Abstract
CKX (cytokinin dehydrogenase) is a flavoprotein that cleaves cytokinins to adenine and the corresponding side-chain aldehyde using a quinone-type electron acceptor. In the present study, reactions of maize (Zea mays) CKX with five different substrates (N6-isopentenyladenine, trans-zeatin, kinetin, p-topolin and N-methyl-isopentenyladenine) were studied. By using stopped-flow analysis of the reductive half-reaction, spectral intermediates were observed indicative of the transient formation of a binary enzyme–product complex between the cytokinin imine and the reduced enzyme. The reduction rate was high for isoprenoid cytokinins that showed formation of a charge-transfer complex of reduced enzyme with bound cytokinin imine. For the other cytokinins, flavin reduction was slow and no charge-transfer intermediates were observed. The binary complex of reduced enzyme and imine product intermediate decays relatively slowly to form an unbound product, cytokinin imine, which accumulates in the reaction mixture. The imine product only very slowly hydrolyses to adenine and an aldehyde derived from the cytokinin N6 side-chain. Mixing of the substrate-reduced enzyme with Cu2+/imidazole as an electron acceptor to monitor the oxidative half-reaction revealed a high rate of electron transfer for this type of electron acceptor when using N6-isopentenyladenine. The stability of the cytokinin imine products allowed their fragmentation analysis and structure assessment by Q-TOF (quadrupole–time-of-flight) MS/MS. Correlations of the kinetic data with the known crystal structure are discussed for reactions with different cytokinins.
Keywords: cytokinin, cytokinin dehydrogenase (CKX), flavoprotein, maize, mass spectrometry (MS), stopped-flow spectrophotometry
Abbreviations: CKX, cytokinin dehydrogenase (EC 1.5.99.12); ZmCKX1, CKX enzyme from Zea mays; Q-TOF, quadrupole–time-of-flight
INTRODUCTION
Flavoproteins fulfil a wide variety of biological functions. Although being recognized for catalysing a range of redox reactions, flavoproteins have also evolved to mediate non-redox processes, e.g. light emission and sensing, transcriptional regulation and DNA repair. They typically contain FAD or FMN as a cofactor covalently or non-covalently bound to the apoprotein. Flavoproteins often also harbour other cofactors, such as haem, [4Fe-4S] clusters or molybdenum [1].
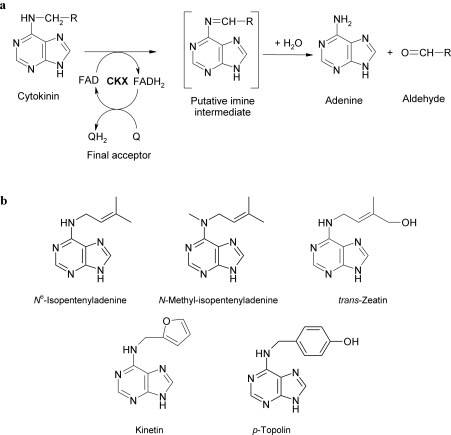
Flavin-containing oxidoreductases mediate the cleavage of a C–H bond with concomitant transfer of two electrons from a donor molecule to oxygen (oxidases) or another electron acceptor (dehydrogenases) [1]. CKX (cytokinin dehydrogenase; EC 1.5.99.12) is a flavoprotein in which the FAD cofactor is covalently linked to a histidine residue. The enzyme catabolizes plant hormones (cytokinins) by irreversible cleavage of their side chain (Figure 1a). This function makes it an important regulator of cytokinin levels in plants [2]. During catalysis a ternary complex composed of cytokinin substrate, enzyme and an electron acceptor leads to conversion of the cytokinin into adenine and the corresponding aldehyde [3]. CKX was originally thought to be an oxidase, with molecular oxygen as the primary electron acceptor. Kinetic data on wheat (Triticum aestivum) and maize (Zea mays) CKXs have shown, however, that oxygen is a poor electron acceptor [4,5], despite a high redox potential (+0.82 V) that favourably accepts electrons from many other covalent flavoproteins [6]. Quinone-type electron acceptors were found to function more efficiently for CKX, indicating that it is a much more effective dehydrogenase than oxidase [4,5,7,8]. A crystal structure of ZmCKX1 (CKX from Z. mays) has been solved recently [5] and has confirmed that ZmCKX1 is a covalent flavoprotein in which FAD is covalently tethered to His105. This monomeric protein of 57 kDa is most active with the isoprenoid cytokinins, trans-zeatin and isopentenyladenine. X-ray analyses revealed that, upon binding, these substrates seal the pore that leads to the catalytic site, leaving no space for electron acceptor entry [9]. So far, the crystallographic data have not revealed potential electron-acceptor-binding sites.
Figure 1. Cytokinin dehydrogenase and cytokinins.
(a) Reaction mechanisms of cytokinin dehydrogenase (EC 1.5.99.12), (b) structures of cytokinins used in the present study.
The present study presents steady-state and pre-steady-state kinetic analyses of the ZmCKX1 reaction with five different substrates (Figure 1b), the typical isoprenoid cytokinins {isopentenyladenine [N6-(2-isopentenyl)adenine] and trans-zeatin [6-(E-4-hydroxy-3-methylbut-2-enylamino)purine]}, aromatic cytokinins {kinetin [N6-(2-furfuryl)adenine)] and p-topolin [N6-(4-hydroxybenzyl)adenine]}, and the cytokinin analogue N-methyl-isopentenyladenine [N6-methyl-N6-(3-methyl-2-butenyl)adenine] that was used previously for mechanistic studies [5]. We also present the first structural identification of a CKX reaction intermediate that was postulated as a cytokinin-derived imine by Laloue and Fox [10]. The structure of the intermediate was determined for all five studied cytokinins using MS fragmentation analysis and accurate mass determination by MS/MS spectroscopy.
MATERIALS AND METHODS
Enzyme and chemicals
The recombinant ZmCKX1 enzyme used in the present study was the same as that described previously [5,9]. The molar absorption coefficient of the ZmCKX1 enzyme was determined spectrophotometrically by comparing the absorption spectrum of the native enzyme with that of the enzyme mixed with SDS. It was found that incubation with 0.1% SDS for 5 min ensured its complete unfolding, as the obtained flavin spectrum was similar to that of free FAD, whereas higher SDS concentrations or longer incubation with SDS did not change the final spectrum. By using ϵFAD=11.3 mM−1·cm−1 at 450 nm [11], the molar absorption coefficient of ZmCKX1 at 450 nm was found to be 11.5 mM−1·cm−1.
Isopentenyladenine, trans-zeatin and kinetin were obtained from Sigma. p-Topolin was from OlChemIm. N-Methyl-isopentenyladenine was synthesized as described previously [12]. Absorption spectra of used cytokinins and corresponding product aldehydes (3-methyl-2-butenal, furfural and 4-hydroxybenzaldehyde; all from Sigma) were measured in 75 mM imidazole/HCl buffer (pH 6.5) and 50 mM Tris/HCl buffer (pH 8.0) respectively.
Stopped-flow kinetics of FAD reduction
Stopped-flow kinetics was performed using an SX17MV stopped-flow instrument equipped with a diode-array detector (Applied Photophysics). Spectral scans were collected either at 2.56 ms or 7.68 ms intervals. To determine the rates for specific processes accurately, the single-wavelength detection mode was used (451 nm for flavin reduction, 360 nm for intermediate decay and 650 nm for monitoring the charge-transfer complex). Anaerobic conditions were accomplished by flushing solutions with nitrogen and by subsequent inclusion of 20 mM glucose and 20 μg/ml glucose oxidase. The ZmCKX1 enzyme (23 μM) was mixed with different concentrations of cytokinin substrates in 75 mM imidazole/HCl (pH 6.5) at 25 °C. The experiment with p-topolin was also performed in 75 mM Tris/HCl buffer (pH range 7.5–8.5).
Monitoring the redox state of the FAD cofactor during catalysis
The experiment was carried out using the stopped-flow apparatus in the single-wavelength detection mode at 451 nm, as above, in 75 mM imidazole/HCl (pH 6.5) at 25 °C. The enzyme (5 μM, final concentration) was first anaerobically reduced with 0.1 mM isopentenyladenine and subsequently mixed with an equal volume of oxygen-saturated buffer [75 mM imidazole/HCl (pH 6.5)] or with anaerobically prepared buffer of the same composition containing 0.5 mM CuCl2.
Spectrophotometric measurements of the reaction intermediate decay
Spectrophotometric measurements were performed using a DU 7500 photodiode array spectrophotometer (Beckman), in the range 250–700 nm, in aerobic mode. The enzyme (5 μM) was incubated at 25 °C with various concentrations of cytokinin substrate in 75 mM imidazole/HCl (pH 6.5) and 75 mM Tris/HCl (pH 8.0) buffers.
Determination of kinetic parameters for the turnover reaction
The assay was performed using methods previously described [13]. An enzyme sample (0.44–120 nM) was incubated in a reaction mixture (total volume of 0.6 ml in an Eppendorf tube) composed of 75 mM imidazole/HCl (pH 6.5) containing 1.25 mM CuCl2 and the appropriate concentration of the substrate at 25 °C and 37 °C. The incubation period varied from 0.5 to 1.5 h depending on the activity. The reaction was stopped by adding 0.3 ml of 40% (w/v) trichloroacetic acid to the tube, which was followed by the addition of 0.2 ml of 4-aminophenol [2% (v/v) solution in 6% (v/v) trichloroacetic acid]. The absorption of the sample was then scanned using a UV-2401PC double-beam spectrophotometer (Shimadzu) in the range 300–700 nm against a blank without the substrate. Concentrations of the reaction products of different cytokinins were then determined from absorptions at specific wavelengths as described by Frébort et al. [13]. All kinetic data were analysed using the program GraFit 4.0.12 (Erithacus Software).
Intermediate identification by MS/MS
The high-resolution MS measurements of cytokinin dehydrogenase reaction intermediates were performed on a hybrid mass analyser Q-TOF micro with MassLynx data system software (Micromass). Cytokinins were diluted from a stock solution of 1 mM in DMSO to 0.05 mM in 30 mM ammonium hydrogen carbonate (pH 8.0). ZmCKX1 [stock solution 87.7 μM in TE buffer (10 mM Tris/HCl, 1mM EDTA, pH 8.0) containing 1 M KCl] was desalted using a Microcon YM-30 filter (Millipore). For the analysis of reaction intermediates, a reaction mixture that contained 1.5 μM ZmCKX1 and 0.05 mM cytokinin in 30 mM ammonium hydrogen carbonate (pH 8.0) was prepared and incubated at 25 °C for 2 min before the measurement. The samples were applied in the form of an in-line spray at an injection rate of 10 μl/min. In the full-scan mode, data acquisition was performed in the range 50–1000 Da, with a cycle time of 47 μs, a scan time of 0.5 s and a collision energy of 5 V. Electrospray ionization in the positive mode was performed using the following parameters: source block/desolvation temperature of 80/150 °C, capillary/cone voltage of 2500/30 V, and spray/cone gas flow (N2) of 50/250 litres/h. For the exact mass determination experiments, the external calibration was performed using lock spray technology with a mixture of 0.1 M NaOH/10% (v/v) formic acid/acetonitrile (1:1:8 by volume) as a reference. For the MS/MS experiments, fragmentation was performed in an argon-gas-filled collision cell with collision energies 15, 20, 25, 30 and 35 V. Other parameters were the same as in the simple MS experiments. Masses of the parent ions and their fragments were calculated and used for the determination of the elementary composition and structure, with accuracy within 5 p.p.m. for full scan and within 15 p.p.m. for MS/MS experiments.
RESULTS
Reductive half-reaction
To study the kinetic properties of ZmCKX1 in more detail, a pre-steady state kinetic study was performed. Five different cytokinins were chosen for the kinetic study: two naturally occurring isoprenoid cytokinins (isopentenyladenine and trans-zeatin), two aromatic cytokinins (kinetin and p-topolin) and a cytokinin analogue (N-methyl-isopentenyladenine) that was shown previously [5] to be a substrate of ZmCKX1 (see Figure 1b for structural formulae). As a reference, the absorption spectra of the cytokinins and available product aldehydes (3-methyl-2-butenal, furfural and 4-hydroxybenzaldehyde) were measured (Supplementary Figure 1 at http://www.BiochemJ.org/bj/398/bj3980113add.htm). None of the compounds had significant absorption above 300 nm, except for 4-hydroxybenzaldehyde (a product of cleavage of p-topolin) that has a typical absorption spectrum with a peak at 330 nm at pH 8.0 (ϵ330 of 17900 M−1·cm−1).
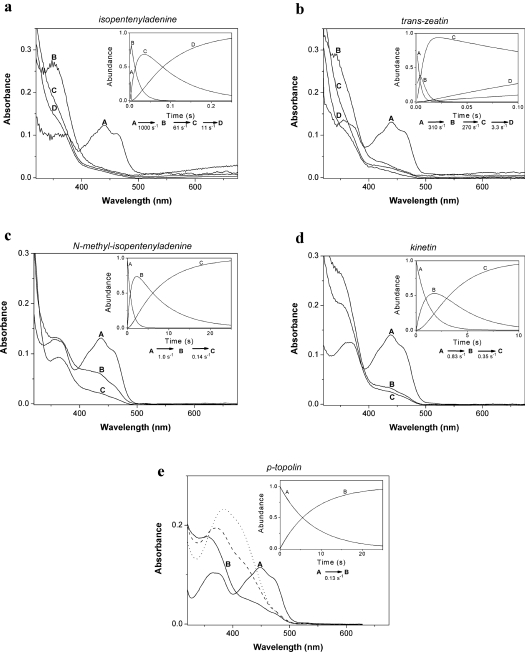
When ZmCKX1 was anaerobically mixed with individual cytokinins using a stopped-flow apparatus equipped with a diode-array detector, the time-resolved spectral analysis (300–670 nm) revealed the presence of at least two distinct kinetic events (Figure 2). During the first process, the FAD cofactor was fully reduced, as evidenced by an absorption decrease around 451 nm. Concomitantly with FAD reduction, the formation of a species characterized by a shoulder at 360 nm was observed with most substrates. This kinetic event was extremely fast for the isoprenoid cytokinins where the rates were 200–1000 s−1 (Figures 2a and 2b), but slow for the aromatic cytokinins and N-methyl-isopentenyladenine (Figures 2c, 2d and 2e) with rates ≤ 1 s−1. For isopentenyladenine and, to some extent, also for trans-zeatin, a broad absorbance band above 550 nm was also observed concomitantly with FAD reduction (Figures 2a and 2b), and this feature was ascribed previously to a charge-transfer complex [5].
Figure 2. Anaerobic reduction of ZmCKX1 with cytokinins.
Using a stopped-flow instrument equipped with a photodiode array detector, spectral data were obtained upon anaerobic mixing of ZmCKX1 with cytokinin substrates. The spectral species shown were derived from global analysis of the data obtained. The data could be fitted using a model in which species A decays in an exponential fashion to form B (A→B). If needed, more exponential decay steps were included in the model. Typically, 23 μM ZmCKX1 was mixed with 0.5 mM of a cytokinin substrate (except for N-methyl-isopentenyladenine and kinetin where 0.25 mM was used) under anaerobic conditions in 75 mM imidazole/HCl (pH 6.5) at 25 °C. The deconvoluted spectra are shown for: (a) isopentenyladenine, (b) trans-zeatin, (c) N-methyl-isopentenyladenine, (d) kinetin, and (e) p-topolin. For p-topolin (e), spectra of species B obtained with 75 mM Tris/HCl (pH 7.5; broken line) and (pH 8.5; dotted line) are shown. Time courses obtained by global analyses for the respective species are shown in the insets. For experimental details see the Materials and methods section.
Although the flavin was fully reduced in this first kinetic process by all natural cytokinins, the cofactor was only partially reduced by N-methyl-isopentenyladenine. Only in a second slow phase was the flavin fully reduced. With most substrates, kinetic processes other than the relatively fast flavin reduction were observed. For the isoprenoid cytokinins, isopentenyladenine and trans-zeatin, the reduction was followed by two relatively slow phases in which the formed intermediates, absorbing at approx. 360 nm, slowly disappeared. In the case of kinetin, the species formed upon flavin reduction also showed a relatively high absorbance at approx. 360 nm and disappeared in a single and slow subsequent step. Unique to p-topolin, the anaerobic reduction of ZmCKX1 revealed a single kinetic event in which the flavin was fully reduced while a species was formed that absorbs at approx. 390 nm. The pH-dependence of this band suggests that it is indeed the phenolic imine of p-topolin, since the aldehyde product 4-hydroxybenzaldehyde would give a peak at 330 nm (Supplementary Figure 1 at http://www.BiochemJ.org/bj/398/bj3980113add.htm). The pKa of the imine is apparently similar to that of the aldehyde, as would be expected.
To elucidate the kinetic behaviour of the individual spectral species as described above further, single wavelength detection modes at 451 nm, 360 nm and 650 nm were applied for higher accuracy. FAD reduction rates were measured as absorbance decay at 451 nm and their dependencies on different concentrations of individual cytokinins were used to determine the reduction rate constants and dissociation constants of the enzyme–substrate complex. The rates of the enzyme–product binary complex decay were measured at 360 nm and the charge-transfer complex was monitored at 650 nm. The resulting data are summarized in Table 1.
Table 1. Kinetic parameters of the catalytic reaction of ZmCKX1 with various cytokinins.
Rate constants of the processes of flavin reduction, FADH2-imine complex decay and charge-transfer formation and decay were monitored under anaerobic conditions at 451, 360 and 650 nm respectively, using single-wavelength stopped flow spectrophotometry for 0.25 mM cytokinin substrates with 10 μM ZmCKX1, in 75 mM imidazole/HCl (pH 6.5) at 25 °C. The enzyme–substrate complex dissociation constants (Kd) were determined at 451 nm with 3 μM ZmCKX1 enzyme reduced with cytokinin substrates in the concentration range 15–250 μM, under the same conditions. Turnover rates and the respective apparent Km′ were determined using the 4-aminophenol method [13], with 0.44–120 nM ZmCKX1 and 1.25 mM CuCl2 as the electron acceptor in 75 mM imidazole/HCl (pH 6.5) at 25 °C. For determining the rate of reduction and the Kd value, the observed reduction rates were measured using at least seven different substrate concentrations. Both the steady-state and the reduction kinetic data could be analysed using a Michaelis–Menten equation, yielding steady-state kinetic parameters (kcat and Km′) and kinetic parameters for the enzyme reduction (kred and Kd). The S.D. obtained upon fitting the data are indicated. For the rates of imine decay and charge transfer decay, the kinetic data were derived from curve fitting using an exponential decay model. The derived values exhibited S.D. of < 5%.
| FAD reduction | FADH2-imine complex decay | Charge-transfer decay | Aldehyde product formation | ||||
|---|---|---|---|---|---|---|---|
| Cytokinin substrate | kred (s−1) | Kd (μM) | Rate constant (s−1) | Amplitude (A650) | Observed rate (s−1) | kcat (s−1) | Km′ (μM) |
| Isopentenyladenine | 1150±50 | 15.1±2.5 | 62 | 0.0115 | 60 | 40.0±2.8 | 1.0±0.6 |
| 7.0* | 0.0028* | 8.0* | |||||
| trans-Zeatin | 244±5 | 13.1±1.1 | 179 | 0.0140 | 222 | –† | 14† |
| 3.1* | 0.0028* | 2.6* | |||||
| N-Methyl-isopentenyladenine | 1.08±0.02 | 25.1±1.4 | 0.16 | 0 | − | 0.13±0.01 | 23.9±1.6 |
| 0.24±0.01* | 13.3±3.2* | 0.024* | |||||
| Kinetin | 0.82±0.02 | 66±3 | 0.013 | 0 | − | 1.90±0.04 | 84.9±6.1 |
| p-Topolin | 0.11±0.01 | 24±3 | 0.009 | 0 | − | 0.23±0.01 | 38.4±3.3 |
*The event was biphasic, producing the indicated second rate constant and amplitude.
Data in Table 1 demonstrate that isopentenyladenine provided very fast and efficient reduction of the enzyme cofactor with an extremely high rate constant of 1150 s−1 and a dissociation constant of 15 μM. In contrast with previous measurements that showed Kd<2 μM [5], the present analysis allowed better fitting for the fast reduction process. Previously [5], only one fast phase was observed, whereas now two phases were present. Along with the fast cofactor reduction, a strong charge-transfer complex absorption band appeared that was relatively stable and decayed in a biphasic process which was synchronized with the binary complex decay. It is supposed that the strong charge-transfer complex is formed due to the tight binding of the intermediate product (imine), positioned next to the isoalloxazine ring system of the flavin cofactor. Repositioning of this product in the binary complex makes the charge-transfer complex weaker, and dissociation of the binary complex leads to its complete loss and to the formation of imine-product (Figure 2a and inset).
trans-Zeatin was also effective in reducing the FAD cofactor, although with a lower rate constant (244 s−1) but with a similar dissociation constant (13 μM) when compared with isopentenyladenine (Table 1). A charge-transfer complex was also observed and decayed in a biphasic process. It seems that product binding in the first binary complex results in less extensive orbital overlap as compared with isopentenyladenine and as observed by a less intense charge-transfer band. This may also explain why the spectral species showing the strong absorption at 360 nm, that was observed for isopentenyladenine (Figure 2a), is less abundant in the trans-zeatin spectral data (Figure 2b).
N-Methyl-isopentenyladenine had a much slower FAD reduction than isopentenyladenine and trans-zeatin, and showed no evidence of the charge-transfer complex formation. The measurements at 451 nm provided a reduction rate fitted by two exponentials, a faster process characterized by kred=1.0 s−1 and Kd=22 μM, and a second reductive event with an apparent rate of 0.2 s−1 and a similar apparent affinity (Kd=12 μM). In the first phase, a relative increase at approx. 360 nm was observed again that disappeared during the second phase. However, the intensity at 360 nm of this intermediate was much lower when compared with the cytokinin substrates mentioned above. For the aromatic cytokinins, the reduction rate constants were very low (0.1–0.8 s−1). The single wavelength stopped-flow kinetic analyses showed that kinetin affinity for the enzyme is somewhat low (Kd=66 μM), whereas p-topolin had relatively stronger binding (Kd=24 μM). The rate constants of the binary complex decay were approx. 0.01 s−1, thus the binary complex was quite stable, but no charge-transfer was observed.
Oxidative half-reaction
To determine the reactivity of reduced ZmCKX1 with molecular oxygen, the oxidative half-reaction was also studied. For pre-steady-state re-oxidation experiments, 20 μM ZmCKX1 was reduced anaerobically by adding 25 μM dithionite. By mixing the dithionite-reduced enzyme with air-saturated buffer, ZmCKX1 was re-oxidized in a slow monophasic process yielding fully oxidized enzyme. The re-oxidation rate was 1.83 s−1. Using a buffer that was saturated with molecular oxygen (0.62 mM oxygen upon mixing), a higher re-oxidation rate was observed, yielding a re-oxidation rate constant of 1.5×104 M−1·s−1. Compared with other flavoprotein oxidases, this re-oxidation rate constant is relatively low, indicating that in vivo the enzyme will not act as an efficient oxidase.
The turnover rate of ZmCKX1, which was previously measured by the formation of the corresponding cytokinin-derived aldehyde product [4,5], was found to be much faster when using alternative electron acceptors such as 2,3-dimethoxy-5-methyl-1,4-benzoquinone and 2,6-dichlophenol indophenol. Although highly effective, these compounds cannot be used for spectrophotometric analysis of FAD re-oxidation due to strong absorption at the wavelength range studied. To overcome this disadvantage, a Cu(II)/imidazole complex was used, since it also functions as an electron acceptor of CKX [4], but does not interfere with FAD absorption. The enzyme was first anaerobically reduced with an excess of isopentenyladenine and then mixed, using the stopped/flow apparatus, with either an oxygen-saturated buffer or an anaerobically prepared Cu(II)/imidazole solution. With oxygen, the enzyme needed approx. 30 s to consume the substrate before starting to revert to the oxidized form in an exponential process (rate of approx. 0.6 s−1). Using Cu(II)/imidazole, the substrate was consumed within 0.7 s and the oxidized form restored with a rate of 28 s−1. These results clearly indicate that the Cu(II)/imidazole complex is approx. 50-fold more effective as an electron acceptor than molecular oxygen. For the remainder of this kinetic study, the steady-state parameters of ZmCKX1 with the cytokinins studied were determined using Cu(II)/imidazole.
The apparent Km′ values and turnover rates measured by cytokinin-derived aldehyde production [13] with Cu(II)/imidazole (pH 6.5) at 25 °C (the temperature used for spectrophotometric measurements and for corresponding MS/MS experiments) are shown in Table 1. The turnover rate was 40 s−1 for isopentenyladenine, but much less for kinetin, N-methyl-isopentenyladenine and p-topolin. The Km′ was as low as 1 μM for isopentenyladenine, whereas with the other cytokinins it ranged from 14–85 μM. At 37 °C, the turnover rates for all cytokinins studied increased approx. 3-fold and the apparent Michaelis constants by approx. 50% (results not shown).
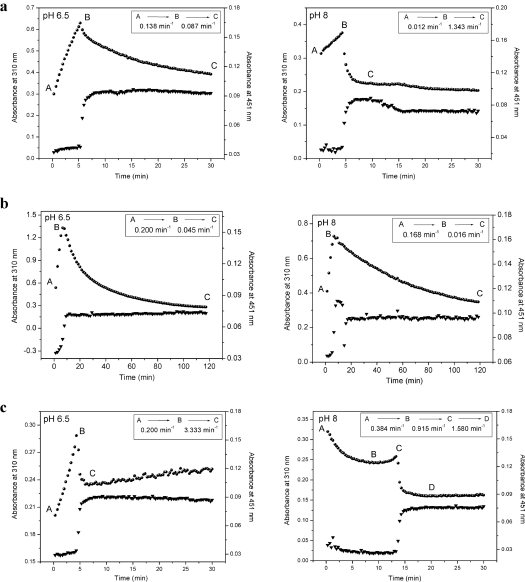
Anaerobic substrate-mediated ZmCKX1 reduction experiments (Figure 2) showed that the binary complex, consisting of reduced enzyme and cytokinin-derived imines, was converted into a species absorbing at 310 nm. This species was proposed previously to be the free imine product [10]. Under aerobic conditions, the enzyme undergoes slow turnover that results in accumulation of the 310-nm species in the reaction mixture [5], before it is hydrolysed to adenine and the corresponding cytokinin-derived aldehyde. In the present study, the accumulation and decay of this spectral species was monitored using diode-array spectrophotometry. The results in Figure 3 show that the 310-nm species derived from isopentenyladenine, trans-zeatin and N-methyl-isopentenyladenine, respectively, accumulated in the reaction mixture until all corresponding substrate was degraded. For isopentenyladenine, this species showed exponential decay, being relatively stable at pH 6.5, but rapidly disappearing at pH 8.0. In the case of N-methyl-isopentenyladenine, this species was also stable, not being significantly affected by pH. In contrast, the 310-nm species derived from trans-zeatin decayed rapidly at both pH values as indicated by a rather sharp decrease in absorption (Figure 3c). The time needed for the substrate consumption and enzyme re-oxidation (detected as absorbance change at 451 nm) was the same in acidic and alkaline conditions when the enzyme was reduced by isopentenyladenine or N-methyl-isopentenyladenine. For trans-zeatin, however, alkaline conditions led to a prolonged re-oxidation phase (3-fold) compared with that at pH 6.5 (Figure 3c), which could be caused by further cyclization of the trans-zeatin-derived aldehyde product, 4-hydroxy-3-methyl-2-butenal.
Figure 3. Cytokinin intermediate formation and decay.
Time courses of the reaction intermediate formation and decay (310 nm; circles) and oxidation changes of the FAD cofactor (451 nm; triangles) observed for ZmCKX1 (5 μM) that was mixed aerobically with 125 μM cytokinins [(a) isopentenyladenine, (b) N-methyl-isopentenyladenine, and (c) trans-zeatin] in 75 mM imidazole/HCl (pH 6.5; left-hand panels) and 75 mM Tris/HCl (pH 8.0; right-hand panels). The data could be fitted using a model in which species A is converted in a hyperbolic fashion into species B that then decays exponentially to form C.
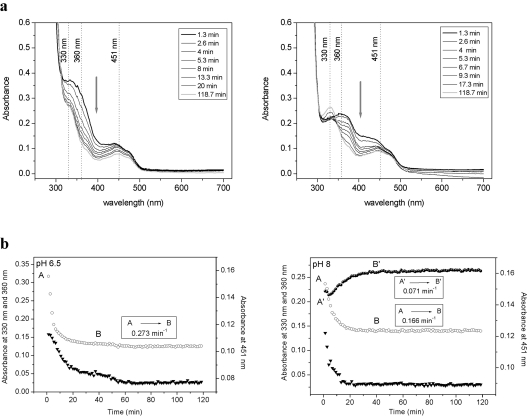
p-Topolin had different behaviour than the other cytokinins. The anaerobic reduction of CKX by this cytokinin already revealed that the enzyme-bound imine product had different spectral properties, with a pH-sensitive absorbance peak at approx. 390 nm (Figure 2e). This imine product had similar stability under acidic and alkaline conditions (Figure 4). Upon prolonged incubation, the imine was hydrolysed to 4-hydroxybenzaldehyde that has a typical absorption spectrum with a peak at 330 nm at pH 8.0 (ϵ330 of 17900 M−1·cm−1), whereas the absorption at pH 6.5 is negligible (ϵ330 of 2750 M−1·cm−1). The striking observation was made that re-oxidation of p-topolin-reduced enzyme (451 nm trace in Figure 4) did not occur even after prolonged incubation (12 h). Only an equimolar amount of the aldehyde was formed before the enzyme was reduced, as can be seen from the 330 nm progress curve at pH 8.0 (Figure 4b, right-hand panel). This observation suggests that the reaction product of p-topolin acts as an inhibitor.
Figure 4. Interaction of ZmCKX1 with p-topolin.
(a) Spectral changes in the range of 250–700 nm, and (b) time courses of the reaction intermediate decay (360 nm, open circles), reduction of the FAD cofactor (451 nm, triangles) and formation of the product 4-hydroxybenzaldehyde (330 nm at pH 8.0, filled circles) observed for ZmCKX1 (5 μM) that was mixed aerobically with 125 μM p-topolin in 75 mM imidazole/HCl (pH 6.5; left-hand panels) and with 13 μM p-topolin in 75 mM Tris/HCl (pH 8.0; right-hand panels). Arrows in (a) indicate the direction of spectral changes as observed from 1.3 min to 118.7 min of the reaction course. The data (b) could be fitted using a model in which species A decays in an exponential fashion to form B. To differentiate the kinetics of the formation of 4-hydroxybenzaldehyde (330 nm) at pH 8, the species are marked as A' and B'.
MS identification of cytokinin-derived imines
Q-TOF (quadrupole–time-of-flight) MS is a powerful tool for analysing the structures of small molecules that allows identification of a structural formula from accurate measurements of the masses of the parent ion and its fragments in the MS/MS mode [14]. Cytokinins were analysed in the direct injection mode and gave parent ions with m/z values that are shown in Table 2. For the analysis of cytokinin intermediates and products in the ZmCKX1 catalytic reaction, a reaction mixture composed of the enzyme and cytokinin was injected into the mass spectrometer shortly after component mixing. For all cytokinins studied, formation of a compound having the m/z of 2 Da lower than the original cytokinin substrate was found (Table 2). The elementary compositions were calculated from accurate masses of the parent ions and the results showed a loss of two hydrogens. This observation is in general agreement with the early proposal of the cytokinin imine formation, a cytokinin[-2H] intermediate of isopentenyladenine [10].
Table 2. MS/MS analysis of cytokinins and cytokinin reaction products.
Accurate masses of the parent ions of studied cytokinins (0.05 mM) in 30 mM ammonium hydrogen carbonate (pH 8.0) and their imine products (cytokinin [-2H]) after conversion by 1.5 μM ZmCKX1 (2 min incubation at 25 °C) were obtained by use of a Q-TOF hybrid mass analyser. For experimental details see Materials and methods section. Experimental values (m/z Exp.) are compared with those calculated for respective ion formulae (m/z Calc.).
| Cytokinin substrate and imine product | m/z Exp. | Formula | m/z Calc. | Difference (p.p.m.) |
|---|---|---|---|---|
| Isopentenyl adenine | 204.1247 | C10H14N5+ | 204.12492 | −1.1 |
| Isopentenyl adenine [-2H] | 202.1090 | C10H12N5+ | 202.10927 | −1.3 |
| trans-Zeatin | 220.1204 | C10H14N5O+ | 220.11984 | 2.6 |
| trans-Zeatin [-2H] | 218.1050 | C10H12N5O+ | 218.10419 | 3.7 |
| N-Methyl-isopentenyladenine | 218.1400 | C11H16N5+ | 218.14057 | −2.5 |
| N-Methyl-isopentenyladenine [-2H] | 216.1254 | C11H14N5+ | 216.12492 | 2.2 |
| Kinetin | 216.0880 | C10H10N5O+ | 216.08854 | −2.5 |
| Kinetin [-2H] | 214.0720 | C10H8N5O+ | 214.07289 | −4.1 |
| p-Topolin | 242.1030 | C12H12N5O+ | 242.10419 | −4.9 |
| p-Topolin [-2H] | 240.0881 | C12H10N5O+ | 240.08854 | −1.8 |
Parent ions of cytokinins were further fragmented to observe and analyse the fragmentation patterns in correlation with known structures. Fragmentation analysis was then applied to cytokinin[-2H] intermediates to analyse their structures. The results of the study are summarized in Figure 5 and Supplementary Figures 2–4 (at http://www.BiochemJ.org/bj/398/bj3980113add.htm).
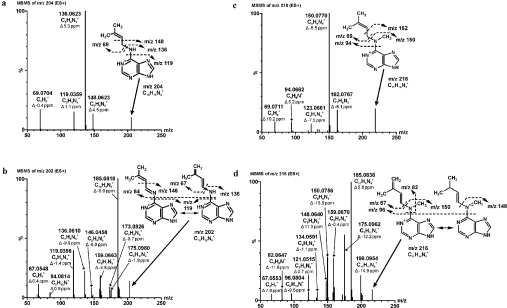
Figure 5. MS/MS analysis of the ZmCKX1 reaction with cytokinins.
Cytokinins [(a) isopentenyladenine (50 μM), (b) isopentenyladenine[-2H] intermediate, (c) N-methyl-isopentenyladenine (50 μM) and (d) N-methyl-isopentenyladenine[-2H] intermediate] were dissolved in 30 mM ammonium hydrogen carbonate (pH 8.0). For the reaction intermediate analysis, a reaction mixture that contained 1.5 μM ZmCKX1 and 50 μM cytokinin in 30 mM ammonium hydrogen carbonate (pH 8.0) was prepared and incubated at 25 °C for 2 min before the measurement. The high-resolution MS/MS experiments were done on a hybrid mass analyser Q-TOF micro with MassLynx data system software. The samples were applied in the form of an in-line spray at an injection rate of 10 μl/min. The data acquisition was performed in the range 50–1000 Da with a cycle time 47 μs, scan time 0.5 s, and collision energy 15, 20, 25, 30 and 35 V. The accurate masses of the parent ions and their fragments were calculated and used for the determination of the elementary composition and structure with fidelity within 15 p.p.m.
Fragmentation of the isopentenyladenine parent ion with a calculated molecular formula C10H14N5+ (Figure 5a) yielded the typical ions of C6H6N5+ (m/z 148.0623), C5H6N5+ (m/z 136.0623) and C5H3N4+ (m/z 119.0359) respectively, and the N6-side-chain ion C5H9+ (m/z 69.0704). The isopentenyladenine[-2H] intermediate ion C10H12N5+ (Figure 5b) had a much more complex fragmentation pattern that was rich in purine ring fragments (m/z 146.0458, 136.0610 and 119.0356) with ions of the side-chain structure, m/z 84.8014 (C10H4N+) and m/z 67.0548 (C5H7+). Besides other detected fragments (m/z 175.0980, 173.0826, 159.0663), a fragment C10H9N4+ (m/z 185.0818) had the maximal relative intensity. The same ion was also present in the MS/MS spectrum of trans-zeatin, although with lower intensity.
The fragmentation pattern of N-methyl-isopentenyladenine ion (C11H16N5+; see Figure 5c), had two ions, C7H8N5+ (m/z 162.0767) and C6H8N5+ (m/z 150.0770), that contained the N-methyladenine ring, and fragments C6H8N+ (m/z 94.0662) and C5H9+ (m/z 69.0711) that originated from a side chain. Other ions with lower intensity (e.g. C5H7N4+, m/z 123.0661) were observed. The N-methyl-isopentenyladenine[-2H] intermediate of the C11H14N5+ ion (Figure 5d) had a more complex MS/MS spectrum than that of N-methyl-isopentenyladenine. The presence of fragment pair C6H8N5+ (m/z 150.0756) and C5H7+ (m/z 67.0553) corresponded with the fragment spectrum of N-methyl-isopentenyladenine. Also, the pattern of purine ring fragments and associates (m/z 185.0838, 175.0962, 159.0670, 148.0640) indicated similarity with the fragmentation of the isopentenyladenine[-2H] intermediate (compare Figure 5b with 5d). However, unique ions of m/z 199.0954 (C11H11N4+), m/z 134.0591 (C6H6N4+) and m/z 121.0515 (C5H5N4+), as well as m/z 96.0804 (C6H10N+) and m/z 82.0647 (C5H8N+) that originated from the side chain, were also observed. The fragmentation pattern of the trans-zeatin C10H14N5O+ ion (Supplementary Figure 2a at http://www.BiochemJ.org/bj/398/bj3980113add.htm) had, besides various purine ring fragments and associates (m/z 185.0815, 148.0614, 136.0618 and 119.0356), loss of water to give rise to the ion C10H12N5+ (m/z 202.1088). Fragments of the trans-zeatin side chain were not found. The trans-zeatin[-2H] intermediate (C10H12N5O+) fragmentation (Supplementary Figure 2b at http://www.BiochemJ.org/bj/398/bj3980113add.htm) had a similar loss of water (C10H10N5+, m/z 200.0909) to the trans-zeatin MS/MS spectrum, one unique (C7H6N5+, m/z 160.0631) and three corresponding purine ring ions C6H6N5+ (148.0604), C5H6N5+ (136.0622) and C5H3N4+ (119.0368). A side-chain fragment, C5H7O+ (m/z 83.0494), was identified as a pair fragment of C5H6N5+.
The fragmentation of the kinetin parent ion with calculated molecular formula C10H10N5O+ (Supplementary Figure 3a at http://www.BiochemJ.org/bj/398/bj3980113add.htm) had the fragments m/z 188.0929 (C9H10N5+), m/z 148.0608 (C6H6N5+) and m/z 119.0357 (C5H3N4+), and the most intense ion m/z 81.0338 (C5H5O+). Only m/z 81.0338 was produced from the N6 side-chain, the other fragments contained the purine ring. The kinetin[-2H] intermediate ion with elementary composition C10H8N5O+ (Supplementary Figure 3b at http://www.BiochemJ.org/bj/398/bj3980113add.htm) underwent the loss of water to give rise to the ion C10H6N5+ (m/z 196.0617) and made the most intensive fragment m/z 119.0345 (C5H3N4+) with a pair fragment of the side chain m/z 96.0453 (C5H6NO+). Both a 3-fold higher intensity of ions m/z 186.0758 (C9H8N5+) and m/z 146.0453 (C6H4N5+) compared with the corresponding kinetin fragments m/z 188.0929 (C9H10N5+) and m/z 148.0608 (C6H6N5+) and an intensity loss of ion m/z 81.0347 (C5H5O+) were observed.
The fragmentation of the p-topolin ion of C12H12N5O+ (Supplementary Figure 4a at http://www.BiochemJ.org/bj/398/bj3980113add.htm) had a major occurrence of ion m/z 136.0608 (C5H6N5+) and also an ion of m/z 107.0499 (C7H7O+) derived from the cytokinin side chain. Fragments C6H6N5+ (m/z 148.0616) and C5H3N4+ (m/z 119.0346) were also observed. On the other hand, the p-topolin[-2H] intermediate of C12H10N5O+ (Supplementary Figure 4b at http://www.BiochemJ.org/bj/398/bj3980113add.htm) produced the most intensive fragment pair of m/z 119.0354 (C5H3N4+) and m/z 122.0598 (C7H8NO+) originating from the side chain. Ions of m/z 146.0460 (C6H4N5+) and m/z 95.0495 (C6H7O+) were the second pair products of fragmentation. The relative intensity of the ion fragment C5H6N5+ decreased from 100% in the p-topolin MS/MS spectrum to 4.3% in the MS/MS spectrum of the p-topolin[-2H] intermediate.
DISCUSSION
CKX is a covalent flavoprotein that plays a crucial role in controlling the level of plant cytokinins. It is capable of catalysing the oxidative side-chain cleavage of a number of cytokinins. A homology model [15] and later the crystal structure [9] revealed a two-domain folding topology similar to members of the vanillyl-alcohol oxidase flavoprotein family [16]. Similar to vanillyl-alcohol oxidase, ZmCKX1 can pass electrons from the covalently bound flavin to molecular oxygen. However, this process is not very efficient, as the reaction turnover of ZmCKX1 can proceed much faster with quinone-type electron acceptors [5]. The active site of ZmCKX1, where cytokinin catabolism takes place, was found to be unique among three-dimensional structures of proteins. A narrow pore, leading to the active site, allows entry only to an aliphatic or aromatic side chain of the cytokinin, the substrate moiety that is oxidatively cleaved. Adenine, the second entity of the cytokinin substrate, protrudes on the protein surface and seals the hydrophobic active site. Such a conformation leaves no space for an electron acceptor to bind in the active site [9], indicating that electron transfer to the protein surface may take place.
Although all cytokinin substrates bind, in principle, in the same structural way to the enzyme, some cytokinins, such as trans-zeatin or isopentenyladenine, react very quickly with the enzyme whereas others, such as N-methyl-isopentenyladenine, p-topolin or kinetin, react slowly [5]. In fact, until recently, aromatic cytokinins were not considered to be substrates of CKX [17]. However, data presented by Frébortová et al. [5] and in the present study show that ZmCKX1 is capable of cleaving aromatic cytokinins, albeit at very low rates. All substrates tested in the present study had a similar affinity for the enzyme, as binding constants only varied to a small extent, with Kd values ranging from 13–66 μM. This indicates that the enzyme mainly recognizes the cytokinins by interacting with the adenine moiety where it can easily accommodate various types of side chains. This is in agreement with the crystallographic data which show that there are several specific hydrogen-bond interactions between the protein and the adenine part of the substrate or product. The Kd values also correspond well with the measured Km′ values. Only the Km′ value observed for isopentenyladenine is relatively low, which can be explained by the fact that the rate of flavin reduction is extremely fast (1150 s−1).
To elucidate the mechanism of CKX-mediated catalysis, including intermediate formation, a pre-steady-state kinetic analysis of ZmCKX1 has been performed in the present study. The advantage of stopped-flow kinetic analysis, employed in the present study, is the ability to measure the reductive and oxidative half-reactions of the flavin-containing oxidoreductase separately. The time-resolved analysis also allows detection of possible spectral intermediates. It is known that isopentenyladenine degradation by CKX is connected with formation of an intermediate reported to be the cytokinin imine product [10] that absorbs at 310 nm. Chemical hydrolysis of this imine would produce adenine and a corresponding aldehyde. The measurements presented in the present study show that catalytic cleavage of all the cytokinins studied proceeds first through the formation of a reaction intermediate detectable at approx. 360 nm (Figure 2). This intermediate is not very stable and dissociates within seconds or even tenths of seconds. It can be ascribed to the formation of a binary complex between the corresponding imine and the reduced enzyme. A similar binary complex was previously observed in the reaction of a flavoprotein homologue, vanillyl-alcohol oxidase, with 4-(methoxymethyl) phenol [18]. In that case, the absorbance increase at 364 nm was ascribed to the formation of a binary complex between the enzyme and the quinone-methide of 4-(methoxymethyl)phenol.
For the cytokinins that rapidly reduce the flavin cofactor, isopentenyladenine and trans-zeatin, the interaction with ZmCKX1 also showed another unique feature that was disclosed by stopped-flow kinetic analysis: a charge-transfer complex is formed resulting in an absorbance band above 550 nm. This complex consists of reduced ZmCKX1 and a tightly bound cytokinin-derived imine positioned near the flavin cofactor. Binding of the imine product in close proximity to the flavin cofactor was also confirmed by X-ray analysis [9]. The crystal structures of ZmCKX1 in complex with the imines of isopentenyladenine and trans-zeatin showed that the imines are bound on top of the isoalloxazine ring of the cofactor. The structure of ZmCKX1 with the imine of trans-zeatin disclosed two possible conformations in which it is bound next to the flavin. In one conformation, charge-transfer is not feasible, which might explain the lower intensity observed for the trans-zeatin-induced charge-transfer absorbance when compared with isopentenyladenine. However, the two conformations of binding might also reflect the transition between the first formed binary complex of reduced ZmCKX1 with the imine and the subsequent conformation (spectra B and C in Figures 2a and 2b). The reductive half-reaction of ZmCKX1 with isopentenyladenine showed that it is undoubtedly the best substrate of the enzyme, being able to reduce the enzyme at a rate of approx. 1150 s−1. An additional hydroxy group present in trans-zeatin (Figure 1) apparently has a negative effect on the flavin reduction reaction, decreasing the reduction rate by approx. 75% (Table 1).
N-Methylation of isopentenyladenine (Figure 1) has a profound effect on the reactivity of this substrate with the enzyme. Although the Km′ value is in the same range as for other substrates, the kcat is low (Table 1). This can be explained by the poor reactivity of the carbon atom that should be oxidized by the flavin. Reduction of ZmCKX1 by N-methyl-isopentenyladenine revealed two reductive phases. Both reductive phases are slow, indicating that flavin reduction is limiting the turnover rate for this substrate. The observed reduction rates for both phases are 1000-fold lower than that with isopentenyladenine. The rates for both reductive phases were strongly dependent on substrate concentration, with similar apparent binding constants (25.1 and 13.3 μM) and relatively slow reduction rates (1.08 and 0.24 s−1, respectively). The extent by which the flavin was reduced in both phases, indicated by the absorbance decrease at approx. 450 nm as analysed by global diode-array data analysis, did not change depending on the substrate concentration. The observed rates obeyed Michaelis–Menten-type kinetic behaviour in which the reduction rate approaches zero when decreasing the substrate concentration. This is indicative of a reduction step that is virtually irreversible. Our results are consistent with a mechanism in which two species of ZmCKX1 exist, one reflecting a conformation that is slightly more efficient at oxidizing N-methyl-isopentenyladenine when compared with the other population. No charge-transfer complex was observed during reduction of the enzyme with N-methyl-isopentenyladenine, indicating that the oxidized cytokinin formed is positioned in a different way when compared with the non-methylated cytokinin isopentenyladenine.
Relatively slow reduction rates were also observed for kinetin and p-topolin. In both cases the reduction of the flavin cofactor involved a one step process. In this first relatively fast phase of flavin reduction, the absorbance at approx. 450 nm decreased significantly suggesting full reduction. This indicates that the reduction is an irreversible process. This was confirmed when analysing the observed substrate concentration-dependent reduction rates: the data could be fitted using a Michaelis–Menten equation. The rate of oxidation for these two cytokinins appears to be limited to the rate of flavin reduction.
Once the binary complex between the reduced enzyme and the cytokinin-derived imine is dissociated, a free cytokinin imine-product is formed. It is noteworthy that for all substrates tested in the present study the binary complex decay is slower by one or two orders of magnitude than flavin reduction (Table 1). This indicates that the binary complex accumulates during the reductive half-reaction. Despite this fact, however, binary complexes are not very stable and are lost within a few seconds or tenths of seconds.
It is known that hydrolysis converts the imine-products into adenine and the corresponding aldehydes that are routinely detected as final products of the enzymatic reaction. As the non-enzymatic hydrolysis is a relatively slow process, the imine accumulates in the reaction mixture as long as a supply of the substrate is available (Figure 3). Cytokinin imines generally accumulate more at slightly acidic rather than at slightly alkaline pH, whereas extreme pH values facilitate instant hydrolysis. For trans-zeatin, an additional reaction occurs at slightly alkaline pH that slows down hydrolytic cleavage and delays enzyme re-oxidation (Figure 3c). The aldehydes derived from isopentenyladenine, N-methyl-isopentenyladenine, trans-zeatin, and kinetin have absorption maxima at approx. or below 300 nm (Supplementary Figure 1 at http://www.BiochemJ.org/bj/398/bj3980113add.htm), and thus were not directly detectable in the presented measurements. On the other hand, the p-topolin-derived aldehyde, 4-hydroxybenzaldehyde, has an absorption maximum at 330 nm and is easily detectable at alkaline pH. When the enzyme was reduced by p-topolin, only the amount of the aldehyde equimolar to the enzyme was formed, and FAD re-oxidation was observed. This indicates that the active site of the enzyme becomes blocked by the p-topolin imine.
Each cytokinin studied gave specific fragmentation in the MS/MS experiments, but a neutral loss of the side-chain amine, resulting in formation of the cation C5H3N4+ that had low relative intensity (2–13%), was conserved. Cytokinins, when converted by ZmCKX1, produced cytokinin[-2H] intermediates that fragmented differently. Fragmentation patterns of these cytokinin-[-2H] intermediates clearly supported the structure of cytokinin imine products. Based on our results, the most likely molecular structures of cytokinin[-2H] intermediates are shown in Figure 5 and Supplementary Figures 2–4 (at http://www.BiochemJ.org/bj/398/bj3980113add.htm). The [-2H] intermediates of kinetin, p-topolin and isopentenyladenine produced the ion C5H3N4+ and the corresponding ions formed from the side chain (C5H6NO+, C7H8NO+ and C5H10N+ respectively) that became one of the major ions in the spectrum. The presence of the ion C6H4N5+ in the MS/MS spectrum of the kinetin[-2H] intermediate (Supplementary Figure 3b at http://www.BiochemJ.org/bj/398/bj3980113add.htm) and the p-topolin[-2H] intermediate (Supplementary Figure 4b at http://www.BiochemJ.org/bj/398/bj3980113add.htm) was indicative of a double bond on the N6 nitrogen of adenine. In contrast, the C6H4N5+ ion and the intensive pair ions (C5H6N5+ and C5H7+) were observed in the isopentenyladenine[-2H] intermediate fragmentation spectrum, indicating two possible locations of the double bond on the N6 side-chain (Figure 5b).
Cleavage of trans-zeatin to the ion C5H3N4+ led to a neutral loss of the side chain, but a side-chain fragment C5H7O+ was observed as a pair fragment to the ion C5H6N5+. The presence of ions C7H6N5+ and C6H6N5+ in the same MS/MS spectrum (Supplementary Figure 2b at http://www.BiochemJ.org/bj/398/bj3980113add.htm) highlighted two possible double bond locations on the N6 side-chain, but neither double bond was observed on the N6.
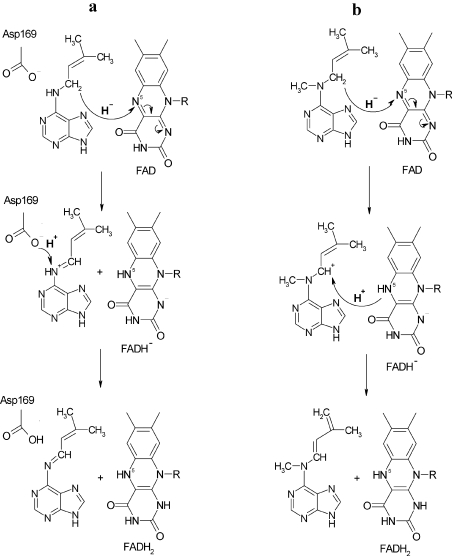
For N-methyl-isopentenyladenine, the fragmentation of the [-2H] intermediate did not show clearly the structure of an imine. The pair fragments (C6H8N5+ and C5H7+) and fragments of the side chain (C6H10N+ and C5H8N+) were observed and indicated a double-bond located on the N6-side chain but not its exact position. On the basis of the results, we propose that the cleavage of N-methyl-isopentenyladenine proceeds via a different mechanism than for isopentenyladenine. Figure 6(a) shows the reductive half-reaction of ZmCKX1 with isopentenyladenine [9], where a hydride ion is transferred from the α-carbon of the cytokinin to the N5 atom of FAD, which is followed by proton abstraction from N6 of the cytokinin by a catalytic base Asp169 to form the imine product. In the case of the N-methyl derivative, we propose again that a hydride is transferred to the flavin N5 atom (Figure 6b). However, the proton abstraction from N6 of the cytokinin substrate is impossible due to the methylation of the amine group. Therefore, in this case, the α-methylene group of the isoprenoid side chain is deprotonated, resulting in the formation and migration of a double bond. The identity of the base in this mechanism is as yet unknown. Asp169 or the reduced flavin cofactor could fulfil this function. Alternatively, they could both be able to act as a base, which would be in line with the observation that reduction of the enzyme with N-methyl-isopentenyladenine appears to follow two kinetic routes (see above). An analysis of an Asp169 mutant enzyme might reveal more details concerning this reaction step.
Figure 6. Reaction mechanisms of cytokinin dehydrogenase for (a) isopentenyladenine, as described by Malito et al. [9], and (b) N-methyl-isopentenyladenine proposed in the present study.
Note that Asp169 could also act as a base, instead of the N5 of the reduced flavin (see Discussion section).
In conclusion, the results of the present paper show that cytokinin cleavage via ZmCKX1 undergoes cascade catalysis and the resulting products, adenine and a side-chain aldehyde, are not directly formed by the enzyme, but result from a hydrolytic cleavage of cytokinin imine products. These products are rather stable and might have some physiological role during the cytokinin degradation in plants. In the case of trans-zeatin, the resulting aldehyde 4-hydroxy-3-methyl-2-butenal has been recognized as an oxidation product of isoprene emitted from vegetation [19]. This aldehyde is likely to undergo a cyclization reaction to form 2-methylfuran as the product. This reaction is facilitated in alkaline pH and slows down the catalytic reaction of ZmCKX1 (see Figure 3c). A similar compound, 4-hydroxy-2-butenal, is known to be a precursor of furan during decomposition of polyunsaturated fatty acids [20]. The possible physiological role of cytokinin imines, resulting aldehydes and their metabolites, remains to be investigated.
Online Data
Acknowledgments
This work was supported by the grant MSM-6198959216 from the Ministry of Education, Youth and Physical Education, Czech Republic. We thank Karel Doležal for helpful discussions.
References
- 1.van Berkel W. J. H., Benen J. A. E., Eppink M. H. M., Fraaije M. W. Flavoprotein kinetics. In: Chapman S. K., Reid G. A., editors. Methods in Molecular Biology, vol. 131, Flavoprotein Protocols. Totowa, NJ: Humana Press; 1999. pp. 61–86. [DOI] [PubMed] [Google Scholar]
- 2.Hare P. D., van Staden J. Cytokinin oxidase: biochemical features and physiological significance. Physiol. Plant. 1994;91:128–136. [Google Scholar]
- 3.Galuszka P., Frébort I., Šebela M., Peč P. Degradation of cytokinins by cytokinin oxidases in plants. Plant Growth Regul. 2000;32:315–327. [Google Scholar]
- 4.Galuszka P., Frébort I., Šebela M., Sauer P., Jacobsen S., Peč P. Cytokinin oxidase or dehydrogenase?. Mechanism of cytokinin degradation in cereals. Eur. J. Biochem. 2001;268:450–461. doi: 10.1046/j.1432-1033.2001.01910.x. [DOI] [PubMed] [Google Scholar]
- 5.Frébortová J., Fraaije M. W., Galuszka P., Šebela M., Peč P., Hrbáč J., Novák O., Bilyeu K. D., English J. T., Frébort I. Catalytic reactions of cytokinin dehydrogenase: preferences for quinones as electron acceptors. Biochem. J. 2004;380:121–130. doi: 10.1042/BJ20031813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fraaije M. W., Van den Heuvel R. H. H., Van Berkel W. J. H., Mattevi A. Covalent flavinylation is essential for efficient redox catalysis in vanillyl-alcohol oxidase. J. Biol. Chem. 1999;274:35514–35520. doi: 10.1074/jbc.274.50.35514. [DOI] [PubMed] [Google Scholar]
- 7.Laskey J. G., Patterson P., Bilyeu K. D., Morris R. O. Rate enhancement of cytokinin oxidase/dehydrogenase using 2,6-dichloroindophenol as an electron acceptor. Plant Growth Regul. 2003;40:189–196. [Google Scholar]
- 8.Kopečný D., Pethe C., Šebela M., Houba-Herin N., Madzak C., Majira A., Laloue M. High-level expression and characterization of Zea mays cytokinin oxidase/dehydrogenase in Yarrowia lipolytica. Biochimie. 2005;87:1011–1022. doi: 10.1016/j.biochi.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Malito E., Coda A., Bilyeu K. D., Fraaije M. W., Mattevi A. Structures of Michaelis and product complexes of plant cytokinin dehydrogenase: implications for flavoenzyme catalysis. J. Mol. Biol. 2004;341:1237–1249. doi: 10.1016/j.jmb.2004.06.083. [DOI] [PubMed] [Google Scholar]
- 10.Laloue M., Fox J. E. Abstracts of 12th International Conference on Plant Growth Substances. In: Bopp M., editor. Berlin, Germany: Springer-Verlag; 1985. p. 23. [Google Scholar]
- 11.de Jong E., van Berkel W. J. H., van der Zwan R. P., de Bont J. A. Purification and characterization of vanillyl-alcohol oxidase from Penicillium simplicissimum. A novel aromatic alcohol oxidase containing covalently bound FAD. Eur. J. Biochem. 1992;208:651–657. doi: 10.1111/j.1432-1033.1992.tb17231.x. [DOI] [PubMed] [Google Scholar]
- 12.Wang J., Letham D. S. Cytokinin oxidase – purification by affinity chromatography and activation by caffeic acid. Plant Sci. 1995;112:161–166. [Google Scholar]
- 13.Frébort I., Šebela M., Galuszka P., Werner T., Schmülling T., Peč P. Cytokinin oxidase/dehydrogenase assay: optimized procedures and applications. Anal. Biochem. 2002;306:1–7. doi: 10.1006/abio.2002.5670. [DOI] [PubMed] [Google Scholar]
- 14.Hopfgartner G., Varesio E., Tschappat V., Grivet C., Bourgogne E., Leuthold L. A. Triple quadrupole linear ion trap mass spectrometer for the analysis of small molecules and macromolecules. J. Mass Spectrom. 2004;39:845–855. doi: 10.1002/jms.659. [DOI] [PubMed] [Google Scholar]
- 15.Popelková H., Galuszka P., Frébortová J., Bilyeu K. D., Frébort I. Cytokinin dehydrogenase: Characterization and structure homology modeling of the flavoprotein catabolizing plant hormones cytokinins. Recent Research Developments in Proteins, vol. 2. In: Pandalai S. G., editor. Kerala: Transworld Research Network; 2003. pp. 63–81. [Google Scholar]
- 16.Fraaije M. W., van Berkel W. J. H., Benen J. A., Visser J., Mattevi A. A novel oxidoreductase family sharing a conserved FAD-binding domain. Trends Biochem. Sci. 1998;23:206–207. doi: 10.1016/s0968-0004(98)01210-9. [DOI] [PubMed] [Google Scholar]
- 17.Mok D. W., Mok M. C. Cytokinin metabolism and action. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:89–118. doi: 10.1146/annurev.arplant.52.1.89. [DOI] [PubMed] [Google Scholar]
- 18.Fraaije M. W., van Berkel W. J. H. Catalytic mechanism of the oxidative demethylation of 4-(methoxymethyl)phenol by vanillyl-alcohol oxidase. Evidence for formation of a p-quinone methide intermediate. J. Biol. Chem. 1997;272:18111–18116. doi: 10.1074/jbc.272.29.18111. [DOI] [PubMed] [Google Scholar]
- 19.Baker J., Arey J., Atkinson R. Formation and reaction of hydroxycarbonyls from the reaction of OH radicals with 1,3-butadiene and isoprene. Environ. Sci. Technol. 2005;39:4091–4099. doi: 10.1021/es047930t. [DOI] [PubMed] [Google Scholar]
- 20.Perez Locas C., Yaylayan V. A. Origin and mechanistic pathways of formation of the parent furan: a food toxicant. J. Agric. Food Chem. 2004;52:6830–6836. doi: 10.1021/jf0490403. [DOI] [PubMed] [Google Scholar]
- 21.Bilyeu K. D., Cole J. L., Laskey J. G., Riekhof W. R., Esparza T. J., Kramer M. D., Morris R. O. Molecular and biochemical characterization of a cytokinin oxidase from maize. Plant Physiol. 2001;125:378–386. doi: 10.1104/pp.125.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.