Abstract
CAR (constitutive active/androstane receptor) regulates both the distal enhancer PBREM (phenobarbital-responsive enhancer module) and the proximal element OARE [OA (okadaic acid) response element] to synergistically up-regulate the endogenous CYP2B6 (where CYP is cytochrome P450) gene in HepG2 cells. In this up-regulation, CAR acts as both a transcription factor and a co-regulator, directly binding to and enhancing PBREM upon activation by xenobiotics such as TCPOBOP {1,4-bis-[2-(3,5-dichloropyridyloxy)]benzene} and indirectly associating with the OARE in response to OA [Swales, Kakizaki, Yamamoto, Inoue, Kobayashi and Negishi (2005) J. Biol. Chem. 280, 3458–3466]. We have now identified the cohesin protein SMC1 (structural maintenance of chromosomes 1) as a CAR-binding protein and characterized it as a negative regulator of OARE activity, thus repressing synergy. Treatment with SMC1 small interfering RNA augmented the synergistic up-regulation of CYP2B6 expression 20-fold in HepG2 cells, while transient co-expression of spliced form of SMC1 abrogated the synergistic activation of a 1.8 kb CYP2B6 promoter. SMC1 indirectly binds to a 19 bp sequence (−236/−217) immediately downstream from the OARE in the CYP2B6 promoter. Both DNA affinity and chromatin immunoprecipitation assays showed that OA treatment dissociates SMC1 from the CYP2B6 promoter, reciprocating the indirect binding of CAR to OARE. These results are consistent with the conclusion that SMC1 binding represses OARE activity and its dissociation allows the recruitment of CAR to the OARE, synergizing PBREM activity and the expression of the CYP2B6 gene.
Keywords: affinity chromatography, constitutive active/androstane receptor (CAR), cytochrome P450 (CYP), phenobarbital-responsive enhancer module, structural maintenance of chromosomes 1 (SMC1), xenobiotic
Abbreviations: ATM, ataxia telangiectasia mutated; CAR, constitutive active/androstane receptor; CBB, Coomassie Brilliant Blue; C-c, C-terminal coiled coil domain; C-g, C-terminal globular domain; ChIP, chromatin immunoprecipitation; CYP, cytochrome P450; DR4 motif, direct repeat 4 motif; DTT, dithiothreitol; ER, oestrogen receptor; GR, glucocorticoid receptor; GST, glutathione S-transferase; hCAR, human CAR; HCC, hepatocellular carcinoma; HRP, horseradish peroxidase; mCAR, mouse CAR; MEM, minimal essential medium; MPG, 3-methyladenine DNA glycosylase; N-c, N-terminal coiled coil domain; N-g, N-terminal globular domain; NP40, Nonidet P40; OA, okadaic acid; OARE, OA response element; PB, phenobarbital; PBREM, PB-responsive enhancer module; PPARα, peroxisome-proliferator-activated receptor α; PXR, pregnane X receptor; RXR, retinoid X receptor; siRNA, small interfering RNA; SMC, structural maintenance of chromosomes; SMC1-NC-del, SMC1 mutant lacking both N- and C-terminal globular domains; TCPOBOP, 1,4-bis-[2-(3,5-dichloropyridyloxy)]benzene
INTRODUCTION
Orphan nuclear receptors such as CAR (constitutive active/androstane receptor), PXR (pregnane X receptor) and PPARα (peroxisome-proliferator-activated receptor α) mediate the xenobiotic-induced transcription of hepatic genes. This induction can result in serious health consequences, since those genes encode enzymes and proteins that are involved in the hepatic metabolisms of compounds such as therapeutic drugs, bilirubin, bile acids, steroid hormones, glucose and fatty acids [1–5]. CAR was the first to be characterized as the receptor that is activated by xenobiotics such as PB (phenobarbital) and the potent PB-type inducer TCPOBOP {1,4-bis-[2-(3,5-dichloropyridyloxy)]benzene}. The activated receptor binds to a DR4 motif (direct repeat 4 motif) within the distal 51-bp PBREM (PB-responsive enhancer module) and induces the transcription of CYP2B genes (where CYP is cytochrome P450) [6–8]. The endogenous Cyp2b gene is not induced by PB in CAR-null mice [9,10]. Moreover, the PBREM-mutated CYP2B gene is not activated by PB in transgenic mice [11]. Thus CAR-mediated activation of PBREM is essential for induction to occur. However, the CAR-mediated activation of the PBREM and CYP2B promoter by PB-type inducers such as TCPOBOP in cell-based assays is far less effective compared with the strong induction of the endogenous Cyp2b gene in mouse livers in vivo. Therefore the high xenobiotic-inducible transcription in livers in vivo is not fully explained simply by the binding of a given receptor to its response element, and requires additional mechanisms to fill the gap.
Using a HepG2 cell line (called Ym17) that stably expresses V5-tagged mCAR (mouse CAR), we found that co-treatment with OA (okadaic acid) synergistically up-regulates the TCPOBOP-activated human CYP2B promoter to levels similar to that of Cyp2b induction in vivo. This synergistic up-regulation, called OA synergy, is regulated by the indirect binding of CAR to the 24 bp element (−256/−233) in the proximal promoter of the human CYP2B6 gene [12]. The 24 bp element, designated OARE (OA response element), appears to be unique to the CYP2B6 gene, since OA synergy is not observed with the bilirubin UDP-glucuronosyltransferase, CYP3A7 and superoxide dismutase 3 genes. These three genes are only moderately induced by PB in livers in vivo. Thus OARE and its activity provide an excellent model to further decipher the CAR-mediated regulatory mechanism that is characteristic of the highly inducible CYP2B gene. In addition, the regulation of OA synergy is independent of the CAR nuclear translocation, since CAR is spontaneously accumulated in the nucleus before OA treatment in Ym17 cells. OA represses the nuclear accumulation of CAR by PB only in the primary mouse hepatocytes in which CAR is retained in the cytoplasm [13]. In the present study, we have now identified SMC1 (structural maintenance of chromosomes 1) as a regulatory factor of the CAR-mediated transcription of the CYP2B6 gene.
SMC1, a member of the SMC family, is a constituent of the cohesin complex [14–18]. DNA damage triggers the activation of the checkpoint protein kinase ATM (ataxia telangiectasia mutated) that phosphorylates various proteins, including SMC1, to form the double-strand break repair complex [18]. In addition to DNA repair, SMC1 may also be involved in cell-cycle checkpoint and gene transcription [15,19]. For example, SMC1 regulates the Cut gene's transcription in Drosophila development by augmenting the interaction between the Wing enhancer and the Cut gene promoter. SMC1 has not yet been implemented in the transcriptional regulation of mammalian genes. Moreover, it has not been explored whether SMC1 can interact with nuclear receptors and, if it does, what the biological consequence from this interaction could be. Here, we have studied the cross-talk mechanism between CAR and SMC1 to regulate the PB-inducible activation of the human CYP2B6 gene, by focusing on OARE and OA synergy. First, we identified SMC1 as a CAR-binding protein. SMC1 indirectly binds to a 19-bp CYP2B6 promoter sequence immediately downstream of OARE and represses OA synergy and, upon OA treatment, dissociates from the promoter. The present study sheds light on the novel molecular mechanism in which CAR, acting as both a transcription factor and co-regulator, co-ordinates multiple elements to regulate this single gene.
MATERIALS AND METHODS
Materials
TCPOBOP and N-dodecanoylsarcosine were purchased from Sigma Chemical Co. (St. Louis, MO, U.S.A.); OA was from Calbiochem (San Diego, CA, U.S.A.); poly(dI-dC)·(dI-dC) was from Amersham Biosciences (Piscataway, NJ, U.S.A.). The plasmids pGL3-basic and pcDNA3.1-V5-His-TOPO were obtained from Promega (Madison, WI, U.S.A.) and Invitrogen (Carlsbad, CA, U.S.A.) respectively. Antibodies were obtained from either Invitrogen or Novus Biological (Littleton, CO, U.S.A.). Normal mouse and rabbit IgGs were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, U.S.A.). ImmunoPure Immobilized Protein A was purchased from Pierce (Rockford, IL, U.S.A.).
Cloning and plasmids
The 1.8-kb 5′-flanking DNA of the CYP2B6 gene and its deletion mutants were cloned into basic firefly luciferase reporter plasmid pGL3-basic as described previously [12]. Two different cDNAs encoding SMC1 were amplified from HepG2 RNAs and were cloned into the pcDNA3.1-V5-His-TOPO plasmid (Invitrogen): full-length SMC1 and an SMC1 variant lacking amino acid residues from positions 313–425 (SMC1Δ313–425). The following mutants were also constructed in pcDNA3.1-V5-His-TOPO; pcDNA3.1-N-g (N-terminal globular domain; SMC1, Met1 to Arg160); pcDNA3.1-N-c (N-terminal coiled coil domain; SMC1, Ser161 to Leu510); pcDNA3.1-hinge (SMC1, Tyr511 to Gly655); pcDNA3.1-C-c (C-terminal coiled coil domain; SMC1, Ala656 to Thr1005); pcDNA3.1-C-g (C-terminal globular domain; SMC1, Leu1006 to Gln1233); pcDNA3.1-SMC1-NC-del (SMC1 mutant lacking both N- and C-terminal globular domains; SMC1, Ser161 to Thr1005). The cDNA of SMC1Δ313–425, N-g, N-c and C-g of SMC1 were also cloned into pGEX4T3 to produce GST (glutathione S-transferase)-fusion proteins. These constructs were verified by their sequences. All other plasmids were previously constructed in this laboratory.
Cell culture
Cells were cultured in MEM (minimal essential medium) supplemented with 10% (v/v) fetal bovine serum and antibiotics (100 units/ml penicillin and 100 μg/ml streptomycin). The Ym17 cell line was produced previously [12].
Real-time PCR
Preparation of total RNA and the subsequent synthesis of first strand cDNA were performed using TRIzol® reagent (Invitrogen) and High Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA, U.S.A.) respectively. With the resulting cDNA, real-time PCR was performed to measure CYP2B6 mRNA using ABI Prism 7700 (Applied Biosystems) as described previously [12]. Primer and probe sets used for PCR analysis were as follows: 5′- and 3′-primers respectively, 5′-AAGCGGATTTGTCTTGGTGAA-3′, 5′-TGGAGGATGGTGGTGAAGAAG-3′, and probe 6FAM (6-carboxyfluorescein)-CATCGCCCGTGCGGAATTGTTC-TAMRA (6-carboxytetramethylrhodamine). The amount of CYP2B6 mRNA was normalized by β-actin mRNA that was measured by using a Pre-Developed Taqman Assay for human β-actin (Applied Biosystems).
Purification of CAR complex
V5-antibody (250 μg) or normal mouse IgG (250 μg) were conjugated with Protein G resin (100 μl) using ImmunoPure Protein G IgG Orientation kit (Pierce). After being treated with TCPOBOP for 1 h at 37 °C, Ym17 cells were harvested from 700 confluent 145 cm2 dishes, from which nuclear extracts and 250 mg of nuclear proteins were prepared as previously described [20]. Nuclear extracts were diluted with the same volume of buffer A [20 mM Tris/HCl, pH 7.5, containing 0.3 M NaCl, 0.2% NP40 (Nonidet P40) and 10% (v/v) glycerol] and were shaken with V5-antibody–Protein G for 2 h. The resin was sequentially eluted with buffer A and 20 mM Hepes buffer (pH 7.5) containing 0.2% N-dodecanoylsarcosine and 0.1 M glycine/HCl (pH 2.8). Finally, the remaining proteins were eluted with 0.1 M glycine/HCl (pH 2.8) containing 0.1% Triton X-100. All procedures were carried out at 4 °C. Purified proteins were freeze-dried, separated on 4–12% NuPage Bis-Tris/polyacrylamide gel, stained with CBB (Coomassie Brilliant Blue) R-250 and subjected to MS analysis.
MS analysis
The MS identification of in-gel digested proteins has been described in detail elsewhere [21–23]. Briefly, individual gel bands were excised and subjected to trypsin proteolysis using a ProGest automated digester (Genomic Solutions). The extracted peptides were analysed on a MALDI–TOF/TOF (matrix-assisted laser-desorption ionization–tandem time-of-flight) mass spectrometer (Voyager 4700) from Applied Biosystems. Data were internally calibrated with trypsin autoproteolysis peaks and submitted to the MASCOT database search engine (Matrix Science) for protein identification by peptide mass ‘fingerprinting’ and sequence tag approaches.
Western blot
Proteins were separated on a 4–12% NuPage Bis-Tris/polyacrylamide gel and transferred on to Immobilon-P membrane (Millipore, Bedford MA, U.S.A.). Subsequently, these membranes were incubated for 1 h at 25 °C or overnight at 4 °C with anti-SMC1, anti-SMC3, anti-V5 or anti-V5–HRP (horseradish peroxidase) (1:5000 dilutions) in 5% (w/v) non-fat milk in Tris-buffered saline/0.2% (v/v) Tween 20. Subsequently, they were incubated with HRP-conjugated anti-rabbit or anti-mouse IgG (1:5000 dilution; Santa Cruz Biotechnology). Protein bands were visualized by ECL® Plus Western blotting detection (Amersham Biosciences).
GST pull-down assays
Recombinant GST and GST-fusion proteins were expressed from their pGEX4T3 plasmids (Amersham Biosciences) in Escherichia coli BL21 (DE3) cells and were purified by standard procedures. pcDNA3.1-SMC1, pcDNA3.1-SMC1Δ313–425, pcDNA3.1-SMC1-NC-del, pcDNA3.1-N-g, pcDNA3.1-N-c, pcDNA3.1-hinge, pcDNA3.1-C-c, pcDNA3.1-C-g, pCR3-mCAR, pCR3-hCAR, pGEM-hRXR and pcDNA3.1-hGR were used to produce in vitro translated proteins in TNT® Quick Coupled Transcription/Translation System (Promega) and [35S]methionine (Amersham Biosciences). GST or GST-fusion proteins were immobilized on glutathione-S-Sepharose beads and were incubated with 5 μl of an in vitro translated protein in 500 μl of HBST (50 mM Hepes, pH 7.5, 0.1 M NaCl and 0.01% Triton X-100) for 30 min at room temperature (25 °C). After three washes in 1 ml of HBST, proteins were eluted in SDS/PAGE sample buffer (NuPAGE LDS Sample buffer; Invitrogen) at 70 °C for 5 min, separated on a 4–12% NuPage Bis-Tris gel (Invitrogen) and stained with CBB. The stained gel was also subjected to autoradiography.
Transfection assays
Ym17 cells in a 24-well plate were transfected with a given pGL3basic luciferase reporter plasmid (0.2 μg/well) and phRL-TK plasmid (Promega) (0.1 μg/well) by Lipofectamine™ 2000 (Invitrogen) according to the manufacturer's instructions, with an additional co-transfection of 0.2 μg/well pcDNA3.1 bearing SMC1 or each of its mutants or pcDNA3.1. After 24 h, the cells were subjected to treatment with chemicals (DMSO, TCPOBOP, OA or TCPOBOP plus OA) for an additional 48 h. Luciferase activity was measured using the Dual-Luciferase reporter assay system (Promega).
siRNA (small interfering RNA) expression
siRNAs were obtained from Dharmacon Research (Lafayette, CO, U.S.A.): SMC1 5′-GCAAUGCCCUUGUCUGUGA-3′, 5′-UCACAGACAAGGGCAUUGCUU-3′. 100 pmol of SMC1 siRNA or control siRNA (siCONTROL Non-Targeting siRNA no. 1) prepared by Dharmacon Research was co-transfected with −1.8-kb-CYP2B6-pGL3basic and hpRL-TK into Ym17 cells in each well of 24-well plates. After 24 h, the cells were treated with chemicals. Reduction of SMC1 protein was confirmed by Western-blot analysis.
DNA affinity chromatography
Dynabeads® M-280 streptavidin (Dynal ASA, Oslo, Norway) was conjugated with multiple copies of wild-type −257/−217-bp oligonucleotide or −257/−217-bp with the −252/−237 region mutated −257/−217mut [12] as described previously [7]. DNA beads were equilibrated with 25 mM Hepes (pH 7.6) containing 0.1 M NaCl, 10% glycerol, 0.5 mM EDTA, 0.5 mM DTT (dithiothreitol), 0.5% NP40, 20 μg/ml herring testes carrier DNA and 10 μg/ml poly(dI-dC)·(dI-dC) and were shaken with 200 μl of nuclear extract (1 mg of protein/ml), then washed with equilibrating buffer three times and then with equilibrating buffer containing 0.3 M NaCl. Subsequently, bound proteins were eluted by increasing the NaCl concentration to 0.5 M.
Immunoprecipitation
The DNA beads were shaken with nuclear extracts and washed with equilibrating buffer (flow-through). The bound proteins were washed with the equilibration buffer containing 0.3 M NaCl (wash) and eluted by increasing NaCl concentration to 0.5 M (eluate). Anti-SMC1 antibody (1 μg) or normal rabbit IgG (1 μg) was mixed with these fractions at 4 °C. After 12–16 h, 20 μl of 50% slurry of Protein A–agarose was added to this incubation mixture and was incubated for an additional 1 h at 4 °C. Agarose was washed by repeated centrifugations in 1 ml of the equilibrating solution for DNA affinity chromatography. Proteins were directly eluted from the agarose in SDS/PAGE sample buffer by incubation at 70 °C for 5 min, separated on a 4–12% NuPage Bis-Tris/polyacrylamide gel and subjected to Western-blot analysis.
ChIP (chromatin immunoprecipitation) assay
Approx. 1.5×107 cells were harvested by trypsinization, for assays using ChIP assay kit (Upstate Biotechnologies, Charlottesville, VA, U.S.A.), at 48 h after treatment with DMSO or TCPOBOP plus OA. Harvested cells were suspended in 10 ml of serum-free MEM and fixed by adding 270 μl of formaldehyde and gently shaking at room temperature for 30 min. A one-twentieth volume of 2.5 M glycine was added to fixed cell suspension with an additional 5 min shaking at room temperature. Fixed cells were collected by centrifugation, washed with cold PBS (pH 7.5), homogenized in 1 ml of 10 mM Hepes (pH 7.5) containing 10 mM KCl, 1.5 mM MgCl2, 1 mM DTT and 0.3% NP40 supplemented with Complete™ Mini (Roche Diagnostics, Mannheim, Germany) and collected by centrifugation. Washed cells were precipitated and resuspended in 500 μl of SDS lysis buffer (ChIP assay kit) supplemented with Complete™ Mini protease inhibitors for sonication by Misonix Microson™ XL 2000 ultrasonic cell disrupter. The sonicated material was centrifuged at 14000 g for 15 min and diluted with dilution buffer (ChIP assay kit) 10 times. The diluted lysates were pre-cleared by incubating with 200 μl of Protein A for 3 h at 4 °C. Cleared lysates (1 ml) were subjected to overnight incubation with either 5 μg of antibody, normal rabbit IgG or no antibody at 4 °C. Immunocomplexes were precipitated by adding 30 μl of Protein A for 1 h at 4 °C with shaking, washed with buffers included in ChIP assay kit, eluted and de-cross-linked in 500 μl of elution buffer (0.1 M NaHCO3, 1% SDS and 0.3 M NaCl) for 4 h at 65 °C. After protease digestion, DNA was purified using QIAquick PCR Purification kit (Qiagen, Valencia, CA, U.S.A.) and suspended in 40 μl of elution buffer. Input chromatin and immunoprecipitated DNA were PCR-amplified in 10 μl of reaction mixture with LA (long and accurate) Taq polymerase (Takara, Otsu Shiga, Japan), resolved on a 1.5% (w/v) agarose gel and visualized by ethidium bromide staining. The primer sequences that were designed to amplify the −338 and −99 region were used for analysis: Forward (5′-AGACAAACAGACAAAGCTAA-3′) and Reverse (5′-AGGAGCATTAGCTTAGAAAA-3′).
RESULTS
Purification and identification of SMC1
Ym17 cells express mCAR–V5-fusion protein [12]. The nuclear proteins prepared from TCPOBOP-treated Ym17 cells were incubated with V5 antibody–agarose or normal IgG–agarose. The proteins that bound to the agarose were eluted with a pH 2.8 buffer containing detergent. CAR was enriched in the eluate from V5 antibody–agarose but not from the normal IgG–agarose (Figures 1A and 1B). After being separated side-by-side on an SDS/polyacrylamide gel, all CBB-stained bands on the gel were subjected to MS analysis. One such band that appeared above the 116.3 kDa marker was only present in the eluate from the V5 antibody–agarose (Figure 1C). Subsequent MS analysis identified this protein as SMC1, while no peptide of SMC1 was detected from the corresponding area of gel after electrophoresis of the proteins eluted from the normal IgG–agarose. A co-purification of SMC1 with CAR indicated that SMC1 is a constituent of the nuclear CAR complex in Ym17 cells.
Figure 1. Purification of CAR complex from nuclear extracts of Ym17 cells.
Protein complexes were purified using a V5 antibody (α-V5) column or normal mouse IgG (nmIgG) column. The nuclear extract (NE), flow-through (FT) fraction and the fraction eluted with 0.2 M glycine/HCl (pH 2.8) containing 0.1% Triton X-100 (E) were applied to an SDS/4–12% polyacrylamide gel. (A) Western-blot analysis was performed using the V5 antibody to detect mCAR. (B) The protein band of mCAR was visualized by silver staining. (C) The band corresponding to SMC1 was stained by CBB.
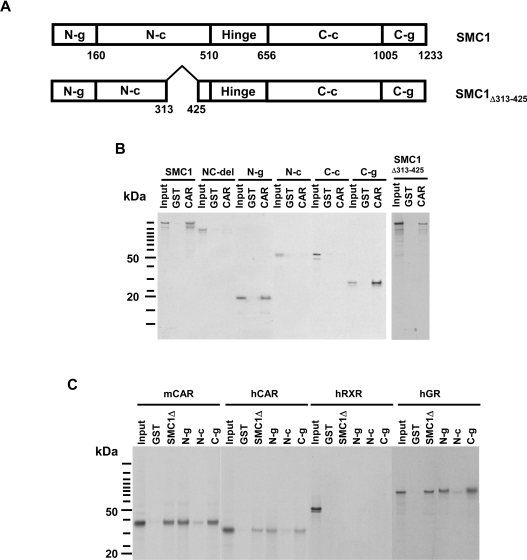
CAR binding region of SMC1
Direct interaction between CAR and SMC1 was analysed by GST pull-down assays. The SMC1 molecule can be divided into the following subdomains: N- and C-terminal globular domains, N- and C-terminal coiled coil regions and a hinge region that connects the two coils (Figure 2A). Both the full-length SMC1 and SMC1Δ313–425 exhibited binding to CAR (Figure 2B). Since the 113-residue deletion resides in the N-terminal coiled coil region, CAR did not appear to bind to this coiled coil region. Therefore the other subdomains (globular domains, coils and hinge) were in vitro translated separately in the presence of 35S-labelled methionine to test their binding to CAR. In binding assays, GST–CAR-fusion protein pulled down both N- and C-terminal globular domains, but neither coiled coil domains (Figure 2B). The assay was also performed using GST-fusion proteins of the SMC1Δ313–425, globular domains and in vitro translated mCAR, substantiating the observation that CAR binds preferentially to the globular domains at both termini of the SMC1 molecule (Figure 2C). Examination of the binding of the hinge region was prevented due to experimental limitations, such as the hinge region not being sufficiently labelled by in vitro translation since it contains only one methionine residue. However, the SMC1-NC-del was not capable of binding to CAR, suggesting that the hinge region is not required for CAR binding. Like mCAR, hCAR (human CAR) and GR (glucocorticoid receptor) bound to globular domains of SMC1 and SMC1Δ313–425 but human RXR (retinoid X receptor) did not (Figure 2C).
Figure 2. GST pull-down assay to identify the CAR-binding region of SMC1.
(A) Schematic diagram of the subdomains of the SMC1 and SMC1Δ313–425 molecule. N-g denotes N-terminal globular domain; N-c, N-terminal coiled coil domain; hinge, hinge domain; C-c, C-terminal coiled coil domain; C-g, C-terminal globular domain; NC-del, SMC1-NC-del. Numbers indicate amino acid residues. (B) GST and GST–mCAR were immobilized on glutathione–Sepharose beads and were incubated with in vitro translated 35S-labelled full-length or mutants of SMC1. Bound proteins were eluted and analysed by SDS/PAGE (4–12% gel) as described in the Materials and methods section. (C) GST, GST–SMC1Δ313–425 (SMC1Δ) and GST–SMC1 subdomains (N-g, N-c or C-g) were incubated with in vitro translated mCAR, hCAR, hRXR or hGR for this assay.
SMC1 as a repressor of the promoter activity
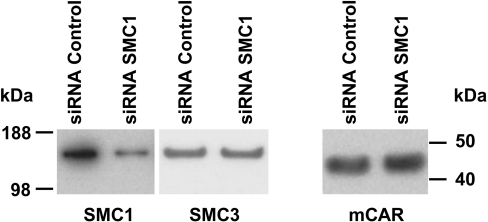
Treatment with OA synergistically up-regulated CAR-mediated induction of the endogenous CYP2B6 gene by TCPOBOP in Ym17 cells, having led us to elucidate the phenomenon called OA synergy [12]. To investigate the role for SMC1 in the synergistic up-regulation of CYP2B6 gene, siRNA was employed to specifically knock down SMC1 in Ym17 cells. Western-blot analysis showed a nearly 70% decrease in SMC1, but not SMC3 or mCAR, in the SMC1 siRNA-transfected Ym17 cells (Figure 3). While the rate of OA synergy (i.e. fold induction of CYP2B6 mRNA by the co-treatment divided by the sum of those by single treatments with TCPOBOP and OA) was 30-fold in control Ym17 cells, OA synergy was increased up to 300-fold in the SMC1 siRNA-transfected Ym17 cells (Figure 4A). Thus SMC1-knocked-down Ym17 cells exhibited OA synergy 10-fold greater than the cells transfected with control siRNA. Both TCPOBOP- and OA-dependent inductions were also increased in SMC1-knocked-down cells (3-fold for TCPOBOP, 4-fold for OA; inset of Figure 4A). A similar effect of SMC1 siRNA was also observed with transcription activity of the −1.8 kb CYP2B6 promoter-luciferase reporter gene; OA synergy was double in SMC1 siRNA-transfected Ym17 cells compared with that observed in cells transfected with control siRNA (Figure 4B). A much stronger effect of SMC1 siRNA was observed with the expression of endogenous CYP2B6 gene, which may suggest that chromatin structure may be involved in SMC1-mediated regulation.
Figure 3. Endogenous SMC1 was depleted by siRNA specific to SMC1.
Nuclear extracts prepared from the cells transfected with control siRNA or SMC1 siRNA were subjected to Western-blot analysis using the SMC1, SMC3 and V5 antibody.
Figure 4. Increase of OA synergy by SMC1 siRNA.
(A) Ym17 cells were transfected with SMC1 siRNA or control siRNA. The transfected cells were treated with TCPOBOP (250 nM), OA (10 nM) or TCPOBOP plus OA (OA, TC) for 48 h. Total cellular RNAs were prepared from the treated cells and subjected to real-time reverse transcriptase–PCR of CYP2B6 mRNA. Fold induction was calculated relative to the levels in DMSO-treated cells. The inset shows enlarged columns for inductions by TCPOBOP and OA. (B) Ym17 cells were transfected with −1.8k-pGL3, phRL-TK plasmids and SMC1 siRNA or control siRNA. The transfected cells were incubated with TCPOBOP (250 nM), OA (10 nM) or TCPOBOP plus OA for 48 h, harvested and assayed for luciferase activity. Fold induction was calculated by taking the control activity (DMSO) as 1.
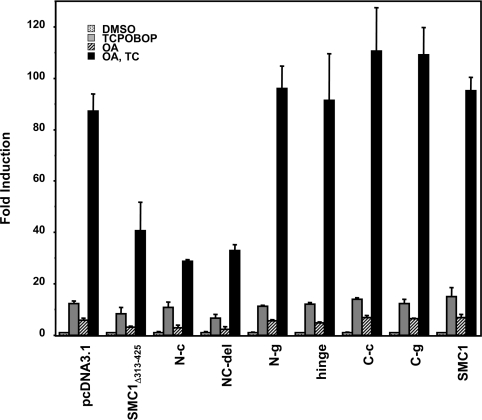
Given the fact that the SMC1 knockdown augmented the synergistic activation of −1.8 kb CYP2B6 promoter, we co-expressed SMC1 with the same promoter in Ym17 cells to investigate whether SMC1 overexpression repressed the promoter. The SMC1Δ313–425 variant was found to effectively repress both the OA-dependent activation as well as OA-dependent synergistic activation of the −1.8 kb CYP2B6 promoter, while the full-length SMC1 did not repress either activation (Figure 5). Since the deletion resides within the N-terminal coiled coil region, the different ability of SMC1Δ313–425 and full-length SMC1 to repress the promoter suggested that this coil region may be responsible for the repression. Therefore various deletion mutants of SMC1 were constructed based on these structural characteristics and were tested for their ability to repress the CYP2B6 promoter. Only the N-terminal coiled coil region (N-c) effectively repressed the OA-dependent activation as well as OA synergy of the −1.8 kb CYP2B6 promoter (Figure 5). No other subdomains (i.e. C-terminal coiled coil, hinge domains and N- and C-terminal globular domains) repressed the promoter activity. This dominant-negative function of the N-terminal coiled coil was further confirmed by using the SMC1-NC-del mutant (Ser161–Thr1005) that contained the coiled coil regions but lacked both globular domains, which also repressed the promoter activity. A similar experiment was performed using V5-His-tagged SMC1, SMC1Δ313–425 and SMC1-NC-del to confirm the expression level of these proteins in transfected cells (see Supplementary data at http://www.BiochemJ.org/bj/398/bj3980125add.htm).
Figure 5. Repression of OA synergy by overexpression of SMC1.
Various SMC1 subdomains, SMC1Δ313–425 and SMC1-NC-del were co-expressed with −1.8k-pGL3 and phRL-TK plasmids in Ym17 cells. The transfected cells were treated with TCPOBOP (250 nM), OA (10 nM) or TCPOBOP plus OA (OA, TC) for 48 h, harvested and assayed for luciferase activity. Fold induction was calculated by taking the control activity (DMSO) as 1.
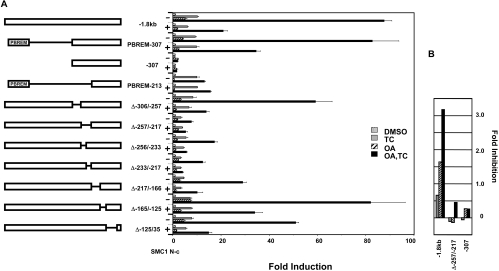
OA synergy occurs when both the PBREM and the −307-bp promoter are present and the OA response activity resides within the −307-bp promoter [12]. To delineate the promoter region responsible for the SMC1-dependent repression of OA synergy, various internal deletions within the −307-bp proximal promoter were constructed in the context of the −1.8-kb CYP2B6 promoter. These deletion constructs were co-expressed with the N-terminal coiled coil of SMC1 (SMC1N-c) in Ym17 cells (Figure 6A). The Δ−306/−257, Δ−217/−166, Δ−165/−125 and Δ−125/−35 deletion constructs retained the OA synergy in the TCPOBOP plus OA-treated Ym17 cells (Figure 6A). Two deletions, namely Δ−256/−233 and Δ−233/−217, significantly decreased OA synergy, while the residual promoter activity was still repressed by SMC1N-c. When both Δ−256/−233 and Δ−233/−217 regions were deleted, the Δ−257/−217 construct lost OA synergy. Additionally, the repression by SMC1N-c also disappeared. Thus the deletion assays delineated the repression of OA synergy by SMC1N-c to the −257/−217 sequence of the CYP2B6 promoter. Noticeably, the degree of repression by SMC1N-c in the TCPOBOP plus OA-treated Ym17 cells was correlated with that in the OA-treated, but not in the TCPOBOP-treated Ym17 cells (Figure 6B). The inhibition by SMC1N-c was not observed with the PBREM-213 construct, underscoring the fact that PBREM is not the direct target of SMC1-mediated repression. In addition, SMC1N-c did not repress the CAR-mediated activation of NR1-luciferase reporter gene, which contains only the DR4 motif (results not shown). SMC1N-c more effectively repressed OA-dependent activation of the −1.8-kb CYP2B6 promoter than the activation by TCPOBOP. Moreover, the OA synergy of the Δ−257/−217 deletion and the −307-bp promoter (used as the negative control that was not regulated by either TCPOBOP or OA) was similarly repressed by SMC1N-c (Figure 6B). These results suggested that SMC1 represses OA synergy primarily by inhibiting the OA response activation of the −251/−217 region of the CYP2B6 promoter, but not TCPOBOP activation of the PBREM.
Figure 6. Delineation of the SMC1-dependent repression to the 41-bp region (−257/−217-bp) of the CYP2B6 promoter.
Various deletion constructs of the −1.8-kb CYP2B6 promoter, shown in the left panel, were co-transfected with phRL-TK and pcDNA3.1 or pcDNA3.1-SMC1 N-coiled coil (N-c) into Ym17 cells. The transfected cells were incubated with TCPOBOP (250 nM), OA (10 nM) or TCPOBOP plus OA for 48 h, harvested and assayed for luciferase activity. Fold induction was calculated by taking the control activity (DMSO) as 1. (B) Fold inhibition of the treatment-dependent promoter activities by the N-coiled coil in the presence or absence of SMC1. The fold inductions in the absence of SMC1 were divided by those in the presence of SMC1. Numbers were calculated by subtracting one from those inhibition rates to make the baseline to zero. The values shown with less than zero indicate no inhibition.
Protein binding to the −257/−217 sequence
First, gel shift assays were performed to show the binding of SMC1 to the −257/−217 region using a bacterially expressed or in vitro translated SMC1 and the 41-bp DNA (−257/−217) as probe. No band was observed with either full-length, N- or C-terminal coiled coil in the gel shift assays (results not shown). Since the recombinant SMC1 did not directly bind to the −257/−217 region, we employed DNA affinity chromatography to investigate the possibility that SMC1 binds to the region in the presence of nuclear proteins. An oligonucleotide of 41 bp spanning the region −257/−217 or the DNA with mutations in the −257/−237 region were conjugated to magnetic beads to produce (−257/−217)-beads and (−257/−217mut)-beads. The nuclear extracts prepared from Ym17 cells treated with DMSO, TCPOBOP, OA and TCPOBOP plus OA were incubated with either (−257/−217)-beads or (−257/−217mut)-beads. After a wash with low-salt buffer [equilibrating buffer (see the DNA affinity chromatography subsection) containing 0.3 M NaCl], proteins were eluted with a buffer containing 0.5 M NaCl, separated on an SDS/polyacrylamide gel and subjected to Western-blot analysis. SMC1 was present in all eluates from the DMSO- or TCPOBOP-treated nuclear extracts, indicating SMC1 being included in the protein complex that bound to the −257/−217 region (Figure 7A). SMC1 was only scarcely recovered in the eluates from the OA-treated or TCPOBOP plus OA-treated Ym17 cells. There was no difference in the SMC1 elution patterns between (−257/−217) and (−257/−217mut), suggesting that the −236/−217 region was critical for the binding of SMC1. Little recovery of SMC1 from DNA affinity chromatography of the OA-treated sample indicated the association of SMC1 with the −236/−217 region and its dissociation from the region after OA treatment.
Figure 7. Treatment-specific interaction of SMC1 with OARRE or CAR.
(A) Nuclear extracts were prepared from the Ym17 cells treated with DMSO, TCPOBOP (250 nM), OA (10 nM) or TCPOBOP plus OA for 48 h and were applied to a DNA affinity column. After a wash with a buffer containing 0.3 M NaCl, nuclear proteins were eluted by 0.5 M NaCl from the (−257/−217)-beads or its mutated version (−257/−217mut)-beads and subjected to Western-blot analysis using SMC1 or V5 antibody. D, T, O and OT denote DMSO, TCPOBOP, OA and TCPOBOP plus OA respectively. Input is percentage of the proteins that were applied on columns. (B) Nuclear extracts prepared from DMSO- or OA-treated Ym17 cells were applied to the (−257/−217)-beads. SMC1 was precipitated from the flow-through (FT) or 0.3 M NaCl-washed fraction by incubation with 1 μg of SMC1 antibody (αSMC1) or normal rabbit IgG (nRb IgG). The resulting immunoprecipitates were resolved on 4–12% polyacrylamide gels and immunoblotted with the SMC1 or V5 antibody. Input is percentage of the proteins that were applied on columns. (C) ChIP assays to show the interaction of SMC1 with the promoter. Ym17 cells were treated with DMSO or TCPOBOP plus OA for 48 h, from which assays were performed for SMC1 antibody (αSMC1), V5 antibody (αV5), normal rabbit IgG (nRb IgG) or no antibody (no Ab). The purified DNA was amplified by PCR using primers specific to the −257/−217 region (amplicon −338 to −99) and resolved on a 1.5% agarose gel. Results shown were generated simultaneously from the same Ym17 cell extracts.
When the DNA affinity-enriched 0.5 M NaCl fractions were subjected to Western-blot analysis using V5 antibody, no CAR was detected in any of these fractions (Figure 7A).
This result was unexpected, since SMC1 was co-purified with CAR from the nuclear extracts (Figure 1). The presence of an SMC1–CAR complex was examined in the other fractions of the nuclear extracts after DNA affinity chromatography with the (−257/−217)-beads: the fraction that did not bind to the beads or was washed out with 0.3 M NaCl (flow-through or wash). Most of the SMC1 was recovered in these two fractions, only 1% of which was obtained in the 0.5 M NaCl eluate. SMC1 antibody co-precipitated CAR from both the flow-through and 0.3 M NaCl wash fractions, indicating that CAR and SMC1 existed as a complex in these unbound fractions to (−257/−217)-beads (Figure 7B). Thus the SMC1 that did not bind to the (−257/−217)-beads is present as a complex with CAR, while the SMC1 bound tightly does not form a complex with CAR. At least two different forms of SMC1 appear to be present in the nucleus of Ym17 cells: complexes with and without CAR. Only the SMC1 without CAR appeared to associate with the −236/−217 region.
To further confirm the interaction of SMC1 with the CYP2B6 promoter, ChIP assays were performed. Ym17 cells were treated with DMSO or TCPOBOP plus OA, from which cross-linked DNA–protein conjugates were precipitated by SMC1 antibody to subsequently amplify the −338/−99 bp region of CYP2B6 promoter that contains the −257/−217 region but not PBREM. Supporting the finding obtained from DNA affinity experiments, ChIP assays clearly showed that SMC1 decreases its interaction with the promoter after drug treatment (Figure 7C, αSMC1), indicating dissociation of SMC1 from the promoter. Background amplification from the sample of no IgG or normal rabbit IgG showed no treatment-specific changes, although these levels of amplification varied from one set of experiments to the next (Figure 7C). Consistent with previous findings [12] and providing an excellent evidence for the specificity of ChIP assays, the interaction of CAR with promoter was greatly enhanced after co-treatment with OA and TCPOBOP (Figure 7C, αV5). All results are consistent with the conclusion that SMC1 binds to the −257/−217 region as a repressor and undergoes OA-dependent dissociation from the promoter.
DISCUSSION
Our previous work characterized OARE as the positive element to which CAR indirectly binds and synergizes the TCPOBOP-dependent PBREM activity following OA treatment [12]. This CAR-mediated synergistic activation, called OA synergy, of the CYP2B6 promoter is unique in which one nuclear receptor, acting as both the transcription factor (i.e. direct binding to PBREM) and co-activator (i.e. indirect association with OARE), regulates multiple distinct elements of a single gene. We have now identified SMC1 as a repressor that negatively regulates the activity of OARE. In response to OA treatment, the SMC1 complex dissociates from the CYP2B6 promoter, thus enabling OARE to recruit CAR as the co-activator and up-regulating synergistically the CYP2B6 promoter.
SMC1 represses OA synergy through its N-terminal coiled coil in the cell-based transfection assays by binding to the −236/−217 region of CYP2B6 promoter, while its binding to CAR in the GST pull-down assays is mediated by the N- and/or C-terminal globular domains. Thus SMC1 spreads these two functions over the different subdomains, complicating the molecular mechanism of how SMC1 represses the OA synergy. Apparently, deletion of the CAR-binding domain may have converted SMC1 into a dominant active form as a repressor, as suggested by the fact that N-c and SMC1-NC-del repressed OA synergy in transient transfection assay. The reason why full-length SMC1 could not repress OA synergy in the cell-base transfection assays remains enigmatic. However, the results from SMC1Δ313/425 variant that could repress OA synergy raises, at least, two possibilities how SMC1 regulates OA synergy. SMC1 may have to be properly regulated to confer its repressive activity, and residue(s) within the 313–425 region may play a regulatory role for SMC1 to acquire this repressive activity. Although it may be artificial, the SMC1Δ313/425 may mimic the native form of SMC1 capable of repressing the synergistic up-regulation of the PBERM enhancer activity by OARE. Alternatively, SMC1Δ313/425, the naturally occurring but minor variant of SMC1, is the true form of SMC1 that is actually involved in the OARE repression in HepG2 cells in vivo. To further decipher the molecular mechanism of how SMC1 represses the OARE, it is important to determine whether the SMC1Δ313/425 is, in fact, the true repressor in future investigations, although developing proper antibodies is required to do this investigation.
SMC1, via the C-terminal globular domain, is known to bind to A-T-rich sequences or DNA fragments that can form secondary structures such as cruciforms [24]. Neither the C-terminal nor the other subdomains bound directly to the CYP2B6 promoter. The N-terminal globular domain contains an ATP-binding site and exerts ATPase activity by interacting with the C-terminal globular domain. This ATPase activity does not appear to be required, at least, for SMC1 binding to CAR, since the receptor can independently bind to the N- or C-terminal globular domain. Although the function of coiled coils is less understood, it is suggested that this region mediates protein–protein interactions [25]. The N-terminal coiled coil may interact with an unknown protein that mediates the indirect binding of SMC1 to cause the repression of OA synergy. SMC1 dissociates from the CYP2B6 promoter in response to OA treatment, in which OA may directly alter the phosphorylation of SMC1. ATM kinase phosphorylates residues Ser957 and Ser966 in the coiled coil region near the C-terminal globular domain [18]. Intriguingly, while these two residues were not phosphorylated in the DMSO- or TCPOBOP-treated Ym17 cells, they were phosphorylated to a certain level in the OA-treated cells (K. Inoue and M. Negishi, unpublished work). Once the DNA-binding protein that mediates the interaction of SMC1 with the −236/−217 region is identified in future investigations, we will be in a better position to answer this question.
The interaction of SMC1 with CAR is not the first demonstration of cross-talk between a nuclear receptor and the DNA repair protein. For example, ERα (oestrogen receptor α) was recently reported to bind to a DNA repair enzyme MPG (3-methyladenine DNA glycosylase) [26]. The binding of MPG attenuates the activity of ERα, down-regulating the transcription of ER response element-bearing genes. In addition to ERα, our study showed that GR also binds to SMC1. Thus the cross-talk of SMC1 with nuclear receptors may be a novel mechanism regulating various receptor-mediated biological functions. CAR activators such as PB and phenytoin are non-genotoxic carcinogens that promote HCC (hepatocellular carcinoma) in rodents [27]. The activation of CAR was recently found to be an essential factor in HCC promotion, as indicated by the fact that PB does not promote HCC in the diethylnitrosamine-treated CAR-null mice [28]. Not only PB and phenytoin, but also the other numerous non-genotoxic carcinogens are, in fact, activators/ligands of the so-called xeno-sensing nuclear receptors including CAR, the PXR and PPARα. The interaction of CAR with SMC1 may provide an experimental model to examine if CAR, or other nuclear receptors, can modulate the activities of SMC1 such as DNA repair, thus opening a new area of biological research into the cross-talk between nuclear receptors and cohesin proteins.
Online Data
Acknowledgments
We thank Dr Karen Swales (William Harvey Research Institute, Queen Mary, University of London, London, U.K.) for providing various constructs and we thank also the members of M.N.'s laboratory for thoughtful discussion.
References
- 1.Swales K., Negishi M. CAR, driving into the future. Mol. Endocrinol. 2004;18:1589–1598. doi: 10.1210/me.2003-0397. [DOI] [PubMed] [Google Scholar]
- 2.Sueyoshi T., Negishi M. Phenobarbital response elements of cytochrome P450 genes and nuclear receptors. Annu. Rev. Pharmacol. Toxicol. 2001;41:123–143. doi: 10.1146/annurev.pharmtox.41.1.123. [DOI] [PubMed] [Google Scholar]
- 3.Huang W., Zhang J., Chua S. S., Qatanani M., Han Y., Granata R., Moore D. D. Induction of bilirubin clearance by the constitutive androstane receptor (CAR) Proc. Natl. Acad. Sci. U.S.A. 2003;100:4156–4161. doi: 10.1073/pnas.0630614100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maglich J. M., Watson J., McMillen P. J., Goodwin B., Willson T. M., Moore J. T. The nuclear receptor CAR is a regulator of thyroid hormone metabolism during caloric restriction. J. Biol. Chem. 2004;279:19832–19838. doi: 10.1074/jbc.M313601200. [DOI] [PubMed] [Google Scholar]
- 5.Qatanani M., Zhang J., Moore D. D. Role of the constitutive androstane receptor in xenobiotic-induced thyroid hormone metabolism. Endocrinology. 2005;146:995–1002. doi: 10.1210/en.2004-1350. [DOI] [PubMed] [Google Scholar]
- 6.Honkakoski P., Moore R., Washburn K. A., Negishi M. Activation by diverse xenochemicals of the 51-base pair phenobarbital-responsive enhancer module in the CYP2B10 gene. Mol. Pharmacol. 1998;53:597–601. doi: 10.1124/mol.53.4.597. [DOI] [PubMed] [Google Scholar]
- 7.Honkakoski P., Zelko I., Sueyoshi T., Negishi M. The nuclear orphan receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-responsive enhancer module of the CYP2B gene. Mol. Cell. Biol. 1998;18:5652–5658. doi: 10.1128/mcb.18.10.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sueyoshi T., Kawamoto T., Zelko I., Honkakoski P., Negishi M. The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J. Biol. Chem. 1999;274:6043–6046. doi: 10.1074/jbc.274.10.6043. [DOI] [PubMed] [Google Scholar]
- 9.Ueda A., Hamadeh H. K., Webb H. K., Yamamoto Y., Sueyoshi T., Afshari C. A., Lehmann J. M., Negishi M. Diverse roles of the nuclear orphan receptor CAR in regulating hepatic genes in response to phenobarbital. Mol. Pharmacol. 2002;61:1–6. doi: 10.1124/mol.61.1.1. [DOI] [PubMed] [Google Scholar]
- 10.Wei P., Zhang J., Egan-Hafley M., Liang S., Moore D. D. The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature (London) 2000;407:920–923. doi: 10.1038/35038112. [DOI] [PubMed] [Google Scholar]
- 11.Ramsden R., Beck N. B., Sommer K. M., Omiecinski C. J. Phenobarbital responsiveness conferred by the 5′-flanking region of the rat CYP2B2 gene in transgenic mice. Gene. 1999;228:169–179. doi: 10.1016/s0378-1119(98)00612-x. [DOI] [PubMed] [Google Scholar]
- 12.Swales K., Kakizaki S., Yamamoto Y., Inoue K., Kobayashi K., Negishi M. Novel CAR-mediated mechanism for synergistic activation of two distinct elements within the human cytochrome P450 2B6 gene in HepG2 cells. J. Biol. Chem. 2005;280:3458–3466. doi: 10.1074/jbc.M411318200. [DOI] [PubMed] [Google Scholar]
- 13.Kawamoto T., Sueyoshi T., Zelko I., Moore R., Washburn K., Negishi M. Phenobarbital-responsive nuclear translocation of the receptor CAR in induction of the CYP2B gene. Mol. Cell. Biol. 1999;19:6318–6322. doi: 10.1128/mcb.19.9.6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jessberger R. SMC proteins at the crossroads of diverse chromosomal processes. IUBMB Life. 2003;55:643–652. doi: 10.1080/15216540310001639661. [DOI] [PubMed] [Google Scholar]
- 15.Hagstrom K. A., Meyer B. J. Condensin and cohesin: more than chromosome compactor and glue. Nat. Rev. Genet. 2003;4:520–534. doi: 10.1038/nrg1110. [DOI] [PubMed] [Google Scholar]
- 16.Harvey S. H., Krien M. J., O'Connell M. J. Structural maintenance of chromosomes (SMC) proteins, a family of conserved ATPases. Genome Biol. 2002;3:REVIEWS 3003.1–3003.5. doi: 10.1186/gb-2002-3-2-reviews3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jessberger R. The many functions of SMC proteins in chromosome dynamics. Nat. Rev. Mol. Cell Biol. 2002;3:767–778. doi: 10.1038/nrm930. [DOI] [PubMed] [Google Scholar]
- 18.Kitagawa R., Bakkenist C. J., McKinnon P. J., Kastan M. B. Phosphorylation of SMC1 is a critical downstream event in the ATM–NBS1–BRCA1 pathway. Genes Dev. 2004;18:1423–1438. doi: 10.1101/gad.1200304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rollins R. A., Morcillo P., Dorsett D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics. 1999;152:577–593. doi: 10.1093/genetics/152.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi K., Sueyoshi T., Inoue K., Moore R., Negishi M. Cytoplasmic accumulation of the nuclear receptor CAR by a tetratricopeptide repeat protein in HepG2 cells. Mol. Pharmacol. 2003;64:1069–1075. doi: 10.1124/mol.64.5.1069. [DOI] [PubMed] [Google Scholar]
- 21.Parker C. E., Warren M. R., Loiselle D. R., Dicheva N. N., Borchers C. H. Identification of components of protein complexes. In: Patterson W. C., Cyr D. M., editors. Ubiquitin-Proteasome Protocols, Methods in Molecular Biology 301. Totowa, NJ: Humana Press; 2005. pp. 117–151. [DOI] [PubMed] [Google Scholar]
- 22.Borchers C., Peter J. F., Hall M. C., Kunkel T. A., Tomer K. B. Identification of in-gel digested proteins by complementary peptide mass fingerprinting and tandem mass spectrometry data obtained on an electrospray ionization quadrupole time-of-flight mass spectrometer. Anal. Chem. 2000;72:1163–1168. doi: 10.1021/ac990937m. [DOI] [PubMed] [Google Scholar]
- 23.Hall M. C., Torres M. P., Schroeder G. K., Borchers C. H. Mnd2 and Swm1 are core subunits of the Saccharomyces cerevisiae anaphase-promoting complex. J. Biol. Chem. 2003;278:16698–16705. doi: 10.1074/jbc.M213109200. [DOI] [PubMed] [Google Scholar]
- 24.Akhmedov A. T., Frei C., Tsai-Pflugfelder M., Kemper B., Gasser S. M., Jessberger R. Structural maintenance of chromosomes protein C-terminal domains bind preferentially to DNA with secondary structure. J. Biol. Chem. 1998;273:24088–24094. doi: 10.1074/jbc.273.37.24088. [DOI] [PubMed] [Google Scholar]
- 25.Anderson D. E., Losada A., Erickson H. P., Hirano T. Condensin and cohesin display different arm conformations with characteristic hinge angles. J. Cell Biol. 2002;156:419–424. doi: 10.1083/jcb.200111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Likhite V. S., Cass E. I., Anderson S. D., Yates J. R., Nardulli A. M. Interaction of estrogen receptor α with 3-methyladenine DNA glycosylase modulates transcription and DNA repair. J. Biol. Chem. 2004;279:16875–16882. doi: 10.1074/jbc.M313155200. [DOI] [PubMed] [Google Scholar]
- 27.Pitot H. C. Altered hepatic foci: their role in murine hepatocarcinogenesis. Annu. Rev. Pharmacol. Toxicol. 1990;30:465–500. doi: 10.1146/annurev.pa.30.040190.002341. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto Y., Moore R., Goldsworthy T. L., Negishi M., Maronpot R. R. The orphan nuclear receptor constitutive active/androstane receptor is essential for liver tumor promotion by phenobarbital in mice. Cancer Res. 2004;64:7197–7200. doi: 10.1158/0008-5472.CAN-04-1459. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.