Abstract
The number of aberrant splicing processes causing human disease is growing exponentially and many recent studies have uncovered some aspects of the unexpectedly complex network of interactions involved in these dysfunctions. As a consequence, our knowledge of the various cis- and trans-acting factors playing a role on both normal and aberrant splicing pathways has been enhanced greatly. However, the resulting information explosion has also uncovered the fact that many splicing systems are not easy to model. In fact we are still unable, with certainty, to predict the outcome of a given genomic variation. Nonetheless, in the midst of all this complexity some hard won lessons have been learned and in this survey we will focus on the importance of the wide sequence context when trying to understand why apparently similar mutations can give rise to different effects. The examples discussed in this summary will highlight the fine ‘balance of power’ that is often present between all the various regulatory elements that define exon boundaries. In the final part, we shall then discuss possible therapeutic targets and strategies to rescue genetic defects of complex splicing systems.
INTRODUCTION
The study of the connections between defective splicing and occurrence of disease has become a central issue in the medical research field (1–7). This is a consequence of the growing list of pathological alterations that can be directly linked with aberrant splicing processes. In fact, for many genes, genomic variations which affect the splicing process may represent up to 50% of all mutations that lead to gene dysfunction (8,9). This awareness has also led to the critical reappraisal of many past observations, and it is now clear that substitutions which had for a long time been regarded as harmless synonymous changes in protein coding regions may have some very severe consequences on splicing processes, and thus on the appearance of disease (10).
The underlying biological reason for such an exuberant growth in the number of diseases derived from splicing derangements is due not only on the intrinsic properties of the exon itself (presence of regulatory elements within the exon, strength of the splice sites etc.) but also on the wider environment within which the exon is found, such as the presence of regulatory elements dispersed throughout the gene at much longer distances (11–14), the splicing events that have taken place before the exon of interest (15,16) or the type of promoter architecture and the consequent polymerase II processivity (17). This means that splicing spoilers can occur virtually everywhere within any given gene and many of the novel types of mutations identified have still poorly characterized functional mechanisms (18). Furthermore, alternative splicing processes (19) are responsible for increasing the functional diversity of the human genome (20–24). Therefore, it is very likely that variations anywhere in the genome may alter the delicate modulation of the splicing process and produce pathological alterations (3).
Numerous methodological developments have also aided researchers recently in the task of building connections between splicing and disease. For example, the refinement of minigene-based technologies for alternative splicing analysis initially described 20 years ago (25) has allowed a relatively fast approach to identify splicing spoilers and to study their underlying functional mechanisms (26,27). In addition, a great boost for the preliminary identification of general splicing regulatory sequences (SREs) has been the development of SELEX methodologies to characterize binding specificities of trans-acting factors involved in splicing regulation (28,29).
Last but not least, scientific interest in this subject has also been steadily growing as potential applications in the molecular therapy field have begun to be appreciated and have started to be tested, such as the recent advances in the use of bi-functional or inhibitory oligonucleotides in rescuing aberrant splicing events (3,30–35), in the engineering of specific compounds by chemical synthesis to target splicing factor protein families (36–38), or in the development of novel vectors based on U7snRNA to rescue protein functionality by exon skipping (39).
The fact that even single nucleotide changes in a pre-mRNA sequence away from the canonical AG/GU nucleotides at the splice junctions are ever more frequently been described as capable of affecting the splicing process (40) was rather unexpected. Indeed, computational analyses confirm that when most of the main determinants of exon definition (3′ss, 5′ ss, branch site and pY tract) are close to the consensus minimal variations may be possible without impairing recognition by the spliceosome (41). This conclusion is also consistent with the rather loose sequence-specificities of many essential splicing factors. For example, in vitro studies have shown that a minimal amount of all the potential base pairings between the 5′ splice site and U1snRNA is sufficient for its correct recognition by U1snRNP (42). Moreover, even in the absence of the 5′ tail of U1snRNA, the U1snRNP particle is capable of selecting a 5′ss-like sequence probably through the U1-C subunit (43). Finally, the observation that over-expression of SR proteins could compensate for absence of U1snRNP in vitro (44,45) provides support to the view that the splicing process can be precise even in the absence of key participants.
Reality, however, has painted a much more complex picture, with the micro-environment having an important role in the ability of a splice site to tolerate a nucleotide transition (46). Furthermore, microarray technology and comparative expressed sequence tags analyses are now vividly depicting a world of widespread differences in alternative splicing levels, both within normal tissues or following pathological alterations (47–49). For this reason, particular effort has been devoted recently to characterize splicing abnormalities in cell transformation and cancer (50,51) as they may provide an additional therapeutic target for this kind of diseases.
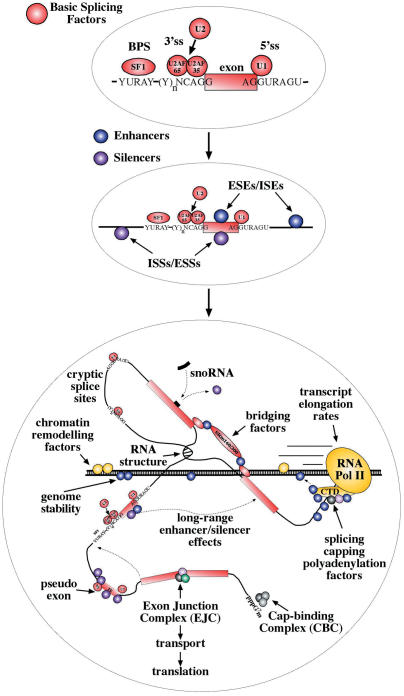
The fact that splicing regulation appears to be a linked network (52–56) means that researchers and clinicians have to consider as a potential splicing mutation virtually every substitution that is encountered within a pre-mRNA sequence (18). The recent evidences that many splicing factors may also be actively involved in other important cellular processes, such as genome stability (57) or mRNA transport and translation (58–61), have just begun to unravel all the connections of the RNA processing system (62,63). In fact, rather than becoming simplified, splicing regulation in the new millenium could be best characterized as right on the edge of chaos. Figure 1 shows a simplified view of how the number of accepted players and functions have grown in recent years. It is for this reason that the connotation of ‘difficult to characterize’ is increasingly associated with many new splicing mutations.
Figure 1.
The three schematic representations show the increase during the last decade of the number of interactions reported to occur during RNA processing. The upper diagram depicts a classic pre-mRNA molecule in which the exon (closed box) is defined by a small number of relatively conserved sequences that bind a well-defined set of basic splicing factors. The complexity of the system begins to increase, as shown in the middle diagram, when the vast and still growing array of enhancer and silencer factors are to be added to the basic picture. These factors can bind particular enhancer and silencer sequences either within the exon itself (ESE and ESS) or in the nearby flanking regions (ISE and ISS) and help increase/decrease exon recognition. As shown in the bottom diagram, in these recent years the complexity of the regulatory elements that participate in splicing control has taken an enormous leap forward by the discovery of the potential connections between splicing, the specific structural features of pre-RNAs and many of the processes that participate in a their life-cycle such as genome stability, RNA processing speed, transport and translation.
The following observations exemplify the problems that the molecular machinery has to resolve to obtain proper splicing:
Potential exon boundaries are particularly abundant in pre-mRNA molecules. For example, in the human hprt gene there are ∼10 times more consensus splice site than those effectively recognized by the splicing machinery (64),
The splicing process occurs in a background of repression, as random human genomic sequences display a higher than average splicing inhibitory activity when tested in minigene systems (65–67),
Positive or negative SREs are abundant in all pre-mRNAs, both within introns and exons regardless if they are constitutively or alternatively spliced (67–69).
All the processes that deal with an RNA life cycle including transcription, splicing, transport, translation and degradation are no longer considered as separated from each other but rather, in line with the literal translation of the Latin word ‘complexus’, appear to be functionally entwined/twisted together (54,70,71). Indeed, just how close relationships within the spliceosome can grow is clearly highlighted by the recent demonstration of extensive molecular tethering between the pre-mRNA being processed and the splicing machinery, allowing processing to occur correctly even when the nucleotide chain of the pre-mRNA molecule is interrupted (72).
As a consequence, the sequence and genomic ‘context’ in which mutations are observed to occur are essential to explain the sometimes widely different outcomes of similar substitutions in different genes (18,55). Several excellent articles have reviewed the individual connections between aberrant splicing and disease in recent years (1–7). Our purpose will be that of using selected examples to highlight the importance of genomic and local context when investigating splicing outcomes. For example, the inactivation of donor site (5′ss) sequences produce a great variety of splicing outcomes that vividly depicts the mind boggling variety of exon/intron definition mechanisms.
WHAT MAY HAPPEN WHEN A DONOR SITE IS INACTIVATED?
The recognition of the donor site is one of the key events of the splicing process (73), and many of its peculiarities are still under investigation (42,43,74–76). Mutations that destroy natural donor sequences are usually observed to cause the skipping of their associated exon (77). However, in a considerable number of cases additional events can also take place, which include cryptic splice site activation and full intron inclusion. Now, whether any of these events takes place or not is often dependent on the individual context in which the substitution occurs. Sometimes evaluating this context may be fairly straightforward; however, in some cases it may also involve long-range genomic context interactions or be dependent on some kind of ‘priming’ effect to the splicing machinery by events happening upstream of the exon in question (see below).
Cryptic splice site activation
Activation of cryptic splice sites is probably the most prominent outcome (after exon skipping) that follows the inactivation of the original donor site. In fact, cryptic splice site activation in the β-globin gene was one of the first medically relevant splicing defects described (78,79). The usage of alternative splice sites following the inactivation of the proper site often leads to a frameshift in the protein coding sequence. It should be noted, however, that cryptic splice site sequences do not simply represent a harmful background to normal splicing pathways. Rather, it is now increasingly clear that they have played a very important role in the still very much debated evolutionary mechanisms of intron gain/loss (80) and recent work has suggested that in some cases they act as preferential sites for intron insertion (81,82).
Following the disruption of the normal donor site it would be convenient to predict beforehand which sites (if any) become activated by simply looking at all the nearby sequences that match the consensus. In practice, however, there is not always a straight correlation between the chosen cryptic site and the calculated strength of the potential sites in the region (83). Contemporary studies on donor (83) and acceptor sites (84) have established that local context features may help the splicing machinery to actively discriminate against the usage of cryptic sites and in favor of natural 5′ and 3′ss. These local context features may play a role in cryptic splice site usage after disruption of the natural site.
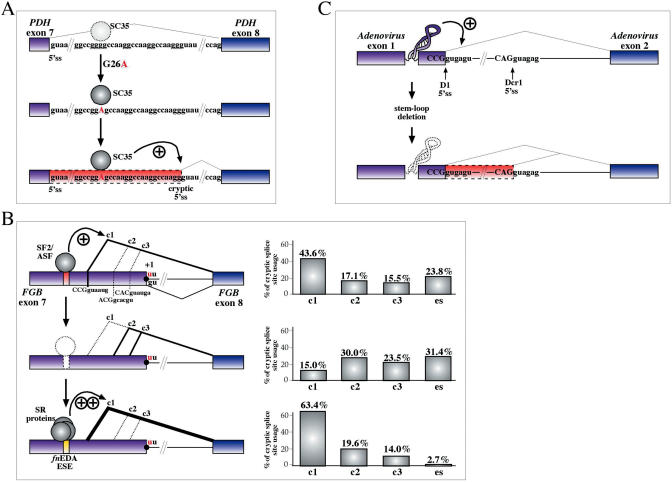
For example, SR protein levels have been long known to affect relative cryptic splice site usage in the human β-globin gene both in vitro and in vivo (85,86). Now, two recent reports have demonstrated a critical role of SR-binding sequences on cryptic splice site selection: in the E1α Pyruvate Dehydrogenase pre-mRNA a single point mutation away from the natural ss strengthens a weak, pre-existing SC35-binding site and has the effect of shifting donor site usage to a previously silent cryptic site (87) (Figure 2A). This mutation has an important biological impact and is linked with mental retardation. In the second case, a strong SF2/ASF-binding region in fibrinogen exon 7 which is not normally needed for constitutive inclusion of this exon in the mature mRNA has the ability to determine an increase in the relative usage levels of a particular cryptic donor site when the natural donor site is inactivated by a IVS7 + 1G > U mutation (88). Interestingly, changing the local context by swapping the native SF2/ASF-binding sequence with a different SR-binding ESE sequence from the fibronectin gene (fnESE) has the ability to change the relative cryptic site usage ratios with respect to the natural fibrinogen sequence (Figure 2B).
Figure 2.
(A) Schematic diagram showing how local context arrangements can influence cryptic splice site activation. For example, in intron 7 of the PDH gene a G26A mutation greatly enhances a preexisting binding site for the SC35 protein (upper and middle panels). The increased binding of this protein causes the shift of donor site usage towards a cryptic donor site in the downstream sequence (lower panel). (B) Another example is represented by a strong SF2/ASF-binding sequence that is not normally needed by the splicing machinery to recognize fibrinogen exon 7 (upper panel). However, if the natural donor site is inactivated by a +1G > U substitution the relative usage of the three cryptic sites which become activated (c1, c2 and c3) and the levels of exon skipping (es) are profoundly influenced by its presence, especially c1 (middle panel). In this case, the importance of local context has also been demonstrated by replacing the natural SF2/ASF-binding sequence with a well-known ESE sequence from the fibronectin gene (fnESE) and observing that the level of c1 usage are substantially increased with respect to the natural sequence (lower panel). (C) Finally, in the case of RNA secondary structure it has been shown that correct utilization of the natural donor site sequence (D1) can be achieved only in the presence of a highly conserved stem-loop structure within Adenovirus type 2 first exon (upper panel). Removal of this sequence has the consequence of activating usage of a cryptic donor site sequence (Dcr1) (lower panel).
Finally, an additional general factor that has been described to affect cryptic splice site usage is represented by RNA secondary structure (12–14,89). For example, in the early region 3 of adenovirus type 2 a stem–loop structure allows the correct and exclusive usage of its D1 natural donor site rather than a cryptic donor site (Dcr1) located 74 nt away (90,91) (Figure 2C). Disruption of this structure results in a shift towards the Dcr1 site.
Intron retention
An intron retention event is identified as the preservation of an entire intronic sequence within some forms of the final processed mRNA. Using this criteria, the frequency of normal intron retention events in the human genome has been estimated recently to be ∼15% in a set of >21 000 annotated genes (92). Most of these events naturally occur in the untranslated regions of the pre-mRNA where the rate of retention is observed to be >10-fold greater with respect to protein coding sequence (92). The biological role of most of these events is currently unknown; however, retained introns within the coding region are significantly shorter than non-retained introns (92,93). Furthermore, their sequences are under selective pressure for coding potential (92), suggesting a potentially regulatory role in protein functions. In recent years, the biological significance of some physiological intron retention events has also been clearly established in several cases such as the generation of the P element and Msl2 transcripts in Drosophila (94,95) or in the developmental regulation of the pro insulin messenger RNA in chicken embryos (96).
Most importantly, aberrant intron retention events have also been shown to be associated with several pathological splicing defects. For example, in the Ret tyrosine kinase gene (97) where it has been connected with the presence of pheocromocytomas, in the HERG gene where it has been identified as being responsible for the development of a long QT syndrome (LQT2 phenotype) in an individual which carried a +6T > C mutation in the donor site of intron 7 of this gene (98) and in a patient affected by Leigh syndrome where the retention of intron 3 in the SURF1 gene follows a single-nucleotide mutation in its donor site (99). As similar mutations in other contexts cause exon skipping it is highly probable that local context features either in the retained intron itself or in the flanking exonic sequences determine this particular outcome. At the moment, very little is known concerning this issue and it is to be hoped that future studies will target these particular issues.
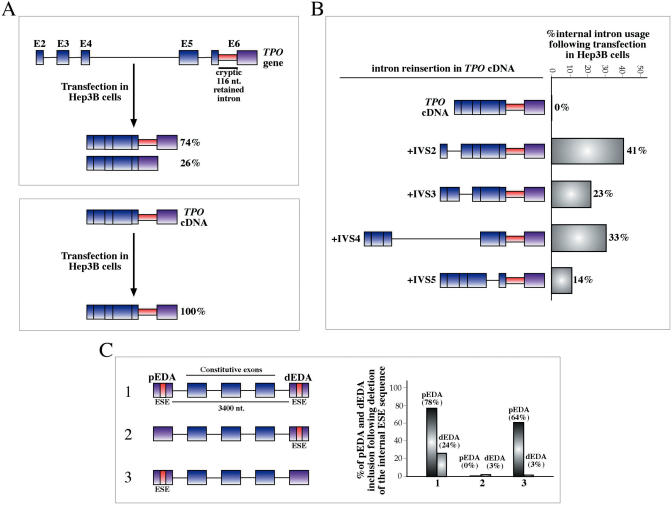
An interesting phenomenon that also deals with intron retention/recognition has been observed by analyzing the last exon of the TPO gene. In 26% of the TPO transcripts a 116 n sequence located in the last exon is removed and hence treated as an ‘alternative intron’ (Figure 3A, upper panel) (15). Strikingly, this short intron behaves as such only if there has been an upstream splicing event. In fact, if the cDNA of the TPO gene is transfected in Hep3B cells removal of the 116 nt intron is completely abolished (Figure 3A, lower panel). The recognition of the small intron 5′ and 3′ss within TPO's last exon seems to require spliceosome assembly on a previous intron, as the effect is obtained not only with the intact intron structure of the TPO gene but also by inserting any single intron in the cDNA sequence (Figure 3B). Although the mechanism responsible for these coordinated intron recognition events is currently unknown, it is clear that some kind of ‘priming’ event from the splicing machinery may be required for the processing of this intron. Intriguingly, a somewhat analogous mechanism, this time on exon recognition, has been proposed recently to exist in the coordination of splicing between two independent alternatively spliced regions within a fibronectin minigene (16) (Figure 3C). In this case, mutations that inhibit inclusion of the proximal exon (pEDA) correlate directly with inclusion levels of the distal exon (dEDA). Furthermore, existence of such a phenomenon has also been described in vivo in a mouse knock-in model (100) between two different alternatively spliced regions of the fibronectin gene (the EDA exon and the V region) (16).
Figure 3.
(A) This figure shows the effect on internal intron retention in exon 6 of the TPO gene. When the TPO gene is transfected in Hep3B cells only two processed mRNAs are produced, a major form lacking all the intervening introns (74%) and a minor form in which there is the removal of an additional 116 nt long sequence (26%) from the processed mRNA (upper panel, boxed in red). On the other hand, lower panel shows that when the TPO cDNA is transfected in Hep3B cells no processing of this internal intron can be observed. (B) The only way of recovering internal intron processing is that of reintroducing at least one of the normally processed introns of the TPO gene. An analogous situation in which the system has to be ‘primed’ before it can function efficiently is provided by the recent observation that coordinated splicing can occur in correspondence of distant alternatively spliced exons. (C) Shows such an experiment where two alternatively spliced EDA exons (pEDA and dEDA) separated by a 3400 nt long spacer containing three constitutively spliced exons are inserted in a minigene systems in either wild type or mutated form (lacking an important ESE sequence). The graph on the left shows that efficient inclusion of the proximal exon strictly correlates with inclusion efficiencies of the distal exon.
These examples, together with the recent description of connections between chromatin factors (101), promoter usage and transcript elongation on the splicing process (17,102) clearly highlight how difficult it is to separate the functional effect of a given nucleotide variation from its wide genomic context, and further examples of connections between the upstream history of the transcription/splicing machinery and downstream exon definition will be discussed below.
Pseudoexon inclusion
The observation that consensus splice site sequences are also very abundant in intronic sequences indicates that introns have the potential ability to encode for ‘false’ exons (commonly referred to as pseudoexons) (64,66). Pseudoexons insertion in mature mRNA have been mainly reported in connection with human diseases and Table 1 shows a comprehensive list of genes in which this kind of events has been described. Nonetheless, and possibly more important from a biological point of view, pseudoexon insertion may well represent a substrate for the evolution of new exons. Indeed, it has been proposed recently that ‘exonization’ of a class of particularly abundant intronic sequences (known as Alu repeats) may well represent a major and frequent source of ‘new’ exons and thus play a very important and useful role in evolution (103–105).
Table 1.
list of pseudoexon inclusion events connected with disease in humans
| Gene | Pseudoexon length (bp) | Activating mutation | Reference |
|---|---|---|---|
| DMD | 58 | 5′ss creation | 182 |
| DMD | 67 | 5′ss creation | 183 |
| DMD | 89 | 5′ss creation | 183 |
| DMD | 90 | 5′ss creation | 183 |
| DMD | 95 | 3′ss strengthening | 182 |
| DMD | 149 | 3′ss creation | 183 |
| DMD | 172/202 | 5′ss creation | 184 |
| CFTR | 49 | 5′ss creation | 185 |
| CFTR | 84 | 5′ss creation | 186 |
| CFTR | 184 | 3′ss deletion | 187 |
| CFTR | 214 | 5′ss creation | 188 |
| CYBB | 56 | 5′ss strengthening | 189 |
| CYBB | 94 | 3′ss creation | 190 |
| PHEX | 50/100/170 | 5′ss creation | 191 |
| α-Gal A | 57 | SRE creation | 107 |
| ATM | 65 | SRE deletion | 108 |
| BRCA1 | 66 | 3′ss creation | 192 |
| GHER | 69 | 5′ss creation | 193 |
| CHM | 98 | 3′ss creation | 194 |
| GHR | 102 | 5′ss creation | 195 |
| ATM | 137 | 5′ss creation | 196 |
| OA1/GPR143 | 165 | 3′ss creation | 197 |
| β-globin | 165 | 5′ss creation | 78 |
| NF-1 | 172 | 3′ss creation | 198 |
Despite the abundance in the genome of potential pseudoexons (intronic sequences between 50 and 200 nt in length with apparently viable 5′ss and 3′ss sites at either end) their inclusion does not seem to be a frequent event during normal pre-mRNA processing. Several different reasons have been put forward to account for this, e.g. it was originally noted that many of these ‘non-spliced’ pseudoexons contain multiple defects in splicing controlling regions despite their apparently good agreement with real exonic sequences (64). In keeping with this, three putative splicing silencer motifs have been found recently to be enriched in these non-spliced pseudoexons compared with their surrounding intronic regions (66). This observation is consistent with previous work which showed that human DNA is particularly rich in sequences which can inhibit splicing (65). Furthermore, an extended computational analysis of 8mer sequences that either promote or inhibit splicing in internal non-coding exons versus non-spliced pseudoexons has revealed that enhancer sequences are highly enriched in real exons whilst silencer sequences tend to cluster within pseudoexons (67). Finally, computational studies have suggested recently that pseudoexons flanking regions have a distinct tendency to form double-stranded structures that include the pseudo exon itself, providing an additional layer that differentiates them from real exons, where flanking regions have the tendency to base pair between themselves (106).
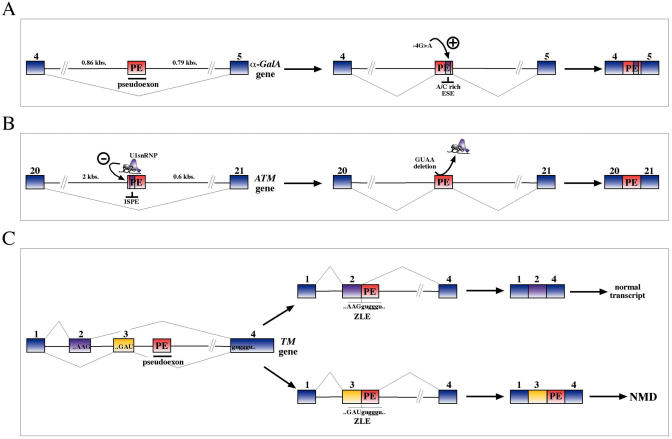
Nonetheless, many intronic sequences seem poised on the brink of becoming exons because, as shown in Table 1, most of these disease-connected pseudoexon inclusion events originate from a single activating mutation. This mutation generally involves the ex novo creation of a strong donor or acceptor site, followed by the subsequent selection of weaker ‘opportunistic’ acceptor or donor site sequences. However, other mechanisms that cause the inclusion of pseudoexon sequences may also take place, and pseudoexon-activating mutations may also involve the creation/deletion of SREs. There are in fact at least two cases, in the α-Gal A (107) (Figure 4A) and the ATM gene (108) (Figure 4B), in which the creation/deletion of SREs has been observed to occur. In the ATM gene the inclusion of a 65 nt long pseudoexon is the consequence of a 4 nt deletion in ATM intron 20 that disrupts a high-affinity U1snRNP-binding site located within the pseudoexon sequence itself. This region was called ISPE (intron splicing processing element) and its disruption resulted in the activation of an immediately upstream cryptic acceptor site and of a downstream cryptic donor site (108). The comparison of this intronic sequence with its homologous region in the mouse has shown the importance of local context also in this case. In fact, an ISPE sequence is also present in the mouse and only a critically missing donor site downstream of this element is the reason why the mouse sequence cannot give rise to an analogous pseudoexon insertion event (109).
Figure 4.
Shows several factors that determine pseudoexon inclusion aside the simple creation of ex novo functional splice sites. In the a-GalA gene (A), a single point mutation G > A observed to occur in position-4 with respect to a cryptic donor site has the effect of creating a novel A/C rich enhancer element that causes inclusion of a 57 bp. pseudoexon in the processed mRNA. In a second example, a 4 nt deletion (GUAA) in the intronic region of the ATM gene between exons 20 and 21 caused the insertion of a 65 nt long pseudoexon in the final mRNA (B). Functional analysis has demonstrated that this deletion abolished binding of an U1snRNP molecule in this position and activated a 3′ss lying 12 nt upstream of this element. Finally, (C) shows the effect of the inclusion of a pseudoexon sequence in the a-tropomyosin pre-mRNA with respect to two mutually exclusive exons 2 and 3. When the pseudoexon sequence (red box) is spliced to exon 2 a 5′ss ZLE sequence is regenerated, thus allowing splicing to occur again giving rise to a normal transcript bearing exon 2 joined to exon 4 (upper panel). On the other hand, when the pseudoexon is joined to exon 3 the ZLE sequence that is regenerated is insufficient to promote joining together of exon 3 to exon 4 and the resulting transcript is thus degraded by NMD (lower panel). This difference in resplicing activity has been proposed to be consistent with the match to consensus of the two ZLE elements (higher for exon 2 than exon 3) and for the higher exon spicing enhancer activity of exon 2.
It is interesting to note that in recent years a lot of effort has gone into deciphering the code that underlies alternatively spliced exon recognition (29,53,69). However, the recent increase in pseudoexon and intron retention events reported in clinical investigations indicates that we should focus also on deciphering splicing codes within introns. In fact, investigations on pseudoexons may well acquire a wider importance in the future, as it has also been proposed recently that many of the these sequences which are thought to be ignored by the splicing machinery also participate in splicing regulation processes (110). The interesting notion is that the functional usage of many of these putative pseudoexon sequences may be often overlooked owing to the fact that their inclusion can lead to premature insertion of a termination codon in the mature mRNA, and its consequently rapid degradation by the nonsense-mediated decay (NMD) pathway (111). This intriguing phenomenon has been described to occur in the rat α-tropomyosin gene in which a pseudoexon sequence localized downstream of the mutually exclusive exons 2 (that is included only in smooth muscle tissue) and 3 (that is included in most cell types) can give rise to different splicing outcomes when it is joined to either exon (110) (Figure 4C). In particular, when joined to exon 2 the formation of an efficient ZLE (zero length exon) sequence allows to resplice exon 2 to the acceptor site of exon 4 (thus giving rise to a normally processed mRNA molecule). On the other hand, when the pseudoexon sequence is joined with exon 3 the resulting ZLE is not sufficient for the resplicing pathway and the pseudoexon 5′ss is used to join with the acceptor site of exon 4 (thus giving rise to a mRNA molecule which is rapidly degraded by NMD.
GENOMIC CONTEXT IS IMPORTANT IN DEFINING EXONIC REGIONS
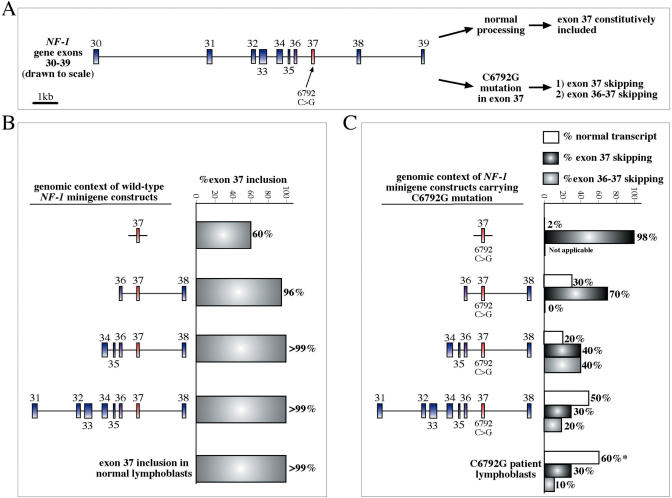
A practical example of the importance of genomic context on the splicing process is represented by the splicing regulation of the neurofibromatosis type-1 (NF-1) exon 37 (112). This exon is localized in an exon-rich genomic cluster (Figure 5A) and mutations within its sequence have been shown to be involved in the development of neurofibromatosis (113). It has been shown recently that this exon is badly recognized by the splicing machinery if it is placed with limited flanking intronic sequences in a minigene with heterologous genomic context. However, it becomes efficiently recognized when located within its natural milieu in the region spanning NF-1 exons 34-38 (Figure 5B). Interestingly, mutations disrupting a SRE sequence within this exon (including some that have been described in neurofibromatosis patients such as C6792G) have genomic context-dependent effects (112). In fact, analysis of patient's lymphoblasts has revealed that this mutation causes both the skipping of exons 36–37 together and the skipping of exon 37 alone. Also in this case, the use of a minigene system to mimick these effects has shown that, after taking in consideration the contribution of the unmutated allele in the patient's lymphoblasts, increasing the genomic context to comprise exons 31–38 allows to duplicate almost exactly the patient's in vivo splicing pattern (Figure 5C).
Figure 5.
(A) Shows a schematic diagram (drawn to scale) of the exon-rich genomic region surrounding exon 37 of the neurofibromatosis (NF-1) gene. This exon is normally included constitutively in the mature mRNA but in the presence of a pathological substitution in position 6792 (a C is replaced by a G) the analysis from patient lymphoblasts reveals both the double skipping of exons 36 and 37 and also skipping of only exon 37. The influence of the genomic context has been tested by engineering a series of minigene constructs that gradually included larger portions of this genomic region, and their functionality compared with the in vivo pattern. (B) Wild-type exon 37 alone with a small portion of flanking intronic regions is not sufficient to yield the observed >99% inclusion in normal lymphoblasts. However, this result can be readily achieved when larger portions of this genomic region are included in the genomic construct. In addition to the normal situation, an increased genomic context also provides a closer correspondence with regards to the exon skipping effects of the C6792G substitution (C). In fact, only the construct carrying most of the NF-1 exons 31 to 38 together with a mutated exon 37 yields a percentage of normal transcript, exon 37 skipping and exon 36–37 skipping that reflects the observed percentage in a patient's lymphoblasts that carry the C6792G substitution (the asterisk indicates the levels of normal mRNA from the C6792G-carrying allele after deducting the estimated contribution of the un mutated allele).
To complicate matters further, it has been observed that whether an exon is recognized as such and to what an extent is also decided by the overall architecture of the pre-mRNA. Two models exist in mRNA processing, the exon definition model and the intron definition model where, as their names suggest, in the former the exon is defined by the acceptor and donor sites that flank it whilst in the latter it is the acceptor and donor sites on the same intron which define its recognition (114,115). Recent experimental evidence has allowed to determine that intron recognition ceases to occur when the intron size increases beyond the 200–250 nt range (116). As splice site recognition across introns of similar size to exons tends to be significantly more efficient than across the exons (116), it follows that exonic sequences with similar exon/intron junction sequences can be recognized with widely different efficiencies depending on the local pre-mRNA architecture where they happen to be present.
SREs ADD A LAYER OF REDUNDANCY FOR THE DEFINITION OF WEAK SPLICE SITES
The first observation that exonic sequences were modulating alternative splicing in a cellular gene was done in 1987 (117). At that time this observation appeared to most researchers as an exceptional case, difficult to interpret. However, in the last 10 years it has been shown that far from being exceptional the presence of SREs in exonic sequences is the rule and their importance in the modulation of pre mRNA processing is now widely recognized.
These SREs reinforce the limited information in the splice sites and, depending on their position of and their effect on splicing, have been generally classified as exonic splicing enhancer and silencer (ESE/ESS) or intronic splicing enhancer and silencer (ISE/ISS) sequences (Figure 1). Although the number of reported proteins recognizing specific enhancer and silencer elements is steadily growing (118), most enhancer sequences have been found to bind members of the SR protein family (119–125) whilst many silencing elements have been found to bind members of the hnRNP protein family (55,126–128). There are, however, several difficulties when trying to evaluate the action and the presence of SREs in many splicing events.
Firstly, it should be noted that there are many exceptions to the rough division stated above. For example, classical SR proteins have also been described to be involved in splicing repression in a few number of cases (129–131) and some members of this protein family (SRp38) act as splicing repressors upon dephosphorylation (132,133). Analogously, some well-represented hnRNP proteins (hnRNP L) may also act as splicing enhancers (134) whilst others, such as hnRNP H, may be involved either in splicing repression or enhancement depending on context (46,135,136). Hence, the simple belonging to a particular family of splicing factors is not a guarantee of function conservation and experimental studies are often required to clarify the role played by even well known splicing factor found in new systems under study.
Secondly, the RNA-binding sequences of many of these splicing factors can be highly degenerate, making it difficult to spot them beforehand on the basis of the primary sequence alone. For example, SELEX analysis of SR protein-binding motifs showed that the major family members recognized fairly degenerate consensus sequences varying from 5 to 7 nt (137) and, although this finding is entirely consistent with the great diversity of ESE sequences mapped within exons (119,138,139), it obviously provides an additional hurdle when trying to identify likely binding sites. To these difficulties it has to be added that much of the same problems apply when investigating common splicing inhibitors, such as hnRNP A1 (140). Moreover, it has to be noted that local context factors, such as RNA secondary structure, may profoundly influence the way that optimal binding motifs are recognized by their specific binding factors (141). Therefore, the simple presence of a high-score-binding motif may not necessarily translate in a strong binding affinity.
In order to obviate to these difficulties several computational approaches have been proposed based on functional SELEX methodologies to identify potential SR protein-binding sites (142), on statistical analysis of genomic exonic sequences to identify splicing-enhancing sequences (67, 68,143,144), or on the information content of the splice site sequences (145). The reason why so much effort is devoted to the development of faster and more efficient ways of predicting SRE presence/action is justified by the clear indication that disruption/creation of these elements has become a very prominent cause of splicing related alterations in disease (4). Indeed, it is probable that many mutations which were previously thought to be harmless polymorphisms may in reality be very powerful modifiers of normal splicing processes (10,146–148).
At present, the accuracy of these methods to successfully predict changes in binding of splicing factors has yielded in some cases encouraging results (143,149,150). However, in several genes such as dystrophin (151), CFTR (147,148) and ATR (152) the predictive power of these in silico approaches has failed to establish a clear connection between splicing and abolishment (or creation) of SREs. Thus, experimental validation of predictions is still a mandatory step in most splicing studies. Nonetheless, because of the realization that accurate in silico approaches will be very useful in the diagnostic and therapeutic field (34,35), studies of this kind are destined to increase. Indeed, statistical approaches have also been aimed recently at characterizing inhibitory sequences (66,69,153) or other kinds of repeated motifs such as UAGG or G-runs which are often found near or within alternatively spliced exons and which may contribute heavily to inclusion/skipping decisions by the splicing machinery (153,154).
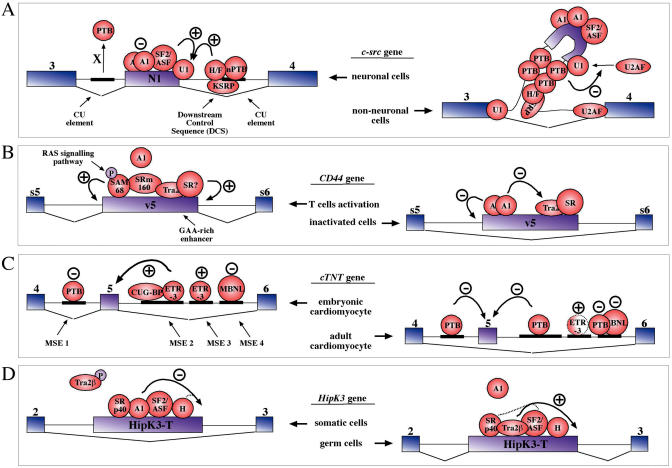
Finally, the mechanisms through which splicing factors bind SRE sequences has been the subject of many studies, especially regarding the interplay between classic antagonistic factors such as ASF/SF2 (a SR protein family member that promotes inclusion) and hnRNP A1 (a classic hnRNP protein that has been historically associated with exon skipping) (155,156). In many instances, however, the number of ‘duelling partners’ increases considerably and a complex number of SRE sequences and binding factors is involved in regulating tissue- or developmentally specific alternatively spliced exons. A schematic diagram of four well-studied regulatory mechanisms is shown in Figure 6. The example given are the c-src exon N1 (157,158) (Figure 6A), the CD44 exon v5 (159–161) (Figure 6B), chicken cTNT exon 5 (162) (Figure 6C) and HipK3 ‘T’ exon (163) (Figure 6D). In all four cases it should be noted that in order to achieve tissue- and/or temporal-precise accuracy a complex arrangement of cis-acting elements (with either positive or negative influence on exon recognition) is complemented by a correspondingly high number of splicing factors.
Figure 6.
Modulation of tissue- and developmental-specific expression of four different exons. (A) Shows that in non-neuronal cells PTB binds to two CU elements which are located both upstream and downstream c-src exon N1 and form a silencing complex that interferes with the interaction between U1snRNP bound at the donor site of this exon and U2AF. This complex does not interfere with binding of U2AF with the U1snRNP present at the donor site of exon 3 and this allows splicing of exon 3 to exon 4, thereby excluding exon N1 from the mature mRNA. On the other hand, in neuronal cells PTB is removed from the CU elements by an unknown factor ‘X’ in the presence of ATP and exon inclusion is promoted by the SF2/ASF protein binding to this exon (which is also bound by its antagonistic molecule, hnRNP A1) and by the DCS complex (which contains hnRNP H/F (H/F), KSRP, plus a neuronal form of PTB known as nPTB or WERI PTB). (B) Shows the Ras signaling pathway-dependent regulation of splicing of the variable v5 exon in the CD44 gene that occurs during T cell activation. In normal situations the enhancer complex in correspondence to a GAA-rich motif near the donor site of this exon is repressed by a strong hnRNP A1-binding sequence. During T cell activation, however, the repression can be overcome by either inhibition or displacement of A1 by phosphorylation of Sam 68 (Src-associated in mitosis) and involvement of powerful splicing coactivators such as SRm160. (C) Represents a model of cTNT exon 5 alternative splicing in the chicken developing heart. In the embryonic cardiomyocite CUG-BP and ETR-3 are expressed at high levels in the nucleus and they bind to the regulatory regions downstream of exon 5 (MSE2 to MSE4) to promote its inclusion. On the other hand, in the adult cardiomyocite CUG-BP and ETR-3 expression is downregulated and skipping is favored by PTB and MBNL proteins that are now free to bind on either side of the exon. Finally, (D) contains a model of HipK3 ‘T’ (testis-specific) differential exon inclusion in both somatic cells and in germ lines. In somatic cells, hnRNP A1 (A1) is principally responsible for the inhibition of the T exon donor site, probably with some help by hnRNP H (H). This is favored by the fact that in these cells the Tra2β protein tends to be in low amounts and is mostly phosphorylated. On the other hand, in germ cells Tra2β expression is upregulated and tends to be in its hypo-phosphorylated form whilst hnRNP A1 is downregulated. This arrangement allows the Tra2β protein to bind in place of A1 and to promote T exon inclusion with the assistance of SF2/ASF and SRp40 which also bind within this exon.
SPLICE SITE STRENGTHS CAN EITHER MASK OR UNCOVER ADDITIONAL SPLICING REGULATORY REGIONS PRESENT IN EXONS
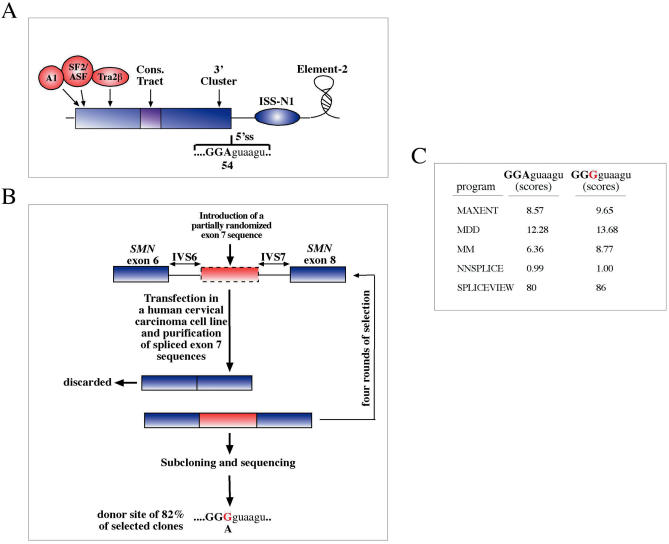
A well-studied working example in which the interplay between splice sites and regulatory sequences has been analyzed in depth is represented by exon 7 in the SMN1/SMN2 genes. This exon plays a key role in a common autosomal recessive disorder, spinal muscular atrophy. In humans, there are two copies of the SMN gene, named SMN1 and SMN2, and inactivation of the SMN1 gene leads to disease because the SMN2 gene cannot compensate for its absence. The reason for this is a critical C to U substitution in exon 7 of the SMN2 gene that does not change the codon but either disrupts an ASF/SF2-binding site (149,164), creates an hnRNP A1-binding site (165), or both (although recent evidence supports the ASF/SF2 model whilst confirming an indirect role for hnRNP A1 in splicing repression). As a result of this substitution, the SMN2 gene produces a transcript where exon 7 is predominantly skipped, leading to the production of a truncated protein and inability to compensate for SMN1 inactivation. In addition to this region, several other SRE sequences both within the exon and in the upstream intron have been identified. These elements include a Tra2-binding site in the region adjacent to the SF2/ASF interaction (166) and a highly structured ISE region in the IVS7 intron a short distance away from the donor site (167) (Figure 7A). Recently, Singh et al. (168) have used a functional selection technique with the aim of identifying additional controlling elements that regulate the splicing of an entire exon. As shown in Figure 7B, they have achieved this purpose by engineering a pre-mRNA template which contained the full sequence of SMN exon 6 and 8 with some intronic sequences and a partially randomized (30%) exon 7 sequences in order to substantially preserve the wild-type characteristics of this exon but at the same time allow mutability of each nucleotide. This pool of pre-mRNA molecules was then transfected in a human cervical carinoma cell line and the spliced products were purified to select the exon 7 sequences that displayed correct recognition by the splicing machinery (Figure 7B). After successive rounds of in vivo selection the different exon 7 sequences were subsequently analyzed to evaluate the range and type of nucleotide substitutions that had occurred with respect to the initial wild-type sequence. Beside confirming previous results with regards to the SF2/ASF and Tra2-binding regions the results of this analysis allowed the identification of two novel regulatory regions, one near the 3′ end of the exon (3′-Cluster) and a conserved CT tract in the middle. Most importantly, however, the weak 5′ss consensus sequence was also found to represent a critical factor in regulating SMN exon 7 recognition, as 82% of the spliced exon 7 clones identified following the functional SELEX contained a A54G substitution (Figure 7B). It is interesting to note that this substitution alone was quite capable of overcoming the negative effect of several mutations within other conserved elements such as the first position of the exon or single mutations within the Tra2-binding site (168).
Figure 7.
(A) Shows a schematic diagram of the splicing regulatory regions that have been described to affect recognition of SMN1/SMN2 exon 7. Recently, a functional SELEX analysis has been applied to this whole exonic sequence. (B) Shows the minigene system based on the human SMN gene used for this assay in which the entire exon 7 is partially randomized. These sequences can then be selected for inclusion in the final spliced products by subjecting them to rounds of selection in vivo. The borders of this randomized exon are dotted in (B) to indicate the fact that these sequences will not always be used as an exon in this technique. After each round of selection the spliced exon 7 sequences are recovered by RT–PCR and reinserted in the minigene splicing cassette. This selection process has allowed the precise mapping, at the single nucleotide level, of all the major determinants involved in the control of exon 7 splicing. Interestingly, 82% of all the final clones contained the same substitution in correspondence to the last position of this exon, and this change was quite capable of overcoming the effects of other negative regulatory mutations. (C) Shows a comparison of the two donor site strengths (the wild type and the one carrying the A54G substitution) using a panel of splice site prediction programs.
It is also interesting to note that several splice site prediction programs such as MaxEnt, MDD and MM (169), NNSplice (170) and Spliceview (171) fail to detect a major improvement in donor site score following the A54G substitution (Figure 7C). As these programs are normally very accurate in detecting splice site strength variations the most likely conclusion is that only the slightly weakening of exon 7 splice site can ‘highlight’ several sequences that would normally not be able to influence its splicing process. This is consistent with the observation that alternatively spliced exons possess splice site sequences which are on average weaker than splice sites belonging to constitutively recognized exons and stresses the importance of the context (172). In addition, it has also to be noted that splice site prediction programs may not pick up additional protein–RNA interactions at the 5′ splice site that may substantially contribute in determining its relative strength. For example, in the NF-1 exon 3 donor site a G-four stretch in correspondence to the donor site has been identified as a binding site of hnRNP H that severely constrained the 5′ss sequences that could vary without disrupting splicing (46).
Nonetheless, the result of SELEX analysis suggests that many skipping events originating from various mutations within the exon may be rescued by even small improvements in splice site quality of the affected exon(s). However, changing the donor site in exons of patients affected by mutations within SRE elements would probably involve some too costly and uncertain investments in gene therapy schemes and, for this reason, alternative targets should be preferred.
INACTIVATION OF STRONG NEGATIVE ELEMENTS CAN RESCUE SPLICING DEFECTS ORIGINATING FROM OTHER NEARBY REGULATORY ELEMENTS
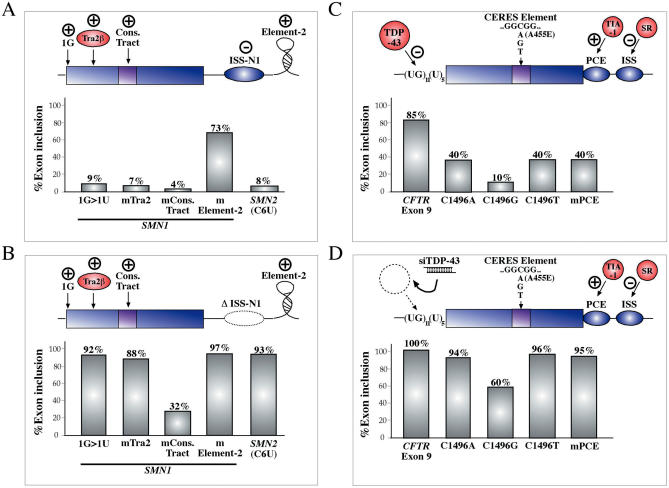
Therefore, in order to optimize therapeutic strategies an important question that needs to be addressed is the existence of hierarchical levels between these splicing regulatory elements, as this may decide which region should be preferentially targeted by the effector molecule of choice. Recently, the presence of a human-specific ISS near to the 5′ss of SMN1/SMN2 exon 7 has been described to have analogous compensatory effects as the A54G substitution (173). Indeed, Figure 8A shows that the effects of mutating three well-characterized SRE elements in SMN1 exon 7 (the first nucleotide, the Tra2 binding sequence, and the conserved tract) can all be overcome by deleting the ISS-N1 sequence (Figure 8B). In keeping with this result, much the same effect can be obtained by the deletion of the ISS-N1 region in the SMN2 sequence that carries the C6U substitution that abrogates SF2/ASF binding.
Figure 8.
(A) Shows a diagrammatic representation of several positive elements that regulate SMN1/SMN2 exon 7 inclusion. The elements analyzed in this experiment are the first nucleotide of exon 7 (1G), the Tra2β-binding site (Tra2), the conserved tract in the middle of the exon (ConsTract) and the intronic splicing enhancer (element-2) in IVS7. Also shown is the dominant ISS element (ISS-N1). The graphical representation underneath shows the levels of exon 7 inclusion following mutation of all of these elements in a suitable minigene construct. Also included are the basal exon 7 inclusion levels from SMN2 exon 7 that carries a natural C6U substitution. (B) Shows the effect of the same mutations/substitutions when the ISS-N1 sequence has been deleted from the minigene constructs. An analogous situation which regards the CFTR exon 9 sequence is schematically represented in (C) in which the (UG)m(U)n repeated motif near the 3′ss of this exon has been shown to bind a powerful negative regulator of splicing, TDP-43. Several other cis-acting elements are also shown in this diagram: a CERES element within the exon, a positive polypyrimidine sequence near the 5′ss (PCE) that binds TIA-1, and a human-specific ISS in IVS9 that binds members of the SR protein family. The graph below shows that mutations within these elements are strongly inhibitory with respect to the basal inclusion level on a TG11T5 background (lane CFTR exon 9). Nonetheless, also in this case the removal of TDP-43 using siRNA technology rescues the effect of all these negative mutations (D).
Interestingly, an analogous situation has been described for exon 9 of the CFTR gene. Skipping of this exon is involved in the occurrence of several forms of cystic fibrosis depending on the presence of particular polymorphisms in a (UG)m(U)n repeated motif near to its 3′ splice site (174). Recently, several studies have confirmed that a nuclear protein, TDP-43, binds to the (UG)m sequence and can inhibit its inclusion in the final mRNA (175–177). However, in addition to this regulatory sequence several other exonic and intronic SREs have been described to take part in splicing regulation of exon 9: a composite exonic regulatory element of splicing (CERES) (147), a positive polypyrimidine rich element near to the 5′ss that binds the TIA-1 splicing factor (PCE) (178), and an ISS further down in IVS9 that binds several members of the SR protein family (129) (Figure 8C). Mutations in all these sequences have modulating effects in CFTR exon inclusion in the mRNA. In particular, the CERES element identified by Pagani et al. (129) lies in correspondence to a previously described C1496A missense mutation (A455E) which should be therefore also considered a splicing mutation. The recent use of siRNA technology to knockdown TDP-43 expression (179) has resulted in a generalized improvement in the inclusion of exon 9 even in the presence of many of these mutations (Figure 8D), demonstrating that removal of this key inhibitory factor may be a useful way of repairing negative mutations also in other controlling elements. In this respect, it is important to note that depletion of this factor has also been described to override the need for several ESEs and ISEs of Apo AII exon 3 (180). Obviously, knowledge of other critical functions of this or any other factor is essential before a general inhibitory approach is used in therapy.
CONCLUDING REMARKS
Taken together, recent results clearly highlight that any pre-mRNA sequence is embedded with regulatory elements, some of which may only be uncovered by disrupting mutations or by other facets of mRNA synthesis (such as transcript elongation rates or secondary structure). The practical consequences of such an arrangement is that many splicing events are the result of a fine ‘balance of power’ between a myriad of controlling factors which can vary from tissue to tissue, during development or in the presence of external stimuli. Moreover, the recent discovery that snoRNAs can also participate actively in the regulation of splicing (181) emphasizes that we are still ignorant of entire classes of splicing regulators. This complexity makes the job of predicting the splicing outcomes of given genomic variations exceedingly difficult for the researcher. Nonetheless, not all these factors possess the same weight when determining splicing outcomes, and the SMN1/SMN2 exon 7 and CFTR exon 9 examples demonstrate that identifying master checkpoints may, if acted upon, compensate for negative effects in several other regulatory elements. This conclusion provides a considerable encouragement towards the search for novel therapeutic strategies aimed at covering as great an amount as possible of splicing mutations. Paradoxically, though, it is very probable that effective cures for many splicing mutations will be developed long before the functional mechanisms that underlie them may be satisfactorily explained.
Acknowledgments
F.E.B. is supported by the Telethon Onlus Foundation (Italy) (grants no. GGP02453 and nos. GGP030261 and GGP06147), by FIRB (RBNE01W9PM) and by a European community grant (EURASNET). The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Nissim-Rafinia M., Kerem B. The splicing machinery is a genetic modifier of disease severity. Trends Genet. 2005;21:480–483. doi: 10.1016/j.tig.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 2.Caceres J.F., Kornblihtt A.R. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 2002;18:186–193. doi: 10.1016/s0168-9525(01)02626-9. [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Blanco M.A., Baraniak A.P., Lasda E.L. Alternative splicing in disease and therapy. Nat. Biotechnol. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- 4.Cartegni L., Chew S.L., Krainer A.R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nature Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 5.Faustino N.A., Cooper T.A. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 6.Tazi J., Durand S., Jeanteur P. The spliceosome: a novel multi-faceted target for therapy. Trends Biochem. Sci. 2005;30:469–478. doi: 10.1016/j.tibs.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Stoilov P., Meshorer E., Gencheva M., Glick D., Soreq H., Stamm S. Defects in pre-mRNA processing as causes of and predisposition to diseases. DNA Cell. Biol. 2002;21:803–818. doi: 10.1089/104454902320908450. [DOI] [PubMed] [Google Scholar]
- 8.Ars E., Serra E., Garcia J., Kruyer H., Gaona A., Lazaro C., Estivill X. Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1. Hum. Mol. Genet. 2000;9:237–247. doi: 10.1093/hmg/9.2.237. [DOI] [PubMed] [Google Scholar]
- 9.Teraoka S.N., Telatar M., Becker-Catania S., Liang T., Onengut S., Tolun A., Chessa L., Sanal O., Bernatowska E., Gatti R.A., et al. Splicing defects in the ataxia-telangiectasia gene, ATM: underlying mutations and consequences. Am. J. Hum. Genet. 1999;64:1617–1631. doi: 10.1086/302418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chamary J.V., Parmley J.L., Hurst L.D. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nature Rev. Genet. 2006;7:98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- 11.Lenasi T., Peterlin B.M., Dovc P. Distal regulation of alternative splicing by splicing enhancer in equine beta-casein intron 1. RNA. 2006;12:498–507. doi: 10.1261/rna.7261206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graveley B.R. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell. 2005;123:65–73. doi: 10.1016/j.cell.2005.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kreahling J.M., Graveley B.R. The iStem, a long-range RNA secondary structure element required for efficient exon inclusion in the Drosophila Dscam pre-mRNA. Mol. Cell. Biol. 2005;25:10251–10260. doi: 10.1128/MCB.25.23.10251-10260.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buratti E., Baralle F.E. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol. Cell. Biol. 2004;24:10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romano M., Marcucci R., Baralle F.E. Splicing of constitutive upstream introns is essential for the recognition of intra-exonic suboptimal splice sites in the thrombopoietin gene. Nucleic Acids Res. 2001;29:886–894. doi: 10.1093/nar/29.4.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fededa J.P., Petrillo E., Gelfand M.S., Neverov A.D., Kadener S., Nogues G., Pelisch F., Baralle F.E., Muro A.F., Kornblihtt A.R. A polar mechanism coordinates different regions of alternative splicing within a single gene. Mol. Cell. 2005;19:393–404. doi: 10.1016/j.molcel.2005.06.035. [DOI] [PubMed] [Google Scholar]
- 17.Kornblihtt A.R. Promoter usage and alternative splicing. Curr. Opin. Cell. Biol. 2005;17:262–268. doi: 10.1016/j.ceb.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 18.Pagani F., Baralle F.E. Genomic variants in exons and introns: identifying the splicing spoilers. Nature Rev. Genet. 2004;5:389–396. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- 19.Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 20.Modrek B., Lee C. A genomic view of alternative splicing. Nature Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 21.Maniatis T., Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 22.Stetefeld J., Ruegg M.A. Structural and functional diversity generated by alternative mRNA splicing. Trends Biochem. Sci. 2005;30:515–521. doi: 10.1016/j.tibs.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 23.Graveley B.R. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 24.Stamm S., Ben-Ari S., Rafalska I., Tang Y., Zhang Z., Toiber D., Thanaraj T.A., Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 25.Vibe-Pedersen K., Kornblihtt A.R., Baralle F.E. Expression of a human alpha-globin/fibronectin gene hybrid generates two mRNAs by alternative splicing. EMBO J. 1984;3:2511–2516. doi: 10.1002/j.1460-2075.1984.tb02165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baralle D., Baralle M. Splicing in action: assessing disease causing sequence changes. J. Med. Genet. 2005;42:737–748. doi: 10.1136/jmg.2004.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooper T.A. Use of minigene systems to dissect alternative splicing elements. Methods. 2005;37:331–340. doi: 10.1016/j.ymeth.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 28.Buratti E., Baralle F.E. Another step forward for SELEXive splicing. Trends Mol. Med. 2005;11:5–9. doi: 10.1016/j.molmed.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 29.Fu X.D. Towards a splicing code. Cell. 2004;119:736–738. doi: 10.1016/j.cell.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 30.Buratti E., Baralle F.E., Pagani F. Can a ‘patch’ in a skipped exon make the pre-mRNA splicing machine run better? Trends Mol. Med. 2003;9:229–232. doi: 10.1016/s1471-4914(03)00072-8. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Blanco M.A. Making antisense of splicing. Curr. Opin. Mol. Ther. 2005;7:476–482. [PubMed] [Google Scholar]
- 32.Wilton S.D., Fletcher S. RNA splicing manipulation: strategies to modify gene expression for a variety of therapeutic outcomes. Curr. Gene Ther. 2005;5:467–483. doi: 10.2174/156652305774329249. [DOI] [PubMed] [Google Scholar]
- 33.Sazani P., Kole R. Therapeutic potential of antisense oligonucleotides as modulators of alternative splicing. J. Clin. Invest. 2003;112:481–486. doi: 10.1172/JCI19547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cartegni L., Krainer A.R. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nature Struct. Biol. 2003;10:120–125. doi: 10.1038/nsb887. [DOI] [PubMed] [Google Scholar]
- 35.Skordis L.A., Dunckley M.G., Yue B., Eperon I.C., Muntoni F. Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc. Natl Acad. Sci. USA. 2003;100:4114–4119. doi: 10.1073/pnas.0633863100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soret J., Bakkour N., Maire S., Durand S., Zekri L., Gabut M., Fic W., Divita G., Rivalle C., Dauzonne D., et al. Selective modification of alternative splicing by indole derivatives that target serine-arginine-rich protein splicing factors. Proc. Natl Acad. Sci. USA. 2005;102:8764–8769. doi: 10.1073/pnas.0409829102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hagiwara M. Alternative splicing: a new drug target of the post-genome era. Biochim. Biophys. Acta. 2005;1754:324–331. doi: 10.1016/j.bbapap.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 38.Graveley B.R. Small molecule control of pre-mRNA splicing. RNA. 2005;11:355–358. doi: 10.1261/rna.7229705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goyenvalle A., Vulin A., Fougerousse F., Leturcq F., Kaplan J.C., Garcia L., Danos O. Rescue of dystrophic muscle through U7 snRNA-mediated exon skipping. Science. 2004;306:1796–1799. doi: 10.1126/science.1104297. [DOI] [PubMed] [Google Scholar]
- 40.Maquat L.E. The power of point mutations. Nature Genet. 2001;27:5–6. doi: 10.1038/83759. [DOI] [PubMed] [Google Scholar]
- 41.Zhang M.Q. Statistical features of human exons and their flanking regions. Hum. Mol. Genet. 1998;7:919–932. doi: 10.1093/hmg/7.5.919. [DOI] [PubMed] [Google Scholar]
- 42.Lund M., Kjems J. Defining a 5′ splice site by functional selection in the presence and absence of U1 snRNA 5′ end. RNA. 2002;8:166–179. doi: 10.1017/s1355838202010786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Du H., Rosbash M. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature. 2002;419:86–90. doi: 10.1038/nature00947. [DOI] [PubMed] [Google Scholar]
- 44.Tarn W.Y., Steitz J.A. SR proteins can compensate for the loss of U1 snRNP functions in vitro. Genes. Dev. 1994;8:2704–2717. doi: 10.1101/gad.8.22.2704. [DOI] [PubMed] [Google Scholar]
- 45.Crispino J.D., Blencowe B.J., Sharp P.A. Complementation by SR proteins of pre-mRNA splicing reactions depleted of U1 snRNP. Science. 1994;265:1866–1869. doi: 10.1126/science.8091213. [DOI] [PubMed] [Google Scholar]
- 46.Buratti E., Baralle M., De Conti L., Baralle D., Romano M., Ayala Y.M., Baralle F.E. hnRNP H binding at the 5′ splice site correlates with the pathological effect of two intronic mutations in the NF-1 and TSHbeta genes. Nucleic Acids Res. 2004;32:4224–4236. doi: 10.1093/nar/gkh752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Relogio A., Ben-Dov C., Baum M., Ruggiu M., Gemund C., Benes V., Darnell R.B., Valcarcel J. Alternative splicing microarrays reveal functional expression of neuron-specific regulators in Hodgkin lymphoma cells. J. Biol. Chem. 2005;280:4779–4784. doi: 10.1074/jbc.M411976200. [DOI] [PubMed] [Google Scholar]
- 48.Pan Q., Shai O., Misquitta C., Zhang W., Saltzman A.L., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 49.Yeo G., Holste D., Kreiman G., Burge C.B. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalnina Z., Zayakin P., Silina K., Line A. Alterations of pre-mRNA splicing in cancer. Genes Chromosomes Cancer. 2005;42:342–357. doi: 10.1002/gcc.20156. [DOI] [PubMed] [Google Scholar]
- 51.Venables J.P. Unbalanced alternative splicing and its significance in cancer. Bioessays. 2006;28:378–386. doi: 10.1002/bies.20390. [DOI] [PubMed] [Google Scholar]
- 52.Smith C.W., Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 53.Matlin A.J., Clark F., Smith C.W. Understanding alternative splicing: towards a cellular code. Nature Rev. Mol. Cell. Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 54.Maniatis T., Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 55.Singh R., Valcarcel J. Building specificity with nonspecific RNA-binding proteins. Nature Struct. Mol. Biol. 2005;12:645–653. doi: 10.1038/nsmb961. [DOI] [PubMed] [Google Scholar]
- 56.Shin C., Manley J.L. Cell signalling and the control of pre-mRNA splicing. Nature Rev. Mol. Cell. Biol. 2004;5:727–738. doi: 10.1038/nrm1467. [DOI] [PubMed] [Google Scholar]
- 57.Li X., Manley J.L. New talents for an old acquaintance: the SR protein splicing factor ASF/SF2 functions in the maintenance of genome stability. Cell Cycle. 2005;4:1706–1708. doi: 10.4161/cc.4.12.2210. [DOI] [PubMed] [Google Scholar]
- 58.Nott A., Le Hir H., Moore M.J. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18:210–222. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanford J.R., Gray N.K., Beckmann K., Caceres J.F. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 2004;18:755–768. doi: 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang Y., Yario T.A., Steitz J.A. A molecular link between SR protein dephosphorylation and mRNA export. Proc. Natl Acad. Sci. USA. 2004;101:9666–9670. doi: 10.1073/pnas.0403533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang Y., Gattoni R., Stevenin J., Steitz J.A. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 62.Graveley B.R. Coordinated control of splicing and translation. Nature Struct. Mol. Biol. 2005;12:1022–1023. doi: 10.1038/nsmb1205-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blaustein M., Pelisch F., Tanos T., Munoz M.J., Wengier D., Quadrana L., Sanford J.R., Muschietti J.P., Kornblihtt A.R., Caceres J.F., et al. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nature Struct. Mol. Biol. 2005;12:1037–1044. doi: 10.1038/nsmb1020. [DOI] [PubMed] [Google Scholar]
- 64.Sun H., Chasin L.A. Multiple splicing defects in an intronic false exon. Mol. Cell. Biol. 2000;20:6414–6425. doi: 10.1128/mcb.20.17.6414-6425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fairbrother W.G., Chasin L.A. Human genomic sequences that inhibit splicing. Mol. Cell. Biol. 2000;20:6816–6825. doi: 10.1128/mcb.20.18.6816-6825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sironi M., Menozzi G., Riva L., Cagliani R., Comi G.P., Bresolin N., Giorda R., Pozzoli U. Silencer elements as possible inhibitors of pseudoexon splicing. Nucleic Acids Res. 2004;32:1783–1791. doi: 10.1093/nar/gkh341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang X.H., Chasin L.A. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang X.H., Kangsamaksin T., Chao M.S., Banerjee J.K., Chasin L.A. Exon inclusion is dependent on predictable exonic splicing enhancers. Mol. Cell. Biol. 2005;25:7323–7332. doi: 10.1128/MCB.25.16.7323-7332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Z., Rolish M.E., Yeo G., Tung V., Mawson M., Burge C.B. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 70.Mendes Soares L.M., Valcarcel J. The expanding transcriptome: the genome as the ‘Book of Sand’. EMBO J. 2006;25:923–931. doi: 10.1038/sj.emboj.7601023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Proudfoot N.J., Furger A., Dye M.J. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 72.Dye M.J., Gromak N., Proudfoot N.J. Exon tethering in transcription by RNA polymerase II. Mol. Cell. 2006;21:849–859. doi: 10.1016/j.molcel.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 73.Horowitz D.S., Krainer A.R. Mechanisms for selecting 5′ splice sites in mammalian pre-mRNA splicing. Trends Genet. 1994;10:100–106. doi: 10.1016/0168-9525(94)90233-x. [DOI] [PubMed] [Google Scholar]
- 74.Freund M., Hicks M.J., Konermann C., Otte M., Hertel K.J., Schaal H. Extended base pair complementarity between U1 snRNA and the 5′ splice site does not inhibit splicing in higher eukaryotes, but rather increases 5′ splice site recognition. Nucleic Acids Res. 2005;33:5112–5119. doi: 10.1093/nar/gki824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roca X., Sachidanandam R., Krainer A.R. Determinants of the inherent strength of human 5′ splice sites. RNA. 2005;11:683–698. doi: 10.1261/rna.2040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Carmel I., Tal S., Vig I., Ast G. Comparative analysis detects dependencies among the 5′ splice-site positions. RNA. 2004;10:828–840. doi: 10.1261/rna.5196404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Krawczak M., Reiss J., Cooper D.N. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum. Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- 78.Treisman R., Orkin S.H., Maniatis T. Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature. 1983;302:591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- 79.Wieringa B., Meyer F., Reiser J., Weissmann C. Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit beta-globin gene. Nature. 1983;301:38–43. doi: 10.1038/301038a0. [DOI] [PubMed] [Google Scholar]
- 80.Roy S.W., Gilbert W. The evolution of spliceosomal introns: patterns, puzzles and progress. Nature Rev. Genet. 2006;7:211–221. doi: 10.1038/nrg1807. [DOI] [PubMed] [Google Scholar]
- 81.Sadusky T., Newman A.J., Dibb N.J. Exon junction sequences as cryptic splice sites: implications for intron origin. Curr. Biol. 2004;14:505–509. doi: 10.1016/j.cub.2004.02.063. [DOI] [PubMed] [Google Scholar]
- 82.Stoltzfus A. Molecular evolution: introns fall into place. Curr. Biol. 2004;14:R351–352. doi: 10.1016/j.cub.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 83.Roca X., Sachidanandam R., Krainer A.R. Intrinsic differences between authentic and cryptic 5′ splice sites. Nucleic Acids Res. 2003;31:6321–6333. doi: 10.1093/nar/gkg830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kralovicova J., Christensen M.B., Vorechovsky I. Biased exon/intron distribution of cryptic and de novo 3′ splice sites. Nucleic Acids Res. 2005;33:4882–4898. doi: 10.1093/nar/gki811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Krainer A.R., Conway G.C., Kozak D. The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell. 1990;62:35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- 86.Caceres J.F., Stamm S., Helfman D.M., Krainer A.R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 87.Gabut M., Mine M., Marsac C., Brivet M., Tazi J., Soret J. The SR protein SC35 is responsible for aberrant splicing of the E1alpha pyruvate dehydrogenase mRNA in a case of mental retardation with lactic acidosis. Mol. Cell. Biol. 2005;25:3286–3294. doi: 10.1128/MCB.25.8.3286-3294.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Spena S., Tenchini M.L., Buratti E. Cryptic splice site usage in exon 7 of the human fibrinogen Bβ-chain gene is regulated by a naturally silent SF2/ASF binding site within this exon. RNA. 2006;12:948–958. doi: 10.1261/rna.2269306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Smith C.W. Alternative splicing—when two's a crowd. Cell. 2005;123:1–3. doi: 10.1016/j.cell.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 90.Domenjoud L., Gallinaro H., Kister L., Meyer S., Jacob M. Identification of a specific exon sequence that is a major determinant in the selection between a natural and a cryptic 5′ splice site. Mol. Cell. Biol. 1991;11:4581–4590. doi: 10.1128/mcb.11.9.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Domenjoud L., Kister L., Gallinaro H., Jacob M. Selection between a natural and a cryptic 5′ splice site: a kinetic study of the effect of upstream exon sequences. Gene Expr. 1993;3:83–94. [PMC free article] [PubMed] [Google Scholar]
- 92.Galante P.A., Sakabe N.J., Kirschbaum-Slager N., de Souza S.J. Detection and evaluation of intron retention events in the human transcriptome. RNA. 2004;10:757–765. doi: 10.1261/rna.5123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stamm S., Zhu J., Nakai K., Stoilov P., Stoss O., Zhang M.Q. An alternative-exon database and its statistical analysis. DNA Cell Biol. 2000;19:739–756. doi: 10.1089/104454900750058107. [DOI] [PubMed] [Google Scholar]
- 94.Laski F.A., Rubin G.M. Analysis of the cis-acting requirements for germ-line-specific splicing of the P-element ORF2-ORF3 intron. Genes Dev. 1989;3:720–728. doi: 10.1101/gad.3.5.720. [DOI] [PubMed] [Google Scholar]
- 95.Gebauer F., Merendino L., Hentze M.W., Valcarcel J. The Drosophila splicing regulator sex-lethal directly inhibits translation of male-specific-lethal 2 mRNA. RNA. 1998;4:142–150. [PMC free article] [PubMed] [Google Scholar]
- 96.Mansilla A., Lopez-Sanchez C., de la Rosa E.J., Garcia-Martinez V., Martinez-Salas E., de Pablo F., Hernandez-Sanchez C. Developmental regulation of a proinsulin messenger RNA generated by intron retention. EMBO Rep. 2005;6:1182–1187. doi: 10.1038/sj.embor.7400539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Le Hir H., Charlet-Berguerand N., de Franciscis V., Thermes C. 5′-End RET splicing: absence of variants in normal tissues and intron retention in pheochromocytomas. Oncology. 2002;63:84–91. doi: 10.1159/000065725. [DOI] [PubMed] [Google Scholar]
- 98.Zhang L., Vincent G.M., Baralle M., Baralle F.E., Anson B.D., Benson D.W., Whiting B., Timothy K.W., Carlquist J., January C.T., et al. An intronic mutation causes long QT syndrome. J. Am. Coll. Cardiol. 2004;44:1283–1291. doi: 10.1016/j.jacc.2004.06.045. [DOI] [PubMed] [Google Scholar]
- 99.Pequignot M.O., Dey R., Zeviani M., Tiranti V., Godinot C., Poyau A., Sue C., Di Mauro S., Abitbol M., Marsac C. Mutations in the SURF1 gene associated with Leigh syndrome and cytochrome C oxidase deficiency. Hum. Mutat. 2001;17:374–381. doi: 10.1002/humu.1112. [DOI] [PubMed] [Google Scholar]
- 100.Muro A.F., Chauhan A.K., Gajovic S., Iaconcig A., Porro F., Stanta G., Baralle F.E. Regulated splicing of the fibronectin EDA exon is essential for proper skin wound healing and normal lifespan. J. Cell. Biol. 2003;162:149–160. doi: 10.1083/jcb.200212079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Batsche E., Yaniv M., Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nature Struct. Mol. Biol. 2006;13:22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- 102.Kornblihtt A.R. Chromatin, transcript elongation and alternative splicing. Nature Struct. Mol. Biol. 2006;13:5–7. doi: 10.1038/nsmb0106-5. [DOI] [PubMed] [Google Scholar]
- 103.Kreahling J., Graveley B.R. The origins and implications of Aluternative splicing. Trends Genet. 2004;20:1–4. doi: 10.1016/j.tig.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 104.Sorek R., Lev-Maor G., Reznik M., Dagan T., Belinky F., Graur D., Ast G. Minimal conditions for exonization of intronic sequences: 5′ splice site formation in alu exons. Mol. Cell. 2004;14:221–231. doi: 10.1016/s1097-2765(04)00181-9. [DOI] [PubMed] [Google Scholar]
- 105.Lei H., Day I.N., Vorechovsky I. Exonization of AluYa5 in the human ACE gene requires mutations in both 3′ and 5′ splice sites and is facilitated by a conserved splicing enhancer. Nucleic Acids Res. 2005;33:3897–3906. doi: 10.1093/nar/gki707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang X.H., Leslie C.S., Chasin L.A. Dichotomous splicing signals in exon flanks. Genome. Res. 2005;15:768–779. doi: 10.1101/gr.3217705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ishii S., Nakao S., Minamikawa-Tachino R., Desnick R.J., Fan J.Q. Alternative splicing in the alpha-galactosidase A gene: increased exon inclusion results in the Fabry cardiac phenotype. Am. J. Hum. Genet. 2002;70:994–1002. doi: 10.1086/339431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pagani F., Buratti E., Stuani C., Bendix R., Dork T., Baralle F.E. A new type of mutation causes a splicing defect in ATM. Nature Genet. 2002;30:426–429. doi: 10.1038/ng858. [DOI] [PubMed] [Google Scholar]
- 109.Lewandowska M.A., Stuani C., Parvizpur A., Baralle F.E., Pagani F. Functional studies on the ATM intronic splicing processing element. Nucleic Acids Res. 2005;33:4007–4015. doi: 10.1093/nar/gki710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Grellscheid S.N., Smith C.W. An apparent pseudo-exon acts both as an alternative exon that leads to nonsense-mediated decay and as a zero-length exon. Mol. Cell. Biol. 2006;26:2237–2246. doi: 10.1128/MCB.26.6.2237-2246.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Maquat L.E. Nonsense-mediated mRNA decay in mammals. J. Cell. Sci. 2005;118:1773–1776. doi: 10.1242/jcs.01701. [DOI] [PubMed] [Google Scholar]
- 112.Baralle M., Skoko N., Knezevich A., De Conti L., Motti D., Baralle D., Buratti E., Baralle F.E. NF1 mRNA biogenesis: effects of the genomic milieu in splicing regulation of the NF1 exon 37 region. FEBS Lett., 2006 doi: 10.1016/j.febslet.2006.07.018. doi:10.1016/j.febslet.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 113.Vandenbroucke I., Callens T., De Paepe A., Messiaen L. Complex splicing pattern generates great diversity in human NF1 transcripts. BMC Genomics. 2002;3:13. doi: 10.1186/1471-2164-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Berget S.M. Exon recognition in vertebrate splicing. J. Biol. Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 115.Ast G. How did alternative splicing evolve? Nature Rev. Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- 116.Fox-Walsh K.L., Dou Y., Lam B.J., Hung S.P., Baldi P.F., Hertel K.J. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc. Natl Acad. Sci. USA. 2005;102:16176–16181. doi: 10.1073/pnas.0508489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mardon H.J., Sebastio G., Baralle F.E. A role for exon sequences in alternative splicing of the human fibronectin gene. Nucleic Acids Res. 1987;15:7725–7733. doi: 10.1093/nar/15.19.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jurica M.S., Moore M.J. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 119.Bourgeois C.F., Lejeune F., Stevenin J. Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:37–88. doi: 10.1016/S0079-6603(04)78002-2. [DOI] [PubMed] [Google Scholar]
- 120.Graveley B.R. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Blencowe B.J. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 2000;25:106–110. doi: 10.1016/s0968-0004(00)01549-8. [DOI] [PubMed] [Google Scholar]
- 122.Fu X.D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 123.Sanford J.R., Ellis J., Caceres J.F. Multiple roles of arginine/serine-rich splicing factors in RNA processing. Biochem. Soc. Trans. 2005;33:443–446. doi: 10.1042/BST0330443. [DOI] [PubMed] [Google Scholar]
- 124.Zhang Z., Krainer A.R. Involvement of SR proteins in mRNA surveillance. Mol. Cell. 2004;16:597–607. doi: 10.1016/j.molcel.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 125.Huang Y., Steitz J.A. SRprises along a messenger's journey. Mol. Cell. 2005;17:613–615. doi: 10.1016/j.molcel.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 126.Krecic A.M., Swanson M.S. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell. Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- 127.Dreyfuss G., Kim V.N., Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nature Rev. Mol. Cell. Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 128.Spellman R., Smith C.W. Novel modes of splicing repression by PTB. Trends. Biochem. Sci. 2006;31:73–76. doi: 10.1016/j.tibs.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 129.Pagani F., Buratti E., Stuani C., Romano M., Zuccato E., Niksic M., Giglio L., Faraguna D., Baralle F.E. Splicing factors induce cystic fibrosis transmembrane regulator exon 9 skipping through a non-evolutionary conserved intronic element. J. Biol. Chem. 2000;275:21041–21047. doi: 10.1074/jbc.M910165199. [DOI] [PubMed] [Google Scholar]
- 130.Wang Y., Wang J., Gao L., Lafyatis R., Stamm S., Andreadis A. Tau exons 2 and 10, which are misregulated in neurodegenerative diseases, are partly regulated by silencers which bind a SRp30c.SRp55 complex that either recruits or antagonizes htra2beta1. J. Biol. Chem. 2005;280:14230–14239. doi: 10.1074/jbc.M413846200. [DOI] [PubMed] [Google Scholar]
- 131.Kanopka A., Muhlemann O., Akusjarvi G. Inhibition by SR proteins of splicing of a regulated adenovirus pre- mRNA. Nature. 1996;381:535–538. doi: 10.1038/381535a0. [DOI] [PubMed] [Google Scholar]
- 132.Nilsen T.W. Too hot to splice. Nature Struct. Mol. Biol. 2004;11:208–209. doi: 10.1038/nsmb0304-208. [DOI] [PubMed] [Google Scholar]
- 133.Shin C., Feng Y., Manley J.L. Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature. 2004;427:553–558. doi: 10.1038/nature02288. [DOI] [PubMed] [Google Scholar]
- 134.Hui J., Stangl K., Lane W.S., Bindereif A. HnRNP L stimulates splicing of the eNOS gene by binding to variable-length CA repeats. Nature Struct. Biol. 2003;10:33–37. doi: 10.1038/nsb875. [DOI] [PubMed] [Google Scholar]
- 135.Martinez-Contreras R., Fisette J.F., Nasim F.U., Madden R., Cordeau M., Chabot B. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol. 2006;4:e21. doi: 10.1371/journal.pbio.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chen C.D., Kobayashi R., Helfman D.M. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat beta-tropomyosin gene. Genes Dev. 1999;13:593–606. doi: 10.1101/gad.13.5.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Liu H.X., Zhang M., Krainer A.R. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ladd A.N., Cooper T.A. Finding signals that regulate alternative splicing in the post-genomic era. Genome. Biol. 2002;3:reviews 0008. doi: 10.1186/gb-2002-3-11-reviews0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zheng Z.M. Regulation of alternative RNA splicing by exon definition and exon sequences in viral and mammalian gene expression. J. Biomed. Sci. 2004;11:278–294. doi: 10.1159/000077096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Abdul-Manan N., Williams K.R. hnRNP A1 binds promiscuously to oligoribonucleotides: utilization of random and homo-oligonucleotides to discriminate sequence from base- specific binding. Nucleic Acids Res. 1996;24:4063–4070. doi: 10.1093/nar/24.20.4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Buratti E., Muro A.F., Giombi M., Gherbassi D., Iaconcig A., Baralle F.E. RNA folding affects the recruitment of SR proteins by mouse and human polypurinic enhancer elements in the fibronectin EDA exon. Mol. Cell. Biol. 2004;24:1387–1400. doi: 10.1128/MCB.24.3.1387-1400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cartegni L., Wang J., Zhu Z., Zhang M.Q., Krainer A.R. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fairbrother W.G., Yeh R.F., Sharp P.A., Burge C.B. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 144.Fairbrother W.G., Yeo G.W., Yeh R., Goldstein P., Mawson M., Sharp P.A., Burge C.B. RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res. 2004;32:W187–W190. doi: 10.1093/nar/gkh393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nalla V.K., Rogan P.K. Automated splicing mutation analysis by information theory. Hum. Mutat. 2005;25:334–342. doi: 10.1002/humu.20151. [DOI] [PubMed] [Google Scholar]
- 146.Pagani F., Raponi M., Baralle F.E. Synonymous mutations in CFTR exon 12 affect splicing and are not neutral in evolution. Proc. Natl Acad. Sci. USA. 2005;102:6368–6372. doi: 10.1073/pnas.0502288102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pagani F., Buratti E., Stuani C., Baralle F.E. Missense, nonsense, and neutral mutations define juxtaposed regulatory elements of splicing in cystic fibrosis transmembrane regulator exon 9. J. Biol. Chem. 2003;278:26580–26588. doi: 10.1074/jbc.M212813200. [DOI] [PubMed] [Google Scholar]
- 148.Pagani F., Stuani C., Tzetis M., Kanavakis E., Efthymiadou A., Doudounakis S., Casals T., Baralle F.E. New type of disease causing mutations: the example of the composite exonic regulatory elements of splicing in CFTR exon 12. Hum. Mol. Genet. 2003;12:1111–1120. doi: 10.1093/hmg/ddg131. [DOI] [PubMed] [Google Scholar]
- 149.Cartegni L., Krainer A.R. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nature Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 150.Liu H.X., Cartegni L., Zhang M.Q., Krainer A.R. A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nature Genet. 2001;27:55–58. doi: 10.1038/83762. [DOI] [PubMed] [Google Scholar]
- 151.Sironi M., Cagliani R., Pozzoli U., Bardoni A., Comi G.P., Giorda R., Bresolin N. The dystrophin gene is alternatively spliced throughout its coding sequence. FEBS Lett. 2002;517:163–166. doi: 10.1016/s0014-5793(02)02613-3. [DOI] [PubMed] [Google Scholar]
- 152.O'Driscoll M., Ruiz-Perez V.L., Woods C.G., Jeggo P.A., Goodship J.A. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nature Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 153.Han K., Yeo G., An P., Burge C.B., Grabowski P.J. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 2005;3:e158. doi: 10.1371/journal.pbio.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kralovicova J., Vorechovsky I. Position-dependent repression and promotion of DQB1 intron 3 splicing by GGGG motifs. J. Immunol. 2006;176:2381–2388. doi: 10.4049/jimmunol.176.4.2381. [DOI] [PubMed] [Google Scholar]
- 155.Zhu J., Mayeda A., Krainer A.R. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell. 2001;8:1351–1361. doi: 10.1016/s1097-2765(01)00409-9. [DOI] [PubMed] [Google Scholar]
- 156.Eperon I.C., Makarova O.V., Mayeda A., Munroe S.H., Caceres J.F., Hayward D.G., Krainer A.R. Selection of alternative 5′ splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol. Cell. Biol. 2000;20:8303–8318. doi: 10.1128/mcb.20.22.8303-8318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sharma S., Falick A.M., Black D.L. Polypyrimidine tract binding protein blocks the 5′ splice site-dependent assembly of U2AF and the prespliceosomal E complex. Mol. Cell. 2005;19:485–496. doi: 10.1016/j.molcel.2005.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chou M.Y., Underwood J.G., Nikolic J., Luu M.H., Black D.L. Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol. Cell. 2000;5:949–957. doi: 10.1016/s1097-2765(00)80260-9. [DOI] [PubMed] [Google Scholar]
- 159.Matter N., Herrlich P., Konig H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 2002;420:691–695. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- 160.Matter N., Marx M., Weg-Remers S., Ponta H., Herrlich P., Konig H. Heterogeneous ribonucleoprotein A1 is part of an exon-specific splice-silencing complex controlled by oncogenic signaling pathways. J. Biol. Chem. 2000;275:35353–35360. doi: 10.1074/jbc.M004692200. [DOI] [PubMed] [Google Scholar]
- 161.Cheng C., Sharp P.A. Regulation of CD44 alternative splicing by SRm160 and its potential role in tumor cell invasion. Mol. Cell. Biol. 2006;26:362–370. doi: 10.1128/MCB.26.1.362-370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ladd A.N., Stenberg M.G., Swanson M.S., Cooper T.A. Dynamic balance between activation and repression regulates pre-mRNA alternative splicing during heart development. Dev. Dyn. 2005;233:783–793. doi: 10.1002/dvdy.20382. [DOI] [PubMed] [Google Scholar]
- 163.Venables J.P., Bourgeois C.F., Dalgliesh C., Kister L., Stevenin J., Elliott D.J. Up-regulation of the ubiquitous alternative splicing factor Tra2beta causes inclusion of a germ cell-specific exon. Hum. Mol. Genet. 2005;14:2289–2303. doi: 10.1093/hmg/ddi233. [DOI] [PubMed] [Google Scholar]
- 164.Cartegni L., Hastings M.L., Calarco J.A., de Stanchina E., Krainer A.R. Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am. J. Hum. Genet. 2006;78:63–77. doi: 10.1086/498853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kashima T., Manley J.L. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nature Genet. 2003;34:460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 166.Hofmann Y., Lorson C.L., Stamm S., Androphy E.J., Wirth B. Htra2-beta 1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2) Proc. Natl Acad. Sci. USA. 2000;97:9618–9623. doi: 10.1073/pnas.160181697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Miyaso H., Okumura M., Kondo S., Higashide S., Miyajima H., Imaizumi K. An intronic splicing enhancer element in survival motor neuron (SMN) pre-mRNA. J. Biol. Chem. 2003;278:15825–15831. doi: 10.1074/jbc.M209271200. [DOI] [PubMed] [Google Scholar]
- 168.Singh N.N., Androphy E.J., Singh R.N. In vivo selection reveals combinatorial controls that define a critical exon in the spinal muscular atrophy genes. RNA. 2004;10:1291–1305. doi: 10.1261/rna.7580704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yeo G., Burge C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 170.Reese M.G., Eeckman F.H., Kulp D., Haussler D. Improved splice site detection in Genie. J. Comput. Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 171.Rogozin I.B., Milanesi L. Analysis of donor splice sites in different eukaryotic organisms. J. Mol. Evol. 1997;45:50–59. doi: 10.1007/pl00006200. [DOI] [PubMed] [Google Scholar]
- 172.Zheng C.L., Fu X.D., Gribskov M. Characteristics and regulatory elements defining constitutive splicing and different modes of alternative splicing in human and mouse. RNA. 2005;11:1777–1787. doi: 10.1261/rna.2660805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Singh N.K., Singh N.N., Androphy E.J., Singh R.N. Splicing of a critical exon of human survival motor neuron is regulated by a unique silencer element located in the last intron. Mol. Cell. Biol. 2006;26:1333–1346. doi: 10.1128/MCB.26.4.1333-1346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Groman J.D., Hefferon T.W., Casals T., Bassas L., Estivill X., Des Georges M., Guittard C., Koudova M., Fallin M.D., Nemeth K., et al. Variation in a repeat sequence determines whether a common variant of the cystic fibrosis transmembrane conductance regulator gene is pathogenic or benign. Am. J. Hum. Genet. 2004;74:176–179. doi: 10.1086/381001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Buratti E., Dork T., Zuccato E., Pagani F., Romano M., Baralle F.E. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 2001;20:1774–1784. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Buratti E., Brindisi A., Giombi M., Tisminetzky S., Ayala Y.M., Baralle F.E. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J. Biol. Chem. 2005;280:37572–37584. doi: 10.1074/jbc.M505557200. [DOI] [PubMed] [Google Scholar]
- 177.Wang H.Y., Wang I.F., Bose J., Shen C.K. Structural diversity and functional implications of the eukaryotic TDP gene family. Genomics. 2004;83:130–139. doi: 10.1016/s0888-7543(03)00214-3. [DOI] [PubMed] [Google Scholar]
- 178.Zuccato E., Buratti E., Stuani C., Baralle F.E., Pagani F. An intronic polypyrimidine-rich element downstream of the donor site modulates cystic fibrosis transmembrane conductance regulator exon 9 alternative splicing. J. Biol. Chem. 2004;279:16980–16988. doi: 10.1074/jbc.M313439200. [DOI] [PubMed] [Google Scholar]
- 179.Ayala Y.M., Pagani F., Baralle F.E. TDP43 depletion rescues aberrant CFTR exon 9 skipping. FEBS Lett. 2006;580:1339–1344. doi: 10.1016/j.febslet.2006.01.052. [DOI] [PubMed] [Google Scholar]
- 180.Mercado P.A., Ayala Y.M., Romano M., Buratti E., Baralle F.E. Depletion of TDP 43 overrides the need for exonic and intronic splicing enhancers in the human apoA-II gene. Nucleic Acids Res. 2005;33:6000–6010. doi: 10.1093/nar/gki897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kishore S., Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 182.Tuffery-Giraud S., Saquet C., Chambert S., Claustres M. Pseudoexon activation in the DMD gene as a novel mechanism for Becker muscular dystrophy. Hum. Mutat. 2003;21:608–614. doi: 10.1002/humu.10214. [DOI] [PubMed] [Google Scholar]
- 183.Beroud C., Carrie A., Beldjord C., Deburgrave N., Llense S., Carelle N., Peccate C., Cuisset J.M., Pandit F., Carre-Pigeon F., et al. Dystrophinopathy caused by mid-intronic substitutions activating cryptic exons in the DMD gene. Neuromuscul. Disord. 2004;14:10–18. doi: 10.1016/s0960-8966(03)00169-x. [DOI] [PubMed] [Google Scholar]
- 184.Ikezawa M., Nishino I., Goto Y., Miike T., Nonaka I. Newly recognized exons induced by a splicing abnormality from an intronic mutation of the dystrophin gene resulting in Duchenne muscular dystrophy. Mutations. in. brief. no. 213. Online. Hum. Mutat. 1999;13:170. doi: 10.1002/(SICI)1098-1004(1999)13:2<170::AID-HUMU12>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 185.Chillon M., Dork T., Casals T., Gimenez J., Fonknechten N., Will K., Ramos D., Nunes V., Estivill X. A novel donor splice site in intron 11 of the CFTR gene, created by mutation 1811+1.6kbA→G, produces a new exon: high frequency in Spanish cystic fibrosis chromosomes and association with severe phenotype. Am. J. Hum. Genet. 1995;56:623–629. [PMC free article] [PubMed] [Google Scholar]
- 186.Highsmith W.E., Burch L.H., Zhou Z., Olsen J.C., Boat T.E., Spock A., Gorvoy J.D., Quittel L., Friedman K.J., Silverman L.M., et al. A novel mutation in the cystic fibrosis gene in patients with pulmonary disease but normal sweat chloride concentrations. N. Engl. J. Med. 1994;331:974–980. doi: 10.1056/NEJM199410133311503. [DOI] [PubMed] [Google Scholar]
- 187.Will K., Dork T., Stuhrmann M., Meitinger T., Bertele-Harms R., Tummler B., Schmidtke J. A novel exon in the cystic fibrosis transmembrane conductance regulator gene activated by the nonsense mutation E92X in airway epithelial cells of patients with cystic fibrosis. J. Clin. Invest. 1994;93:1852–1859. doi: 10.1172/JCI117172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Monnier N., Gout J.P., Pin I., Gauthier G., Lunardi J. A novel 3600+11.5 kb C>G homozygous splicing mutation in a black African, consanguineous CF family. J. Med. Genet. 2001;38:E4. doi: 10.1136/jmg.38.1.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Rump A., Rosen-Wolff A., Gahr M., Seidenberg J., Roos C., Walter L., Gunther V., Roesler J. A splice-supporting intronic mutation in the last bp position of a cryptic exon within intron 6 of the CYBB gene induces its incorporation into the mRNA causing chronic granulomatous disease (CGD) Gene. 2006;371:174–181. doi: 10.1016/j.gene.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 190.Noack D., Heyworth P.G., Newburger P.E., Cross A.R. An unusual intronic mutation in the CYBB gene giving rise to chronic granulomatous disease. Biochim. Biophys. Acta. 2001;1537:125–131. doi: 10.1016/s0925-4439(01)00065-5. [DOI] [PubMed] [Google Scholar]
- 191.Christie P.T., Harding B., Nesbit M.A., Whyte M.P., Thakker R.V. X-linked hypophosphatemia attributable to pseudoexons of the PHEX gene. J. Clin. Endocrinol. Metab. 2001;86:3840–3844. doi: 10.1210/jcem.86.8.7730. [DOI] [PubMed] [Google Scholar]
- 192.Balz V., Prisack H.B., Bier H., Bojar H. Analysis of BRCA1, TP53, and TSG101 germline mutations in German breast and/or ovarian cancer families. Cancer. Genet. Cytogenet. 2002;138:120–127. doi: 10.1016/s0165-4608(02)00601-5. [DOI] [PubMed] [Google Scholar]
- 193.Wang M., Dotzlaw H., Fuqua S.A., Murphy L.C. A point mutation in the human estrogen receptor gene is associated with the expression of an abnormal estrogen receptor mRNA containing a 69 novel nucleotide insertion. Breast Cancer Res. Treat. 1997;44:145–151. doi: 10.1023/a:1005753117205. [DOI] [PubMed] [Google Scholar]
- 194.van den Hurk J.A., van de Pol D.J., Wissinger B., van Driel M.A., Hoefsloot L.H., de Wijs I.J., van den Born L.I., Heckenlively J.R., Brunner H.G., Zrenner E., et al. Novel types of mutation in the choroideremia (CHM) gene: a full-length L1 insertion and an intronic mutation activating a cryptic exon. Hum. Genet. 2003;113:268–275. doi: 10.1007/s00439-003-0970-0. [DOI] [PubMed] [Google Scholar]
- 195.Metherell L.A., Akker S.A., Munroe P.B., Rose S.J., Caulfield M., Savage M.O., Chew S.L., Clark A.J. Pseudoexon activation as a novel mechanism for disease resulting in atypical growth-hormone insensitivity. Am. J. Hum. Genet. 2001;69:641–646. doi: 10.1086/323266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.McConville C.M., Stankovic T., Byrd P.J., McGuire G.M., Yao Q.Y., Lennox G.G., Taylor M.R. Mutations associated with variant phenotypes in ataxia-telangiectasia. Am. J. Hum. Genet. 1996;59:320–330. [PMC free article] [PubMed] [Google Scholar]
- 197.Vetrini F., Tammaro R., Bondanza S., Surace E.M., Auricchio A., De Luca M., Ballabio A., Marigo V. Aberrant splicing in the ocular albinism type 1 gene (OA1/GPR143) is corrected in vitro by morpholino antisense oligonucleotides. Hum. Mutat. 2006;27:420–426. doi: 10.1002/humu.20303. [DOI] [PubMed] [Google Scholar]
- 198.Raponi M., Upadhyaya M., Baralle D. Functional splicing assay shows a pathogenic intronic mutation in neurofibromatosis type 11 (NF1) due to intronic sequence exonization. Hum. Mutat. 2006;27:294–295. doi: 10.1002/humu.9412. [DOI] [PubMed] [Google Scholar]