Abstract
Blood stages of Plasmodium falciparum export proteins into their erythrocyte host, thereby inducing extensive host cell modifications that become apparent after the first half of the asexual development cycle (ring stage). This is responsible for a major part of parasite virulence. Export of many parasite proteins depends on a sequence motif termed Plasmodium export element (PEXEL) or vacuolar transport signal (VTS). This motif has allowed the prediction of the Plasmodium exportome. Using published genome sequence, we redetermined the boundaries of a previously studied region linked to P. falciparum virulence, reducing the number of candidate genes in this region to 13. Among these, we identified a cluster of four ring stage-specific genes, one of which is known to encode an exported protein. We demonstrate that all four genes code for proteins exported into the host cell, although only two genes contain an obvious PEXEL/VTS motif. We propose that the systematic analysis of ring stage-specific genes will reveal a cohort of exported proteins not present in the currently predicted exportome. Moreover, this provides further evidence that host cell remodeling is a major task of this developmental stage. Biochemical and photobleaching studies using these proteins reveal new properties of the parasite-induced membrane compartments in the host cell. This has important implications for the biogenesis and connectivity of these structures.
INTRODUCTION
Plasmodium falciparum is the causative agent of the most severe form of human malaria, killing 1–2 million people each year (Snow et al., 2005). The symptoms of this disease are associated with the continuous asexual multiplication of P. falciparum parasites within red blood cells (RBCs) (for review, see Miller et al., 2002). In this part of the life cycle, the parasite develops, within 48 h from the ring stage, to the trophozoite stage and finally to the schizont stage after which the infected RBCs (IRBCs) rupture, releasing up to 32 merozoite stage parasites that infect new RBCs.
The ring stage lasts for almost half of this cycle (∼0-20 h after invasion) and shows low metabolic activity with little change in size and morphology (Zolg et al., 1984; de Rojas and Wasserman, 1985). P. falciparum ring forms are the only stages apparent in the blood circulation of infected humans, because more mature parasites adhere to the endothelium of various organs to avoid clearance by the spleen (Langreth and Peterson, 1985). The subsequent accumulation of infected RBCs in the microvasculature is largely responsible for malaria-associated morbidity and mortality. This sequestration of IRBCs is mediated by the parasite protein P. falciparum erythrocyte membrane protein 1 (PfEMP-1), which is exported to the surface of the IRBCs and is encoded by a family of variant antigens (Baruch et al., 1995; Smith et al., 1995; Su et al., 1995). Because human RBCs are devoid of a functional protein trafficking system, the parasite has to establish a protein export system in the host cell before PfEMP-1 can be displayed on the surface of the infected cell. This occurs concomitant with other extensive host cell modifications and involves export of a large number of parasite proteins into the RBC, causing dramatic changes in the topology, permeability, deformability and adhesiveness of the host cell (for reviews, see Cooke et al., 2001, 2004; Marti et al., 2005). PfEMP-1 occurs on the RBC surface ∼16-20 h after invasion (Gardner et al., 1996; Kriek et al., 2003), coinciding with the end of the ring stage. Hence, it seems that the survival of the rapidly growing later stages critically depends on successful host cell modifications during the ring stage, and it can be hypothesized that the slow growth of ring stages represents a “lag” phase during which the parasite installs these host cell modifications.
The first systematic screen for ring stage-specific genes in P. falciparum revealed 14 genes of which only two genes showed significant homology to known genes in other organisms (Spielmann and Beck, 2000). In contrast, >50% of trophozoite-specific genes had homologues in other organisms (Spielmann and Beck, 2000). Several of the identified genes have now been characterized and have all been shown to encode exported proteins. Early transcribed membrane proteins (ETRAMPs) localize to the parasitophorous vacuole membrane (PVM), which surrounds the parasite inside the host cell (Spielmann et al., 2003, 2006). Skeleton binding protein 1 (SBP1) and membrane-associated protein (MAHRP)1 localize to the Maurer’s clefts (Blisnick et al., 2000; Spycher et al., 2003), parasite-induced vesicular structures in the host cell that are involved in the export of cytoadherence determinants to the RBC surface (Wickham et al., 2001; Knuepfer et al., 2005b). Interestingly, SBP1 has now been shown to be essential for export of PfEMP-1 to the RBC surface (Cooke et al., 2006). Thus, it seems that many ring stage-specific genes code for proteins that 1) show no homologies to proteins in other organisms and 2) are exported into the host cell and therefore may be involved in host cell modifications. The lack of homology of these proteins to known proteins in other organisms indicates that this is at least in part a parasite-specific process with no precedent in mammalian biology and may represent a potential target for chemotherapeutic intervention.
A short sequence motif necessary for the export of Plasmodium proteins into the host cell has recently been identified (Hiller et al., 2004; Marti et al., 2004). This motif, termed Plasmodium export element (PEXEL) or vacuolar transport signal (VTS), was used to predict the P. falciparum exportome (Hiller et al., 2004; Marti et al., 2004; Sargeant et al., 2006). Most of the genes predicted to encode exported proteins show maximal transcription between the late schizont and late ring stage, peaking in early to mid-ring stage (Marti et al., 2004). P. falciparum seems to have a greatly expanded exportome compared with other Plasmodia, and this was attributed to the requirement for trafficking of cytoadherence determinants to the host cell surface (Sargeant et al., 2006).
Early studies showed that a region of ∼55 kb on the right arm of chromosome 9 was linked to cytoadherence in P. falciparum (Day et al., 1993; Barnes et al., 1994; Bourke et al., 1996). Because this region did not seem to encode PfEMP-1, its effect on cytoadherence must be indirect such as by impairing export or presentation of PfEMP1 (Bourke et al., 1996; Trenholme et al., 2000). Here, we report a more accurate mapping of the region linked to cytoadherence on chromosome 9, reducing the candidate genes to 13. We find that this region contains a cluster of four ring stage-specific genes. We have previously shown that one of these genes codes for a Maurer’s cleft protein (Hawthorne et al., 2004). Here, we report the characterization of the remaining ring stage-specific genes in that locus. In support of a major role of ring stages proteins in modification of the host cell, we found that all analyzed genes encode exported proteins. However, only two of the four ring stage-specific proteins encoded by this region have an obvious PEXEL motif. Because the ring stage-specific proteins, SBP1 and MAHRP1, also do not possess an obvious PEXEL, we propose that the analysis of genes exclusively transcribed in ring stages might add valuable components to the currently predicted exportome of P. falciparum and shed light on how proteins without an obvious PEXEL are exported into the host cell.
MATERIALS AND METHODS
PCR Analysis of Genomic B8, 3D7, and D10 DNA
Primers pairs PFI1715wF/PFI1715wR, PFI1755cF/PFI1755cR, PFI1770wF/PFI1770wR, PFI1775wF/PFI1775wR, PFI17580cF/PFI1780cR, and PFI1785wF/PFI1785wR (listed in Supplemental Table S1) and primers for the etramp2 control (Spielmann et al., 2006) were used on genomic DNA prepared from B8, 3D7, and D10 parasites.
Plasmid Construction
For the construction of plasmids for recombinant protein expression, the sequence corresponding to the rex2, rex3, and rex4 C termini were amplified from P. falciparum 3D7 genomic DNA using primers 40cfw and 40crv (corresponding to amino acids 61–94 of ring-exported protein 2 [REX2]), 55cfw and 55crv (corresponding to amino acids 48–326 of REX3), and 60wfw and 60wrv (corresponding to amino acids 44–214 of REX4) and cloned into the SmaI site of pGEX-6P-2 (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). Primers are listed in Supplemental Table S1.
Plasmids for P. falciparum transfection, containing the complete coding sequence for each of the genes, were obtained as follows. For the transgenic expression of REX4-GFP under the control of the crt promoter, rex4 was amplified from 3D7 cDNA using primers 60wfwAflII and 60wrvApaI and cloned into vector pARL (Wickham et al., 2001) modified to contain AflII/ApaI restriction sites between the crt promoter region and the GFP (pARL1mGFPmT; kindly generated by A. Adisa, La Trobe University, Melbourne, Victoria, Australia). For the expression of REX4myc in P. falciparum, we used a vector based on pHHC*/DR0.28 (O’Donnell et al., 2002) containing etramp4myc (Spielmann et al., 2006) cloned into the XhoI cleaved vector using XhoI/SalI to destroy the second XhoI site (Spielmann, unpublished data). This vector was cleaved with XhoI/StuI to replace etramp4 before the myc tag with full-length rex4, which was amplified from 3D7 cDNA using primers 60cfwfXhoI and 60crv. For the transgenic expression of REX2-GFP and REX3-GFP the corresponding coding sequences were amplified from 3D7 cDNA using primers 40cBamHI/40cPstI and 55cBamHI/55cPstI, respectively and cloned into pHGB Gateway Entry vector (a kind gift of Dr. Chris Tonkin, University of Melbourne) containing the crt promoter region (derived from vector pARL) and modified to contain BglII/PstI sites between the promoter and the green fluorescent protein (GFP). This vector containing either the rex2 or rex3 sequences was used in clonase reactions (Invitrogen, Carlsbad, CA) with a pHHC*/DR0.28 vector containing a destination cassette to obtain vectors mediating expression of REX2 and REX3 GFP fusion proteins under the crt promoter. Inserts of all constructs were confirmed by sequencing.
Production of Recombinant Proteins and Immunization of Mice
Glutathione S-transferase (GST) fusion proteins were expressed in Escherichia coli BL21 and purified as described previously (Spielmann et al., 2003). Mice were immunized according to standard procedures (Gardiner et al., 2004) using 30–50 μg of recombinant protein per injection.
Parasite Culture and Transfection
Plasmodium falciparum was cultured in RPMI 1640 medium containing 10% human serum as described previously (Trager and Jensen, 1976). Parasites used were 3D7, a clone of NF54 with full-length chromosome 9 (Walliker et al., 1987); D10, a noncytoadherent clone of FC27 lacking the right end of chromosome 9 (Anders et al., 1983); and B8, a cytoadherent clone of ITG2F6 with an intermediate deletion on the right arm of chromosome 9 (Biggs et al., 1989). Both D10 and B8 also have chromosome 2 deletion, which has removed both Knob-associated histidine-rich protein (kahrp) and erythrocyte membrane protein 3 (pfemp3). Plasmids were transfected into P. falciparum using standard electroporation conditions (Wu et al., 1996).
Immunofluorescence Assays and Fluorescence Recovery after Photobleaching (FRAP) Analysis
Immunofluorescence assays (IFAs) were carried out essentially as described previously (Spielmann et al., 2006) using air-dried IRBCs fixed with acetone (for REX2) or parasites coated onto wells using concanavalin A (for REX3 and REX4). Because fixing of the coated cells with 4% formaldehyde alone did not prevent the release of the RBC cytoplasm, we included 0.005% glutaraldehyde into the fixing solution (Tonkin et al., 2004). However, this protocol was not suitable for REX2, because only a poor signal was obtained (although staining pattern was comparable with the signal in acetone-fixed cells), irrespective of whether cells were permeabilized with 0.1% Triton X-100 after fixation. No signal was obtained with these sera when methanol fixation protocols (which retain the host cell cytoplasm) were used.
Antibody dilutions used were mouse anti-REX2 (1/1500), mouse anti-REX3 (1/1000), mouse anti-REX4 (1/400), rabbit anti-REX1 (1/2000), and rabbit anti-myc (1/150; all Cell Signaling Technology, Beverly, MA). Secondary antibodies were Cy2 and Texas Red-conjugated goat AffiniPure antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA). Samples were viewed on an Axioscope 2 Mot + (Carl Zeiss, Jena, Germany) equipped with a Zeiss 63×/1.4 Plan Apochromat lens. Images were captured with an Axiocam MRm camera (Carl Zeiss) using Axiovision AC software (Carl Zeiss).
FRAP was performed as described previously (Klonis et al., 2002; Knuepfer et al., 2005a) using an inverted Leica TCS SP2 confocal microscope equipped with a 100×/1.4 oil immersion lens. Samples were mounted in culture medium under a coverslip on to a glass slide. Photobleaching studies were performed using the time-lapse feature of the Leica TCS SP2 software and images were processed using NIH ImageJ (http://rsb.info.nih.gov/ij).
Selective Permeabilization of IRBCs, Proteinase K Protection Assay, and Triton X-114 Phase Separation
IRBCs were enriched from 20 ml of parasite culture (5% hematocrit, 5–10% parasitemia) using Percoll (Aley et al., 1986), washed in RPMI 1640 medium without serum, and equally split into three parts. Part 1 was dissolved in 100 μl of SDS-PAGE sample buffer (total IRBCs). Part 2 was treated with 70 μl of 0.03% saponin in phosphate-buffered saline (PBS) for 10 min on ice and centrifuged at 16,000 × g for 5 min to separate particulate fraction from the supernatant. The supernatant was brought to 100 μl with SDS-PAGE sample buffer, and the pellet was washed in PBS and dissolved in 100 μl of SDS-PAGE sample buffer. Part 3 was treated with 100 μl of PBS containing 100 U of activated streptolysin O (SLO; Sigma-Aldrich, St. Louis, MO) at 37°C for 15 min, centrifuged, and resuspended in SDS-PAGE sample buffer as described for the saponin lysed IRBCs. Equivalent fractional amounts were analyzed by immunoblotting.
For proteinase K protection assays, washed SLO pellets (∼2-5 × 108 parasites) were split into three fractions and resuspended in 100 μl of PBS or 100 μl of PBS containing 1 mg/ml proteinase K or 100 μl of 0.03% saponin in PBS containing 1 mg/ml proteinase K and stored on ice for 30 min. Reactions were stopped, and protein was precipitated by adding trichloroacetic acid to 10%. Pellets were resuspended in 1× SDS-PAGE sample buffer, and equal amounts were subjected to Western analysis.
Triton X-114 phase separation was carried out as described previously (Spielmann et al., 2003), except that protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany) was added to the protein extracts instead of phenylmethylsulfonyl fluoride.
Western Analysis
Protein extracts were resolved on reducing SDS-PAGE gels (12 or 15% [for REX2] acrylamide) and transferred to 0.2-μm polyvinylidene difluoride membranes (Macherey-Nagel, Aston, PA) using 10 mM 3-cyclohexylamino-1-propanesulfonic acid, pH 11.2, without methanol as described previously (Spielmann et al., 2006). Immunoreactions were carried out using the following primary antibody dilutions: anti-REX2 (1/2000), anti-REX3 (1/2000), anti-REX4 (1/400), anti-REX1 (1/5000), anti-ETRAMP4 (1/650), and monoclonal anti-GFP (Roche Diagnostics) (1/2000). Anti-SBP1C and anti-SBP1N (1/1000) were a kind gift of Dr. Brian Cooke (Monash University, Melbourne, Australia) and were used at 1/2000 and 1/1000, respectively. Monoclonal anti-P. falciparum glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies were a kind gift of Dr. C. Daubenberger (Swiss Tropical Institute, Basel, Switzerland) and were used at 1/2000. The rabbit anti-serine–rich protein (SERP) serum was a kind gift of Prof. K. Lingelbach (Philipps-University, Marburg, Germany) and was used at 1/1000. Sheep horseradish peroxidase-conjugated secondary antibodies (Chemicon International, Temecula, CA) were used at 1/5000, and detection was carried out using enhanced chemiluminescence (GE Healthcare).
RESULTS
A Cluster of Ring Stage-specific Genes in a Region on the Right Arm of Chromosome 9 Linked to Parasite Virulence
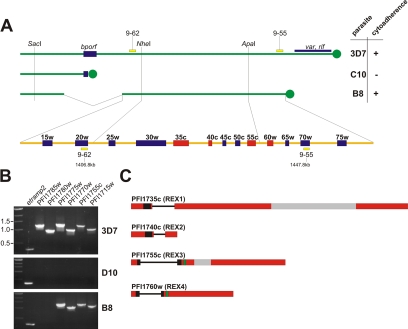
An ∼55-kb region on the right arm of chromosome 9 is known to be linked to cytoadherence and gametocytogenesis of IRBCs (Day et al., 1993; Barnes et al., 1994; Bourke et al., 1996). This region was mapped experimentally by Southern blotting with a number of sequence markers (Bourke et al., 1996; Holt et al., 1998). To identify the genes in this region, we used these sequence markers in BLAST searches (http://www.PlasmoDB.org) to locate the corresponding region in the completed P. falciparum genome (Figure 1). The outmost markers available comprise a region of 41 kb, including the genes PFI1720w–PFI1770w. The remaining ∼14 kb of the estimated 55-kb sequence linked to the observed phenotypes must therefore border this region on both sides. Toward the centromere this can only include gene PFI1715w, because this is the only gene located between PFI1720w and the common break point region associated with the loss of cytoadherence (Figure 1). If all of the remaining ∼14 kb were located toward the telomere, it would add the genes PFI1775w and PFI1780w. PCR analysis on genomic DNA of the parasite line B8 (which is cytoadherent) suggested that PFI1715w and PFI1775w were still present in the linked region, whereas PFI1780w was not (Figure 1B). Thus, the region linked to cytoadherence and gametocytogenesis contains 13 genes (PFI1715w–PFI1775w) (Figure 1A). These results confirmed the previous conclusion that this region does not contain any var genes and that the loss of cytoadherence must be due to an indirect effect on PfEMP-1, such as preventing its export/presentation on the RBC surface by impairing host cell modifications (Bourke et al., 1996; Trenholme et al., 2000).
Figure 1.
Remapping of the region on the right arm of chromosome 9 linked to cytoadherence. (A) The right arm of chromosome 9 is shown for the P. falciparum isolates 3D7 (cytoadherent), C10 (noncytoadherent), and B8 (cytoadherent) (modified from Bourke et al., 1996). Single restriction sites determined by Southern analysis (Barnes et al., 1994) and the previously published markers 9-62 and 9-55 (Holt et al., 1998) were compared with the published 3D7 genome (www.PlasmoDB.org) using BLAST and virtual digests (NEBcutter, version 2.0; http://tools.neb.com/NEBcutter2/index.php). The region linked to cytoadherence and the predicted genes therein (as determined by the Plasmodium Sequencing Project and available at http://www.PlasmoDB.org) are indicated below (not drawn to scale). Genes PFI1715w–PFI1775w are indicated as 15w–75w. bporf, breakpoint open reading frame; telomeres are indicated as filled circles; the positions of the markers 9-62 and 9-55 on chromosome 9 are indicated in kilobase pairs; ring stage-specific genes are shown as red boxes. (B) PCR on genomic DNA of 3D7, D10 (which contains a deletion on the right arm of chromosome 9 similar to C10), and B8 showed that B8 still contains gene PFI1775w but not PFI1780w. Thus, the region linked to cytoadherence comprises the genes PFI1715w–PFI1775w. etramp2 was used as a positive control. (C) Gene structure of the four ring stage-specific genes in this region. Exons are indicated as boxes, introns as lines; hydrophobic stretches are indicated in black; PEXEL motifs in green; and areas encoding repeats are in gray.
To identify candidate genes coding for proteins involved in host cell modifications, we looked for ring stage-specific genes in the region defined above. Strikingly, analysis of the published transcriptome data (LeRoch et al., 2003) revealed that four of the 13 genes in this region were ring stage-specific (PFI1735c, PFI1740c, PFI1755c, and PFI1760w). PFI1740c has previously been identified in a screen for ring stage-specific genes (Spielmann and Beck, 2000), and we previously characterized PFI1735c (Hawthorne et al., 2004), which we termed REX1. By analogy, we tentatively named PFI1740c, PFI1755c, and PFI1760w REX2, REX3, and REX4, respectively.
The transcriptome data (LeRoch et al., 2003) also indicated an induction of PFI1765c and PFI1770w in ring stages; however, transcription levels are comparably low and increase significantly in gametocytes, indicating that the increased transcription in ring stages might arise from parasites committed to gametocytogenesis, which at that stage cannot be distinguished from other ring stages. Therefore, these genes were not analyzed further.
REX2, REX3, and REX4 Are Encoded by Two Exon Genes and Show No Homology to Known Proteins in Other Organisms
REX2, REX3, and REX4 as well as the previously described REX1 (PFI1735c), are encoded by two exons (as predicted by the Plasmodium Sequencing Project; http://www.Plasmodb.org). Sequencing of cDNA confirmed the predicted gene structure for rex3 and rex4, whereas the gene structure of rex2 differed from the prediction in PlasmoDB but was in accordance with a previously published cDNA fragment (Spielmann and Beck, 2000; GenBank accession no. AJ290921) (our unpublished data). All four genes encode proteins containing a single hydrophobic stretch (Figure 1C). Both REX3 and REX4 contain a PEXEL motif after the hydrophobic stretch (Marti et al., 2004), suggesting that they might be exported into the host cell. REX1 and REX2 do not have a discernible PEXEL motif. REX2, REX3, and REX4 show no similarity to known proteins in other organisms. However, REX3 and REX4 share 31% identity over 154 amino acid residues with a single gap in REX3 (our unpublished data). Their adjacent location on the chromosome indicates that they may have arisen from a gene duplication event. It is noteworthy that the domain homologous between REX3 and REX4 is also found in proteins in P. vivax (PlasmoDB accession no. Pv_6869, Pv_6571) and P. knowlsei (PlasmoDB accession no. Pk112f03plc) but not in rodent malaria parasites (our unpublished data).
REX2 and REX3 Are Exported Proteins
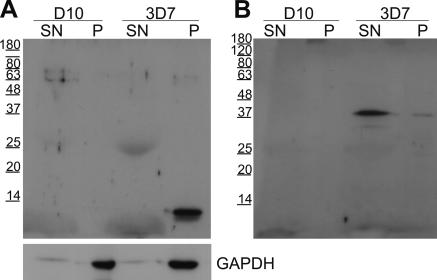
To analyze whether REX2, REX3, and REX4 were exported proteins, we raised antisera in mice against the recombinantly expressed C termini of these proteins. The sera raised against REX2 and REX3 each reacted with a single band in extracts of 3D7 parasites but not in extracts of D10 parasites (which do not possess the right arm of chromosome 9) (Figure 2). This indicated that these sera were specific for the respective proteins. REX2 migrated at 13 kDa (predicted full-length molecular mass, 10.7 kDa) and REX3 at 39 kDa (predicted full-length molecular mass, 37.6 kDa). Taking into account the possible cleavage of the hydrophobic stretch as a signal peptide, these proteins migrate slower in SDS-PAGE gels than expected. This is probably due to nonuniform binding of SDS to these proteins rich in charged residues and has previously been noted for P. falciparum proteins (Coppel et al., 1994). The serum raised against REX4 did not detect any protein in 3D7 parasite extracts but detected a single band in transgenic parasites expressing GFP- or myc-tagged REX4 (see below). This suggested that endogenous REX4 was not made in sufficient levels to be detected by this serum (although the serum was specific and was sufficiently sensitive to detect chimeric REX4 in total parasite extracts).
Figure 2.
Sera raised against the C terminus of REX2 and REX3 recognize a single band in Western blots with 3D7 parasite extracts but not with D10 parasites. D10 and 3D7 parasites were saponin lysed, and the pellet and the supernatant fraction were analyzed by Immunoblotting. When probed with the serum raised against REX2, a single band of 13 kDa was detected in the saponin pellet of 3D7 but not D10 parasites (A). A single band of 39 kDa was observed in the saponin supernatant when probed with the serum raised against REX3 (B). Western analysis of the same samples using a serum against GAPDH demonstrated equal amounts of protein present in D10 and 3D7 parasite and also showed that saponin treatment did not release the contents of the parasite cytoplasm. Size markers are indicated in kilodaltons on the left of each blot.
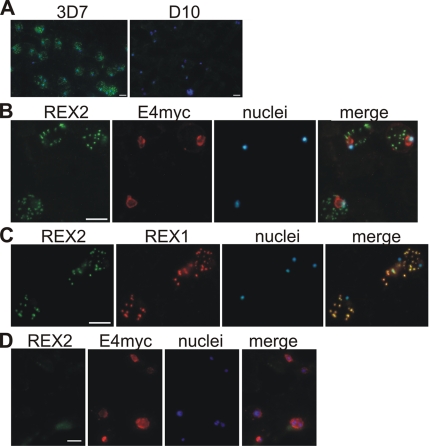
In IFAs, REX2 was detected in a punctuate pattern in 3D7 IRBCs but not in D10 parasites, confirming the specificity of this serum in IFA (Figure 3A). We then used transgenic 3D7 and D10 parasites expressing a myc-tagged ETRAMP4, a marker for the parasite PVM that can be detected by commercial rabbit anti-myc serum (Spielmann et al., 2006). Colocation studies with the mouse anti-REX2 serum showed that the dots labeled with this serum were situated outside of the parasite PVM in 3D7-ETRAMP4myc parasites (Figure 3B). No signal was detected in D10-ETRAMP4myc parasites (Figure 3D). Thus, REX2 is exported into the host cell. The punctate staining pattern in the host cell cytoplasm is typical of Maurer’s cleft staining. Colocation with the previously described Maurer’s cleft protein REX1 in 3D7 parasites confirmed that REX2 localizes to these structures (Figure 3C).
Figure 3.
Location of REX2 in acetone fixed IRBCs. (A) Specificity of the anti-REX2 serum: anti-REX2 serum reacted only with 3D7 (left) but not D10 parasites (right). Merges with nuclear stain (Hoechst) are shown. (B) IFAs with 3D7 parasites expressing a myc-tagged ETRAMP4 (Spielmann et al., 2006) showed that REX2 (green; Cy2) has a dotty appearance outside of the PVM (ETRAMP4myc in the PVM detected by a rabbit anti-myc antibody; red, Texas Red). No REX2 signal was detectable in D10 parasites expressing ETRAMP4myc (D). (C) REX2 (green; Cy2) colocalized with the Maurer’s cleft protein REX1 (Hawthorne et al., 2004) (red; Texas Red) in 3D7 parasites. Bars, 5 μm.
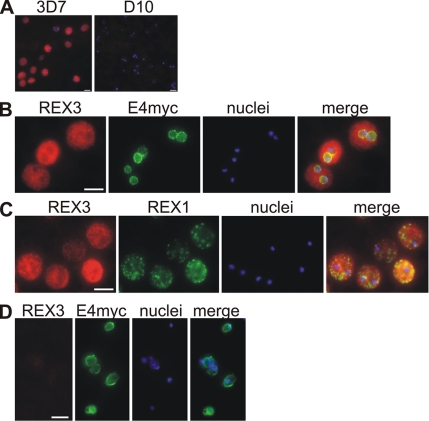
The serum raised against REX3 also reacted only with 3D7 but not D10 parasites (Figure 4A). Similar to REX2, it stained predominantly the area outside of the PVM as judged by colocation with ETRAMP4myc in transgenic parasites (Figure 4B). However, the staining was more uniform than seen with Maurer’s cleft proteins and colocation IFAs with REX1 in 3D7 parasites showed that REX3 staining was not restricted to the clefts (Figure 4C). This indicated that REX3 is exported into the cytoplasm of the host cell but does not accumulate at any particular structure therein. Thus, both REX2 and REX3 seem to be exported proteins but localize to different compartments in the infected host cell.
Figure 4.
Location of REX3 in IRBCs fixed with 4% formaldehyde/0.005% glutaraldehyde. Acetone fixation was not suitable to detect REX3 (see Materials and Methods). (A) Specificity of the anti-REX3 serum: anti-REX3 serum reacted only with 3D7 (left) but not D10 parasites (right). Merges with nuclear stain (Hoechst) are shown. (B) IFAs with 3D7 parasites expressing a myc-tagged ETRAMP4 (Spielmann et al., 2006) showed that REX3 (red; Texas Red) occurs as a uniform stain outside of the PVM (ETRAMP4myc in the PVM detected by a rabbit anti-myc antibody; green, Cy2). The fainter red stain inside the area delineated by the PVM is likely to represent REX3 in the host cell cytoplasm from different focal planes above/below the parasite. No REX3 signal was detectable in D10 parasites expressing ETRAMP4myc (D). (C) REX3 (red; Texas Red) did not colocalize with the Maurer’s cleft protein REX1 (Hawthorne et al., 2004) (green; Cy2) in 3D7 parasites. Bars, 5 μm.
REX2 Is an Integral Maurer’s Cleft Protein with its C Terminus Facing the Host Cell Cytoplasm
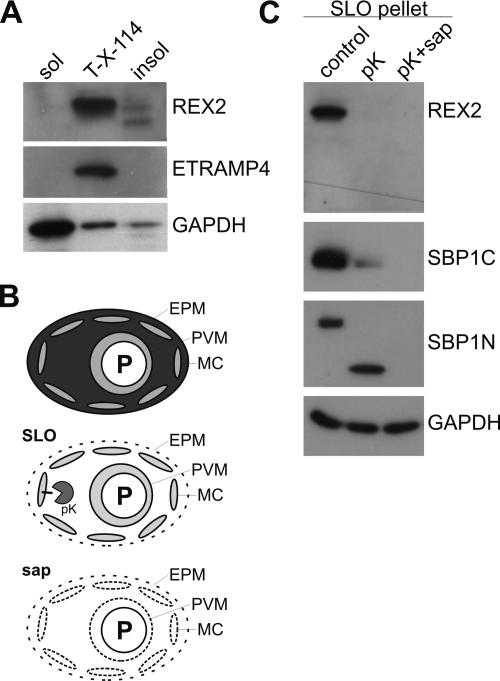
To test the nature of the association of REX2 with the Maurer’s clefts, we studied the solubility of REX2. Saponin treatment lyses the erythrocyte membrane and the PVM of IRBCs (Figure 5B). The parasites and the ghosts of infected cells (including the Maurer’s clefts) can then be pelleted to separate them from the soluble content of the IRBCs. Western analysis showed that REX2 was found in the pellet of saponin lysed IRBCs (Figure 2A). This indicated that REX2 is tightly associated with the Maurer’s clefts. To test whether REX2 is a Maurer’s cleft integral membrane protein and whether its hydrophobic stretch acts as a transmembrane domain, we performed Triton X-114 partitioning of saponin-released parasites. The integral PVM protein ETRAMP4 (Spielmann et al., 2003) and the predominantly soluble cytoplasmic parasite protein GAPDH (Daubenberger et al., 2003) were used as controls (Figure 5A). REX2 was predominantly associated with the membrane fraction, but some REX2 occurred as two bands of slightly smaller size in the Triton X-114–insoluble fraction (Figure 5A). This suggested that REX2 is an integral membrane protein of the Maurer’s clefts and that a smaller fraction of REX2 molecules is Triton X-114 insoluble. Because the smaller bands present in the insoluble fraction were not visible in total extracts, it is possible that these smaller species resulted from nonphysiological limited proteolysis in this fraction (which must have occurred despite the presence of protease inhibitors).
Figure 5.
Biochemical confirmation of REX2 location and analysis of its topology at the Maurer’s clefts. (A) Triton X-114 partitioning of 3D7 parasite extracts. REX2 was predominantly found in the membrane fraction (lane T-X-114). A smaller proportion of REX2 molecules were found in the insoluble fraction (lane insol). No REX2 was present in the soluble fraction (lane sol). The integral PVM protein ETRAMP4 was used as a positive control on the same samples. Reprobing of the membrane with a monoclonal anti-GAPDH antibody showed the expected distribution for this protein, which is known to be predominately soluble in the parasite cytoplasm with a smaller fraction associated with particulate material after parasite lysis (Daubenberger et al., 2003). (B) Schematic depiction of the treatment of IRBCs (top cell) with SLO (second cell from top) or saponin (sap, bottom cell). SLO treatment releases only the cytoplasm of the IRBC (Ansorge et al., 1996; Kriek et al., 2003). Saponin treatment also permeabilizes the PVM (Ansorge et al., 1996) and the Maurer’s clefts (this study). Addition of proteinase K (pK) will digest protein domains not protected by the Maurer’s cleft membrane or the PVM. Black bar, integral Maurer’s cleft protein. (C) Proteinase K protection assay showed that the C terminus of REX2 faces the host cell cytoplasm and confirmed that REX2 is an exported protein. SLO-treated parasites were either incubated with pK or proteinase K in the presence of saponin (pK + sap) or mock treated (control). Western analysis showed that REX2 was digested both in presence and absence of saponin, demonstrating that its C terminus faces the host cell cytoplasm. The integral Maurer’s cleft protein SBP1 served as a control for the integrity of the clefts and completeness of the SLO lysis. SBP1 is located in the Maurer’s clefts membrane with its C terminus facing the red cell cytoplasm (Blisnick et al., 2000). Analysis of this protein in these samples showed that its C terminus was digested in the presence of proteinase K (using a serum against its C terminus) and that digestion left a smaller N-terminal fragment (using a serum against its N terminus) that was protected by the cleft membrane. In the presence of saponin, SBP1 was completely digested, demonstrating that the Maurer’s clefts were permeabilized by this treatment. Membranes were reprobed for GAPDH to show equal loading and that parasite internal proteins were not digested.
To biochemically confirm the IFA results and to analyze the orientation of REX2 in the Maurer’s clefts, we treated IRBCs with SLO, which permeabilizes the RBC membrane of IRBCs but leaves the PVM (Ansorge et al., 1996) and the Maurer’s clefts intact (Kriek et al., 2003; Figure 5B). These cells were then subjected to digestion with proteinase K in the presence or absence of saponin and analyzed by immunoblotting using the serum raised against REX2, which is directed against its C-terminal region. The C-terminal region of REX2 was not protected from digestion, both in presence and absence of saponin (Figure 5C), indicating that this domain faces the host cell cytoplasm (if REX2 had the opposite orientation in the cleft membrane, a smaller band would have been expected after proteinase K treatment without saponin). These results also confirmed that REX2 is an exported protein, because it otherwise would not have been accessible for digestion. To confirm the integrity of the clefts after SLO treatment and to check the completeness of the SLO lysis, extracts from each experiment were tested for the integral Maurer’s cleft protein SBP1 (Blisnick et al., 2000). SBP1 is a single-pass transmembrane protein with its C-terminal region facing the RBC cytoplasm and its N terminus buried inside the clefts. After SLO treatment, the C-terminal region of SBP1 was digested upon proteinase K treatment, whereas the Maurer’s cleft internal N-terminal region was protected and occurred as a smaller fragment (Figure 5C). In the presence of saponin, both the N- and C-terminal regions of SBP1 were digested, but the parasite internal protein GAPDH was protected (Figure 5C). These data show that proteinase K had access to digest only exported proteins and that the Maurer’s clefts were still intact after SLO treatment. Moreover, because the Maurer’s cleft lumenal part of SBP1 was digested in the presence of saponin, these results suggested that the Maurer’s cleft membrane has similar properties to the PVM, which is known to be permeabilized by this reagent.
REX3 Is a Soluble Protein Exported into the Host Cell Cytoplasm
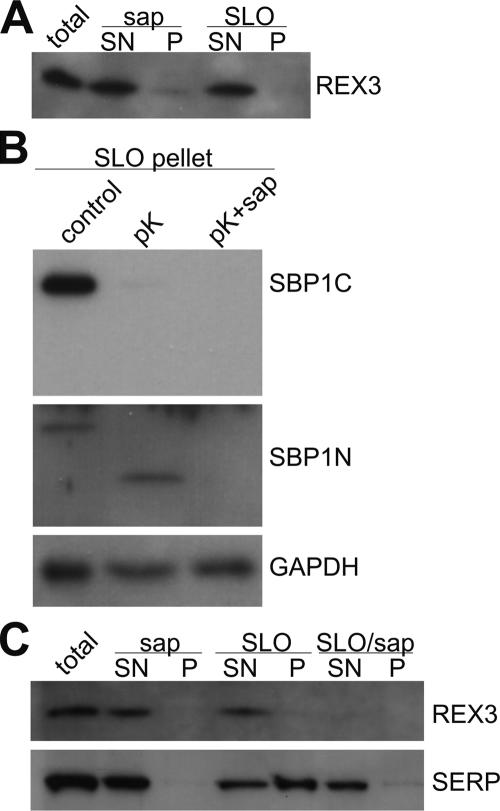
In IFAs, REX3 showed distribution throughout the host cell cytoplasm (Figure 4) and Western analysis indicated that it was released by saponin treatment of IRBCs, indicating that it is a soluble exported protein (Figure 2). However, because saponin also permeabilizes the PVM and the Maurer’s clefts, a location in the parasitophorous vacuole (PV) or inside of the Maurer’s clefts in addition to the host cell cytoplasm cannot be excluded. For example, glycophorin-binding protein is found both in the PV and the host cell cytoplasm (Ansorge et al., 1996). We therefore treated IRBCs with SLO, which does not release the contents of the PV and the Maurer’s clefts. Nevertheless, REX3 was still quantitatively released under these conditions (Figure 6A). To confirm that the SLO treatment did only permeabilize the RBC plasma membrane, we treated the lysed cells with proteinase K and analyzed SBP1. Because the SBP1 C terminus was digested but the N terminus protected, this showed that the SLO lysis was quantitative without breaching the Maurer’s clefts (Figure 6B). This excludes that the presence of REX3 in the clefts lumen. When we analyzed the PV soluble protein SERP (Delplace et al., 1987), we noticed that the PVM was partly breached under these SLO concentrations, as apparent from the release of some of this protein into the supernatant (Figure 6C). However, although treatment of the SLO pellet with saponin released the remaining SERP, no REX3 was released. Thus, REX3 was already quantitatively released by the SLO treatment, indicating that no REX3 was present in the PV (Figure 6C). From these results it can be concluded that REX3 is located exclusively in the host cell cytoplasm. Moreover, these results suggest that although the PVM and the Maurer’s cleft membrane are similarly permeabilized by the saponin concentrations used, the PVM seems to be more susceptible to SLO treatment than the Maurer’s clefts.
Figure 6.
Biochemical confirmation of the location of REX3. (A) IRBCs (total) were treated with saponin (sap) or SLO and separated into pellet (P) and supernatant (SN). These fractions were analyzed for the presence of REX3 by immunoblotting. REX3 was found in the supernatant after each treatment, demonstrating that it is exported into the host cell cytoplasm. To confirm that the SLO treatment did not breach the Maurer’s clefts or the parasite plasma membrane, the SLO pellet was subjected to proteinase K treatment and analyzed for SBP1 and GAPDH (B) as described in Figure 5. (C) To confirm that no REX3 was present in the PV, parasites were lysed as described in A, but the SLO pellet (containing the parasite inside the PVM) was also subjected to saponin treatment (SLO/sap) and separated into supernatant and pellet. These samples were analyzed for REX3 and the PV marker SERP on Western blots. Although the PVM of some cells was breached by the SLO concentration used (as apparent from partial release of SERP), a significant part of SERP was only released after saponin treatment of the SLO pellet. This fraction did not contain any detectable REX3, excluding the presence of this protein in the PV.
Analysis of Transgenic REX2 and REX3 in Live Parasites
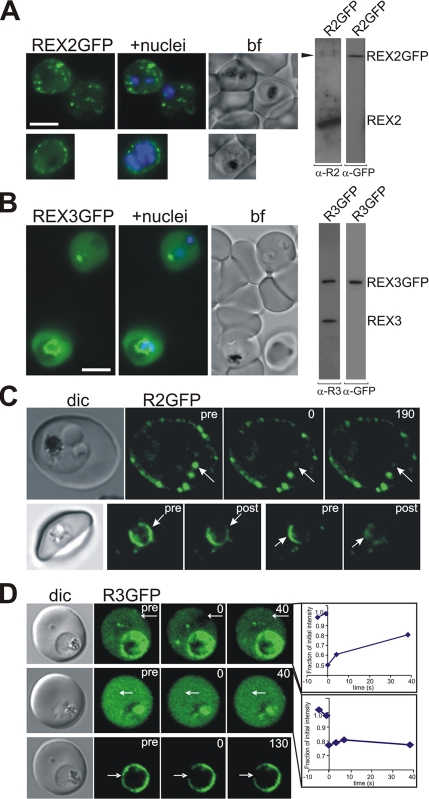
To analyze REX proteins in live parasites, we transfected 3D7 parasites with constructs mediating expression of full-length GFP-tagged REX2 and REX3, respectively. Both GFP-tagged proteins were successfully expressed in the presence of the endogenous protein, and the integrity of the chimeric proteins was maintained as judged by Western analysis (Figure 7, A and B). Single additional bands with molecular masses of 39 and 66 kDa were observed for REX2-GFP and REX3-GFP, respectively. This is in good agreement with the expected molecular mass of these chimeric proteins. The location of the transgenic fusion proteins matched that of the endogenous protein. REX2-GFP occurred as puncta of fluorescence in the host cell cytoplasm, which is typical for a Maurer’s cleft location (Figure 7A). A three-dimensional reconstruction (see Supplemental Movie 1) indicates that these puncta lie close to the RBC membrane, as previously reported for Maurer’s clefts (Wickham et al., 2001; Spycher et al., 2003). A time-lapse movie (see Supplemental Movie 2) shows that these puncta are not mobile, which is consistent with tethering of the Maurer’s clefts to the RBC cytoskeleton (Blisnick et al., 2000). Interestingly, younger stage transfectants were occasionally observed (<5% of cells) in which REX2-GFP was associated partly with the PV and partly with structures that were mobile in the host cell cytoplasm (see Supplemental Movie 3). This may represent the formation and budding of nascent Maurer’s clefts (Spycher et al., 2006), although other explanations for this phenomena are possible. These data show that a full-length PEXEL-negative protein fused to GFP can be exported into the host cell and correctly trafficked to the Maurer’s clefts.
Figure 7.
Analysis of GFP-tagged REX2 and REX3 in live parasites. (A) Live 3D7 parasites expressing REX2-GFP show a punctuate pattern of fluorescence in the host cell cytoplasm when analyzed by fluorescence microscopy. Some chimeric REX2 remains within the parasite. The top panels show two trophozoite-infected cells, and the bottom panel shows a schizont-stage parasite. Right, Western blot analysis of extracts of 3D7 parasites expressing GFP-tagged REX2 (R2GFP) showing integrity of the chimeric protein. REX2-GFP was only poorly detected by the specific REX2 serum, possibly due to masking of the short C-terminal domain by the GFP or due to comparably low expression levels of the chimeric protein. (B) Live 3D7 parasites expressing REX3-GFP show a more homogeneous fluorescence in the host cell cytoplasm. Some of the REX3-GFP protein remains within the parasite. The top RBC is infected with two ring-stage parasites and the lower cell with a trophozoite stage parasite. Nuclei were stained with Hoechst; bf, bright field image. Bars, 5 μm. Right, Western blot analysis of extracts of 3D7 parasites expressing GFP-tagged REX3 (R3GFP) showing integrity of the chimeric protein. (C and D) Photobleaching analysis of REX2-GFP (R2GFP) and REX3-GFP (R3GFP) parasites using confocal microscopy. Each series consists of a differential interference contrast image (dic), and images showing prebleach fluorescence (pre), fluorescence after the bleach pulse (time point 0), and postbleach images (post) were acquired after the times indicated (in seconds). The position of the bleach pulse is indicated by a white arrow. (C) Top row, after a 250-ms bleach pulse of an individual focal region of REX2-GFP fluorescence, no recovery is seen over the time frame analyzed (190 s). Bottom row, two different regions of the PV were subjected to bleach pulses (3 × 30 s each). Localized bleaching of the PV-associated REX2-GFP is observed which does not affect the regions distal to the bleach pulse. (D) Top, after a 1-s laser pulse, some localized bleaching of REX3-GFP fluorescence was observed in the host cell cytoplasm with recovery of the signal over 40 s. Middle, in a different infected red blood cell, no localized bleaching of REX3-GFP in the host cell cytoplasm is seen after a 1-s laser pulse. The graphs show the fluorescence intensity over time in the bleached region relative to the prebleach fluorescence levels. The bottom panels show localized bleaching of PV-associated REX3-GFP after a 750-ms laser pulse. No recovery of fluorescence is observed over the time frame analyzed (130 s).
By contrast, REX3-GFP showed a more homogenous distribution in the host cell (Figure 7B). Some of the population of the chimeric proteins was also observed in the PV or inside of the parasite, presumably in the endoplasmic reticulum (Figure 7). This indicates a reduced efficiency of export due to the GFP-tag. A time-lapse movie of a REX3-GFP transfectant shows apparent diffusion of the labeled protein within the host cell cytosol and indicates the presence of some larger aggregates of protein (see Supplemental Movie 4).
To analyze the interconnectivity of the Maurer’s clefts and the molecular organization of the chimeric proteins, we performed FRAP on individual REX2-GFP foci in REX2-GFP transfectants. After a short bleach pulse to ablate the signal associated with an individual cleft, no recovery of fluorescence was observed over 2 min (Figure 7C, top). This indicates that there are no connections between individual clefts or that REX2-GFP is restricted in its ability to diffuse in the cleft membrane by a strong attachment to fixed structures. In some early stage parasites, a population of REX2-GFP was observed in association with the PV confirming that this is an intermediate compartment en route to the Maurer’s clefts. Repeated bleaching of one region of the PV depleted the fluorescence signal from that region but not from other regions (Figure 7C, bottom). This suggests that PV-associated REX2-GFP is contained within subcompartments of the PV as has been reported for other exported proteins (Wickham et al., 2001; Adisa et al., 2003).
REX3-GFP in the host cell cytoplasm was also subjected to a photobleach analysis (Figure 7D). In some cells, localized bleaching was observed with recovery within a few seconds (Figure 7D, top). Recovery on this time scale is consistent with the predicted mobility for a protein aggregate (Klonis et al., 2002) and suggests that REX3-GFP is present in the IRBC cytosol as part of a complex with itself or with other proteins. Similar results have been observed for other GFP fusion proteins in the IRBC cytosol (Knuepfer et al., 2005a, b). In some cells, application of a bleach pulse caused a small, uniform decrease in fluorescence intensity with no obvious local bleaching and no recovery (Figure 7D, middle). This indicates a very rapidly diffusing species with a half-time for recovery that is quicker than the time taken (∼1 s) to collect the first postbleach image and is consistent with a monomeric protein or a small aggregate. Together, these data suggest that chimeric REX3 protein is present as a component of complexes of variable size throughout the host cell cytoplasm. The PV-associated population of REX3 was also examined by photobleach analysis. Again, bleaching of one region of the PV depleted the fluorescence signal from that region but not from other regions (Figure 7D, bottom), indicating the presence of PV subcompartments.
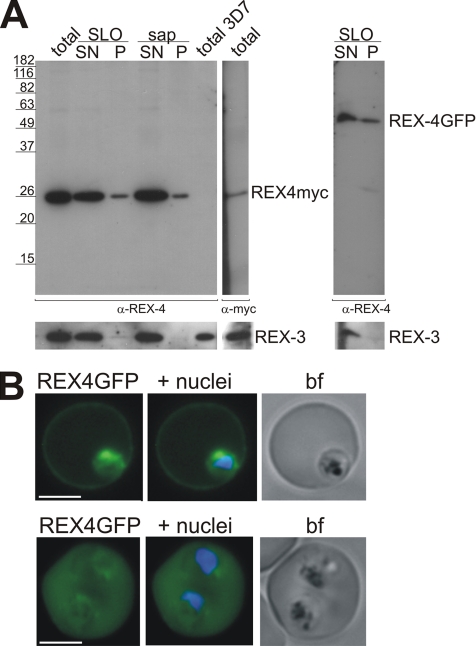
Transgenic REX4 Is Exported into the Host Cell
Despite the clear evidence for transcription of REX4 (LeRoch et al., 2003; our unpublished reverse transcription-PCR results), we were unable to detect the corresponding protein by IFA or immunoblots of asexual stage parasites of a number of different strains (our unpublished data). REX4 was also not detectable in cultures that were stressed to induce gametocytogenesis. However, our serum specifically detected GFP-tagged or myc-tagged REX4 in transgenic parasites, demonstrating that it is sufficiently sensitive to detect this protein in total parasite extracts (Figure 8A). At present, it is not clear whether REX4 is not translated or whether it is simply present at levels too low to be detected by our antiserum. However, the transgenic parasites clearly show that REX4 can be exported by the parasite (Figure 8, A and B). GFP-tagged REX4 is located in the host cell cytoplasm with some protein remaining inside the parasite, similar to the location of GFP-tagged REX3 (Figure 7B). Western analysis confirmed these results and indicated that the export of the myc-tagged REX4 was more efficient than that of REX4-GFP (Figure 8A; our unpublished data). These results demonstrated that REX4 contains all the features required for an exported protein and that GFP interfered with export to some extent.
Figure 8.
Transgenically expressed REX4 is exported into the host cell. (A) Western analysis of 3D7 parasites expressing myc-tagged (left) and GFP-tagged REX4. IRBCs (total) expressing REX4myc were treated with either saponin (sap) or SLO and separated into pellet (P) and supernatant (SN). Immunoblots were analyzed using a specific REX4 serum. REX4myc was mostly soluble and exported into the host cell, with a small fraction of REX4myc molecules remaining with the parasite pellet. No signal was observed with the anti-REX4 serum in untransfected 3D7 (total 3D7). REX4-GFP (right) was also exported into the host cell, but more of the chimeric protein remained within the parasite. REX3 was used as a control to show efficient release of the host cell cytoplasm. (B) Parasites expressing REX4-GFP showed uniform fluorescence in the host cell cytoplasm with some of the chimeric protein remaining inside of the parasite. Top, a ring stage infected RBC. Bottom, a RBC infected with two trophozoite stage parasites. Nuclei were stained with Hoechst; bf, bright field image. Bars, 5 μm.
DISCUSSION
The right end of chromosome 9 has long been known to be linked to cytoadherence and gametocytogenesis in P. falciparum (Day et al., 1993). In this work, we established that the region linked to these phenotypes contains only 13 genes, and we showed that the ring stage-specific genes therein encode exported proteins, although only two of them are part of the currently predicted exportome. This was supported by immunofluorescence assays and transgenic expression of tagged versions of these proteins. These results were confirmed by biochemical analysis using subcellular fractionation of IRBCs, and these studies also allowed us to gain novel insight into the properties of the different membrane compartments in IRBCs. Importantly, the availability of parasites lacking the right arm of chromosome 9 provided a convenient control to support the specificity of our antisera. This is especially important, because cross-reactivity of antisera to Maurer’s clefts is common (our unpublished observation), and studies to establish export of PEXEL-negative proteins have to be rigorously controlled. To gain insight into PEXEL-independent export, it is crucial to establish the exact location of these proteins, which might help to uncover potential differences to PEXEL-mediated export.
Although REX2 and REX3 were readily detected in 3D7 parasites, no endogenous REX4 could be detected. This was puzzling, because transcription levels of rex4 are comparably high (LeRoch et al., 2003) and its sequence similarity to rex3 is too low to make cross-reactivity of microarray probes a likely explanation for the measured transcription levels. However, it is possible that this gene encodes a protein for which the mRNA is stored and translated in a different life cycle stage or under certain conditions only. Discrepancies between transcription and translation in P. falciparum have been noted previously (Coulson et al., 2004). Alternatively, our sera might not be sensitive enough to detect REX4, which might be considerably less abundant than the transgenically expressed REX4 proteins. Nevertheless, our data clearly show that REX4 contains all the features required for an exported protein, and it is likely to be exported if translated.
To date, only a few P. falciparum proteins have been shown to be soluble proteins resident in the host cell cytoplasm. In this respect, it is interesting to note that our initial IFA experiments using acetone or formaldehyde fixation of IRBCs revealed that these reagents did not prevent solubilization and release of the host cell cytoplasm (as judged by loss of hemoglobin after fixing). For soluble antigens exported into the host cell, this led to either a lack of signal or a weak Maurer’s clefts stain. The cleft staining was possibly due to fixing of the antigen to these structures. Adding 0.005% glutaraldehyde to the formaldehyde fixing solution solved this problem. This method also enabled us to visualize other proteins soluble in the RBC cytoplasm currently under investigation in our laboratory, which we previously failed to detect using other fixation methods (our unpublished data). Thus, proteins soluble in the host cell cytoplasm as well as the cytoplasmic fractions of proteins associated with structures in the host cell might be missed using the common acetone or formaldehyde fixation methods. This highlights the need to verify IFA results with Western data from selectively permeabilized and fractionated IRBCs when studying exported proteins in malaria parasites. Interestingly, a recent proteomics study on ghost fractions of IRBCs reported the absence of a signal in IFAs for some of the identified proteins that were clearly detected on Western blots (Vincensini et al., 2005). Two of these proteins (Pf10_0013 and Mal7P1.174) contain a PEXEL motif but no predicted transmembrane domain, similar to REX3 and REX4, and thus might be soluble proteins resident in the host cell cytoplasm.
Our topology analysis indicated that REX2 is an integral Maurer’s cleft protein with its N terminus inside the cleft and the C terminus facing the RBC cytoplasm. This orientation is similar to that of SBP1 (Blisnick et al., 2000), which we used as a control in this study. Using saponin instead of a harsher detergent in our proteinase K assay, we were able to selectively permeabilize the clefts and the PVM only. To our knowledge, this is the first report demonstrating that the cleft membrane is permeabilized by saponin. The susceptibility of the Maurer’s cleft membrane to saponin indicates that it shares similar properties to that of the PVM. However, under the conditions used in this study, the clefts membrane seemed to be less susceptible to SLO than the PVM. It has recently been suggested that the clefts and the PVM are connected by a continuous membrane network (Wickert et al., 2003). Our data suggest, however, that the properties of the membranes of these two compartments are not identical and thus may not be continuous. Our FRAP analysis using REX2-GFP support previous FRAP studies using proteins transiently associating with Maurer’s clefts (Knuepfer et al., 2005b) or the resident integral cleft protein MAHRP1 (Spycher et al., 2006). In each case, the data indicate that Maurer’s clefts are separate entities. FRAP data cannot exclude the possibility that the proteins analyzed are simply restricted in their diffusion at the clefts where they might be associated with fixed structures. However, it seems unlikely that this is the case for all of the increasing number of cleft associated proteins analyzed by FRAP.
REX2-GFP seems to traffic through the PV en route to the Maurer’s clefts. Occasionally, REX2-GFP–containing structures that seemed to have budded from the PVM but not yet docked at the RBC membrane were observed in the IRBC cytosol.
FRAP analysis has previously been used to look at the dynamics of GFP chimeras of KAHRP, PfEMP3, and PfEMP1, which are present transiently as a soluble population in the cytoplasm of IRBCs before binding to sites at the RBC membrane (Wickham et al., 2001; Knuepfer et al., 2005a, b). For all of these proteins, the recovery time after bleaching was slower than expected for monomeric species and suggested the presence of large aggregates. In some cells, REX3-GFP diffused too rapidly to be monitored using spot photobleaching protocols, whereas in other cells the diffusion rate was at the limit of detection. This suggests that REX3 is present in a range of complexes of variable size and might be monomeric in some cells. The difference in mobilities of different GFP-fusion proteins suggests that there may be a range of complexes with different compositions diffusing within the IRBC cytosol.
Discovery of the PEXEL/VTS motif has not only provided insight into the mechanism by which exported proteins cross the barrier of the PVM but also, importantly, allowed prediction of the P. falciparum exportome, which presumably represents a complement of parasite proteins involved in host cell remodeling (Hiller et al., 2004; Marti et al., 2004). However, there are exceptions to the rule that a PEXEL/VTS motif is needed to cross the PVM, because some PEXEL-negative exported proteins have been documented (Blisnick et al., 2000; Spycher et al., 2003). Our analysis of the cluster of ring stage-specific genes on chromosome 9 revealed that only two of the 4 exported proteins possess an obvious PEXEL. There is further evidence that this proportion of PEXEL-negative exported proteins is maintained among other ring stage-specific proteins. Several of a set of genes exclusively transcribed in ring stages (Spielmann and Beck, 2000) have now been further characterized. Of the 14 genes identified, two genes have homologues in other organisms and three genes are etramps, which encode integral PVM proteins (Spielmann et al., 2003). Of the remaining nine genes, four genes encode PEXEL-positive proteins and can be assumed to be exported, although this remains to be experimentally demonstrated. The remaining five genes code for PEXEL-negative proteins. The locations of three of these (now called SBP1, MAHRP1, and REX2) have since been determined, and they are all exported to the Maurer’s clefts (Blisnick et al., 2000; Spycher et al., 2003; this study). Hence, apart from the ETRAMPs (which are exported but do not need a PEXEL, because they remain in the PVM), >50% of the identified proteins do not have a PEXEL, and all three tested so far are exported. It is likely that the analysis of further ring stage-specific genes that show no homology to known proteins in other organisms will reveal more exported proteins without an obvious PEXEL. This might add a substantial new cohort to the currently predicted P. falciparum exportome.
If PEXEL-negative exported proteins are common, the question arises how they are exported. It is possible that these proteins are either escorted by other proteins or contain a PEXEL that is too divergent to be easily identified. However, that the PEXEL-negative proteins seem to be membrane associated may point to a third possibility. Assuming that clefts form from the PVM, which has yet to be experimentally verified, during the ring stage, membrane-associated PEXEL-negative proteins would not need a sequence to pass through the PVM. It would be sufficient for these proteins to accumulate in regions that will become cleft material. In this respect, it is interesting to note that evidence for the presence of different domains in the PVM has recently been provided (Spielmann et al., 2006). Although several transmembrane proteins, such as members of the RIFIN (Kyes et al., 1999), STEVOR (Kaviratne et al., 2002), and PfMC-2TM (Sam-Yellowe et al., 2004) families, are exported in a PEXEL-dependent manner (Marti et al., 2004; Przyborski et al., 2005), these proteins are expressed at the trophozoite stage when the Maurer’s clefts have already formed. This would not allow them to be exported during cleft biogenesis. However, this remains to be experimentally demonstrated.
PEXEL-independent export to the clefts might be possible or even required for membrane-associated early proteins, explaining the high representation of PEXEL-negative proteins among ring stage-specific genes. It is possible that these proteins are involved in building some of the export components and structures used by proteins expressed in later stages. Moreover, the lack of nonexported proteins encoded by the ring stage-specific genes analyzed so far further highlights host cell remodeling as a major task of this parasite stage.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. B. Cooke, K. Lingelbach, and C. Daubenberger for antibodies and Drs. A. Cowman, B. Crabb, A. Adisa, and C. Tonkin for plasmid vectors. T. S. gratefully acknowledges the Swiss National Science Foundation, the Novartis Foundation, the Freiwillige Akademische Gesellschaft Basel for support during this work. D. K., D. G., and K. T. gratefully acknowledge support from Australian National Health and Medical Research Council (Program Grant 290208) and Mark Nicholson, Alice Hill, and The Tudor Foundation.
Abbreviations used:
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- IRBC
infected red blood cell
- KAHRP
Knob-associated histidine-rich protein
- MAHRP
membrane-associated histidine-rich protein
- PEXEL
Plasmodium export element
- PfEMP
Plasmodium falciparum erythrocyte membrane protein
- PV
parasitophorous vacuole
- PVM
parasitophorous vacuole membrane
- RBC
red blood cell
- REX
ring-exported protein
- SBP
skeleton-binding protein
- SERP
serine-rich protein
- VTS
vacuolar transport signal.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-04-0291) on June 7, 2006.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Adisa A., Rug M., Klonis N., Foley M., Cowman A. F., Tilley L. The signal sequence of exported protein-1 directs the green fluorescent protein to the parasitophorous vacuole of transfected malaria parasites. J. Biol. Chem. 2003;278:6532–6542. doi: 10.1074/jbc.M207039200. [DOI] [PubMed] [Google Scholar]
- Aley S. B., Sherwood J. A., Marsh K., Eidelman O., Howard R. J. Identification of isolate-specific proteins on sorbitol-enriched Plasmodium falciparum infected erythrocytes from Gambian patients. Parasitology. 1986;92:511–525. doi: 10.1017/s0031182000065410. [DOI] [PubMed] [Google Scholar]
- Anders R. F., Brown G. V., Edwards A. Characterization of an S antigen synthesized by several isolates of Plasmodium falciparum. Proc. Natl. Acad. Sci. USA. 1983;80:6652–6656. doi: 10.1073/pnas.80.21.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansorge I., Benting J., Bhakdi S., Lingelbach K. Protein sorting in Plasmodium falciparum-infected red blood cells permeabilized with the pore-forming protein streptolysin O. Biochem. J. 1996;315:307–314. doi: 10.1042/bj3150307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes D. A., Thompson J., Triglia T., Day K., Kemp D. J. Mapping the genetic locus implicated in cytoadherence of Plasmodium falciparum to melanoma cells. Mol. Biochem. Parasitol. 1994;66:21–29. doi: 10.1016/0166-6851(94)90032-9. [DOI] [PubMed] [Google Scholar]
- Baruch D. I., Pasloske B. L., Singh H. B., Bi X., Ma X. C., Feldman M., Taraschi T. F., Howard R. J. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- Biggs B. A., Culvenor J. G., Ng J. S., Kemp D. J., Brown G. V. Plasmodium falciparum: cytoadherence of a knobless clone. Exp. Parasitol. 1989;69:189–197. doi: 10.1016/0014-4894(89)90187-2. [DOI] [PubMed] [Google Scholar]
- Blisnick T., Morales Betoulle M. E., Barale J. C., Uzureau P., Berry L., Desroses S., Fujioka H., Mattei D., Braun Breton C. Pfsbp1, a Maurer’s cleft Plasmodium falciparum protein, is associated with the erythrocyte skeleton. Mol. Biochem. Parasitol. 2000;111:107–121. doi: 10.1016/s0166-6851(00)00301-7. [DOI] [PubMed] [Google Scholar]
- Bourke P. F., Holt D. C., Sutherland C. J., Kemp D. J. Disruption of a novel open reading frame of Plasmodium falciparum chromosome 9 by subtelomeric and internal deletions can lead to loss or maintenance of cytoadherence. Mol. Biochem. Parasitol. 1996;82:25–36. doi: 10.1016/0166-6851(96)02715-6. [DOI] [PubMed] [Google Scholar]
- Cooke B. M., Buckingham D. W., Glenister F. K., Fernandez K. M., Bannister L. H., Marti M., Mohandas N., Coppel R. L. A Maurer’s cleft-associated protein is essential for expression of the major malaria virulence antigen on the surface of infected red blood cells. J. Cell Biol. 2006;172:899–908. doi: 10.1083/jcb.200509122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke B. M., Lingelbach K., Bannister L. H., Tilley L. Protein trafficking in Plasmodium falciparum-infected red blood cells. Trends Parasitol. 2004;20:581–589. doi: 10.1016/j.pt.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Cooke B. M., Mohandas N., Coppel R. L. The malaria-infected red blood cell: structural and functional changes. Adv. Parasitol. 2001;50:1–86. doi: 10.1016/S0065-308X(01)50029-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppel R. L., Davern K. M., McConville M. J. Immunochemistry of parasite antigens. In: van Oss C. J., van Regenmortel M.H.V., editors. Immunochemistry. New York: Marcel Dekker; 1994. pp. 475–532. [Google Scholar]
- Coulson R. M., Hall N., Ouzounis C. A. Comparative genomics of transcriptional control in the human malaria parasite, Plasmodium falciparum. Genome Res. 2004;14:1548–1554. doi: 10.1101/gr.2218604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubenberger C. A., Tisdale E. J., Curcic M., Diaz D., Silvie O., Mazier D., Eling W., Bohrmann B., Matile H., Pluschke G. The N′-terminal domain of glyceraldehyde-3-phosphate dehydrogenase of the apicomplexan Plasmodium falciparum mediates GTPase Rab2-dependent recruitment to membranes. Biol. Chem. 2003;384:1227–1237. doi: 10.1515/BC.2003.135. [DOI] [PubMed] [Google Scholar]
- Day K. P., Karamalis F., Thompson J., Barnes D. A., Peterson C., Brown H., Brown G. V., Kemp D. J. Genes necessary for expression of a virulence determinant and for transmission of Plasmodium falciparum are located on a 0.3-megabase region of chromosome 9. Proc. Natl. Acad. Sci. USA. 1993;90:8292–8296. doi: 10.1073/pnas.90.17.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rojas M. O., Wasserman M. Temporal relationships on macromolecular synthesis during the asexual cell cycle of Plasmodium falciparum. Trans. R. Soc. Trop. Med. Hyg. 1985;79:792–796. doi: 10.1016/0035-9203(85)90119-1. [DOI] [PubMed] [Google Scholar]
- Delplace P., Fortier B., Tronchin G., Dubremetz J., Vernes A. Localization, biosynthesis, processing and isolation of a major 126 kDa antigen of the parasitophorous vacuole of Plasmodium falciparum. Mol. Biochem. Parasitol. 1987;23:193–201. doi: 10.1016/0166-6851(87)90026-0. [DOI] [PubMed] [Google Scholar]
- Gardiner D. L., Spielmann T., Dixon M. W., Hawthorne P. L., Ortega M. R., Anderson K. L., Skinner-Adams T. S., Kemp D. J., Trenholme K. R. CLAG 9 is located in the rhoptries of Plasmodium falciparum. Parasitol. Res. 2004;93:64–67. doi: 10.1007/s00436-004-1098-4. [DOI] [PubMed] [Google Scholar]
- Gardner J. P., Pinches R. A., Roberts D. J., Newbold C. I. Variant antigens and endothelial receptor adhesion in Plasmodium falciparum. Proc. Natl. Acad. Sci. USA. 1996;93:3503–3508. doi: 10.1073/pnas.93.8.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawthorne P. L., Trenholme K. R., Skinner-Adams T. S., Spielmann T., Fischer K., Dixon M. W., Ortega M. R., Anderson K. L., Kemp D. J., Gardiner D. L. A novel Plasmodium falciparum ring stage protein, REX, is located in Maurer’s clefts. Mol. Biochem. Parasitol. 2004;136:181–189. doi: 10.1016/j.molbiopara.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Hiller N. L., Bhattacharjee S., van Ooij C., Liolios K., Harrison T., Lopez-Estrano C., Haldar K. A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science. 2004;306:1934–1937. doi: 10.1126/science.1102737. [DOI] [PubMed] [Google Scholar]
- Holt D. C., Bourke P. F., Mayo M., Kemp D. J. A high resolution map of chromosome 9 of Plasmodium falciparum. Mol. Biochem. Parasitol. 1998;97:229–233. doi: 10.1016/s0166-6851(98)00123-6. [DOI] [PubMed] [Google Scholar]
- Kaviratne M., Khan S. M., Jarra W., Preiser P. R. Small variant STEVOR antigen is uniquely located within Maurer’s clefts in Plasmodium falciparum-infected red blood cells. Eukaryot. Cell. 2002;1:926–935. doi: 10.1128/EC.1.6.926-935.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klonis N., Rug M., Wickham M., Harper I., Cowman A. F., Tilley L. Fluorescence photobleaching analysis for the study of cellular dynamics. Eur. Biophys. J. 2002;31:36–51. doi: 10.1007/s00249-001-0202-2. [DOI] [PubMed] [Google Scholar]
- Knuepfer E., Rug M., Klonis N., Tilley L., Cowman A. F. Trafficking determinants for PfEMP3 export and assembly under the Plasmodium falciparum-infected red blood cell membrane. Mol. Microbiol. 2005a;58:1039–1053. doi: 10.1111/j.1365-2958.2005.04895.x. [DOI] [PubMed] [Google Scholar]
- Knuepfer E., Rug M., Klonis N., Tilley L., Cowman A. F. Trafficking of the major virulence factor to the surface of transfected P. falciparum-infected erythrocytes. Blood. 2005b;105:4078–4087. doi: 10.1182/blood-2004-12-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriek N., Tilley L., Horrocks P., Pinches R., Elford B. C., Ferguson D. J., Lingelbach K., Newbold C. I. Characterization of the pathway for transport of the cytoadherence-mediating protein, PfEMP1, to the host cell surface in malaria parasite-infected erythrocytes. Mol. Microbiol. 2003;50:1215–1227. doi: 10.1046/j.1365-2958.2003.03784.x. [DOI] [PubMed] [Google Scholar]
- Kyes S. A., Rowe J. A., Kriek N., Newbold C. I. Rifins: a second family of clonally variant proteins expressed on the surface of red cells infected with Plasmodium falciparum. Proc. Natl. Acad. Sci. USA. 1999;96:9333–9338. doi: 10.1073/pnas.96.16.9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langreth S. G., Peterson E. Pathogenicity, stability, and immunogenicity of a knobless clone of Plasmodium falciparum in Colombian owl monkeys. Infect. Immun. 1985;47:760–766. doi: 10.1128/iai.47.3.760-766.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeRoch K. G., et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- Marti M., Baum J., Rug M., Tilley L., Cowman A. F. Signal-mediated export of proteins from the malaria parasite to the host erythrocyte. J. Cell Biol. 2005;171:587–592. doi: 10.1083/jcb.200508051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti M., Good R. T., Rug M., Knuepfer E., Cowman A. F. Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science. 2004;306:1930–1933. doi: 10.1126/science.1102452. [DOI] [PubMed] [Google Scholar]
- Miller L. H., Baruch D. I., Marsh K., Doumbo O. K. The pathogenic basis of malaria. Nature. 2002;415:673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- O’Donnell R. A., Freitas-Junior L. H., Preiser P. R., Williamson D. H., Duraisingh M., McElwain T. F., Scherf A., Cowman A. F., Crabb B. S. A genetic screen for improved plasmid segregation reveals a role for Rep20 in the interaction of Plasmodium falciparum chromosomes. EMBO J. 2002;21:1231–1239. doi: 10.1093/emboj/21.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przyborski J. M., Miller S. K., Pfahler J. M., Henrich P. P., Rohrbach P., Crabb B. S., Lanzer M. Trafficking of STEVOR to the Maurer’s clefts in Plasmodium falciparum-infected erythrocytes. EMBO J. 2005;24:2306–2317. doi: 10.1038/sj.emboj.7600720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sam-Yellowe T. Y., Florens L., Johnson J. R., Wang T., Drazba J. A., LeRoch K. G., Zhou Y., Batalov S., Carucci D. J., Winzeler E .A., Yates J. R., 3rd. A Plasmodium gene family encoding Maurer’s cleft membrane proteins: structural properties and expression profiling. Genome Res. 2004;14:1052–1059. doi: 10.1101/gr.2126104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargeant T. J., Marti M., Caler E., Carlton J. M., Simpson K., Speed T. P., Cowman A. F. Lineage-specific expansion of proteins exported to erythrocytes in malaria parasites. Genome Biol. 2006;7:R12. doi: 10.1186/gb-2006-7-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. D., Chitnis C. E., Craig A. G., Roberts D. J., Hudson-Taylor D. E., Peterson D. S., Pinches R., Newbold C. I., Miller L. H. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell. 1995;82:101–110. doi: 10.1016/0092-8674(95)90056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow R. W., Guerra C. A., Noor A. M., Myint H. Y., Hay S. I. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmann T., Beck H. P. Analysis of stage-specific transcription in Plasmodium falciparum reveals a set of genes exclusively transcribed in ring stage parasites. Mol. Biochem. Parasitol. 2000;111:453–458. doi: 10.1016/s0166-6851(00)00333-9. [DOI] [PubMed] [Google Scholar]
- Spielmann T., Fergusen D. J., Beck H. P. etramps, a new Plasmodium falciparum gene family coding for developmentally regulated and highly charged membrane proteins located at the parasite-host cell interface. Mol. Biol. Cell. 2003;14:1529–1544. doi: 10.1091/mbc.E02-04-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmann T., Gardiner D. L., Beck H. P., Trenholme K. R., Kemp D. J. Organization of ETRAMPs and EXP-1 at the parasite-host cell interface of malaria parasites. Mol. Microbiol. 2006;59:779–794. doi: 10.1111/j.1365-2958.2005.04983.x. [DOI] [PubMed] [Google Scholar]
- Spycher C., Klonis N., Spielmann T., Kump E., Steiger S., Tilley L., Beck H. P. MAHRP-1, a novel Plasmodium falciparum histidine-rich protein, binds ferriprotoporphyrin IX and localizes to the Maurer’s clefts. J. Biol. Chem. 2003;278:35373–35383. doi: 10.1074/jbc.M305851200. [DOI] [PubMed] [Google Scholar]
- Spycher C., Rug M., Klonis N., David J. P., Ferguson D.J.P., Cowman A. F., Beck H. P., Tilley L. Genesis of and trafficking to the Maurer’s clefts of P. falciparum-infected erythrocytes. Mol. Cell. Biol. 2006;26:4074–4085. doi: 10.1128/MCB.00095-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X. Z., Heatwole V. M., Wertheimer S. P., Guinet F., Herrfeldt J. A., Peterson D. S., Ravetch J. A., Wellems T. E. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- Tonkin C. J., van Dooren G. G., Spurck T. P., Struck N. S., Good R. T., Handman E., Cowman A. F., McFadden G. I. Localization of organellar proteins in Plasmodium falciparum using a novel set of transfection vectors and a new immunofluorescence fixation method. Mol. Biochem. Parasitol. 2004;137:13–21. doi: 10.1016/j.molbiopara.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Trager W., Jensen J. B. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- Trenholme K. R., Gardiner D. L., Holt D. C., Thomas E. A., Cowman A. F., Kemp D. J. clag 9, A cytoadherence gene in Plasmodium falciparum essential for binding of parasitized erythrocytes to CD36. Proc. Natl. Acad. Sci. USA. 2000;97:4029–4033. doi: 10.1073/pnas.040561197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincensini L., Richert S., Blisnick T., Van Dorsselaer A., Leize-Wagner E., Rabilloud T., Braun Breton C. Proteomic analysis identifies novel proteins of the Maurer’s clefts, a secretory compartment delivering Plasmodium falciparum proteins to the surface of its host cell. Mol. Cell Proteomics. 2005;4:582–593. doi: 10.1074/mcp.M400176-MCP200. [DOI] [PubMed] [Google Scholar]
- Walliker D., Quakyi I. A., Wellems T. E., McCutchan T. F., Szarfman A., London W. T., Corcoran L. M., Burkot T. R., Carter R. Genetic analysis of the human malaria parasite Plasmodium falciparum. Science. 1987;236:1661–1666. doi: 10.1126/science.3299700. [DOI] [PubMed] [Google Scholar]
- Wickham M. E., Rug M., Ralph S. A., Klonis N., McFadden G. I., Tilley L., Cowman A. F. Trafficking and assembly of the cytoadherence complex in Plasmodium falciparum-infected human erythrocytes. EMBO J. 2001;20:5636–5649. doi: 10.1093/emboj/20.20.5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickert H., Wissing F., Andrews K. T., Stich A., Krohne G., Lanzer M. Evidence for trafficking of PfEMP1 to the surface of P. falciparum-infected erythrocytes via a complex membrane network. Eur. J. Cell Biol. 2003;82:271–284. doi: 10.1078/0171-9335-00319. [DOI] [PubMed] [Google Scholar]
- Wu Y., Kirkman L. A., Wellems T. E. Transformation of Plasmodium falciparum malaria parasites by homologous integration of plasmids that confer resistance to pyrimethamine. Proc. Natl. Acad. Sci. USA. 1996;93:1130–1134. doi: 10.1073/pnas.93.3.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolg J. W., Macleod A. J., Scaife J. G., Beaudoin R. L. The accumulation of lactic acid and its influence on the growth of Plasmodium falciparum in synchronized cultures. In Vitro. 1984;20:205–215. doi: 10.1007/BF02618189. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.