Abstract
RNA–protein interactions are vitally important in a wide range of biological processes, including regulation of gene expression, protein synthesis, and replication and assembly of many viruses. We have developed a computational tool for predicting which amino acids of an RNA binding protein participate in RNA–protein interactions, using only the protein sequence as input. RNABindR was developed using machine learning on a validated nonredundant data set of interfaces from known RNA–protein complexes in the Protein Data Bank. It generates a classifier that captures primary sequence signals sufficient for predicting which amino acids in a given protein are located in the RNA–protein interface. In leave-one-out cross-validation experiments, RNABindR identifies interface residues with >85% overall accuracy. It can be calibrated by the user to obtain either high specificity or high sensitivity for interface residues. RNABindR, implementing a Naive Bayes classifier, performs as well as a more complex neural network classifier (to our knowledge, the only previously published sequence-based method for RNA binding site prediction) and offers the advantages of speed, simplicity and interpretability of results. RNABindR predictions on the human telomerase protein hTERT are in good agreement with experimental data. The availability of computational tools for predicting which residues in an RNA binding protein are likely to contact RNA should facilitate design of experiments to directly test RNA binding function and contribute to our understanding of the diversity, mechanisms, and regulation of RNA–protein complexes in biological systems. (RNABindR is available as a Web tool from http://bindr.gdcb.iastate.edu.)
Keywords: bioinformatics, RNA binding site, RNA–protein interactions, RNABindR, telomerase, prediction
INTRODUCTION
Understanding the molecular mechanisms by which proteins recognize and discriminate between specific RNA molecules is critical for comprehending the functional implications of these interactions in cells. RNA–protein interactions, in addition to their importance in protein synthesis, mRNA processing, and viral replication, have recently been shown to play critical roles in cellular defense and developmental regulation (Hall 2002; Tian et al. 2004), underscoring the importance of understanding the molecular determinants of RNA–protein interactions.
At least nine families of RNA binding proteins have been identified using sequence-based analyses of RNA binding proteins, together with functional characterization of mutations that affect the specificity or affinity of RNA binding (for review, see Chen and Varani 2005). In contrast, the number of experimentally determined structures for RNA–protein complexes is still relatively small and heavily biased (ribosomal proteins represent ∼50% of all RNA binding proteins in the Protein Data Bank [PDB]). Nevertheless, several computational analyses of RNA–protein complexes have generated databases of RNA–protein contacts and provided valuable insights into the biophysical basis of interaction patterns between ribonucleotides and amino acids (Cusack 1999; Draper 1999; Jones et al. 2001; Kim et al. 2003; Hoffman et al. 2004; Jeong et al. 2004; Jeong and Miyano 2006).
Because of the importance of RNA–protein interactions in biological regulation and the considerable effort required to identify RNA binding residues through biophysical analyses of RNA–protein complexes or in vitro binding studies, there is an urgent need for computational methods to identify RNA binding sites based on primary amino acid sequence alone. Machine learning techniques offer an attractive approach to construction of classifiers for this task, using data sets of experimentally well-characterized RNA–protein complexes. Three recent studies have reported the use of support vector machines (SVMs) to identify RNA binding proteins and assign them to functional classes (e.g., rRNA binding, mRNA binding, tRNA binding, viral RNA binding, etc.) using only the amino acid sequence (Han et al. 2004), a combination of sequence and pseudo-amino acid composition as input (Cai and Lin 2003), or a variety of sequence-based information, including predicted solvent accessibility and predicted secondary structure (Yu et al. 2006). Our previous work (Yan et al. 2004a,b, 2006) has demonstrated the feasibility of constructing classifiers for protein–protein and protein–DNA binding site identification using machine learning approaches. However, there has been little work using machine learning approaches to construct classifiers for identifying RNA binding sites from primary amino acid sequence.
In this article, we present RNABindR, a fast and simple tool for predicting RNA binding sites. In its current implementation, RNABindR requires only protein sequence information as input; no information regarding the structure of the protein or the sequence or structure of the RNA is required. Although inclusion of structure-derived information, when available, can improve predictions, we focus here on sequence-based prediction to provide a broadly applicable tool. To demonstrate the utility of RNABindR, we make predictions on the telomerase protein TERT, for which the structure of the protein–RNA complex has not been determined. The predictions are in good agreement with the experimentally characterized RNA binding regions of TERT.
The only previously published sequence-based method for predicting interface residues, to our knowledge, is a neural network classifier reported by Miyano's group (Jeong et al. 2004; Jeong and Miyano 2006). The results of our experiments demonstrate that the performance of RNABindR, using a Naive Bayes classifier trained and tested on the same data set, is comparable to that of the neural network classifier. Unlike the neural network classifier, which requires multiple passes through the data during training, the Naive Bayes classifier requires only one pass through the training data, is easily updatable, and is rather straightforward to interpret.
RESULTS
Sequence characteristics of RNA binding sites
Arginine-rich motifs (Weiss and Narayana 1998) are abundant in RNA binding sites, and other strong biases in the types of amino acids present in RNA–protein interfaces have been reported in several previous studies (Lustig et al. 1997; Jones et al. 2001; Kim et al. 2003; Jeong et al. 2004; Jeong and Miyano 2006). To evaluate whether these primary sequence biases can be effectively exploited in a machine learning approach to identify amino acid sequence correlates of RNA binding sites, we generated a nonredundant data set of 109 RNA binding proteins (see Materials and Methods) to estimate the interface propensity for each amino acid type as follows:
 |
An interface propensity value greater than 0 indicates that an amino acid is overrepresented in RNA–protein interfaces relative to the protein sequence as a whole. Figure 1 shows the interface propensity (solid bars) for each of the 20 amino acids, as well as the frequency with which that amino acid occurs in each of the two positions immediately flanking a known interface residue (cross-hatched bars). Interface propensities, estimated from a smaller data set of 55 ribosomal protein chains (data not shown) did not differ significantly from those estimated using the larger data set of 109 RNA binding proteins, and our results using both data sets are consistent with previously published data (Jones et al. 2001).
FIGURE 1.
Certain amino acids are highly favored in RNA–protein interfaces. Interface propensities for the indicated amino acids are shown as solid bars; the hatched bars to the left and right of the solid bar are the propensities for the amino acid to occur in the position immediately before or after an interface residue, respectively. The residues are placed in the order of increasing hydrophobicity based on the (Kyte and Doolittle 1982) hydropathy index.
As expected, the positively charged amino acids arginine and lysine show the highest interface propensities, 1.29 and 1.17, respectively, consistent with their ability to participate in interactions both with bases and with the negatively charged phosphate backbone of RNA. Together, arginine and lysine account for 32% of the interface residues in our data set. While this is a significant fraction of interface residues, it also shows that one cannot focus solely on positively charged amino acids to discover how RNA–protein recognition occurs. Another favored residue, histidine (0.60), also can be positively charged and can participate in stacking interactions with RNA bases through its imidazole ring. Tryptophan and tyrosine are slightly preferred, with propensities of 0.21 and 0.18, respectively. In contrast, phenylalanine (−0.60) and negatively charged amino acids glutamate (−1.13) and aspartate (−0.62) are significantly underrepresented in interfaces, as are hydrophobic residues such as leucine, isoleucine, valine, and alanine (all below −0.84). Importantly, there are significant biases in the types of amino acids that tend to be “sequence neighbors” of interface residues. For instance, glycine is highly preferred on either side of an interface residue (0.50 and 0.47); its small size may enhance flexibility, allowing protein domains to adopt conformations that facilitate RNA binding.
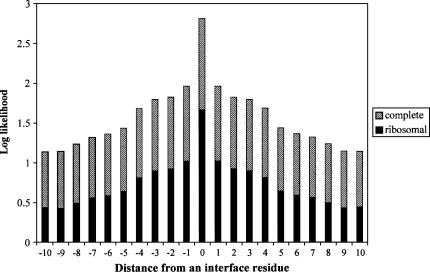
If the biases in amino acid propensities noted above are frequently accompanied by clustering of interface residues within the primary sequence of an RNA binding protein, a machine learning algorithm should be able to “learn” sequence composition characteristics or other signals in the neighborhood surrounding interface residues, based on a validated training data set, and generate a classifier for predicting likely interface residues in test sequences. The tendency of protein–protein interface residues to be clustered along the primary sequence of proteins has been noted previously (Jones et al. 2001; Ofran and Rost 2003; Yan et al. 2004b). We examined the tendency of RNA–protein interface residues to be similarly clustered in our data set of RNA binding proteins by calculating the log-likelihood that a residue is an interface residue, given that it is at a certain distance from another interface residue (Fig. 2). The log-likelihood is given by log2(P observed/P background) where P observed is the observed probability that a given neighbor of an interface residue is also an interface residue and Pbackground is the probability that the position is an interface residue by chance (∼0.14 for our data set, because ∼14% of the residues in our data set are interface residues).
FIGURE 2.
RNA binding residues tend to occur in clusters within primary sequence. The log likelihood that a position neighboring an interface residue also contains an interface residue based on the nonredundant data set of 109 RNA binding proteins. The hatched portion of the bars represents the log likelihood for the entire data set of 109 proteins. The solid portion of the bars represents the log likelihood for the ribosomal protein subset of 55 proteins. Likelihood values >0 mean that the position has higher probability than random of also being an interface residue.
This analysis revealed that 95% of interface residues in the data set of 109 RNA binding proteins have at least one additional interface residue among the four amino acids on either side, and 49% have at least four. The tendency of interface residues to be clustered within the primary sequence is more pronounced in the subset of 55 ribosomal proteins: 97% of interface residues in the ribosomal data set have at least one additional interface residue within four amino acids on either side and 63% have at least four neighboring interface residues. For the data set of 54 nonribosomal proteins, the corresponding values are 90% and 23%, respectively. Thus, this tendency of interface residues to cluster in primary sequence, together with the distinct interface propensities of individual amino acids, suggests that it should be possible to capture functionally relevant sequence signals in the neighborhood of interface residues and to exploit these using a machine learning approach to predict RNA binding sites in proteins.
Using a Naive Bayes classifier, RNABindR, reliably predicts RNA–protein interface residues using only amino acid sequence information
The performance of RNABindR, using a Naive Bayes classifier, was evaluated in leave-one-out cross-validation experiments as described in Materials and Methods. Table 1 summarizes an example of the results obtained using four different input window sizes and a threshold, θ, that was empirically determined to provide an optimal correlation coefficient on the training set. Using an input window of 25 amino acids, the classifier achieved an overall accuracy of 85% with a correlation coefficient of 0.35, specificity+ of 0.51 and sensitivity+ of 0.38 (see Materials and Methods for definitions). Adding information such as secondary structure, relative accessible surface area, sequence entropy, hydrophobicity, and electrostatic potential to the amino acid sequence inputs did not improve RNABindR performance. Performance on the ribosomal subset was better than the average performance over the entire data set (data not shown). However, performance on the ribosomal subset was the same whether the training set used was the ribosomal subset or the entire data set.
TABLE 1.
Interface residue prediction performance of RNABindR

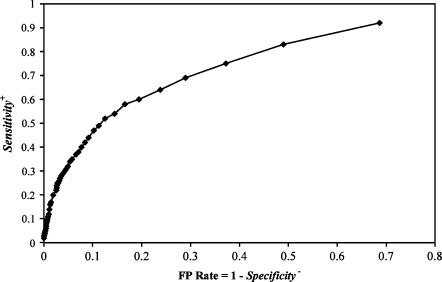
In specific biological applications, such as identifying critical residues for site-specific mutagenesis, it may be more important to predict interface residues with high specificity (i.e., to produce a smaller number of “positive” interface residue predictions with high confidence) than to obtain a high correlation coefficient. We report results obtained with classifiers trained to obtain an optimal correlation coefficient (CC) because CC is a more meaningful measure than specificity or sensitivity for comparing different classifiers (see Materials and Methods; Baldi et al. 2000). With a Naive Bayes classifier, it is straightforward to vary the threshold θ to increase specificity + at the expense of a decrease in sensitivity +. This is illustrated in Figure 3, which shows a receiver operating characteristic (ROC) plot of sensitivity + against false positive rate, defined as (1-specificity −). At the expense of lower sensitivity, a very low false positive rate can be achieved.
FIGURE 3.
Receiver operating characteristic (ROC) curve for RNABindR predictions. The ROC curve illustrates how varying the cutoff threshold θ determines the trade-off between sensitivity+ and false positive rate (1-specificity−), where specificity− is defined as FP/(FP + TN). Results shown are for an input window of 25 amino acids.
While these statistics allow evaluation of the performance of RNABindR in identifying RNA–protein interface amino acids on a per residue basis, an important criterion for evaluating its utility in practice is whether it correctly identifies a significant fraction of the total interface residues in individual RNA binding proteins. For the complete data set, RNABindR effectively recognized binding sites in 59% of proteins by correctly identifying at least 20% of the interface residues (Fig. 5A, see below).
FIGURE 5.
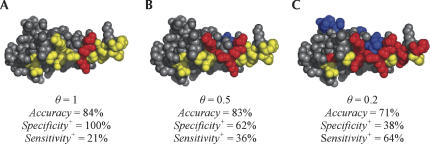
RNABindR sensitivity and specificity trade-off. Changing the value of the threshold parameter θ causes a trade-off between specificity and sensitivity in predicting RNA binding residues. The example shown here is the double-stranded RNA binding protein from Xenopus, PDB ID 1DI2:A, also shown in Figure 4B. The color scheme in this figure is the same as in Figure 4.
Evaluating RNABindR predictions in the context of three-dimensional structures
In developing RNABindR, we have not taken advantage of available structural information regarding the target protein or its cognate RNA because it is much more common to have the sequence of a protein without a structure. Nevertheless, it is informative to evaluate RNABindR results by visualizing them in the context of three-dimensional structures of known RNA–protein complexes. Figure 4 shows examples in which RNABindR was tested on one protein chosen from each of the four different categories of complexes in the complete data set (see Table 2): (1) rRNA; (2) mRNA, snRNA, dsRNA, and siRNA; (3) tRNA; and (4) viral RNA. For each protein, the predicted versus actual interface residues, shown in red, are mapped onto surface plots of PDB structures (Fig. 4, cf. left and middle panels). In the panels on the far right, a different coloring scheme is used to illustrate the performance of RNABindR on individual residues in each protein (see below).
FIGURE 4.
Predictions mapped onto three-dimensional structures of RNA binding proteins. Examples of RNABindR results for four different types of RNA–protein complexes are shown: (A) ribosomal protein L15, PDB 1JJ2:K (Klein et al. 2001); (B) Xenopus dsRNA binding protein, PDB 1DI2:A (Ryter and Schultz 1998); (C) Ebola virus Vp40, PDB 1H2C:A (Gomis-Ruth et al. 2003); (D) tRNA pseudouridine synthase, PDB 1R3E:A (Pan et al. 2003). Predicted RNA binding sites, with predicted interface residues shown in red and predicted noninterface residues in gray (left panels). Actual RNA binding sites, with actual interface residues in red and actual noninterface residues in gray (middle panels). The performance of RNABindR for individual residues, with true positives (TPs) shown in red, false positives (FPs) in blue, false negatives (FNs) in yellow, and true negatives (TNs) in gray (right panels). Thus, in this representation, red + yellow residues correspond to the actual interface (derived from the PDB structure), red + gray residues correspond to correctly predicted residues (both interface and noninterface), and blue + yellow residues correspond to misclassified residues. Results shown were predicted with RNABindR using an input window of 25 amino acids and θ = 0.5. All structure diagrams were generated using PyMol (http://www.pymol.org).
TABLE 2.
RNA binding proteins in the nonredundant training data set
Results obtained for ribosomal protein L15 (PDB 1JJ2:K), a structural component of the large ribosomal subunit from the archaebacterium Haloarcula marismortui (Klein et al. 2001) are shown in Figure 4A. This was the “best” prediction (ranked #1 out of 109) based on correlation coefficient (0.63). For clarity and because of its large size, the RNA partner is not included in this example. In L15, one of the two RNA binding sites was detected with very high specificity (Fig. 4A, cf. red residues representing the predicted interface in the left panel and the actual interface in the middle panel). In the rightmost panel (Fig. 4A), interface residues of L15 that were correctly identified as such (true positives, TPs) are shown in red: 40 out of 42 predicted interface residues are, in fact, interface residues (specificity + = 95%). There were only two false positive (FP) predictions, shown in blue. True negatives (TNs), in gray, and false negatives (FNs), in yellow, are also shown. Note that although the specificity for interface residues in this example is high (95%), the accuracy is relatively low (80%) compared with the average over the complete data set (85%), largely due to failure of RNABindR to detect any interface residues in one of two RNA binding sites on the L15 protein. As described below, sensitivity (for the training data set) can be enhanced by choosing a lower value for θ. In the case of L15, this results in better coverage (i.e., higher sensitivity), allowing the second RNA binding domain to be detected, but at the loss of specificity (data not shown).
Results of similar analyses for a protein from each of the other three classes of RNA–protein complexes are shown in Figure 4B–D. Figure 4B shows results for the double-stranded RNA binding motif (dsRBM) domain of the Xenopus dsRNA binding protein A bound to RNA (in green wire frame). The prediction for this protein ranked 23rd (CC = 0.38) with an overall accuracy of 83%. A simple search for RNA binding motifs on this protein reveals that the entire 69 amino acid sequence included in the crystal structure is the canonical dsRBM. However, there are only 13 actual interface residues within this motif, all clustered on one face of the protein shown. RNABindR correctly identified 5 of these 13 interface residues. Figure 5 illustrates how lowering the threshold θ significantly improves identification of the interface residue class. The interface residue predictions for the dsRNA binding protein shown in Figure 4 are shown for three different values of θ. In Figure 5A when θ is relatively high, a small number of interface residues are predicted with high specificity. Figure 5B shows the predictions using the value of θ obtained by optimizing RNABindR on the training set. In Figure 5C when θ is low, many more interface residues are predicted, but we sacrifice specificity to do so.
The Ebola virus matrix protein, Vp40, bound to a 3-nt RNA ligand (accuracy 95%) is shown in Figure 4C, and a tRNA pseudouridine synthase bound to a tRNA ligand (51 nt) is shown in Figure 4D. These predictions were ranked 19th (CC = 0.42) and 34th (CC = 0.29) out of 109, respectively. Performance statistics provided in the figure captions illustrate that the specificity and sensitivity for noninterface residues are much higher than for interface residues in both cases.
Comparison of RNABindR predictions with mapped RNA binding sites in the telomerase protein, TERT
The primary motivation for developing RNABindR (which does not require structural information) was to provide a tool for identifying potential RNA binding sites in proteins when information regarding the RNA–protein complex or its interface is not available. To demonstrate the utility of RNABindR in such cases, we have applied it to the prediction of RNA binding residues in the human telomerase protein hTERT. Telomerase is the ribonucleoprotein complex responsible for maintaining telomere length by adding short repeated sequences to the ends of chromosomes (for reviews, see Blackburn 2005; Autexier and Lue 2006). TERT is the reverse transcriptase component of telomerase and binds to the essential telomerase RNA subunit (TR), which serves as the template for synthesis of telomeric DNA repeats. The C-terminal half of hTERT contains the reverse transcriptase domain (RT), and two RNA interaction domains (RIDs) have been mapped to the N-terminal half of the protein (Bachand and Autexier 2001; Lai et al. 2001; Moriarty et al. 2002, 2005). RID2 is a relatively high affinity RNA binding domain and RID1 is a lower affinity RNA binding domain (for review, see Autexier and Lue 2006). RID1 and RID2 each contain several elements that are conserved at the primary sequence level and, in some cases, have been shown to be important for RNA binding based on mutagenesis and in vitro binding experiments (Bachand and Autexier 2001; Lai et al. 2001; Moriarty et al. 2002, 2005).
Figure 6A shows the RNA interface residues predicted by RNABindR mapped onto functional domains of hTERT defined by in vitro catalytic activity and/or RNA binding assays (Bachand and Autexier 2001; Lai et al. 2001; Moriarty et al. 2002, 2005). The prediction that most residues involved in hTERT RNA binding lie outside the RT domain is in agreement with experimental results that have demonstrated that the RT and RNA binding domains of hTERT are separable (Lai et al. 2001; Moriarty et al. 2004). Most clusters of predicted RNA binding residues are located within the experimentally mapped RNA binding domains, RID1 and RID2, or correspond to arginine-rich portions of the variable “linker” region between them, which has been shown to contribute to hTERT RNA binding in vitro (Moriarty et al. 2002).
FIGURE 6.
RNABindR predictions on telomerase reverse transcriptase (TERT). Mapped functional domains and conserved motifs of TERT are shown at the top. Shaded boxes on lines labeled “Predictions” show clusters of predicted RNA interface residues. (A) Human telomerase reverse transcriptase (hTERT). Boundaries of two major RNA interaction domains (RIDs) indicated by open boxes (Moriarty et al. 2005). The amino acid sequence that includes one of the clusters of predicted RNA-interface residues, located in RID2, is shown at the bottom. Two boxed regions, amino acids 481–490 and amino acids 508–517, correspond to deletion mutations that have been shown to decrease hTERT RNA binding activity by 60% and 70%, respectively (Moriarty et al. 2002). Individual interface residues predicted by RNABindR are indicated by + below the sequence. (B) Tetrahymena thermophila telomerase reverse transcriptase (tTERT). The two RNA binding domains are indicated by open boxes. The amino acid sequence of the C-terminal end of the TEN RNA binding domain is shown, with individual interface residues predicted by RNABindR indicated by + below the sequence. Removing residues 1–12 and 182–191 (boxed in the sequence view) abolished RNA binding of the TEN domain construct (Jacobs et al. 2005, 2006). RNABindR predicts a cluster of interface residues in residues 182–191, but no interface residues are predicted in residues 1–12. (N) N terminus, (TEN) telomerase essential N-terminal domain, (GQ, CP, QFP, and T) conserved sequence motifs, (RT) reverse transcriptase domain.
The amino acid sequence of a conserved portion of RID2 containing a cluster of predicted RNA binding residues is shown in the lower portion of Figure 6A. This predicted cluster lies within the “QFP” motif in RID2 and encompasses amino acids whose deletion results in a 60% reduction in RNA binding (amino acids 481–490, in box) (Moriarty et al. 2002). Another cluster of interface residues within RID2 (but outside the region whose sequence is shown) also overlaps with sequences within the “T” motif required for full RNA binding activity based on deletion studies (Lai et al. 2001). Three clusters of predicted interface residues lie within or overlap the boundaries of RID1, which appears to comprise a lower affinity binding domain that contributes to, but is not absolutely required for, RNA binding (Moriarty et al. 2002). An example of a case in which RNABindR does not predict interface residues corresponding to amino acids whose deletion results in reduced RNA binding activity is also shown in the lower portion of Figure 6A (amino acids 508–517, in box). It is important to note, however, that loss of RNA binding activity in these experiments could be due either to deletion of residues that directly contact RNA or to loss of binding due to an indirect effect on the overall structure or stability of hTERT. Moreover, experimental data that provide evidence for or against the role of specific amino acids in the hTERT–TR interaction are not available for most residues within the mapped RNA binding domains. Overall, the RNABindR predictions are in very good agreement with currently available experimental data and identify several additional amino acids that could potentially contribute to hTERT RNA binding activity.
Tetrahymena TERT also has two RNA binding domains. The higher affinity domain, residues 195–516, is essential for telomerase RNA binding (Lai et al. 2001), and mutagenesis experiments have demonstrated that specific residues within the CP and T motifs are important for RNA binding (Bryan et al. 2000; Lai et al. 2002). Figure 6B shows RNABindR predictions mapped onto the functional regions of Tetrahymena TERT. RNABindR predicts two clusters of interface residues, one near the T motif, but none in the CP motif. A lower affinity RNA binding domain, referred to as the TEN domain, contributes to telomerase RNA binding (O'Connor et al. 2005). Residues 1–12 and 182–191 within this domain are especially important for RNA binding; they are susceptible to digestion by Lys-C in the absence of RNA and protected in the presence of RNA (Jacobs et al. 2005, 2006). Also, the deletion of these two short segments abolishes RNA binding (Jacobs et al. 2006). Notably, the only interface residues predicted by RNABindR in this domain are a cluster from 185–191. Thus, RNABindR predictions for Tetrahymena TERT agree well with the available experimental data.
The performance of RNABindR, implementing a Naive Bayes classifier, is comparable to that of a more complex neural network classifier
To our knowledge, there is only one other published successful application of a machine learning approach to sequence-based prediction of interface residues in RNA–protein complexes. Using a data set of 96 chains from RNA–protein complexes and a total of 4782 interface residues, Miyano's group (Jeong et al. 2004) used a neural network to predict interface residues in RNA binding proteins. Miyano's group reported a CC = 0.59 obtained using filtering and state-shifting. Both filtering and state-shifting take advantage of the fact that interface residues are clustered along the primary sequence. Filtering removes incorrect interface predictions that are isolated, i.e., if a residue is predicted to be an interface residue, but no neighboring residues are predicted as interface residues, the prediction is changed to noninterface. State-shifting corrects predictions for residues that were misclassified as noninterface residues by changing the prediction to interface if a neighboring residue is predicted to be an interface residue. Both filtering and state-shifting use information that is generally unavailable to the classifier, i.e., there is no a priori way to determine the false positive and false negative predictions in a test sequence that is not part of the training set. Hence, we do not attempt a comparison of results obtained by filtering and state-shifting with our results. To facilitate direct comparison of RNABindR with the published neural network classifier, we trained and tested RNABindR using a Naive Bayes classifier on the same data set used in Miyano's study. Table 3 shows the best results reported by Miyano's group (Jeong et al. 2004) using a neural network, without filtering and state-shifting, compared with the best results (in terms of correlation coefficient) obtained using RNABindR. Notably, the overall results are comparable, but the Naive Bayes method is considerably faster and easier to implement.
TABLE 3.
Comparison of RNABindR (Naive Bayes classifier) with a neural network classifier

RNABindR detects known PROSITE RNA binding motifs
To compare the motifs picked out by RNABindR with known RNA binding motifs, we identified all PROSITE motifs (Hulo et al. 2004) annotated as nucleic acid binding in the proteins from our data set. PROSITE contains a collection of sequence patterns that are known to be associated with a particular protein family or function. By identifying all of the PROSITE motifs that are involved in nucleic acid binding in our nonredundant data set, we can compare RNABindR performance with simply searching for known RNA binding sequence motifs. RNABindR identified 104 out of 109 proteins (95%) in the nonredundant data set as RNA binding proteins, whereas PROSITE identified RNA binding motifs in only 17 out of 109 chains (15.6%). The interface residues predicted by RNABindR lie within the boundaries of the PROSITE motifs for 16 out of these 17 chains, demonstrating that RNABindR does identify known RNA binding motifs. Furthermore, the fact that RNABindR detected RNA binding sites in 88 proteins that do not contain any PROSITE motif whose annotation indicates a role in RNA binding suggests that RNABindR could be used to identify novel RNA binding motifs.
The tRNA pseudouridine synthase protein shown in Figure 4D contains the PROSITE PUA domain (PS50890), which is predicted to be an RNA binding domain. The PROSITE PUA domain contains 77 amino acids, only six of which contact RNA. RNABindR predicted a cluster of three interface residues in this region, shifted relative to the cluster of three actual interface residues in the complex structure, but precisely overlapping one actual interface residue. This example illustrates that RNABindR is able to identify specific interface residues, while a search for sequence motifs, such as a PROSITE search, may only identify larger RNA binding domains.
DISCUSSION
In this article, we have presented RNABindR, a machine-learning-based tool for identifying RNA binding sites in proteins. To generate a widely applicable tool, we developed RNABindR to use only protein sequence information as input. This achievement is significant because the results presented here were obtained using a relatively small training set of nonredundant RNA–protein complexes chosen from the PDB to allow rigorous evaluation of classification performance. On this data set of 109 diverse proteins (sequence identity below 30%), RNABindR performs well enough to be useful, with 85% accuracy, 0.35 CC, 0.51 specificity+, and 0.38 sensitivity+. Higher specificity values (with lower sensitivity) can be obtained in practice, if required, because RNABindR uses a Naive Bayes classifier, which allows the user to trade off sensitivity against specificity by tuning the classification threshold.
To evaluate RNABindR's ability to identify potential RNA binding sites in proteins for which structural information is not available, we predicted RNA binding residues in the telomerase TERT protein. To date, there is no high-resolution structure of the hTERT–TR complex, primarily because it has not been possible to obtain sufficient quantities of soluble full-length hTERT for detailed biophysical studies (Jacobs et al. 2005). Thus, we compared RNABindR predictions with available experimental data regarding conserved motifs and RNA binding domains in hTERT. The fact that RNABindR correctly predicted clusters of interface residues within known RNA binding domains of hTERT and, in several cases, precisely identified interface residues defined by mutagenesis experiments for hTERT suggests that RNABindR could be valuable in designing experiments to identify RNA binding sites in other experimentally recalcitrant RNA–protein complexes (for an example of this, see Terribilini et al. 2006). Although there is still no experimental structure for any TERT–RNA complex, the recent determination of the structure of a domain of Tetrahymena TERT prompted us to evaluate RNABindR predictions on Tetrahymena TERT. A cluster of predicted interface residues from 185–191 in Tetrahymena TERT is confirmed by the available experimental evidence for RNA binding in this region of the protein. Several residues in both hTERT and Tetrahymena TERT predicted by RNABindR are located outside the boundaries of the essential RNA binding and RT catalytic domains so far defined by experiments. It will be interesting to determine whether these predicted RNA binding residues may, in fact, contact RNA to stabilize the complex or to assist in other functions, such as subnuclear localization of TERT (Blackburn 2005).
We found that the performance of RNABindR, using a Naive Bayes classifier, was comparable to that of the only previously published sequence-based tool for predicting RNA binding sites, a neural network classifier developed by Miyano's group (Jeong et al. 2004). An advantage of RNABindR over the neural network classifier is that the latter method requires the exploration of several alternative neural network architectures (number of layers between the input and output layers, the number of neurons in such intermediate layers, and the connectivity between layers) before settling on an optimal network structure. In contrast, a Naive Bayes classifier does not require such hand-tuning. A Naive Bayes classifier requires significantly less computational effort (a single pass through the training data) to train than a neural network classifier (which requires multiple passes through the training data), making it especially well suited for use with large data sets or in settings that call for incremental update of the classifier as new training data become available.
Several classes of RNA binding domains and motifs that mediate the recognition of RNA by proteins have been very well characterized (Draper 1999). Two abundant and structurally defined RNA binding motifs are the RDB or RNA-recognition motif (RRM), which is the most common single-stranded RNA binding motif, and the double-stranded RNA binding motif (dsRBM) (Hall 2002), which recently has been shown to play important roles in regulatory interactions mediated by siRNAs and miRNAs (Tian et al. 2004). Shorter sequence motifs, including the arginine-rich-motif (ARM) motif and Arg-Gly-Gly (RGG) box are also found in a large number of proteins (Mulder et al. 2003). Within the nonredundant data set of 109 validated RNA binding proteins, only 17 PROSITE RNA binding motifs were identified. RNABindR predicted RNA binding residues in 104 of the 109 proteins and predicted interface residues within 16 of the 17 PROSITE RNA binding motifs. Additionally, most of the sequences “hit” by the 17 PROSITE motifs consist of relatively long stretches of amino acids that contain very few actual interface residues. Because the PROSITE motifs were not generated for the purpose of identifying interface residues, this comparison is not intended to prove “better performance” of RNABindR but instead to indicate that RNABindR may also be valuable for identifying novel RNA binding motifs.
A major challenge in post-genomics research is the functional annotation of novel proteins of known sequence (and, increasingly, known structure) but unknown function. For example, ORFans, orphan open reading frames that share no significant sequence similarity with any ORFs outside the genome in which they reside, represent 20%–30% of genes in sequenced genomes, but their origins and functions are largely mysterious (Fischer and Eisenberg 1999; Siew and Fischer 2004). Recently, several groups have demonstrated success in automatic prediction of protein functional interactions and intermolecular interfaces based on primary sequence information (Rost et al. 2003; Pang et al. 2004). However, when additional types of information are available (e.g., structural motifs, physical interactions, expression profiles, cellular localization, phylogenetic relationships), they can be incorporated to improve the accuracy of functional annotation. For example, for DNA binding proteins, the use of structure-derived features such as small binding motifs, solvent accessibility, and positive electrostatic potential have been shown to improve detection of HTH, HhH, and HLH DNA binding motifs (Shanahan et al. 2004). The prediction of protein–protein interface residues is also significantly improved by incorporating diverse types of information (Bradford and Westhead 2004; Neuvirth et al. 2004; Nissink and Taylor 2004; Sen et al. 2004; de Vries et al. 2006; Hoskins et al. 2006).
In experiments not reported here, we did not obtain significant improvement in classifier performance by incorporating sequence conservation information derived from multiple sequence alignments or residue solvent accessibility information derived from known structures of proteins in the training data set (see Materials and Methods; data not shown). This was unexpected because including sequence entropy or relative solvent accessibility of the target residue along with the input of amino acid identities does, in fact, enhance performance when a similar Naive Bayes classifier is used to predict interface residues in DNA–protein binding sites (Yan et al. 2006). Current experiments are directed at investigating the basis for this difference between DNA and RNA binding site classifiers. We are also exploring different encodings that may result in classifiers that more effectively exploit additional types of information.
Even without using information derived from structure, it should be possible to enhance prediction of RNA–protein interface residues. Recent results from Jeong and Miyano (2006) have shown that using position-specific scoring matrices (PSSMs) derived from PSI-BLAST searches can improve prediction performance of neural network classifiers. Recent preliminary experiments using PSSMs as inputs for RNABindR resulted in improved prediction performance comparable with that of Jeong and Miyano (data not shown). Other methods to improve prediction of interface residues may include, for example, adding “filters” that eliminate false positives based on the estimated probability that a particular interface residue should be located near other interface residues within the primary sequence, as has been done to improve performance of classifiers for identifying protein–protein interface residues (Ofran and Rost 2003; Yan et al. 2004b). Alternatively, training on larger data sets of structurally or functionally related RNA binding proteins, generated by relaxing the redundancy criterion, may generate higher accuracy predictions for specific subclasses of RNA binding proteins.
The RNABindR results reported here, together with results of previous studies published by Jeong and Miyano (Jeong et al. 2004; Jeong and Miyano 2006), demonstrate that computational approaches can successfully identify RNA–protein interface residues using only amino acid sequence as input. For many proteins—notably, the ORFans, mentioned above—the deduced amino acid sequence is often the only information available. The approach we propose here requires only the primary sequence of the protein partner, implying that many structural determinants of RNA binding sites can be captured by local sequence characteristics. The simplicity of RNABindR, together with the fact that a relatively high level of accuracy can be achieved using only protein sequence information (and no information about the identity, sequence, or structure of the RNA partner), suggests that it may prove valuable for functional annotation of putative RNA binding proteins and for genomewide identification of RNA binding residues in protein. RNABindR is available at http://bindr.gdcb.iastate.edu.
MATERIALS AND METHODS
Data set
A data set of RNA–protein interactions was extracted from structures of known RNA–protein complexes solved by X-ray crystallography in the PDB (Berman et al. 2000). Proteins with >30% sequence identity or structures with resolution worse than 3.5Å were removed using PISCES (Wang and Dunbrack 2003). This resulted in a set of 109 nonredundant protein chains containing a total of 25,118 amino acids. Amino acids in the RNA–protein interface were identified using ENTANGLE (Allers and Shamoo 2001). Using default parameters, 3518 (14%) of the amino acids in the data set are defined as interface residues (positive examples). Table 2 lists the PDB identifiers for all 109 proteins in the nonredundant data set, which includes four major classes of RNA–protein complexes. A smaller data set extracted from only ribosomal proteins (55 chains) was used in some experiments. The ribosomal protein data set comprises a total of 7522 amino acids, 2363 (31%) of which are defined as interface residues. These data sets and others are available at http://bindr.gdcb.iastate.edu.
Naive Bayes classifier using only amino acid sequence information as input
The results reported in this article were obtained using RNABindR implementing a Naive Bayes classifier (Mitchell 1997), which was chosen based on exploration of several different machine learning algorithms, including support vector machines, decision trees, and Bayesian networks. The performance of the Naive Bayes classifier was comparable to or better than that of all other methods tested (data not shown). The Naive Bayes classifier assumes the independence of attributes. This assumption greatly reduces the complexity of the classifier and improves the reliability of the estimated parameters when the dimensionality of the input is high relative to the size of the available training set. Despite its simplicity and the fact that the independence assumption may not apply in certain cases, the Naive Bayes classifier often performs at least as well as more sophisticated methods for many problems (Buntine 1991). We used the Naive Bayes classifier from the Weka package (Witten and Frank 2005). In RNABindR, the input to a Naive Bayes classifier is a window x = (x −n,x −n+1,…x T−1,x T,x T+1 …x n−1,x n) of 2n + 1 contiguous amino acid identities, with n amino acid sequence residues on either side of the target residue xT. The output is an instance c ε {+,−} , where + indicates that the target residue x T at the center of the window is an interface residue and − indicates x T is a noninterface residue. A training example is an ordered pair (x, c) where x = (x −n,x −n+1,…x T−1,x T,x T+1 …x n−1,x n) and c is the corresponding class label (interface or noninterface). A training data set D is simply a collection of labeled training examples. In our experiments, several values of n from 2 to 14 (corresponding to windows of width 5–29) were tried.
Let X = (X −n,…X T,…X n) denote the random variable corresponding to the input to the classifier and C denote the binary random variable corresponding to the output of the classifier. The Naive Bayes classifier assigns input x the class label + (interface) if:
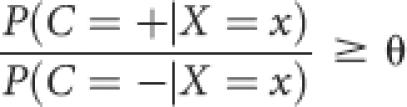 |
and the class label − (noninterface) otherwise. The choice of θ = 1 corresponds to assigning the most probable class label. The desired trade-off of sensitivity against specificity can be achieved by varying θ.
Because the inputs are assumed to be independent given the class, we have:
 |
The relevant probabilities are estimated from the training set using the Laplace estimator (Mitchell 1997). The resulting Naive Bayes classifier classifies a target amino acid residue x T as an interface residue or as a noninterface residue based on the identities of the n amino acid residues on either side.
Naive Bayes classifiers using sequence plus additional information
We experimented with adding relative accessible surface area (rASA), sequence entropy, hydrophobicity, secondary structure, or electrostatic potential to the sequence-based classifier described above. rASA for each residue in the absence of RNA was computed using the program Naccess (http://wolf.bms.umist.ac.uk/naccess/). Each training and test example for the Naive Bayes classifier with rASA added is as follows: x = (x −n,x −n+1,…x T−1,x T,x T+1 …x n−1,x n r T) , where x i is as defined above and r T is the rASA of the target residue. Inputs are encoded similarly for all features. Sequence entropy was encoded using the relative entropy for each residue from the HSSP database (http://www.cmbi.kun.nl/gv/hssp/). Hydrophobicity of each residue was obtained from the consensus normalized hydrophobicity scale derived by Eisenberg et al. (1984). The secondary structure of each residue was extracted from the PDB (http://www.rcsb.org/pdb/). Electrostatic potentials were calculated using the program APBS (http://agave.wustl.edu/apbs/). The electrostatic potential for each residue is the average over all its atoms.
Performance evaluation
The performance of RNABindR was evaluated using leave-one-out cross-validation experiments. That is, in each of the 109 experiments, the Naive Bayes classifier was trained using data from 108 chains and evaluated on the 109th chain. The threshold θ was chosen to maximize the correlation coefficient on the training set. The performance measures reported represent averages over the 109 experiments. The performance of a classifier designed to classify protein residues into interface and noninterface residues is completely summarized by TP (true positives), i.e., the number of interface residues correctly identified as such by the classifier; FP (false positives), i.e., the number of noninterface residues misclassified as interface residues by the classifier; FN (false negatives), i.e., the number of interface residues that are misclassified as noninterface residues by the classifier; and TN (true negatives), i.e., the number of noninterface residues that are correctly identified as such by the classifier. Note that N, the total number of instances used for evaluation of the classifier is given by N = TP + FP + FN + TN.
Commonly used performance measures include accuracy, correlation coefficient (CC), specificity+, sensitivity+, specificity−, and sensitivity− (Baldi et al. 2000). Specificity+ is the fraction of positive predictions (residues predicted to be RNA binding residues) that are actually RNA binding residues. For example, if 100 interface residues are predicted to be RNA binding residues by RNABindR and 50 of them are actually interface residues, specificity+ is 0.5. Sensitivity+ is the fraction of RNA binding residues that are predicted to be RNA binding residues by RNABindR. For example, if a protein contains 20 actual interface residues and RNABindR predicts that 15 of these 20 are interface residues, sensitivity+ is 0.75.
 |
 |
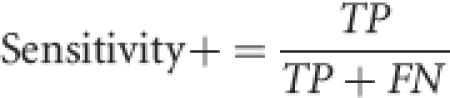 |
Specificity − and Sensitivity − are similarly defined.
Each of these performance measures summarizes the information contained in the four numbers (TP, FP, FN, TN) with a single number (e.g., accuracy), with inevitable loss of information. In the case of data sets in which there is a large difference between the number of instances belonging to the two classes, using accuracy alone to evaluate the classifier can be misleading (Baldi et al. 2000; Yan et al. 2004a,b). The RNA binding site data set contains 14% interface and 86% noninterface examples. A classifier that simply predicts each residue to be noninterface would have an accuracy of 0.86, but such a classifier would be completely useless in correctly identifying the interface residues. Thus, it is desirable to consider multiple performance measures collectively to evaluate the performance of a classifier and compare its performance with other classifiers (Baldi et al. 2000; Yan et al. 2004b).
As noted earlier, it is possible (and in many settings desirable), to trade off the sensitivity of the classifier against its false positive rate. The Receiver Operating Characteristic curve (ROC curve), a plot of the sensitivity + or the “hit rate” versus the false positive rate (1-specificity −) characterizes such trade-off for a classifier. We used the Weka package (Witten and Frank 2005) to obtain the ROC plot for RNABindR.
ACKNOWLEDGMENTS
This research was supported in part by grants from the National Institutes of Health (GM066387) to V.H., D.D., and R.J., research assistantships from USDA IFAFS Multidisciplinary Graduate Education Training Grant (2001-52100-11506) for M.T., and from Iowa State University Biotechnology Council and Center for Integrated Animal Genomics for J.L. We thank Satoru Miyano, Euna Jeong, and I-Fang Chung for kindly providing their data set and members of our research groups for ideas and discussions.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2197306.
REFERENCES
- Allers J., Shamoo Y. Structure-based analysis of protein–RNA interactions using the program ENTANGLE. J. Mol. Biol. 2001;311:75–86. doi: 10.1006/jmbi.2001.4857. [DOI] [PubMed] [Google Scholar]
- Autexier C., Lue N.F. The structure and function of telomerase reverse transcriptase. Annu. Rev. Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- Bachand F., Autexier C. Functional regions of human telomerase reverse transcriptase and human telomerase RNA required for telomerase activity and RNA–protein interactions. Mol. Cell. Biol. 2001;21:1888–1897. doi: 10.1128/MCB.21.5.1888-1897.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi P., Brunak S., Chauvin Y., Andersen C.A., Nielsen H. Assessing the accuracy of prediction algorithms for classification: An overview. Bioinformatics. 2000;16:412–424. doi: 10.1093/bioinformatics/16.5.412. [DOI] [PubMed] [Google Scholar]
- Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn E.H. Telomeres and telomerase: Their mechanisms of action and the effects of altering their functions. FEBS Lett. 2005;579:859–862. doi: 10.1016/j.febslet.2004.11.036. [DOI] [PubMed] [Google Scholar]
- Bradford J.R., Westhead D.R. Improved prediction of protein–protein binding sites using a support vector machines approach. Bioinformatics. 2004;21:1487–1494. doi: 10.1093/bioinformatics/bti242. [DOI] [PubMed] [Google Scholar]
- Bryan T.M., Goodrich K.J., Cech T.R. Telomerase RNA bound by protein motifs specific to telomerase reverse transcriptase. Mol. Cell. 2000;6:493–499. doi: 10.1016/s1097-2765(00)00048-4. [DOI] [PubMed] [Google Scholar]
- Buntine W. Theory refinement on Bayesian networks. Morgan-Kaufmann; San Mateo, CA: 1991. [Google Scholar]
- Cai Y.D., Lin S.L. Support vector machines for predicting rRNA-, RNA-, and DNA-binding proteins from amino acid sequence. Biochim. Biophys. Acta. 2003;1648:127–133. doi: 10.1016/s1570-9639(03)00112-2. [DOI] [PubMed] [Google Scholar]
- Chen Y., Varani G. Protein families and RNA recognition. FEBS J. 2005;272:2088–2097. doi: 10.1111/j.1742-4658.2005.04650.x. [DOI] [PubMed] [Google Scholar]
- Cusack S. RNA–protein complexes. Curr. Opin. Struct . Biol. 1999;9:66–73. doi: 10.1016/s0959-440x(99)80009-8. [DOI] [PubMed] [Google Scholar]
- de Vries S.J., van Dijk A.D.J., Bonvin A.M.J.J. WHISCY: What information does surface conservation yield? Application to data-driven docking. Proteins. 2006;63:479–489. doi: 10.1002/prot.20842. [DOI] [PubMed] [Google Scholar]
- Draper D.E. Themes in RNA–protein recognition. J. Mol. Biol. 1999;293:255–270. doi: 10.1006/jmbi.1999.2991. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Weiss R.M., Terwilliger T.C. The hydrophobic moment detects periodicity in protein hydrophobicity. Proc. Natl. Acad. Sci. 1984;81:140–144. doi: 10.1073/pnas.81.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer D., Eisenberg D. Finding families for genomic ORFans. Bioinformatics. 1999;15:759–762. doi: 10.1093/bioinformatics/15.9.759. [DOI] [PubMed] [Google Scholar]
- Gomis-Ruth F.X., Dessen A., Timmins J., Bracher A., Kolesnikowa L., Becker S., Klenk H.D., Weissenhorn W. The matrix protein VP40 from Ebola virus octamerizes into pore-like structures with specific RNA binding properties. Structure. 2003;11:423–433. doi: 10.1016/S0969-2126(03)00050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall K.B. RNA–protein interactions. Curr. Opin. Struct. Biol. 2002;12:283–288. doi: 10.1016/s0959-440x(02)00323-8. [DOI] [PubMed] [Google Scholar]
- Han L.Y., Cai C.Z., Lo S.L., Chung M.C., Chen Y.Z. Prediction of RNA-binding proteins from primary sequence by a support vector machine approach. RNA. 2004;10:355–368. doi: 10.1261/rna.5890304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman M.M., Khrapov M.A., Cox J.C., Yao J., Tong L., Ellington A.D. AANT: The amino acid–nucleotide interaction database. Nucleic Acids Res. 2004;32:D174–D181. doi: 10.1093/nar/gkh128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins J., Lovell S., Blundell T.L. An algorithm for predicting protein–protein interaction sites: Abnormally exposed amino acid residues and secondary structure elements. Protein Sci. 2006;15:1017–1029. doi: 10.1110/ps.051589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulo N., Sigrist C.J., Le Saux V., Langendijk-Genevaux P.S., Bordoli L., Gattiker A., De Castro E., Bucher P., Bairoch A. Recent improvements to the PROSITE database. Nucleic Acids Res. 2004;32:D134–D137. doi: 10.1093/nar/gkh044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs S.A., Podell E.R., Wuttke D.S., Cech T.R. Soluble domains of telomerase reverse transcriptase identified by high-throughput screening. Protein Sci. 2005;14:2051–2058. doi: 10.1110/ps.051532105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs S.A., Podell E.R., Cech T.R. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat. Struct. Mol. Biol. 2006;13:218–225. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- Jeong E., Miyano S. A weighted profile method for protein–RNA interacting residue prediction. Trans. Comput. Syst. Biol. 2006;IV:123–139. [Google Scholar]
- Jeong E., Chung I., Miyano S. A neural network method for identification of RNA-interacting residues in protein. Genome Inform. Ser. Workshop Genome Inform. 2004;15:105–116. [PubMed] [Google Scholar]
- Jones S., Daley D.T., Luscombe N.M., Berman H.M., Thornton J.M. Protein–RNA interactions: A structural analysis. Nucleic Acids Res. 2001;29:943–954. doi: 10.1093/nar/29.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Jeong E., Lee S.W., Han K. Computational analysis of hydrogen bonds in protein–RNA complexes for interaction patterns. FEBS Lett. 2003;552:231–239. doi: 10.1016/s0014-5793(03)00930-x. [DOI] [PubMed] [Google Scholar]
- Klein D.J., Schmeing T.M., Moore P.B., Steitz T.A. The kink-turn: A new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lai C.K., Mitchell J.R., Collins K. RNA binding domain of telomerase reverse transcriptase. Mol. Cell. Biol. 2001;21:990–1000. doi: 10.1128/MCB.21.4.990-1000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.K., Miller M.C., Collins K. Template boundary definition in Tetrahymena telomerase. Genes & Dev. 2002;16:415–420. doi: 10.1101/gad.962602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig B., Arora S., Jernigan R.L. RNA base–amino acid interaction strengths derived from structures and sequences. Nucleic Acids Res. 1997;25:2562–2565. doi: 10.1093/nar/25.13.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell T. Machine learning. McGraw-Hill; Boston, MA: 1997. [Google Scholar]
- Moriarty T.J., Huard S., Dupuis S., Autexier C. Functional multimerization of human telomerase requires an RNA interaction domain in the N terminus of the catalytic subunit. Mol. Cell. Biol. 2002;22:1253–1265. doi: 10.1128/MCB.22.4.1253-1265.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty T.J., Marie-Egyptienne D.T., Autexier C. Functional organization of repeat addition processivity and DNA synthesis determinants in the human telomerase multimer. Mol. Cell. Biol. 2004;24:3720–3733. doi: 10.1128/MCB.24.9.3720-3733.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty T.J., Ward R.J., Taboski M.A., Autexier C. An anchor site-type defect in human telomerase that disrupts telomere length maintenance and cellular immortalization. Mol. Biol. Cell. 2005;16:3152–3161. doi: 10.1091/mbc.E05-02-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder N.J., Apweiler R., Attwood T.K., Bairoch A., Barrell D., Bateman A., Binns D., Biswas M., Bradley P., Bork P., et al. The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res. 2003;31:315–318. doi: 10.1093/nar/gkg046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuvirth H., Raz R., Schreiber G. ProMate: A structure based prediction program to identify the location of protein–protein binding sites. J. Mol. Biol. 2004;338:181–199. doi: 10.1016/j.jmb.2004.02.040. [DOI] [PubMed] [Google Scholar]
- Nissink J.W., Taylor R. Combined use of physicochemical data and small-molecule crystallographic contact propensities to predict interactions in protein binding sites. Org. Biomol. Chem. 2004;2:3238–3249. doi: 10.1039/B405205F. [DOI] [PubMed] [Google Scholar]
- O'Connor C.M., Lai C.K., Collins K. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J. Biol. Chem. 2005;280:17533–17539. doi: 10.1074/jbc.M501211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofran Y., Rost B. Predicted protein–protein interaction sites from local sequence information. FEBS Lett. 2003;544:236–239. doi: 10.1016/s0014-5793(03)00456-3. [DOI] [PubMed] [Google Scholar]
- Pan H., Agarwalla S., Moustakas D.T., Finer-Moore J., Stroud R.M. Structure of tRNA pseudouridine synthase TruB and its RNA complex: RNA recognition through a combination of rigid docking and induced fit. Proc. Natl. Acad. Sci. 2003;100:12648–12653. doi: 10.1073/pnas.2135585100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang P.S., Jankowsky E., Wadley L.M., Pyle A.M. Prediction of functional tertiary interactions and intermolecular interfaces from primary sequence data. J. Exp. Zoolog. B Mol. Dev. Evol. 2004;304:50–63. doi: 10.1002/jez.b.21024. [DOI] [PubMed] [Google Scholar]
- Rost B., Liu J., Nair R., Wrzeszczynski K.O., Ofran Y. Automatic prediction of protein function. Cell. Mol. Life Sci. 2003;60:2637–2650. doi: 10.1007/s00018-003-3114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryter J.M., Schultz S.C. Molecular basis of double-stranded RNA–protein interactions: Structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen T.Z., Kloczkowski A., Jernigan R.L., Yan C., Honavar V., Ho K.M., Wang C.Z., Ihm Y., Cao H., Gu X., et al. Predicting binding sites of hydrolase-inhibitor complexes by combining several methods. BMC Bioinformatics. 2004;5 doi: 10.1186/1471-2105-5-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanahan H.P., Garcia M.A., Jones S., Thornton J.M. Identifying DNA-binding proteins using structural motifs and the electrostatic potential. Nucleic Acids Res. 2004;32:4732–4741. doi: 10.1093/nar/gkh803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siew N., Fischer D. Structural biology sheds light on the puzzle of genomic ORFans. J. Mol. Biol. 2004;342:369–373. doi: 10.1016/j.jmb.2004.06.073. [DOI] [PubMed] [Google Scholar]
- Terribilini M., Lee J.H., Yan C., Jernigan R.L., Carpenter S., Honavar V., Dobbs D. Identifying interaction sites in “recalcitrant” proteins: Predicted protein and RNA binding sites in Rev proteins of HIV-1 and EIAV agree with experimental data. Pac. Symp. Biocomput. 2006;2006:415–426. [PMC free article] [PubMed] [Google Scholar]
- Tian B., Bevilacqua P.C., Diegelman-Parente A., Mathews M.B. The double-stranded-RNA-binding motif: Interference and much more. Nat. Rev. Mol. Cell Biol. 2004;5:1013–1023. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- Wang G., Dunbrack R.L., Jr. PISCES: A protein sequence culling server. Bioinformatics. 2003;19:1589–1591. doi: 10.1093/bioinformatics/btg224. [DOI] [PubMed] [Google Scholar]
- Weiss M.A., Narayana N. RNA recognition by arginine-rich peptide motifs. Biopolymers. 1998;48:167–180. doi: 10.1002/(SICI)1097-0282(1998)48:2<167::AID-BIP6>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Witten I.H., Frank E. Data mining: Practical machine learning tools and techniques. Morgan Kaufmann; San Francisco: 2005. [Google Scholar]
- Yan C., Dobbs D., Honavar V. Identification of interface residues in protease-inhibitor and antigen-antibody complexes: A support vector machine approach. Neural Comput. Appl. 2004a;13:123–129. doi: 10.1007/s00521-004-0414-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., Dobbs D., Honavar V. A two-stage classifier for identification of protein–protein interface residues. Bioinformatics. 2004b;20:I371–I378. doi: 10.1093/bioinformatics/bth920. (Suppl 1) [DOI] [PubMed] [Google Scholar]
- Yan C., Terribilini M., Wu F., Jernigan R.L., Dobbs D., Honavar V. Predicting DNA-binding sites in proteins from amino acid sequence. BMC Bioinformatics. 2006;7 doi: 10.1186/1471-2105-7-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Cao J., Cai Y., Shi T., Li Y. Predicting rRNA-, RNA-, and DNA-binding proteins from primary structure with support vector machines. J. Theor. Biol. 2006;240:175–184. doi: 10.1016/j.jtbi.2005.09.018. [DOI] [PubMed] [Google Scholar]