced by various metals in root cultures of Rubia tinctorum. Plant Physiol. 110:1145-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mejare, M., and L. Bulow. 2001. Metal-binding proteins and peptides in bioremediation and phytoremediation of heavy metals. Trends Biotechnol. 19:67-73. [DOI] [PubMed] [Google Scholar]
- 15.Mejare, M., S. Ljung, and L. Bulow. 1998. Selection of cadmium specific hexapeptides and their expression as OmpA fusion proteins in Escherichia coli. Protein Eng. 11:489-494. [DOI] [PubMed] [Google Scholar]
- 16.Nriagu, J. O., and J. M. Pacyna. 1988. Quantitative assessment of worldwide contamination of air, water and soils by trace metals. Nature 333:134-139. [DOI] [PubMed] [Google Scholar]
- 17.Ortiz, D. F., L. Kreppel, D. M. Speiser, G. Schell, G. McDonald, and D. W. Ow. 1992. Heavy metal tolerance in the fission yeast requires an ATP-binding cassette-type vacuolar membrane transporter. EMBO J. 11:3491-3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ortiz, D. F., T. Ruscitti, K. F. McCue, and D. W. Ow. 1995. Transport of metal-binding peptides by HMT1, a fission yeast ABC-type vacuolar membrane protein. J. Biol. Chem. 270:4721-4728. [DOI] [PubMed] [Google Scholar]
- 19.Pazirandeh, M., L. A. Chrisey, J. M. Mauro, J. R. Campbell, and B. P. Gaber. 1995. Expression of the Neurospora crassa metallothionein gene in Escherichia coli and its effect on heavy-metal uptake. Appl. Microbiol. Biotechnol. 43:1112-1117. [DOI] [PubMed] [Google Scholar]
- 20.Rijstenbil, J. W., and J. A. Wijnholds. 1996. HPLC analysis of nonprotein thiols in planktonic diatoms: pool size, redox state and response to copper and cadmium exposure. Mar. Biol. 127:45-54. [Google Scholar]
- 21.Schmoger, M. E. V., M. Oven, and E. Grill. 2000. Detoxification of arsenic by phytochelatins in plants. Plant Physiol. 122:793-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silver, S. 1996. Bacterial resistances to toxic metal ions—a review. Gene 179:9-19. [DOI] [PubMed] [Google Scholar]
- 23.Sousa, C., A. Cebolla, and V. de Lorenzo. 1996. Enhanced metalloadsorption of bacterial cells displaying poly-His peptides. Nat. Biotechnol. 14:1017-1020. [DOI] [PubMed] [Google Scholar]
- 24.Sousa, C., P. Kotrba, T. Ruml, A. Cebolla, and V. De Lorenzo. 1998. Metalloadsorption by Escherichia coli cells displaying yeast and mammalian metallothioneins anchored to the outer membrane protein LamB. J. Bacteriol. 180:2280-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valls, M., S. Atrian, V. de Lorenzo, and L. A. Fernandez. 2000. Engineering a mouse metallothionein on the cell surface of Ralstonia eutropha CH34 for immobilization of heavy metals in soil. Nat. Biotechnol. 18:661-665. [DOI] [PubMed] [Google Scholar]
- 26.Vatamaniuk, O. K., E. A. Bucher, J. T. Ward, and P. A. Rea. 2001. A new pathway for heavy metal detoxification in animals: phytochelatin synthase is required for cadmium tolerance in Caenorhabditis elegans. J. Biol. Chem. 276:20817-20820. [DOI] [PubMed] [Google Scholar]
- 27.Vatamaniuk, O. K., S. Mari, Y. P. Lu, and P. A. Rea. 1999. AtPCS1, a phytochelatin synthase from Arabidopsis: Isolation and in vitro reconstitution. Proc. Natl. Acad. Sci. USA 96:7110-7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vatamaniuk, O. K., S. Mari, Y. P. Lu, and P. A. Rea. 2000. Mechanism of heavy metal ion activation of phytochelatin (PC) synthase: blocked thiols are sufficient for PC synthase-catalyzed transpeptidation of glutathione and related thiol peptides. J. Biol. Chem. 275:31451-31459. [DOI] [PubMed] [Google Scholar]