Abstract
The evolution of marine microbes over billions of years predicts that the composition of microbial communities should be much greater than the published estimates of a few thousand distinct kinds of microbes per liter of seawater. By adopting a massively parallel tag sequencing strategy, we show that bacterial communities of deep water masses of the North Atlantic and diffuse flow hydrothermal vents are one to two orders of magnitude more complex than previously reported for any microbial environment. A relatively small number of different populations dominate all samples, but thousands of low-abundance populations account for most of the observed phylogenetic diversity. This “rare biosphere” is very ancient and may represent a nearly inexhaustible source of genomic innovation. Members of the rare biosphere are highly divergent from each other and, at different times in earth's history, may have had a profound impact on shaping planetary processes.
Keywords: biodiversity, low abundance, marine, microbes, rarefaction
The world's oceans are teeming with microscopic life forms. Nominal cell counts of >105 cells per ml in surface sea water (1, 2) predict that the oceans harbor 3.6 × 1029 microbial cells with a total cellular carbon content of ≈3 × 1017 g (3). Communities of bacteria, archaea, protists, and unicellular fungi account for most of the oceanic biomass. These microscopic factories are responsible for 98% of primary production (3, 4) and mediate all biogeochemical cycles in the oceans (4). Given the enormous number of microbes and their vast metabolic diversity, the accumulation of mutations during the past 3.5 billion years should have led to very high levels of genetic and phenotypic variation.
Direct interrogation of microbial genomes based on comparisons of orthologous gene sequences have shown that, in addition to enormous phylogenetic diversity, the complexity of microbial life (the number of different kinds or “species” of microbes) is at least 100 times greater than estimates based on cultivation-dependent surveys (5). With each new survey, this window on the microbial world increases in size. There have been spectacular discoveries of previously unknown microorganisms, many of which have major impacts on oceanic processes (6–8). At the genomic level, comparisons of chromosomal size sequences from cultivars, marine microbial metagenomic analyses of bacterial artificial chromosomes (9), and shotgun small-insert libraries (10) reveal unanticipated levels of metabolic diversity and extensive horizontal gene transfer. Recurrent discoveries of novel genetic information suggest that cryptic “genetic reservoirs” reshape genomic architecture through lateral gene transfer processes (11). There is evidence of hitherto unrecognized physiological groups among the planktonic microbes (12, 13). Two inescapable conclusions emerge from these phylogenetic, genomic, and metagenomic analyses: (i) microbes account for the majority of genetic and metabolic variation in the oceans and (ii) the genetic diversity, community composition, relative abundance, and distribution of microbes in the sea remain undersampled and essentially uncharted.
Gene sequences, most commonly those encoding rRNAs, provide a basis for estimating microbial phylogenetic diversity (5, 7, 14–18) and generating taxonomic inventories of marine microbial populations (5, 7, 14–18). Evolutionary distances between orthologous sequences (19) or similarities to database entries identified through BLAST (20), FASTA (21), or Bayesian classifiers (22) identify operational taxonomic units (OTUs) that correspond to species or kinds of organisms. A variety of parametric and nonparametric methods extrapolate information from observed frequencies of OTUs or species abundance curves to predict the number of different microbial taxa in a local sample (23–26). Richness estimates of marine microbial communities through comparisons of rRNAs range from a few hundred phylotypes per ml in the water column (19) to as many as 3,000 from marine sediments (27, 28). One of the largest water column surveys (1,000 PCR amplicons) described the presence of only 516 unique sequences and estimated occurrence of ≈1,600 coexisting ribotypes in a coastal bacterioplankton community (29). Using data from metagenomic surveys of the Sargasso Sea, nonparametric treatments of rRNA sequences from marine systems argue that the oceans might contain as many as 106 different kinds of microbes (26). Yet, all of these inferences suffer from a paucity of data points (a small number of homologous sequences used to document the presence of individual microbes in a sample) relative to the very large number of organisms (generally 105 to 106 per ml) in oceanic waters. The detection of organisms that correspond to the most abundant OTUs requires minimal sampling, whereas the recovery of sequences from minor components (those present only a few times in a liter of seawater) demands surveys that are many orders of magnitude larger than those reported in the literature.
Insufficient detail about the relative numbers of individuals that represent both major and minor populations constrains the accuracy of both log-normal distribution and nonparametric estimators of taxonomic richness for microbial communities. This information is necessary for meaningful comparisons between community compositions from different environments. Recognizing the undersampled nature of single-cell organisms in the sea, the International Census of Marine Microbes has mounted an effort to increase the efficiency of molecular-based surveys of microbial taxa in both open ocean waters and the benthos. As an alternative to analyzing sequences of nearly full-length PCR amplicons of homologous genes from environmental DNA samples, sequence tags (30) from hypervariable regions in rRNAs can provide measures of richness and relative abundance for OTUs in microbial communities. This “tag sequencing” strategy is analogous to the Bar Code of Life initiative (31), which relies on evolutionary similarities and differences between mitochondrial-encoded cytochrome oxidases to distinguish closely related genera and species. Nearly unique rRNA tag sequences correspond to individual OTUs. Enumerating the number of different rRNA tags provides a first-order description of the relative occurrence of specific microbes in a sample. The highly variable nature of the tag sequences and paucity of positions do not allow direct inference of phylogenetic frameworks. However, when tag sequences serve as a query against a comprehensive reference database of hypervariable regions within the context of full-length rRNA sequences of known phylotypes, it is possible to extract information about taxonomic identity and previously undocumented microbial diversity.
Results and Discussion
To develop a global, in-depth description of the diversity of microbes and their relative abundance in the sea, we exploited the massively parallel DNA sequencing capacity of 454 Life Sciences (Branford, CT) technology (32) to economically increase the number of sampled PCR amplicons in environmental surveys by orders of magnitude. We sequenced ≈118,000 PCR amplicons that span the V6 hypervariable region of ribosomal RNAs from environmental DNA preparations. We examined bacterial community compositions for six paired samples (sample pairs collected at the same coordinates but from different depths) from the meso- and bathypelagic realms at different locations in the North Atlantic Deep Water loop of the ocean conveyor belt and two samples of basalt-hosted diffuse hydrothermal vent fluids collected from the 1998 eruption zone of Axial Seamount (latitude, 45°58′N; longitude, −130°00′W), an active submarine volcano located on the Juan de Fuca Ridge in the northeast Pacific Ocean (Table 1). The number of reads per sample ranged from 6,505 to nearly 23,000 sequences (Table 2). To minimize effects of random sequencing errors, we used a systematic trimming procedure to eliminate sequences with multiple undetermined residues or mismatches to the PCR primers at the beginning of a read. On average, this stringent trimming procedure reduced the size of a data set by 24%. This approach is a conservative one that asserts that sequences with multiple undetermined residues or incorrect primers will display higher random error rates.
Table 1.
Environmental DNA samples used for sequence tag analyses (collected from North Atlantic Deep Water and Axial Seamount, Juan de Fuca Ridge)
| Sample ID | Site | Lat °N, Long °W | Depth, m | Temperature, °C | Cells per ml of water |
|---|---|---|---|---|---|
| 53R | Labrador seawater | 58.300, −29.133 | 1,400 | 3.5 | 6.4 × 104 |
| 55R | Oxygen minimum | 58.300, −29.133 | 500 | 7.1 | 1.8 × 105 |
| 112R | Lower deep water | 50.400, −25.000 | 4,121 | 2.3 | 3.9 × 104 |
| 115R | Oxygen minimum | 50.400, −25.000 | 550 | 7 | 1.5 × 105 |
| 137 | Labrador seawater | 60.900, −38.516 | 1,710 | 3 | 3.3 × 104 |
| 138 | Labrador seawater | 60.900, −38.516 | 710 | 3.5 | 5.2 × 104 |
| FS312 | Bag City | 45.916, −129.983 | 1,529 | 31.2 | 1.2 × 105 |
| FS396 | Marker 52 | 45.943, −129.985 | 1,537 | 24.4 | 1.6 × 105 |
Lat, latitude; Long, longitude.
Table 2.
Data summary and phylotype OTUs
| Sample ID | Total reads | Trimmed tags | Unique tags | OTUs |
|---|---|---|---|---|
| 53R | 6,505 | 5,000 | 2,656 | 1,184 |
| 55R | 18,439 | 13,902 | 7,187 | 2,555 |
| 112R | 12,916 | 9,282 | 5,752 | 2,135 |
| 115R | 14,731 | 11,005 | 5,777 | 2,049 |
| 137 | 18,137 | 13,907 | 6,752 | 2,480 |
| 138 | 18,451 | 14,374 | 7,168 | 2,550 |
| FS312 | 6,605 | 4,835 | 2,769 | 1,362 |
| FS396 | 22,994 | 17,666 | 8,699 | 3,290 |
Value under trimmed tags are the numbers of reads remaining after the removal of primers and low-quality data. Values under unique tags are the numbers of distinct sequences within a set of trimmed tags. OTUs were calculated by comparing each tag to the V6RefDB database and combining tags with the nearest identical reference.
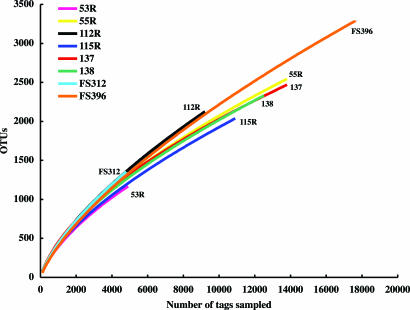
To assess taxonomic diversity, each trimmed 454 read (tag sequence) served as a query to identify its closest match in a reference database (V6RefDB) of ≈40,000 unique V6 sequences extracted from the nearly 120,000 published rRNA genes for the bacteria domain (22, 33–35). In most cases, the information content of V6 sequences is sufficient to identify phylogenetic affinity with full-length sequences in a reference database. Within the 120,000 published rRNA genes, 99.3% of V6 sequences that occur two or more times correctly identify the major bacterial phyla for each query. Random sampling of 5 × 106 (of a possible 1.44 × 1010) pairwise distances for V6 and full-length sequences shows that the differences between full-length sequences containing identical V6 hypervariable regions ranges from 0% to 5% for >90% of the random comparisons. Table 2 shows that there are a very large number of tags from each sampling site that we define as separate OTUs according to their matches to different sequences in the V6RefDB database. Fig. 1 is a rarefaction analysis based on best matches for each tag to sequences in V6RefDB and their frequency of recovery. These rarefaction curves describe unprecedented levels of bacterial complexity for marine samples, yet none has reached the curvilinear or plateau phase. The likelihood that they represent underestimates of the number of different kinds of bacteria in each sample is supported by observations of significant variation among tags with closest matches to the same sequence in V6RefDB. For example, the analysis of sample FS396 identified 4,288 tag sequences that most closely matched the V6 from the epsilon-proteobacterium Wolinella spp., but multiple alignment of these sequences reveals 45 clusters that are minimally 10% divergent from each other.
Fig. 1.
Rarefaction analysis for each sample based on best matches against the V6RefDB database. The frequency of observed best matches to V6RefDB (OTUs) for each site was used to calculate rarefaction curves with the program Analytic Rarefaction 1.3.
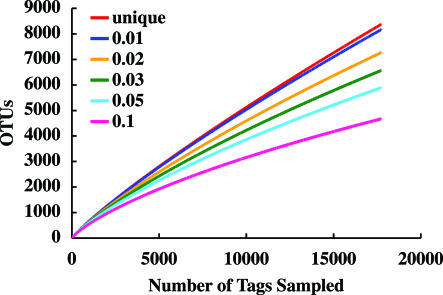
As an alternative to defining OTUs by their best matches against V6RefDB, we clustered sequence tags into groups of defined sequence variation that ranged from unique sequences (no variation) to 10% differences by using DOTUR (19). These clusters served as OTUs for generating rarefaction curves and for making calculations with the abundance-based coverage estimator ACE (24, 25) and the Chao1 (23) estimator of species diversity. Table 3 shows that the species diversity estimates obtained with ACE and Chao1 are at least an order of magnitude greater than published analyses of any microbial community. The rarefaction curves in Fig. 2 are from the analysis of the diffuse flow sample FS396. Even when relatively large genetic distances (5% or 10% difference) define similarity groups, the rarefaction curves predict that additional sampling will lead to significantly increased estimates of total diversity.
Table 3.
Similarity-based OTUs and species richness estimates
| Sample ID | Reads | Cluster distance |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0.03 |
0.05 |
0.10 |
||||||||
| OTU | ACE | Chao1 | OTU | ACE | Chao1 | OTU | ACE | Chao1 | ||
| 53R | 5,000 | 1,946 | 7,247 | 6,997 | 1,732 | 5,616 | 5,288 | 1,316 | 3,351 | 3,018 |
| 55R | 13,902 | 5,266 | 19,235 | 18,191 | 4,673 | 14,959 | 14,209 | 3,644 | 9,618 | 9,080 |
| 112R | 9,282 | 4,241 | 16,002 | 13,772 | 3,770 | 12,341 | 10,870 | 2,958 | 8,011 | 7,108 |
| 115R | 11,005 | 4,000 | 12,767 | 11,296 | 3,413 | 9,189 | 8,585 | 2,457 | 5,553 | 5,448 |
| 137 | 13,907 | 4,554 | 13,698 | 11,991 | 3,866 | 9,734 | 8,705 | 2,708 | 5,353 | 5,181 |
| 138 | 14,374 | 5,136 | 18,656 | 16,600 | 4,508 | 13,852 | 12,424 | 3,293 | 7,596 | 6,941 |
| FS312 | 4,835 | 1,941 | 5,599 | 5,482 | 1,681 | 4,233 | 4,080 | 1,227 | 2,466 | 2,346 |
| FS396 | 17,666 | 6,326 | 23,315 | 20,949 | 5,573 | 18,003 | 16,889 | 4,291 | 11,520 | 10,567 |
The species richness estimates were determined by using the program DOTUR as described in Methods.
Fig. 2.
Rarefaction analysis for sample FS396 based on pairwise distance. Rarefaction is shown for OTUs that contain unique sequences and OTUs with differences that do not exceed 1%, 2%, 3%, 5%, or 10%. Rarefaction of the other seven samples showed curves with similar slopes.
Our analyses of tag sequence composition both by querying V6RefDB and by cluster analyses show that bacterial diversity estimates for the diffuse flow vents of Axial Seamount and the deep water masses of the North Atlantic are much greater than any published description of marine microbial diversity. An elevated rate of random sequencing errors is an unlikely explanation for observations of high diversity in these tag sequencing studies. The recovery of the predicted sequence of the proximal PCR primer is better than 99.5% for the 118,000 unprocessed reads. A second estimate of sequencing error comes from comparisons of sequences that are similar but not identical to tags that occur at high frequency as exact matches to each other. Clusters that contained >10 identical sequence tags were assumed to represent real low-frequency biological variants; clusters with <10 tags were assumed to have occurred from random sequencing error. For the combined data set of processed tags from all samples, a perfectly identical tag sequence occurred 6,550 times. It corresponds to the nearly full-length rRNA sequence from an uncultured marine delta-proteobacterium (GenBank accession no. AF355040). Comparison of this tag to sequences that are no more than 10% divergent identified 140 tags with only 162 differences from the high-frequency sequence over a total of 388,020 positions, indicating an accuracy rate of 99.96%.
The extraordinary estimates of diversity for the tag sequences more likely reflect the small size of the PCR amplicons (<120 bp) and sequencing protocols that do not require construction of recombinant libraries. Analyses of different size PCR products by conventional cloning and capillary sequencing strategies reveal an inverse relationship between the size of the PCR amplicons and the diversity of recovered sequences (data not shown). With smaller PCR targets, the polymerases will encounter fewer higher order structures that might retard procession along the template. A difference in PCR efficiency of <15% will lead to an ≈100-fold difference in the representation of templates in an amplicon library after 30 cycles. By eliminating this bias, the relative number of amplicons will more accurately reflect the proportional abundance of different organisms in a microbial community.
The relative abundance of different OTUs in the tag sequence data set varies by more than three orders of magnitude. Inferences of the taxonomic affinity for each tag according to its best match in V6RefDB reveal a small number of dominant bacterial populations in samples from the diffuse flow and deep waters of the North Atlantic. For example, although there is a high diversity of both indigenous subseafloor and deep-sea microbes in the diffuse fluids, nearly 50% of the population in FS396 corresponds to divergent epsilon-proteobacteria, a group known to have a widespread distribution and dominance in most deep-sea vent habitats (36–38). Within the epsilon-proteobacteria, 125 different taxa represent anywhere from 0.1% to as much as 15% of the phylotypes. Epsilon-proteobacteria were only recently cultured from hydrothermal vents, and those isolates characterized are either mesophilic or moderately thermophilic sulfur reducers that grow autotrophically with hydrogen under anaerobic conditions and many use the reverse tricarboxylic acid cycle to fix carbon (36, 39–41). These organisms likely serve as an important member of the microbial community in both the shallow and deep subseafloor biosphere. In the deep waters of the North Atlantic, the major populations (alpha-, gamma-, and delta-proteobacteria) are similar but not identical, and these major populations in turn are markedly different from bacterial populations in the diffuse water samples.
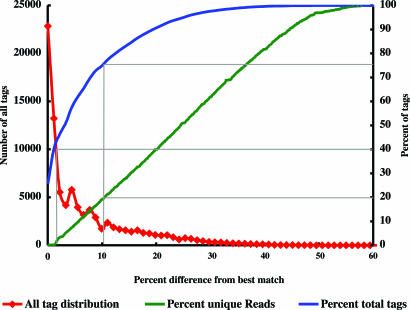
Underlying the major populations are broad distributions of distinct bacterial taxa that represent extraordinary diversity. Fig. 3 plots the percentage difference of “total tags” and “unique tags” to their best match in V6RefDB. For the collection of total tags, 25% are identical to a sequence in our V6 reference database, 40% are no more than 3% different, and ≈75% are no more than 10% different from a sequence in the reference database. On the other hand, analyses of “unique reads” reveal that 80% are >10% different from a closest match in V6RefDB, and, as indicated by the distribution of “all tags,” the most divergent tag sequences represent very low-abundance OTUs. The range of sequence variation between the low-abundance OTUs is comparable to their percentage differences with V6RefDB. Based on their apparent low levels of sequence similarity, these tags most likely represent microbial lineages that have been evolving over extended evolutionary time scales. The detection and enumeration of these low-abundance OTUs requires the sampling of many more PCR amplicons than can be economically achieved by using conventional cloning and capillary sequencing technology. At the same time, the 454 tag sequence strategy circumvents potential bias that cloning procedures might introduce. It is extendable to archaea and protists, offering the microbial oceanography community an opportunity to capture data about the numbers and kinds of microbes in all provinces of the oceans.
Fig. 3.
Similarity of 454 sequence tags from FS396 to the V6RefDB database. “All tag distribution” plots the number of tag sequences for all samples versus the percentage difference from the best-matching sequence in V6RefDB. “Percent unique reads” from all samples shows the percentage difference between each distinct tag sequence and its best match in V6RefDB. “Percent total tags” plots the cumulative percentage of reads in all samples at or below a given percentage difference from best matches in V6RefDB.
Microbial diversity in the oceans (and likely elsewhere) is much greater than previous estimates that are based on conventional molecular techniques. In traditional molecular studies, dominant populations have masked the detection of low-abundance OTUs, their overwhelming genetic diversity, and their individual distribution patterns in marine environments. The large number of highly diverse, low-abundance OTUs constitutes a “rare biosphere” that is largely unexplored. Some of its members might serve as keystone species within complex consortia; others might simply be the products of historical ecological change with the potential to become dominant in response to shifts in environmental conditions (e.g., when local or global change favors their growth). Because we know so little about the global distribution of members of the rare biosphere, it is not yet possible to know whether they represent specific biogeographical distributions of bacterial taxa, functional selection by particular marine environments, or cosmopolitan distribution of all microbial taxa (the “everything is everywhere” hypothesis).
The rare biosphere has temporal and spatial dimensions that impact our perceptions of known microbial diversity. Low-abundance populations at a sample site might eventually become dominant in response to environmental change. On the other hand, at a single point in time, dominant populations at one site can correspond to low-abundance populations at a second site. The large diversity of low-abundance taxa relative to V6RefDB (Fig. 3) reflects the sparse distribution in nature of microbial populations that constitute the rare biosphere. For distinct environmental conditions (the North Atlantic Deep Waters versus diffuse flows), new dominant populations emerge from the rare biosphere. Differences in major populations under dissimilar biogeochemical regimes are hardly unexpected, but the idea that underrepresented populations define such enormous diversity with potential to take over a particular ecological niche has profound implications. Several ecological models that account for frequency-dependent mechanisms predict a survival advantage for rare species, which are less prone to predation and direct competition with dominant community members. The concept of a rare biosphere forces us to rethink the potential feedback mechanisms between shifts in extremely complex microbial populations and how the genomes of their constituents change over evolutionary time scales. The rare biosphere may serve as a potentially inexhaustible reservoir of genomic innovation, which could explain how microbial communities recover from environmental catastrophe and why every previously uncharacterized microbial genome offers so much genetic novelty even when compared with closely related taxa.
By necessity, microbial oceanographers have focused their efforts on dominant components of microbial communities that mediate biogeochemical processes. What they have not tackled are very low-abundance members of microbial populations. The extreme phylogenetic diversity of the rare biosphere suggests these minor populations have persisted over geological time scales and that they may episodically reshape planetary processes.
Methods
Study Sites and Sample Collection.
The TRANSAT-1 and TRANSAT-2 cruises (TRANSAT-1, September 2002 to October 2002; TRANSAT-2, May 2003 to June 2003) collected a total of 344 samples from different depths (80–4,500 m) and locations (latitude, 63°N to 35°N) in the oceanic conveyor belt following the western and eastern branches of the North Atlantic Deep Water from near its source of formation in the Greenland–Iceland Norwegian Sea to the Azores and Bermuda and deep water masses of the North Atlantic. Samples were collected by filtering 1 liter of water onto 0.2-μm HCl-rinsed polycarbonate filters, shock-freezing in liquid nitrogen, and storing at −80°C until processing in the laboratory.
The National Oceanic and Atmospheric Administration New Millennium Observatory Program collected basalt-hosted diffuse hydrothermal vent fluids from the 1998 eruption zone of Axial Seamount on the Juan de Fuca Ridge in the northeast Pacific Ocean. Using the remotely operated vehicle ROPOS, fluids were collected in 2003 from Bag City Vent (FS312) and in 2004 from Marker 52 (FS396) at Axial Seamount. A complete description of the study site and the diffuse vents along the eruptive fissure can be found in ref. 42. Filtered and unfiltered fluids were sampled by using the hydrothermal fluid and particle sampler. Once a stable temperature was reached on the intake probe, fluids were pumped through a Sterivex-GP filter (0.22-μm pore size; Millipore, Billerica, MA), and the temperature and volume of the fluid collected were monitored throughout the 10- to 20-min sampling time required to obtain 1 liter (FS312) or 2 liters (FS396) of fluid. All filters for DNA extraction were placed in 50-ml sterile Falcon tubes and stored at −80°C.
DNA Extraction.
DNA was extracted according to the method described in ref. 16 with minor modifications. These modifications included the addition of phenol to the chloroform:isoamyl alcohol step (25:24:1), the addition of lysozyme (200 μl; 50 mg/ml) once along with SDS and proteinase K, and resuspension of the DNA in 95 μl of TE buffer (10 mM Tris/1 mM EDTA, pH 8.0). DNA was visualized by using pulse field gel electrophoresis and quantified both spectrophotometrically and with PicoGreen (Molecular Probes, Carlsbad, CA).
PCR Amplicon Library Construction and Sequencing of Environmental Samples.
We used the ARB database program (34) to design PCR primers that flank the V6 hypervariable region of bacterial 16S rRNAs (Escherichia coli positions 967-1046). The oligonucleotide design included 454 Life Science's A or B sequencing adapter (shown in lowercase in the following) fused to the 5′ end of primer A-967F, 5′-gcctccctcgcgccatcag-CAACGCGAAGAACCTTACC-3′, and B-1046R, 5′-gccttgccagcccgctcag-CGACAGCCATGCANCACCT-3′. We generated PCR amplicon libraries for each environmental DNA sample. The amplification mix contained 5 units of Pfu Turbo polymerase (Stratagene, La Jolla, CA), 1× Pfu reaction buffer, 200 μM dNTPs (Pierce Nucleic Acid Technologies, Milwaukee, WI), and a 0.2 μM concentration of each primer in a volume of 100 μl. Genomic DNA (3–10 ng) was added to three separate 30-μl amplification mixes. Cycling conditions were an initial denaturation at 94°C for 3 min; 30 cycles of 94°C 30 s, 57°C for 45 s, and 72°C for 1 min; and a final 2-min extension at 72°C. The products were pooled after cycling and cleaned by using the MinElute PCR purification kit (Qiagen, Valencia, CA). The quality of the product was assessed on a Bioanalyzer 2100 (Agilent, Palo Alto, CA) using a DNA1000 LabChip. Only sharp, distinct amplification products with a total yield of >200 ng were used for 454 sequencing. The fragments in the amplicon libraries were bound to beads under conditions that favor one fragment per bead. The beads were emulsified in a PCR mixture in oil, and PCR amplification occurred in each droplet, generating ≈10 million copies of a unique DNA template. After breaking the emulsion, the DNA strands were denatured, and beads carrying single-stranded DNA clones were deposited into wells on a PicoTiterPlate (454 Life Sciences) for pyrosequencing on a Genome Sequencer 20 system (Roche, Basel, Switzerland) at 454 Life Sciences (Branford, CT). By masking the 454 Life Sciences PicoTiterPlate (where the pyrosequencing occurs) into 16 zones, we recovered 118,778 sequence reads that represented forward and reverse reads of the eight sample sites.
Removal of Low-Quality Sequence Tags.
To minimize effects of random sequencing errors, we eliminated (i) sequences that did not perfectly match the PCR primer at the beginning of a read, (ii) sequence reads with <50 bp after the proximal PCR primer if they terminated before reaching the distal primer, and (iii) sequences that contained more than one undetermined nucleotide (N). We included only the first 100 bp after the proximal PCR primer, because 454 DNA pyrosequencing quality degrades beyond this point. The procedure used a combination of BLASTN (20) (with nondefault parameters −q −1 and G 1) and the EMBOSS program fuzznuc (43) (with nondefault parameter − mismatch = 3) to identify complete, mismatched, and partial distal primers that arise from early sequence termination. Both the proximal and distal primers were trimmed from high-quality reads before database searches and similarity calculations.
Construction of V6RefDB and Assignment of Phylotype OTUs.
To assign phylotypes to the 454 tag sequences, we built a reference database of 44,011 nonidentical V6 sequences extracted from 119,480 bacterial rRNAs derived from multiple sources (22, 34, 35) and curated unannotated environmental RNA sequences by using the Ribosomal Database Project II's (22) classifier (22). Each unique tag sequence serves as a BLAST query to the reference database. Because BLAST uses a local alignment algorithm, we collected the top BLAST hits (to a maximum of 250) and aligned them with the query sequence by using the program MUSCLE (44) (with parameters −diags and −maxiters 2). We have empirically determined that two iterations are sufficient to identify high-quality alignments for sequences that are no more than 10% divergent. Phylotype assignments are made according to the V6 reference sequence(s) that display the minimum distance to the query. The frequency of observed best matches to V6RefDB (OTUs) for each site was used to calculate rarefaction curves with the program Analytic Rarefaction 1.3.
Assignment of Similarity-Based OTUs and Species Richness Estimators.
Before distance-based analyses, we used MUSCLE as above to align all of the tag sequences from a single sample. To calculate many thousands of pairwise distances (where pairwise distance equals mismatches, including indels, divided by sequence length) we developed a program called quickdist, based on code modified from QuickTree (45). To avoid overestimating distances between 454 tag sequences from the rapidly diverging variable regions, we ignore terminal gaps and treat gaps of any length as a single evolutionary event or mismatch. These pairwise distances served as input to DOTUR (19) for clustering tags into OTUs, generating rarefaction curves, and calculating the species richness estimator ACE and Chao1 values by using sampling without replacement, a parameter of 100 for precision of distance, jumbled input order, and 50,000 iterations.
Identification of Closely Related Tag Sequence Clusters for Estimating Sequencing Error Rates.
We identified the set of most frequently occurring identical tags from FS396 (994 sequences) and from all samples combined (6,550 sequences) by using the unix shell commands “sort ∣ uniq −c ∣ sort −nr −k 1.” We identified other tags that were no more than 15% divergent from the collection of 994 or 6,550 identical tags by using the shell command “agrep −8” and assembled all divergence tags into clusters with at least 90% identity by using Sequencher (Gene Codes, Ann Arbor, MI).
Data Availability.
The supplemental.zip file available at http://jbpc.mbl.edu/research_supplements/g454/20060412-private provides access to unprocessed and trimmed tag sequence files for each sample. The V6RefDB database is also available at this web site.
Acknowledgments
We thank the captain and crew of R/V Pelagia for their help during work at sea in the North Atlantic; David Butterfield, Sheryl Bolton, the remotely operated vehicle ROPOS, and the National Oceanic and Atmospheric Administration/Pacific Marine Environmental Laboratory Vents Program for the collection of diffuse flow samples; and Steven Holland for providing access to the program Analytic Rarefaction 1.3. This work was supported by National Aeronautics and Space Administration Astrobiology Institute Cooperative Agreement NNA04CC04A (to M.L.S.), a National Research Council Astrobiology Research Associateship Award (to J.A.H.), the Alfred P. Sloan Foundation's ICoMM field project, and subcontracts from the Woods Hole Center for Oceans and Human Health from the National Institutes of Health and National Science Foundation (NIH/NIEHS 1 P50 ES012742-01 and NSF/OCE 0430724-J. Stegeman PI to H.G.M. and M.L.S.). Support for the TRANSAT cruises was provided by a grant from the Earth and Life Science Division of the Dutch Science Foundation (Netherlands Organization for Scientific Research Project 811.33.004) to G.J.H.
Abbreviation
- OTU
operational taxonomic unit
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (trace archive accession nos. TI1369949070–TI1370073576).
References
- 1.Porter K. G., Feig Y. S. Limnol. Oceanogr. 1980;25:943–948. [Google Scholar]
- 2.Hobbie J. E., Daley R. J., Jasper S. Appl. Environ. Microbiol. 1977;33:1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitman W. B., Coleman D. C., Wiebe W. J. Proc. Natl. Acad. Sci. USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atlas R. M., Bartha R. Microbial Ecology: Fundamentals and Applications. Redwood City, CA: Benjamin/Cummings; 1993. [Google Scholar]
- 5.Pace N. R. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 6.Fuhrman J. A., McCallum K., Davis A. A. Appl. Environ. Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giovannoni S. J., Britschgi T. B., Moyer C. L., Field K. G. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 8.Hallam S. J., Mincer T. J., Schleper C., Preston C. M., Roberts K., Richardson P. M., Delong E. F. PLoS Biol. 2006;4:e95. doi: 10.1371/journal.pbio.0040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Béjà O., Suzuki M. T., Koonin E. V., Aravind L., Hadd A., Nguyen L. P., Vilacorta R., Amjadi M., Garrigues C., Jovanovich S. B., et al. Environ. Microbiol. 2000;2:516–529. doi: 10.1046/j.1462-2920.2000.00133.x. [DOI] [PubMed] [Google Scholar]
- 10.Tringe S. G., von Mering C., Kobayashi A., Salamov A. A., Chen K., Chang H. W., Podar M., Short J. M., Mathur E. J., Detter J. C., et al. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 11.Frigaard N. U., Martinez A., Mincer T. J., DeLong E. F. Nature. 2006;439:847–850. doi: 10.1038/nature04435. [DOI] [PubMed] [Google Scholar]
- 12.DeLong E. F., Preston C. M., Mincer T., Rich V., Hallam S. J., Frigaard N. U., Martinez A., Sullivan M. B., Edwards R., Brito B. R., et al. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 13.Venter J. C., Remington K., Heidelberg J. F., Halpern A. L., Rusch D., Eisen J. A., Wu D., Paulsen I., Nelson K. E., Nelson W., et al. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 14.DeLong E. F. Proc. Natl. Acad. Sci. USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hewson I., Vargo G. A., Fuhrman J. A. Microb. Ecol. 2003;46:322–336. doi: 10.1007/s00248-002-1067-3. [DOI] [PubMed] [Google Scholar]
- 16.Huber J. A., Butterfield D. A., Baross J. A. Appl. Environ. Microbiol. 2002;68:1585–1594. doi: 10.1128/AEM.68.4.1585-1594.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knittel K., Losekann T., Boetius A., Kort R., Amann R. Appl. Environ. Microbiol. 2005;71:467–479. doi: 10.1128/AEM.71.1.467-479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rappe M. S., Giovannoni S. J. Annu. Rev. Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 19.Schloss P. D., Handelsman J. Appl. Environ. Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearson W. R. Methods Mol. Biol. 1994;25:365–389. doi: 10.1385/0-89603-276-0:365. [DOI] [PubMed] [Google Scholar]
- 22.Cole J. R., Chai B., Farris R. J., Wang Q., Kulam S. A., McGarrell D. M., Garrity G. M., Tiedje J. M. Nucleic Acids Res. 2005;33:D294–D296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chao A. Scand. J. Stat. 1984;11:265–270. [Google Scholar]
- 24.Chao A. J. Am. Stat. Assoc. 1992;87:210–217. [Google Scholar]
- 25.Chao A., Ma M. C., Yang K. Biometrika. 1993;80:193–201. [Google Scholar]
- 26.Curtis T. P., Sloan W. T., Scannell J. W. Proc. Natl. Acad. Sci. USA. 2002;99:10494–10499. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kemp P. F., Aller J. Y. GEMS Microb. Ecol. 2004;47:161–177. doi: 10.1016/S0168-6496(03)00257-5. [DOI] [PubMed] [Google Scholar]
- 28.Ravenschlag K., Sahm K., Pernthaler J., Amann R. Appl. Environ. Microbiol. 1999;65:3982–3989. doi: 10.1128/aem.65.9.3982-3989.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Acinas S. G., Klepac-Ceraj V., Hunt D. E., Pharino C., Ceraj I., Distel D. L., Polz M. F. Nature. 2004;430:551–554. doi: 10.1038/nature02649. [DOI] [PubMed] [Google Scholar]
- 30.Kysela D. T., Palacios C., Sogin M. L. Environ. Microbiol. 2005;7:356–364. doi: 10.1111/j.1462-2920.2004.00712.x. [DOI] [PubMed] [Google Scholar]
- 31.Hebert P. D., Cywinska A., Ball S. L., deWaard J. R. Proc. Biol. Sci.; 2003. pp. 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Margulies M., Egholm M., Altman W. E., Attiya S., Bader J. S., Bemben L. A., Berka J., Braverman M. S., Chen Y. J., Chen Z., et al. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeSantis T. Z., Dubosarskiy I., Murray S. R., Andersen G. L. Bioinformatics. 2003;19:1461–1468. doi: 10.1093/bioinformatics/btg200. [DOI] [PubMed] [Google Scholar]
- 34.Ludwig W., Strunk O., Westram R., Richter L., Meier H., Yadhukumar, Buchner A., Lai T., Steppi S., Jobb G., et al. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wheeler D. L., Barrett T., Benson D. A., Bryant S. H., Canese K., Chetvernin V., Church D. M., DiCuccio M., Edgar R., Federhen S., et al. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campbell B. J., Jeanthon C., Kostka J. E., Luther G. W., III, Cary S. C. Appl. Environ. Microbiol. 2001;67:4566–4572. doi: 10.1128/AEM.67.10.4566-4572.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huber J. A., Butterfield D. A., Baross J. A. FEMS Microbiol. Ecol. 2003;43:393–409. doi: 10.1111/j.1574-6941.2003.tb01080.x. [DOI] [PubMed] [Google Scholar]
- 38.Reysenbach A. L., Longnecker K., Kirshtein J. Appl. Environ. Microbiol. 2000;66:3798–3806. doi: 10.1128/aem.66.9.3798-3806.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alain K., Querellou J., Lesongeur F., Pignet P., Crassous P., Raguenes G., Cueff V., Cambon-Bonavita M. A. Int. J. Syst. Evol. Microbiol. 2002;52:1317–1323. doi: 10.1099/00207713-52-4-1317. [DOI] [PubMed] [Google Scholar]
- 40.Hugler M., Wirsen C. O., Fuchs G., Taylor C. D., Sievert S. M. J. Bacteriol. 2005;187:3020–3027. doi: 10.1128/JB.187.9.3020-3027.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takai K., Campbell B. J., Cary S. C., Suzuki M., Oida H., Nunoura T., Hirayama H., Nakagawa S., Suzuki Y., Inagaki F., Horikoshi K. Appl. Environ. Microbiol. 2005;71:7310–7320. doi: 10.1128/AEM.71.11.7310-7320.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Butterfield D. A., Lilley M. D., Huber J. A., Roe K. K., Embley R. W., Baross J. A., Massoth G. J. In: The Subseafloor Biosphere at Mid-Ocean Ridges, Wilcock W. S. D., DeLong E. F., Kelley D. S., Baross J. A., Cary S. C., editors. Washington, DC: Am. Geophys. Union; 2004. pp. 269–289. Vol. Geophysical Monograph 144. [Google Scholar]
- 43.Rice P., Longden I., Bleasby A. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 44.Edgar R. C. BMC Bioinformatics. 2004;19:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howe K., Bateman A., Durbin R. Bioinformatics. 2002;18:1546–1547. doi: 10.1093/bioinformatics/18.11.1546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The supplemental.zip file available at http://jbpc.mbl.edu/research_supplements/g454/20060412-private provides access to unprocessed and trimmed tag sequence files for each sample. The V6RefDB database is also available at this web site.