Abstract
CDP/Cux (CCAAT-displacement protein/cut homeobox) contains four DNA binding domains, namely, three Cut repeats (CR1, CR2, and CR3) and a Cut homeodomain. CCAAT-displacement activity involves rapid but transient interaction with DNA. More stable DNA binding activity is up-regulated at the G1/S transition and was previously shown to involve an N-terminally truncated isoform, CDP/Cux p110, that is generated by proteolytic processing. CDP/Cux has been previously characterized as a transcriptional repressor. However, here we show that expression of reporter plasmids containing promoter sequences from the human DNA polymerase α (pol α), CAD, and cyclin A genes is stimulated in cotransfections with N-terminally truncated CDP/Cux proteins but not with full-length CDP/Cux. Moreover, expression of the endogenous DNA pol α gene was stimulated following the infection of cells with a retrovirus expressing a truncated CDP/Cux protein. Chromatin immunoprecipitation (ChIP) assays revealed that CDP/Cux was associated with the DNA pol α gene promoter specifically in the S phase. Using linker scanning analyses, in vitro DNA binding, and ChIP assays, we established a correlation between binding of CDP/Cux to the DNA pol α promoter and the stimulation of gene expression. Although we cannot exclude the possibility that stimulation of gene expression by CDP/Cux involved the repression of a repressor, our data support the notion that CDP/Cux participates in transcriptional activation. Notwithstanding its mechanism of action, these results establish CDP/Cux as an important transcriptional regulator in the S phase.
CDP/Cux (CCAAT-displacement protein/Cut homeobox) belongs to a family of transcription factors present in all metazoans and is involved in the control of proliferation and differentiation (reviewed in reference 49). In Drosophila melanogaster, the gene was named cut, after the cut-wing phenotype, more than 50 years ago (27). Overall, genetic studies of Drosophila suggested that cut plays a role, late in development, in determining cell type specificity in several tissues (8-10, 13, 19, 31, 32, 41, 42, 46). In higher vertebrates, there are two CDP/Cux genes, called the CDP-1 and CDP-2 genes in humans and the Cux-1 and Cux-2 gene in mice and chickens (50, 57, 66). The cux-1 knockout mice displayed phenotypes in various organs, including phenotypes for curly whiskers, growth retardation, delayed differentiation of lung epithelia, altered hair follicle morphogenesis, male infertility, a deficit in T and B cells, and a surplus of myeloid cells (21, 44, 59, 65). In contrast to the small size of the cux-1 knockout mice, transgenic mice expressing Cux-1 under the control of the cytomegalovirus enhancer-promoter displayed multiorgan hyperplasia and organomegaly (38). Thus, from genetic studies both of Drosophila and the mouse, it is clear that the CDP/Cux/Cut gene plays an important role in the development and homeostasis of several tissues.
At the molecular level, CDP/Cux is a complex protein with four evolutionarily conserved DNA binding domains, namely, three Cut repeats (CR1, CR2, and CR3) and a Cut homeodomain (HD) (1, 2, 26, 50). The full-length protein, which we refer to as CDP/Cux p200, was found to be proteolytically processed at the G1/S transition of the cell cycle, thereby generating the CDP/Cux p110 isoform that contains three DNA binding domains, CR2, CR3, and HD (48). In addition, two alternate, tissue-specific mRNA species were found to code for a CDP/Cux p75 isoform that contains only two DNA binding domains: CR3 and HD (23, 68). Despite early claims made by us and others on the basis of results obtained with glutathione S-transferase (GST) fusion proteins (1, 2, 25, 26), individual Cut repeats cannot bind to DNA on their own but need to cooperate with a second Cut repeat or with the Cut homeodomain (47). CR1CR2 was found to make a rapid but transient interaction with DNA, whereas CR2CR3HD and CR3HD bound more slowly, but stably, to DNA (47). Predictably, CDP/Cux p110 and p75 exhibited DNA binding properties similar to that of CR2CR3HD and CR3HD. However, somewhat surprisingly, CDP/Cux p200 behaved like CR1CR2 and made an unstable interaction with DNA, suggesting that DNA binding by CR3HD is inhibited in the context of the full-length protein (47, 48). The carboxy-terminal domain (CTD) of the protein was found to contain two active repression domains, and the CTD was reported to recruit histone deacetylase 1 (39, 45). The protein was shown to repress transcription by at least two mechanisms: competition for binding site occupancy and active repression (45). CDP/Cux was reported to repress a large number of genes, in particular those genes expressed in precursor cells prior to terminal differentiation (6, 17, 20, 28, 33-35, 37, 39, 43, 45, 51, 53, 55, 60, 63, 64, 69, 78). In addition, the binding of CDP/Cux to a number of matrix attachment regions raises the possibility that the protein is involved in higher-order chromatin organization or may be able to target certain regulatory loci to specific regions of the nucleus (5, 16, 40, 63, 75).
A number of studies demonstrated that CDP/Cux is regulated in a cell cycle-dependent manner and may have a specific function in S phase. The histone nuclear factor D (HiNF-D), which was later found to include CDP/Cux as its DNA binding partner, was shown to be up-regulated in S phase in normal cells (29, 70, 72, 73, 77). The up-regulation of stable DNA binding at the G1/S transition was shown to involve at least two posttranslational modifications: dephosphorylation of the Cut homeodomain by the Cdc25A phosphatase and proteolytic cleavage of CDP/Cux p200 between CR1 and CR2 to generate CDP/Cux p110 (17, 48). Later in the cell cycle, DNA binding was found to decrease in G2 following the phosphorylation by cyclin A or Cdk1 of two serine residues, S1237 and S1270, in the region of the Cut homeodomain (58). The rise and decline of stable DNA binding activity of CDP/Cux at the beginning and at the end of S phase suggest that the CDP/Cux p110 isoform plays a role in S phase. We reported that CDP/Cux was able to repress a reporter plasmid carrying the promoter of the p21Waf1/Cip1 gene (17). Moreover, inhibition of CDP/Cux expression in S phase, by way of an antisense vector, restored expression of the p21Waf1/Cip1 reporter to the higher level observed in G1 (17). Interestingly, expression of another cyclin kinase inhibitor, p27, was shown to be down-regulated in the CMV/Cux-1 transgenic mice (38).
The binding of HiNF-D to the promoters of several S phase-specific histone genes at the same time in the cell cycle when these genes are induced is consistent with the notion that HiNF-D functions as a transcriptional activator (3, 4, 36, 72, 73, 77). However, cotransfection of CDP/Cux with a reporter containing the promoter of the FO108-H4 histone gene did not lead to the activation of this reporter but rather to its repression (72). In contrast, in another study, cotransfection of CDP/Cux with the ITF2 transcription factor led to the activation of a reporter containing the tyrosine hydroxylase gene promoter (79). These results may indicate that CDP/Cux needs to cooperate with other proteins in order to mediate activation. Another difference that might explain the discrepancy between these results was that the latter study utilized an incomplete rat CDP/Cux cDNA clone that expressed an N-terminally truncated CDP/Cux protein equivalent to the p110 processed isoform (79). The latter two possibilities are not mutually exclusive. Indeed, one can envision a complex interdependent mechanism in which transcriptional activation requires both stable DNA binding by CDP/Cux p110 and cooperation with other proteins.
In the present study, we show that CDP/Cux p110, but not CDP/Cux p200, was capable of stimulating expression of a reporter containing the promoter of the DNA polymerase α (DNA pol α) gene. Moreover, the introduction of a truncated CDP/Cux protein by retroviral infection led to an increase in DNA pol α mRNA level. Using in vitro mutagenesis and DNA binding assays, we were able to establish a correlation between the binding of CDP/Cux to DNA pol α promoter sequences and the stimulation of the DNA pol α reporter plasmid. The potential mechanisms by which CDP/Cux may stimulate expression of the DNA pol α reporter plasmid are discussed.
MATERIALS AND METHODS
Plasmid construction.
The DNA pol α −1561/+47 reporter plasmid was constructed as previously described (48). In the following manner, 5′ deletion constructs were made. −1158/+47 was inserted into pGL3-Basic, and both were digested with HindIII and NcoI. −402/+47 was constructed via insertion of the BssHII/NcoI promoter fragment into MluI/NcoI of pGL3-Basic; the promoter was digested with SphI, and the overhang was removed with T4 DNA polymerase, followed by digestion with NcoI, yielding −248/+47, which was cloned into SmaI/NcoI of pGL3-Basic. −116/+47 was made by digestion of the promoter with SacII/NcoI and was ligated with pGL3-Basic digested with SacI/NcoI. −65/+47 was made by digestion of DNA pol α with EagI/NcoI and ligation into SmaI/NcoI sites of pGL3-Basic. Linker scanning mutants were made by PCR with pGL3-pol α (−65/+47) as a template, with the 5′-most primer being 5′AGGTACGGGAGGTACTTGGAGCGG3′ and the 3′-most primer being 5′ATGTCGTTCGCGGGCGCAACTGCAACTC3′. The sequences of the inner primers can be made available upon request. Briefly, inner primers used to generate the upstream fragments had the tail sequence 5′GACTTGAAGCTTTC. Inner primers for the downstream fragments had the tail sequence 5′GACTGAAAGCTTCA. Upstream fragments were digested with NotI/HindIII, and downstream fragments were digested with HindIII/BstBI. Upstream and downstream fragments were ligated together with pGL3-Basic digested with NotI/BstBI. Constructs were sequenced to verify the absence of mutations. Sequences and/or maps will be provided upon request for CDP/Cux 831-1505, 831-1505, 659-1192+NLS, and CR2CR3HD constructs. All other CDP/Cux constructs have been described in previous studies (48, 58).
Expression and purification of CDP/Cux fusion proteins.
The full-length CDP/Cux protein was expressed in SF9 insect cells by using a baculovirus vector as previously described (47). All truncated CDP/Cux proteins were otherwise expressed in bacteria by using the pET-15b vector (Novagen). The expression plasmids were introduced into the BL21 (DE3) strain of Escherichia coli and induced with 1 mM isopropyl-β-d-thiogalactopyranoside for 1.5 h. Proteins were purified on nickel-nitrilotriacetic acid-agarose (Qiagen) in accordance with the manufacturer's instructions.
Cell culture, transfection, and synchronization.
HeLa and C33A are human epithelial cell lines derived from cervical carcinomas (18). HS578T, T47D, and MCF-7 are human epithelial cell lines derived from breast carcinomas (11, 24, 76). T98G is a human fibroblastic cell line derived from a gliobastoma multiform tumor (62). NIH 3T3, C33A, and HeLa cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). HS578T, T47D, and MCF-7 cells were grown in DMEM supplemented with 5% FBS. T98G cells were grown in EMEM supplemented with 10% FBS. Synchronization in G1/S was performed by two methods, namely, serum starvation-stimulation (posttransfection, cells were maintained in DMEM plus 0.4% FBS for 36 h, followed by 18 h in DMEM plus 10% FBS) and thymidine block (posttransfection, cells were cultured overnight in DMEM plus 10% FBS supplemented with 2 mM thymidine and harvested). Transient transfections were performed with ExGen500 (MBI Fermentas) in accordance with the manufacturer's instructions.
Luciferase assay.
Cells were plated in 12-well plates to be approximately 50% confluent on the day of transfection. A total of 750 ng of DNA (250 ng of reporter DNA and 500 ng of effector DNA) was transfected. Cells were either synchronized (see above) or harvested 24 to 48 h later. Luciferase assays were performed as described previously (48). Because the internal control plasmid is itself often repressed by CDP/Cux, as a control for transfection efficiency, the purified β-galactosidase protein (Sigma) was included in the transfection mix, as previously described (30). The luciferase activity was then normalized based on β-galactosidase activity.
CDP/Cux antibodies and Western blot analysis.
Antibodies 861 and 1300 have previously been described (23, 48). To generate polyclonal antibodies against various regions of CDP/Cux (1,505 amino acids [aa]), rabbits were injected with 500 μg of purified bacterial fusion protein containing various regions of CDP/Cux in Freund's complete adjuvant. Antibodies were raised against the following regions: α23, aa 23 to 50; α403, aa 403 to 449; α510, aa 510 to 541; α861, aa 861 to 936; and α1300, aa 1300 to 1402. The animals were boosted twice with 250 μg of protein, and serum was collected 10 days after the last boost. Polyclonal antibodies were purified by affinity chromatography. The serum was passed through two GST affinity columns, and the flowthrough was then applied to a GST-CDP/Cux affinity column to isolate antibodies.
Preparation of nuclear extracts and Western blot analysis.
Nuclear extracts were prepared as described previously (48). For Western blotting, indicated quantities of nuclear extracts were recovered as described above and were resuspended in Laemmli buffer. Proteins were then boiled for 5 min and loaded on a sodium dodecyl sulfate-polyacrylamide gel. The gels were equilibrated for 10 min in 0.1 M Tris-0.192 M glycine-20% (vol/vol) methanol, and the proteins were electrotransferred to polyvinylidene difluoride membranes overnight at 40 V at 4°C. The membranes were then washed with Tris-buffered saline (TBS; 10 mM Tris [pH 8], 150 mM NaCl) and blocked in TBS supplemented with 5% milk and 2% bovine serum albumin for 2 h at room temperature. The indicated antibodies were diluted 1:1,000 in TBS supplemented with 0.1% Tween (TBST), and membranes were incubated for 1 h at room temperature. Following five washes with TBST, horseradish peroxidase-conjugated secondary antibodies (Santa Cruz) were diluted 1:4,000 in TBST, and membranes were incubated for 40 min at room temperature. Membranes were washed five times with TBST followed by two washes with TBS. Proteins were detected with the ECL kit from Amersham Pharmacia Biotech.
EMSA.
Electrophoretic mobility shift assays (EMSAs) were performed with indicated quantities of purified protein. Samples were incubated at room temperature for 20 min in a final volume of 30 μl of 25 mM NaCl, 10 mM Tris (pH 7.5), 1 mM MgCl2, 5 mM EDTA (pH 8.0), 5% glycerol, 1 mM dithiothreitol, and 3 μg of bovine serum albumin with 0.2 pmol of radiolabeled oligonucleotides. Samples were loaded on a 5% polyacrylamide (29:1)-0.5× Tris-borate-EDTA gel and separated by electrophoresis at 8 V/cm in 0.5× Tris-borate-EDTA. Gels were dried and visualized by autoradiography.
Calculation of the DNA binding affinity.
To determine the dissociation constant (KD), EMSAs were performed essentially as described above, but with a fixed concentration of DNA (≤10 pM) and a wide range of protein concentrations and with the following modifications. Less than 10 pM DNA was used, and protein and DNA were incubated for 15 min at room temperature. The binding affinity (KD) was calculated using the method described by Janet Carey (14, 15). The amount of free and bound DNA was quantitated by scanning of the autoradiograms on a phosphorimager (Amersham Pharmacia Biotech Typhoon 8600) and verified by scintillation counting of the excised bands in an independent experiment. The data were plotted as the fraction of free DNA versus the log of protein concentration. Since the protein concentrations did not take into account the fraction of inactive proteins, our data are referred to as the apparent dissociation constants [KD(app)].
Oligonucleotides.
The sequences of oligonucleotides used in this study are as follows: TCGAGACGATATCGATAAGCTTCTTTTC (universal CDP/Cux consensus binding site); TCGAGACGGTATCGATAGCTTCTTTTC (ATCGAT); GGGCCGCTGATTGGCTTTCAGGCTGGCGCCTCGA (DNApol α −40/−14); GGGCCGCTGAAAGCTTCACAGGCTGGCGCCTCGA (DNA pol α −40/−14 mut −35/−26). Underlined sequences represent mutations introduced in linker-scanning analysis.
DNase I footprinting.
The DNA pol α fragment −116/+47 was used for this analysis. The plasmid was 32P labeled at the NcoI site with the Klenow fragment of DNA polymerase I and cleaved with SacII. After electrophoresis through a 5% polyacrylamide gel, the labeled fragments were purified by passive elution in 10 mM Tris-HCl (pH 7.5)-1 mM EDTA. DNase footprinting analysis was performed as previously described (25). End-labeled DNA (8,000 cpm per tube) was incubated with purified bacterially expressed fusion proteins for 15 min at room temperature in a final volume of 75 μl in 10 mM Tris (pH 7.5), 25 mM NaCl, 1 mM MgCl2, 1 mM dithiothreitol, 5% glycerol, and 4% (wt/vol) polyvinyl alcohol. Two hundred twenty-five microliters of 10 mM MgCl2-5 mM CaCl2 was added for 90 s. Various dilutions of DNase I were added, and samples were then incubated for 90 s. At that time, 270 μl of DNase stop solution (20 mM EDTA, 1% sodium dodecyl sulfate, 0.2 M NaCl) was added, and the solution was mixed by vortexing. Following phenol-chloroform extraction and ethanol precipitation, samples were electrophoresed through a 6% denaturing polyacrylamide (40:1) gel in 1× Tris-borate-EDTA. Gels were dried and visualized by autoradiography.
Chromatin immunoprecipitation.
Chromatin was prepared as described previously (52) with the following exception. Sonication was performed for 5 s followed by a 2-min incubation on ice, and this was repeated a total of four to six times. A single aliquot was retained for chromatin quality assessment and chromatin quality control. We estimated that 15 × 10 6 to 20 × 10 6 cells per aliquot would ensure similar titers of chromatin in all subsequent immunoprecipitations. To ensure quality control between experiments, protein A-agarose beads from the chromatin immunoprecipitation (ChIP) assay kit (no. 17-295; Upstate Biotechnology) were used for the immunoprecipitations. The extract was first incubated with 30 μl of beads for 1 h. After centrifugation, the supernatant was transferred to a new tube and incubated overnight at 4°C with 2 μg of purified CDP/Cux antibody. The next day, 25 μl of beads was added, and incubation was continued for at least 1 h at 4°C. The supernatant was removed, and the beads were washed two times with low-salt buffer, two times with high-salt buffer, and then once with Tris-EDTA. Elution and DNA purification followed the protocol of Nissen and Yamamoto, with the addition of an additional phenol-chloroform extraction (52). The presence of binding to three different regions of the DNA pol α gene promoter in the immunoprecipitated chromatins was analyzed by PCR with specific sets of oligonucleotides: region 1 (PCR 1), 5′CCCTCAGCTCTAGCTTTTCCCTAAGGGG3′ and 5′CATGGTCCCGAATCTCCCGATTCC3′; region 2 (PCR 2), 5′GGTTCTCTCCTGGTTGGAAAAGCTTG3′ and 5′TTGCCCACATGCTTATTGATCCCTTC3′; region 3 (PCR 3), 5′GGTGCCTTATTGCTCTGTTCTCACATGG3′ and 5′CAGCTGATTACTTCCCACATGCCCG3′. PCRs, in 50 μl, were done with Taq polymerase (MBI Fermentas) for a total of 37 cycles. The temperature of hybridization corresponded to the value of the melting temperature (Tm) of the oligonucleotides for the first 6 cycles. The hybridization temperatures were decreased until 5°C below the Tm for the rest of the PCR.
In vivo DNA binding to transfected reporter plasmids.
HS578T cells were transfected with pGL3-Pol α (−65/+47), pGL3-Pol α (−65/+47) mut (−35/−26), and either pXJ42 or pXJ42/CDP/Cux CR2CR3HD. DNA was extracted approximately 24 h posttransfection. The chromatin immunoprecipitation protocol described above was used, except that samples of extracted DNA were not sonicated. Primers used in PCR are as follows: 5′CCGAGCCGCTGATTGGCTTT3′ (WT) or 5′CCGAGCCGCTGAAAGCTTCA3′ (mut −35/−26) was used with 5′AGCGGTTCCATCTTCCAGCGGATAGA3′.
Retroviral infections and RT-PCR.
HS578T cells were infected by the addition of virus-containing supernatant from 293VSV producer cells (54). Cells were harvested 48 h postinfection. To minimize the extent of proteolytic processing of the full-length CDP/Cux protein, cells had been plated so as to reach near-confluence 48 h postinfection. Real-time reverse transcriptase (RT) PCR was performed with a LightCycler with the Fast Start DNA Master SYBR Green I kit (Roche) and the following primers for DNA pol α: sense primer, 5′GCTTCACCGAATCCTTTCTCTGTG3′ (mRNA position 581 to 604); antisense primer, 5′TTCCTCATCTGCCCCTTTTACC3′ (1030 to 1009). DNA pol α RNA was normalized to the amount of glyceraldehyde phospho-dehydrogenase (GAPDH) (see reference 23 for the primer sequence) RNA amplified.
RESULTS
CDP/Cux stimulates the DNA pol α gene promoter during S phase in NIH 3T3 cells.
A search of the promoter database with the CDP/Cux consensus binding site revealed that the proximal promoter sequences of the DNA pol α gene contained several putative CDP/Cux binding sites in both Drosophila melanogaster and humans (Fig. 1). The DNA pol α gene was previously shown to be up-regulated at the transcriptional level in S phase (56). Using RT-PCR, we confirmed that DNA pol α mRNA expression was up-regulated in S phase following reentry of NIH 3T3 cells into the cell cycle (data not shown). To determine whether CDP/Cux could regulate the human DNA pol α gene promoter, NIH 3T3 cells were cotransfected with a luciferase reporter plasmid containing the sequence from −1561 to +47 of the human DNA pol α gene and either an empty vector or a vector expressing CDP/Cux 817-1505 (Fig. 1). This recombinant protein corresponds to the 110-kDa isoform, CDP/Cux p110, that is generated by proteolytic processing in S phase of the cell cycle (48). CDP/Cux p110 had little or no effect on the expression of the DNA pol α reporter when transfected NIH 3T3 cells were allowed to grow asynchronously (Fig. 1A and B). In contrast, expression of the DNA pol α reporter was stimulated in the presence of CDP/Cux p110 when NIH 3T3 cells were synchronized in S phase either by thymidine block (Fig. 1A) or by serum starvation and restimulation (Fig. 1B). The same assay was repeated with a panel of transformed cell lines that were allowed to grow asynchronously. Significant stimulation of the DNA pol α reporter was observed in HS578T, T47D, and T98G cells, and moderate levels of stimulation were observed in C33A, MCF-7, and HeLa cells (Fig. 1C). The levels of activation may vary due to differences in transfection efficiency or levels of endogenous CDP/Cux, which may affect the response to its overexpression. In summary, CDP/Cux p110 was able to stimulate expression of the DNA pol α reporter in several cell lines; however, in NIH 3T3 cells, this stimulatory effect was dependent upon the cells being synchronized in S phase.
FIG. 1.
CDP/Cux stimulates the DNA pol α gene promoter. (A and B) NIH 3T3 cells were cotransfected with a DNA pol α reporter construct (pGL3-pol α [−1561/+47]) and either an empty vector or a vector expressing a CDP/Cux protein containing aa 817 to 1505 (pXM/HSCDP-817-1505). Cells were either left unsynchronized or synchronized in S phase by thymidine block (A) or by serum starvation-restimulation (B), as described in Materials and Methods. Cytoplasmic extracts were prepared and processed to measure luciferase activity. Results are expressed as relative light units (RLU) normalized to β-galactosidase activity from an internal control and are representative of the mean of results from a minimum of two separate experiments. (C) Various tumor cell lines were cotransfected as described for panel A and were left unsynchronized. Results are expressed as activation when CDP/Cux is transfected relative to activation with transfection of empty vector. (D) HS578T cells were cotransfected with the DNA pol α −1561/+47 reporter construct and either an empty vector or increasing amounts of the pXM/HSCDP-817-1505 vector. Luciferase activity was expressed as relative light units normalized to β-galactosidase activity from an internal control and is representative of the mean of results of three separate experiments. A diagrammatic representation of the reporter and effector plasmids is shown at the bottom. The bars within the DNA pol α promoter sequences represent putative CDP/Cux binding sites.
N-terminal truncation of CDP/Cux is necessary for stimulation of the DNA pol α reporter.
To investigate the mechanism by which CDP/Cux was able to stimulate expression of the DNA pol α reporter, the reporter assay was repeated with effector plasmids expressing CDP/Cux recombinant proteins with progressive N-terminal truncations. HS578T cells were utilized for these assays, as our preliminary experiments revealed that this line consistently displayed the highest level of stimulation by CDP/Cux. Full-length CDP/Cux protein was unable to stimulate expression and, in fact, produced what appeared to be weak repression (Fig. 2A). Yet, the protein expressed from this plasmid was functional, since it was able to repress a reporter plasmid carrying the promoter of the p21Waf1/Cip1 gene, as previously published (Fig. 2B) (17). In contrast to full-length CDP/Cux, all N-terminally truncated CDP/Cux proteins were able to stimulate expression of the DNA pol α reporter (Fig. 2A). This stimulatory effect correlated well with the ability of CDP/Cux proteins to bind to a consensus binding site that is specific for CR3HD or CR2CR3HD (Fig. 2D, lanes 3 to 6). In contrast, the full-length CDP/Cux protein, whose expression was confirmed by Western blotting (Fig. 2C, lane 1), interacted only weakly with this sequence (Fig. 2C, lane 2). These data are in agreement with previous studies showing that full-length CDP/Cux binds to DNA only transiently and exhibits a preference for sequences containing not just one but two CAAT or CGAT motifs (47, 48). Thus, we conclude that p110, but not p200, is able to stimulate expression of the DNA pol α reporter.
FIG. 2.
Amino-terminal truncation of CDP/Cux is necessary for stimulation of DNA pol α gene expression. (A) HS578T cells were cotransfected with the DNA pol α −1561/+47 reporter construct (see Fig. 1) and vectors expressing CDP/Cux proteins with progressive N-terminal truncations, as indicated. At 48 h posttransfection, cells were harvested and cytoplasmic and nuclear extracts were prepared. Cytoplasmic extracts were analyzed for luciferase activity as described in the legend to Fig. 1A. (B) HS578T cells were cotransfected with a reporter plasmid carrying the promoter of the p21Waf1/Cip1 gene and a vector expressing the full-length CDP/Cux proteins. At 48 h posttransfection, cytoplasmic extracts were prepared and analyzed for luciferase activity as described in the legend to Fig. 1A. (C) Nuclear extracts were separated on an 8% sodium dodecyl sulfate-polyacrylamide gel. Proteins were transferred to polyvinylidene difluoride membrane and analyzed by Western blotting with an anti-HA antibody that recognizes the C terminus. (D) Nuclear extracts were analyzed by EMSA with oligonucleotides encoding a CDP/Cux consensus binding site.
The CTD is expendable, but the Cut homeodomain is required, for stimulation.
Removal of the CTD of CDP/Cux had no effect on the reporter assay, whereas removal of the Cut homeodomain prevented stimulation of the DNA pol α reporter (Fig. 3A). All CDP/Cux proteins were expressed at high levels, as observed by Western blot analysis (Fig. 3B). All recombinant proteins bound to DNA efficiently with the exception of CDP/Cux 659-1192, in which the Cut homeodomain was deleted (Fig. 3C, compare lane 5 with lanes 2 to 4). We conclude that the Cut homeodomain is required for the stimulation of the DNA pol α reporter. In summary, results from mapping analysis demonstrated that aa 1 to 1061 and 1301 to 1505 are dispensable. Moreover, the presence of the N-terminal portion of the protein, from aa 1 to 659, and the absence of the Cut homeodomain will prevent the stimulatory function of CDP/Cux. While the mapping data presented were obtained in the HS578T cell line, similar results were obtained in NIH 3T3 cells (data not shown). These results are consistent with the notion that CDP/Cux must be able to make a stable interaction with DNA in order to stimulate expression from the DNA pol α reporter.
FIG.3.
The Cut homeodomain, but not the carboxy-terminal domain of CDP/Cux, is required for stimulation. HS578T cells were cotransfected with the DNA pol α −65/+47 reporter construct (see Fig. 5) and vectors expressing N- and C-terminally truncated CDP/Cux proteins, as indicated. At 48 h posttransfection, cells were harvested and cytoplasmic and nuclear extracts were prepared. (A) Cytoplasmic extracts were analyzed for luciferase activity as described in the legend to Fig. 1A. The means of results of three transfections are shown. (B) Nuclear extracts were subjected to Western blot analysis with the anti-861 antibody. (C) Nuclear extracts were analyzed by EMSA with oligonucleotides encoding a CDP/Cux consensus binding site as described in the legend to Fig. 2C. Complexes containing CDP/Cux were supershifted with antibody 861 (lanes 6 to 10) but not with a nonspecific antibody (lanes 11 to 15). A diagrammatic representation of the effector plasmids and the region recognized by antibody 861 is shown at the bottom.
CDP/Cux can stimulate the expression of reporter plasmids containing the promoter sequences of other S phase-specific genes.
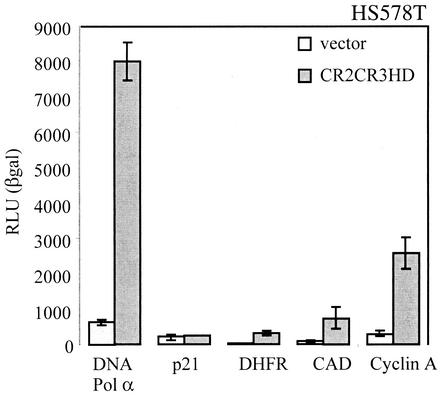
To verify whether stimulation of gene expression by CDP/Cux was unique to the DNA pol α promoter, we tested promoter sequences from a number of genes whose expression is up-regulated in S phase. As a control, we also tested a reporter containing the core promoter of the p21Waf1/Cip1gene. This reporter was previously shown to be repressed by CDP/Cux in cotransfection assays (17, 58). Cotransfection with CDP/Cux CR2CR3HD did not affect the expression of the p21Waf1/Cip1 reporter. It is likely that repression of the p21Waf1/Cip1 promoter by CDP/Cux requires the action of the active repression domains present in the CTD. In contrast, reporter plasmids carrying the promoters from the dihydrofolate reductase (DHFR), carbamoyl-phosphate synthase-aspartate carbamoyltransferase-dihydroorotase (CAD), and cyclin A genes were stimulated by CDP/Cux CR2CR3HD, albeit to a lesser extent than what was observed with the DNA pol α reporter (Fig. 4).
FIG. 4.
CDP/Cux CR2CR3HD stimulates other S phase-specific gene promoters. HS578T cells were cotransfected with the indicated reporter constructs and either empty vector or a vector expressing CDP/Cux CR2CR3HD. Luciferase assays were performed as described in the legend to Fig. 1A. Average levels of activation for DNA pol α, p21, DHFR, CAD, and cyclin A are 12-, 1-, 12-, 8-, and 9-fold, respectively.
CDP/Cux can stimulate the core promoter of the DNA pol α gene.
To identify the DNA pol α promoter sequences that are required for stimulation by CDP/Cux, a series of reporter plasmids with progressive 5′ deletions were tested in the cotransfection assays. CDP/Cux stimulated the expression of all reporter plasmids, including one plasmid carrying DNA pol α sequences from −65 to +47 (Fig. 5A). We conclude that the core promoter of the DNA pol α gene contains sequences that allow its stimulation in the presence of CDP/Cux.
FIG. 5.
CDP/Cux can stimulate the core DNA pol α gene promoter. (A) HS578T cells were cotransfected with DNA pol α reporter constructs with progressive 5′ deletions and either an empty vector or a vector expressing CDP/Cux CR2CR3HD. Luciferase assays were performed as described. Results are expressed as activation relative to that of the −1561/+47 reporter, which was assigned a value of 100%. (B) Linker scanning mutations were introduced at 10-bp intervals within the reporter construct containing the core DNA pol α gene promoter. Mutations consisted of the replacement of the indicated sequence with the sequence GAAAGCTTCA. HS578T cells were cotransfected with the indicated reporter constructs and either an empty vector or a vector expressing CDP/Cux CR2CR3HD. Luciferase assays were performed as described in the legend to Fig. 1A. Results are expressed as activation relative to that of the wild-type −65/+47 reporter, which was assigned a value of 100%.
Linker scanning mutations were introduced into the −65/+47 DNA pol α reporter plasmid. These mutations consisted of the serial replacement of 10-bp sequences with the sequence GAAAGCTTCA. Two replacement mutations, at positions −35 to −26 and −25 to −16, significantly reduced the ability of CDP/Cux CR2CR3HD to stimulate gene expression (Fig. 5B).
CDP/Cux CR2CR3HD can bind to the core DNA pol α gene promoter in vitro.
DNase footprinting analysis was performed to verify whether CDP/Cux proteins containing the two DNA binding domains CR2 and CR3 along with the Cut homeodomain (CR2CR3HD), or those containing just CR3HD, would interact with the core DNA pol α gene promoter. A DNA fragment was end labeled at position +47, incubated with purified bacterially expressed His-CR2CR3HD or His-CR3HD protein, and treated with DNase I. A protected region was observed between nucleotides (nt) −14 and −40 of the coding strand (Fig. 6A, lanes 2 and 6 to 7). Interestingly, an inverted CCAAT motif resides within this interval, at position −30 to −34. EMSA with double-stranded oligonucleotides corresponding to nt −40 to −14 confirmed that the purified His-CR2CR3HD protein could form a strong retarded complex with this sequence (Fig. 6B, lane 2). In contrast, the full-length CDP/Cux protein was unable to make a stable interaction with the DNA pol α sequence (Fig. 6D, lanes 6 to 9).
FIG.6.
A truncated CDP/Cux protein can bind to the core DNA pol α gene promoter. (A) DNase I footprinting analysis of the core DNA pol α promoter. A DNA fragment including sequences from −116 to +47 of the DNA pol α promoter was end labeled at position +47 and was incubated with the purified bacterially expressed His-CR2CR3HD (lane 2) or His-CR3HD protein (lanes 5 to 7). A sequencing reaction was run in parallel. (B) EMSA was performed with radiolabeled oligonucleotides containing nt −40 to −14 of the DNA pol α gene promoter, either of the wild type (left panel) or with the linker replacement mutation at position −35 to −26 (right panel), and 20 ng of purified bacterially expressed His-CDP/Cux CR2CR3HD protein. As competitors, the same cold oligonucleotides representing the wild type and mutated sequence were added as indicated. (C) EMSA was performed with the same radiolabeled oligonucleotides as described in the legend to panel B with increasing amounts of His-CDP/Cux CR2CR3HD. The KD(app) was determined as described in Materials and Methods. (D) EMSA was performed with oligonucleotides containing nt −40 to −14 of the DNA pol α gene promoter and various amounts of the purified histidine-tagged CDP/Cux recombinant proteins, 831-1336 (CR2CR3HD) and 1-1505.
A mutation that reduces stimulation of expression in vivo also reduces DNA binding in vitro.
Two assays were performed to verify whether the replacement mutation at position −35/−26 would reduce the affinity of His-CR2CR3HD for this sequence. In the first assay, wild-type or mutated −40/−14 oligonucleotides were used as cold competitors in EMSA with the wild-type −40/−14 probe. Whereas a 100-fold excess of the wild-type oligonucleotides completely eliminated the retarded complex, the oligonucleotides with the −35/−26 replacement mutation did not compete as efficiently (Fig. 6B, lanes 3 and 4). In the second assay, oligonucleotides with the −35/−26 replacement mutation were used as an EMSA probe. The His-CR2CR3HD protein was able to generate a complex, although the intensity of the complex appeared weaker than that of the wild-type sequence (Fig. 6B, compare lanes 2 and 6). Moreover, as we had seen previously, the wild-type oligonucleotides were more efficient competitors than the mutated ones (Fig. 6B, lanes 7 and 8).
The DNA binding affinity for the wild-type and mutated oligonucleotides was assessed by performing EMSA with a fixed concentration of DNA (≤10 pM) and a wide range of protein concentrations (Fig. 6C). The KD(app) values were measured as described in Materials and Methods. His-CR2CR3HD exhibited KD(app) values of 5.8 × 10−8 M and 1.3 × 10−7 M for the wild-type and mutated oligonucleotides, respectively (Fig. 6C). Thus, the −35/−26 replacement mutation reduced the affinity of CR2CR3HD for the core DNA pol α promoter by a factor of approximately 2.2-fold. In other experiments, we found that the −25/−16 replacement mutation that also reduced the stimulatory effect of CDP/Cux on the DNA pol α reporter did not affect the interaction of CDP/Cux with the DNA pol α promoter sequences in vitro (data not shown). We postulate that this mutation interferes with the binding of another protein that participates in the transcriptional activation of the DNA pol α promoter and is required for the stimulatory effect of CDP/Cux.
In summary, a CDP/Cux protein containing CR2CR3HD was able to stimulate expression of a reporter containing the sequence −65 to +47 from the DNA pol α gene promoter. This stimulatory effect was abolished by the replacement of the sequence from −35 to −26 or −25 to −16. In vitro, a purified CDP/Cux protein containing CR2CR3HD was able to interact with the core DNA pol α promoter sequences; however, the replacement of the sequence from −35 to −26 diminished the affinity of CDP/Cux for the DNA pol α promoter. Thus, a correlation was established between the stimulation of the core DNA pol α gene promoter in vivo and the interaction of CDP/Cux with DNA pol α promoter sequences in vitro.
CDP/Cux binds the DNA pol α gene promoter in vivo, specifically during the S phase of the cell cycle.
ChIP assays were performed to investigate whether endogenous CDP/Cux proteins bind to the promoter of the DNA pol α gene in vivo. Primers were designed to amplify three different regions of the DNA pol α gene promoter and one region upstream of the GAPDH gene promoter. With total chromatin as a template, each pair of primers amplified a DNA fragment of the expected molecular weight (Fig. 7A, lane 3). With chromatin obtained after immunoprecipitation with the anti-CDP/Cux antibody 861, an amplified fragment was observed for the regions from −1179 to −843 and −173 to +47 (Fig. 7A, lane 2, PCR 1 and 2). In contrast, no fragment was obtained with primers for the GAPDH gene promoter or the upstream region of the DNA pol α gene promoter between nt −1505 and −1229 (Fig. 7A, lane 2, GAPDH and PCR3). Controls consisted of template chromatin that was obtained following immunoprecipitation with either anti-immunoglobulin G (IgG) or anti-hemagglutinin (HA) antibodies. No amplified fragment was observed with any of the primer pairs (Fig. 7A, lanes 4 and 5). We conclude that CDP/Cux can interact with the DNA pol α gene promoter in vivo. We note also that a more intense signal was obtained in the region from −1179 to −843 than in the proximal promoter region (Fig. 7A, lane 2, compare PCR 1 and 2). The reason for this is not entirely clear but may involve the fact that this region of the promoter contains a higher concentration of sequence motifs that match the CDP/Cux consensus binding site (see the map in Fig. 1). To verify whether the association of CDP/Cux with the DNA pol α gene promoter is regulated during the cell cycle, we performed ChIP assays on synchronized HS578T cells. Binding to PCR regions 1 and 2 was detected specifically in S phase (Fig. 7B, lane 2). CDP/Cux did not interact with the DNA pol α gene promoter in G2/M or G1 (Fig. 7B, lanes 3 and 4).
FIG. 7.
CDP/Cux binds the DNA pol α gene promoter in vivo, specifically during the S phase of the cell cycle. (A) CDP/Cux binds to the DNA pol α gene promoter in vivo. ChIP was performed with HS578T cells and either of the following antibodies: anti-861 CDP/Cux antibody (lane 2), anti-IgG (secondary) antibody (lane 4), or anti-HA antibody (lane 5). The immunoprecipitated DNA was used as the template in PCR with the indicated primers from the DNA pol α gene promoter or from the GAPDH promoter (lane 2). As control, the PCRs were performed in parallel with total chromatin (lane 3). Shown below is a map of the DNA pol α gene promoter indicating the positions of primers used in PCRs 1, 2, and 3. (B) Binding of CDP/Cux to the DNA pol α gene promoter occurs in S phase only in HS578T cells. HS578T cells were synchronized by double thymidine block, as detailed in Materials and Methods. To obtain populations of cells enriched in either S, G2/M, or G1 phase, cells were grown for 0, 4, or 10 h, respectively, following the second thymidine block. Cells were harvested and processed for ChIP assay and cell cycle analysis. Cell cycle distribution was monitored by fluorescence-activated cell sorting analysis after staining of the DNA with propidium iodide. The fluorescence-activated cell sorting profiles are shown at the bottom, together with the calculated proportion of cells in each phase of the cell cycle. (C) Chromatin immunoprecipitations (ChIP) were performed using HS578T cells and the indicated antibodies. The immunoprecipitated DNA was used as template in real-time PCR using either PCR 1 or PCR 2 primers as indicated. The results are expressed as levels of activation using as a control the anti-IgG sample which was given a value of 1. In the rightmost panel, nuclear extracts from HS578T cells were subjected to immunoprecipitation with the indicated antibodies, followed by immunoblotting with the 1300 antibody. Below is a schematic representation of CDP/Cux isoforms and the regions recognized by the respective antibodies. The evolutionarily conserved domains are indicated (CC, coiled-coil; CR1, CR2 and CR3, Cut repeats 1, 2 and 3; HD, homeodomain).
To verify whether the full-length CDP/Cux protein can interact with the DNA pol α gene promoter in vivo, ChIP assays were performed with a panel of antibodies that recognize various regions of CDP/Cux (see the diagram in Fig. 7C). Three of these antibodies, namely, antibodies 23, 403, and 510, recognize only the full-length protein, while antibodies 861 and 1300 can bind to both the full-length and the processed isoforms (Fig. 7C, rightmost panel). As a control, ChIP assays were performed with IgG. The signal obtained with this sample was taken as background and attributed with a value of 1. Significantly stronger signals were observed with samples obtained with antibodies 861 and 1300. In contrast, samples obtained with antibodies that recognize only the full-length protein did not generate a stronger signal than background. We cannot exclude the possibility that the epitope recognized by an antibody is masked when the protein is bound to DNA. However, it becomes difficult to evoke this possibility to explain the absence of amplification with three different antibodies. The results rather suggest that the full-length CDP/Cux protein does not make a stable interaction with the DNA pol α gene promoter.
Reduction in stimulation of gene expression correlates with a decrease in DNA binding in vivo.
Since we observed a correlation between a decrease in DNA binding in vitro and a reduction in the stimulation of gene expression in transfection assays with the linker scanning mutant −35/−26, we considered the possibility that the decreased affinity of CDP/Cux for this mutant precluded the recruitment of CDP/Cux to the reporter plasmid following transfection into cells. To test this hypothesis, we designed oligonucleotide primers that would specifically amplify sequences from either the wild type or the −35/−26 mutant reporter plasmid. In preliminary experiments, the wild-type primers were found to efficiently amplify the wild-type, but not the mutated, sequence, whereas the mutated primers amplified the mutated, but not the wild-type, sequence (Fig. 8A). HS578T cells were cotransfected with both the wild-type and the mutated reporter plasmids together with either an empty vector or a vector coding for CDP/Cux CR2CR3HD. With total chromatin, a fragment of the expected molecular weight was amplified with each pair of primers to verify that the cells had each received the two plasmids as expected (Fig. 8B, lanes 2 to 3 and 8 to 9). Faint amplified fragments were observed when chromatin obtained by immunoprecipitation with the preimmune serum or chromatin derived from cells that had received the empty effector plasmid was used as a template (Fig. 8, lanes 4 to 7 and 10 to 11). We think this is due to the fact that a small, background amount of CDP/Cux is immunoprecipitated with beads alone, regardless of the buffer used. However, when the chromatin was first subjected to immunoprecipitation with the anti-CDP/Cux antibody, a fragment of strong intensity was amplified by the wild-type primers but not by the mutated primers (Fig. 8, lanes 12 and 13). This result indicates that in cells containing both the wild-type and the mutated reporter plasmids, the recombinant CDP/Cux protein was able to interact efficiently with the wild-type reporter plasmid but not with the mutated plasmid.
FIG. 8.
A linker scanning mutation at positions −35 to −26 of the DNA pol α gene promoter prevents the binding of CDP/Cux to the reporter plasmid in vivo. (A) PCR primers (indicated by arrows in the diagram) were designed to specifically amplify either the wild-type DNA pol α gene promoter or the mutant promoter containing the linker scanning mutation at positions −35 to −26 (see Fig. 7). PCRs were performed with either wild-type or mutated plasmid DNA template and the corresponding primers, as indicated. (B) CDP/Cux in vivo binds to the wild-type DNA pol α reporter but not to the −35/−26 mutant reporter. HS578T cells were cotransfected with the wild type and mutant −35/−26 reporter construct and either an empty vector or a vector expressing CDP/Cux CR2CR3HD. After 2 days, ChIP assays were performed with the indicated primers and antibodies. As control, the PCRs were performed in parallel using total chromatin (lane 3). Abbreviations: Pre-I, preimmune serum; CDP, antibody 1300, a CDP/Cux-specific antibody; wt, wild type; mut, mutant.
CDP/Cux can regulate the endogenous DNA pol α gene.
The experiments described above demonstrated that expression of a DNA pol α reporter could be stimulated in the presence of N-terminally truncated CDP/Cux protein. These experiments, however, did not establish whether CDP/Cux could regulate the endogenous DNA pol α gene. To address this question, we infected HS578T cells with high-titer retroviral vectors expressing either the full-length or a truncated CDP/Cux protein. Cells were harvested 48 h following infection. RNA and proteins were purified from 70 and 30% of the cells, respectively. Expression of the recombinant CDP/Cux proteins was verified by Western blot analysis (Fig. 9B). RT-PCR analysis was used to quantitate DNA pol α mRNA (Fig. 9A). Compared with the amount in cells infected with an empty retrovirus, the amount of DNA pol α mRNA was increased fourfold on average in cells infected with the retrovirus expressing the truncated CDP/Cux protein. However, no increase in DNA pol α expression was observed in cells infected with the full-length CDP/Cux vector. Again, the stimulation in gene expression by the truncated CDP/Cux protein could result from true activation or repression of a repressor. Nonetheless, these results clearly demonstrate the endogenous DNA pol α gene can be regulated in response to CDP/Cux.
FIG. 9.
CDP/Cux stimulates the endogenous DNA pol α gene promoter. (A) HS578T cells were infected with pREV retroviral vectors expressing a full-length or a truncated CDP/Cux protein. To minimize the extent of proteolytic processing of the full-length CDP/Cux protein, cells had been plated so as to reach near confluence 48 h postinfection, at which time total RNA and whole-cell extracts were prepared. Expression of DNA pol α RNA was determined by real-time PCR and was normalized for GAPDH RNA expression. DNA pol α expression in infected cells was compared with that in uninfected cells and is expressed as fold activation relative to expression in uninfected cells. Values represent the means of results of three separate infections. (B) Nuclear extracts were subjected to Western blot analysis with the anti-1300 antibody.
DISCUSSION
The CDP/Cux transcription factor was originally characterized as a transcriptional repressor (6, 17, 20, 28, 33-35, 37, 39, 43, 45, 51, 53, 55, 60, 63, 64, 69, 78). Recently, it was shown that an N-terminally truncated isoform, CDP/Cux p110, is generated by proteolytic processing at the G1/S transition of the cell cycle. Results from the present study revealed that CDP/Cux p110 is able to stimulate transcription from a reporter plasmid containing the DNA pol α promoter (Fig. 1 to 5). Moreover, expression of the endogenous DNA pol α gene was stimulated in a population of cells infected with a retrovirus expressing a truncated CDP/Cux protein (Fig. 9). These results suggest, yet do not demonstrate, that CDP/Cux p110 can function as a transcriptional activator. Stimulation of transcription, whether of a reporter or an endogenous gene, is consistent with a number of possible mechanisms. CDP/Cux p110 might directly activate transcription. It is unlikely that CDP/Cux p110 functions like a classical transcriptional activator with a DNA binding domain and an activation domain. No region of CDP/Cux was found to function as an activation domain in the Gal4 fusion assay, in which various regions of a protein are fused to the DNA binding domain of the Gal4 transcription factor and are assayed together with a Gal4 reporter plasmid (45). Therefore, we envision that CDP/Cux p110 might contribute, perhaps as an architectural factor, to the formation of a larger complex or enhanceosome (22). This mode of action would be consistent with what we know of the HiNF-D complex, which includes CDP/Cux as well as a number of other proteins, and whose presence on the promoter of histone genes coincides with their induction in S phase (3, 4, 36, 67, 70-73).
In light of the well-characterized role of CDP/Cux as a transcriptional repressor, we cannot exclude the possibility that CDP/Cux p110 represses the expression of another repressor that down-modulates the DNA pol α promoter. Indeed, as cells were harvested 48 h posttransfection or -infection, any regulatory effect might be direct or indirect. However, we did not obtain any evidence in support of a repression mechanism. DNA fragments containing sequences from −65 to +47 of the DNA pol α promoter were tested in EMSA and DNase footprinting analyses. Using protein extracts from cells overexpressing CDP/Cux, we did not observe a decrease or disappearance of a retarded complex (data not shown). It still remains possible that CDP/Cux p110 interferes with the binding of a repressor that could not be detected in unfractionated nuclear extracts. In contrast to the lack of evidence in favor of a repression mechanism, we were able to establish a correlation between transcriptional stimulation and the ability of CDP/Cux p110 to bind to the DNA pol α promoter sequences in EMSA and ChIP assays. These results strongly suggest, yet do not prove, that CDP/Cux p110 functioned as a true activator. More direct evidence that CDP/Cux p110 may function as a transcriptional activator would be provided if the addition of CDP/Cux p110 to an in vitro transcription system led to transcriptional activation. Again, however, in a crude in vitro system we could not exclude the possibility that CDP/Cux p110 stimulates transcription by competing with, and displacing, a repressor. Therefore, we do not think that experimental evidence from a single assay will be sufficient to demonstrate one mechanism of action at the expense of another. Instead, the accumulation of evidence from a panoply of assays and experimental conditions will gradually build a case in favor of one mechanism.
A recombinant CDP/Cux protein corresponding to CDP/Cux p110 was able to stimulate transcription, but the full-length CDP/Cux isoform was not (Fig. 2 and 9). These results indicate that stimulation of transcription is a specific property of CDP/Cux p110 that is not shared with CDP/Cux p200. As CDP/Cux p110 is capable of making a stable interaction with DNA, whereas CDP/Cux p200 is not, the need for N-terminal truncation is likely to reflect the requirement for stable DNA binding. This would be compatible with the two mechanisms cited above: direct activation and repression of a repressor. CDP/Cux was previously shown to repress by two mechanisms: active repression and competition for binding site occupancy (45). While competition can be accomplished via transient or stable DNA binding, active repression was shown to involve the recruitment of a histone deacetylase (39). Similarly, transcriptional activation has been associated with the recruitment of histone acetyltransferases, chromatin-remodeling machines, general transcription factors, and/or the stabilization of other site-specific transcription factors (7, 12, 22, 61, 74). Although this has not been formally tested, it is generally assumed that a DNA-binding transcription factor must be able to make a stable interaction with DNA in order to participate in transcriptional activation or active repression. In the case of CDP/Cux, it is clear that proteolytic processing modifies its DNA binding properties. It is possible that another consequence of processing is to change the ability of CDP/Cux to interact with other proteins.
One linker mutation between nt −25 and −16 abolished transcriptional stimulation by CDP/Cux but did not affect its affinity for the core promoter (Fig. 5B and data not shown). It is likely that this mutation interferes with the binding of another protein that participates in the transcriptional activation of the DNA pol α promoter and is required for the stimulatory effect of CDP/Cux. This protein could be another transcription factor or one of the components of the preinitiation complex.
Results from ChIP assays showed that two regions of the DNA pol α gene promoter could be immunoprecipitated with CDP/Cux, the core promoter and a region approximately 1 kbp upstream. The latter was immunoprecipitated more efficiently, yet we found that the core promoter of the DNA pol α gene was sufficient to allow its stimulation in reporter assays. This result does not exclude the possibility that the upstream sequences may also contribute to the recruitment of CDP/Cux to the DNA pol α gene promoter. We envision that the core promoter, which contains a low-affinity binding site for CDP/Cux p110, was able to recruit CDP/Cux p110 when the protein was overexpressed in transfected cells, but it is possible that the upstream sequences play an important role in the recruitment of CDP/Cux p110 when the protein is expressed at physiological levels. In agreement with this notion, we found that a substantial fraction of purified CDP/Cux p110 elutes as a multimeric complex on a size exclusion column (L. Leduy and A. Nepveu, unpublished observations). Whether CDP/Cux p110 can multimerize in vivo and how this process is regulated should be addressed in future studies.
Earlier studies on the HiNF-D factor pointed to a role of CDP/Cux in the S phase of the cell cycle (29, 70, 72, 73, 77). More recently, various posttranslational modifications of CDP/Cux were shown to regulate its DNA binding activity in a cell cycle-dependent manner (17, 48, 58). Results presented herein have revealed a novel activity of this transcription factor that pertains to its role in cell cycle progression: CDP/Cux p110 is capable of stimulating, directly or indirectly, the promoters of DNA pol α and other genes that are induced in S phase. Future studies should ascertain the role of CDP/Cux p110 as a transcriptional activator. Another important issue will be to evaluate the role of CDP/Cux in the regulation of the cell cycle in various cell types. Unless the cux-1 and cux-2 genes are partially redundant, the fact that cux homozygous knock-out mice did not exhibit embryonic lethality indicates that cux-1 is not an essential gene (21, 44, 59). Yet, some phenotypes of the cux-1 homozygous knockout mice, like the smaller size of the mice, their defect in hair growth, and their reduced numbers of B and T cells, are compatible with a role of cux-1 in the proliferation of at least certain cells. Interestingly, these phenotypes are in striking contrast with the multiorgan hyperplasia and organomegaly displayed by a cux-1 transgenic mouse (38). The identity of the cells in which CDP/Cux contributes to proliferation and the developmental signals to which CDP/Cux responds should be the subject of intense investigation in the future.
Acknowledgments
We are grateful to P. J. Farnham and J. M. Blanchard for the generous gifts of DHFR and CAD and of cyclin A reporter constructs, respectively.
A.N. is the recipient of a scholarship from the Fonds de la Recherche en Santé du Québec. M.T. is the recipient of studentships from the Royal Victoria Hospital Research Institute (2000-2001) and the Alexander McGee Memorial Fellowship (2001-2002). This research was supported by grant no. MT-11590 from the Canadian Institute of Health Research of Canada to A.N.
REFERENCES
- 1.Andres, V., M. D. Chiara, and V. Mahdavi. 1994. A new bipartite DNA-binding domain: cooperative interaction between the cut repeat and homeo domain of the cut homeo proteins. Genes Dev. 8:245-257. [DOI] [PubMed] [Google Scholar]
- 2.Aufiero, B., E. J. Neufeld, and S. H. Orkin. 1994. Sequence-specific DNA binding of individual Cut repeats of the human CCAAT displacement/Cut homeodomain protein. Proc. Natl. Acad. Sci. USA 91:7757-7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aziz, F., A. J. van Wijnen, J. L. Stein, and G. S. Stein. 1998. HiNF-D (CDP-cut/CDC2/cyclin A/pRB-complex) influences the timing of IRF-2-dependent cell cycle activation of human histone H4 gene transcription at the G1/S phase transition. J. Cell. Physiol. 177:453-464. [DOI] [PubMed] [Google Scholar]
- 4.Aziz, F., A. J. Vanwijnen, P. S. Vaughan, S. J. Wu, A. R. Shakoori, J. B. Lian, K. J. Soprano, J. L. Stein, and G. S. Stein. 1998. The integrated activities of IRF-2 (HiNF-M), CDP/Cut (HiNF-D) and H4TF-2 (HiNF-P) regulate transcription of a cell cycle controlled human histone H4 gene: mechanistic differences between distinct H4 genes. Mol. Biol. Rep. 25:1-12. [DOI] [PubMed] [Google Scholar]
- 5.Banan, M., I. C. Rojas, W. H. Lee, H. L. King, J. V. Harriss, R. Kobayashi, C. F. Webb, and P. D. Gottlieb. 1997. Interaction of the nuclear matrix-associated region (MAR)-binding proteins, SATB1 and CDP/Cux, with a MAR element (L2a) in an upstream regulatory region of the mouse CD8a gene. J. Biol. Chem. 272:18440-18452. [DOI] [PubMed] [Google Scholar]
- 6.Barberis, A., G. Superti-Furga, and M. Busslinger. 1987. Mutually exclusive interaction of the CCAAT-binding factor and of a displacement protein with overlapping sequences of a histone gene promoter. Cell 50:347-359. [DOI] [PubMed] [Google Scholar]
- 7.Bjorklund, S., and Y. J. Kim. 1996. Mediator of transcriptional regulation. Trends Biochem. Sci. 21:335-337. [DOI] [PubMed] [Google Scholar]
- 8.Blochlinger, K., R. Bodmer, L. Y. Jan, and Y. N. Jan. 1990. Patterns of expression of cut, a protein required for external sensory organ development in wild-type and cut mutant Drosophila embryos. Genes Dev. 4:1322-1331. [DOI] [PubMed] [Google Scholar]
- 9.Blochlinger, K., L. Y. Jan, and Y. N. Jan. 1991. Transformation of sensory organ identity by ectopic expression of Cut in Drosophila. Genes Dev. 5:1124-1135. [DOI] [PubMed] [Google Scholar]
- 10.Bodmer, R., S. Barbel, S. Shepherd, J. W. Jack, L. Y. Jan, and Y. N. Jan. 1987. Transformation of sensory organs by mutations of the cut locus of D. melanogaster. Cell 51:293-307. [DOI] [PubMed] [Google Scholar]
- 11.Brooks, S. C., E. R. Locke, and H. D. Soule. 1973. Estrogen receptor in a human cell line (MCF-7) from breast carcinoma. J. Biol. Chem. 248:6251-6253. [PubMed] [Google Scholar]
- 12.Brown, C. E., T. Lechner, L. Howe, and J. L. Workman. 2000. The many HATs of transcription coactivators. Trends Biochem. Sci. 25:15-19. [DOI] [PubMed] [Google Scholar]
- 13.Cai, H. N., and M. Levine. 1997. The gypsy insulator can function as a promoter-specific silencer in the Drosophila embryo. EMBO J. 16:1732-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carey, J. 1991. Gel retardation. Methods Enzymol. 208:103-117. [DOI] [PubMed] [Google Scholar]
- 15.Carey, J. 1988. Gel retardation at low pH resolves trp repressor-DNA complexes for quantitative study. Proc. Natl. Acad. Sci. USA 85:975-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chattopadhyay, S., C. E. Whitehurst, and J. Chen. 1998. A nuclear matrix attachment region upstream of the T cell receptor beta gene enhancer binds Cux/CDP and SATB1 and modulates enhancer-dependent reporter gene expression but not endogenous gene expression. J. Biol. Chem. 273:29838-29846. [DOI] [PubMed] [Google Scholar]
- 17.Coqueret, O., G. Berube, and A. Nepveu. 1998. The mammalian Cut homeodomain protein functions as a cell-cycle dependent transcriptional repressor which downmodulates p21WAF1/CIP1/SDI1 in S phase. EMBO J. 17:4680-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crook, T., D. Wrede, and K. H. Vousden. 1991. p53 point mutation in HPV negative human cervical carcinoma cell lines. Oncogene 6:873-875. [PubMed] [Google Scholar]
- 19.Dorsett, D. 1993. Distance-independent inactivation of an enhancer by the suppressor of Hairy-wing DNA-binding protein of Drosophila. Genetics 134:1135-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dufort, D., and A. Nepveu. 1994. The human Cut homeodomain protein represses transcription from the c-myc promoter. Mol. Cell. Biol. 14:4251-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellis, T., L. Gambardella, M. Horcher, S. Tschanz, J. Capol, P. Bertram, W. Jochum, Y. Barrandon, and M. Busslinger. 2001. The transcriptional repressor CDP (Cutl1) is essential for epithelial cell differentiation of the lung and the hair follicle. Genes Dev. 15:2307-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fry, C. J., and P. J. Farnham. 1999. Context-dependent transcriptional regulation. J. Biol. Chem. 274:29583-29586. [DOI] [PubMed] [Google Scholar]
- 23.Goulet, B., P. Watson, M. Poirier, L. Leduy, G. Berube, S. Meterissian, P. Jolicoeur, and A. Nepveu. 2002. Characterization of a tissue-specific CDP/Cux isoform, p75, activated in breast tumor cells. Cancer Res. 62:6625-6633. [PubMed] [Google Scholar]
- 24.Hackett, A. J., H. S. Smith, E. L. Springer, R. B. Owens, W. A. Nelson-Rees, J. L. Riggs, and M. B. Gardner. 1977. Two syngeneic cell lines from human breast tissue: the aneuploid mammary epithelial (Hs578T) and the diploid myoepithelial (Hs578Bst) cell lines. J. Natl. Cancer Inst. 58:1795-1806. [DOI] [PubMed] [Google Scholar]
- 25.Harada, R., G. Bérubé, O. J. Tamplin, C. Denis-Larose, and A. Nepveu. 1995. DNA-binding specificity of the Cut repeats from the human Cut-like protein. Mol. Cell. Biol. 15:129-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harada, R., D. Dufort, C. Denis-Larose, and A. Nepveu. 1994. Conserved cut repeats in the human cut homeodomain protein function as DNA binding domains. J. Biol. Chem. 269:2062-2067. [PubMed] [Google Scholar]
- 27.Hertweck, H. 1931. Anatomie und variabilitaet des nerven-systems und der sinneorgane von Drosophila melanogaster. J. Exp. Zool. 139:559-663. [Google Scholar]
- 28.Higgy, N. A., H. A. Tarnasky, I. Valarche, A. Nepveu, and F. A. Vanderhoorn. 1997. Cux/CDP homeodomain protein binds to an enhancer in the rat c-Mos locus and represses its activity. Biochim. Biophys. Acta 1351:313-324. [DOI] [PubMed] [Google Scholar]
- 29.Holthuis, J., T. A. Owen, A. J. van Wijnen, K. L. Wright, A. Ramsey-Ewing, M. B. Kennedy, R. Carter, S. C. Cosenza, K. J. Soprano, J. B. Lian, J. L. Stein, and G. S. Stein. 1990. Tumor cells exhibit deregulation of the cell cycle histone gene promoter factor HiNF-D. Science 247:1454-1457. [DOI] [PubMed] [Google Scholar]
- 30.Howcroft, T. K., S. L. Kirshner, and D. S. Singer. 1997. Measure of transient transfection efficiency using beta-galactosidase protein. Anal. Biochem. 244:22-27. [DOI] [PubMed] [Google Scholar]
- 31.Jack, J., and Y. DeLotto. 1995. Structure and regulation of a complex locus: the cut gene of Drosophila. Genetics 139:1689-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jack, J., D. Dorsett, Y. Delotto, and S. Liu. 1991. Expression of the cut locus in the Drosophila wing margin is required for cell type specification and is regulated by a distant enhancer. Development 113:735-747. [DOI] [PubMed] [Google Scholar]
- 33.Jackson, R. J., S. J. Antonia, K. L. Wright, N. S. Moon, A. Nepveu, and T. Munoz-Antonia. 1999. Human cut-like repressor protein binds TGFbeta type II receptor gene promoter. Arch. Biochem. Biophys. 371:290-300. [DOI] [PubMed] [Google Scholar]
- 34.Kim, E. C., J. S. Lau, S. Rawlings, and A. S. Lee. 1997. Positive and negative regulation of the human thymidine kinase promoter mediated by CCAAT binding transcription factors NF-Y/CBF, DBPA, and CDP/Cut. Cell Growth Differ. 8:1329-1338. [PubMed] [Google Scholar]
- 35.Last, T. J., M. Birnbaum, A. J. van Wijnen, G. S. Stein, and J. L. Stein. 1998. Repressor elements in the coding region of the human histone H4 gene interact with the transcription factor CDP/cut. Gene 221:267-277. [DOI] [PubMed] [Google Scholar]
- 36.Last, T. J., A. J. van Wijnen, M. C. de Ridder, G. S. Stein, and J. L. Stein. 1999. The homeodomain transcription factor CDP/Cut interacts with the cell cycle regulatory element of histone H4 genes packaged into nucleosomes. Mol. Biol. Rep. 26:185-194. [DOI] [PubMed] [Google Scholar]
- 37.Lawson, N. D., A. Khannagupta, and N. Berliner. 1998. Isolation and characterization of the cDNA for mouse neutrophil collagenase: demonstration of shared negative regulatory pathways for neutrophil secondary granule protein gene expression. Blood 91:2517-2524. [PubMed] [Google Scholar]
- 38.Ledford, A. W., J. G. Brantley, G. Kemeny, T. L. Foreman, S. E. Quaggin, P. Igarashi, S. M. Oberhaus, M. Rodova, J. P. Calvet, and G. B. Vanden Heuvel. 2002. Deregulated expression of the homeobox gene Cux-1 in transgenic mice results in downregulation of p27(kip1) expression during nephrogenesis, glomerular abnormalities, and multiorgan hyperplasia. Dev. Biol. 245:157-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, S., L. Moy, N. Pittman, G. Shue, B. Aufiero, E. J. Neufeld, N. S. LeLeiko, and M. J. Walsh. 1999. Transcriptional repression of the cystic fibrosis transmembrane conductance regulator gene, mediated by CCAAT displacement protein/Cut homolog, is associated with histone deacetylation. J. Biol. Chem. 274:7803-7815. [DOI] [PubMed] [Google Scholar]
- 40.Liu, J., A. Barnett, E. J. Neufeld, and J. P. Dudley. 1999. Homeoproteins CDP and SATB1 interact: potential for tissue-specific regulation. Mol. Cell. Biol. 19:4918-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, S., and J. Jack. 1992. Regulatory interactions and role in cell type specification of the Malpighian tubules by the cut, kruppel, and caudal genes of Drosophila. Dev. Biol. 150:133-143. [DOI] [PubMed] [Google Scholar]
- 42.Liu, S., E. McLeod, and J. Jack. 1991. Four distinct regulatory regions of the cut locus and their effect on cell type specification in Drosophila. Genetics 127:151-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo, W., and D. G. Skalnik. 1996. CCAAT displacement protein competes with multiple transcriptional activators for binding to four sites in the proximal gp91(phox) promoter. J. Biol. Chem. 271:18203-18210. [DOI] [PubMed] [Google Scholar]
- 44.Luong, M. X., C. M. van der Meijden, D. Xing, R. Hesselton, E. S. Monuki, S. N. Jones, J. B. Lian, J. L. Stein, G. S. Stein, E. J. Neufeld, and A. J. van Wijnen. 2002. Genetic ablation of the CDP/Cux protein C terminus results in hair cycle defects and reduced male fertility. Mol. Cell. Biol. 22:1424-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mailly, F., G. Berube, R. Harada, P. L. Mao, S. Phillips, and A. Nepveu. 1996. The human Cut homeodomain protein can repress gene expression by two distinct mechanisms: active repression and competition for binding site occupancy. Mol. Cell. Biol. 16:5346-5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Modolell, J., W. Bender, and M. Meselson. 1983. Drosophila melanogaster mutations suppressible by the suppressor of Hairy-wing are insertions of a 7.3-kilobase mobile element. Proc. Natl. Acad. Sci. USA 80:1678-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moon, N. S., G. Berube, and A. Nepveu. 2000. CCAAT displacement activity involves Cut repeats 1 and 2, not the Cut homeodomain. J. Biol. Chem. 275:31325-31334. [DOI] [PubMed] [Google Scholar]
- 48.Moon, N. S., P. Premdas, M. Truscott, L. Leduy, G. Berube, and A. Nepveu. 2001. S phase-specific proteolytic cleavage is required to activate stable DNA binding by the CDP/Cut homeodomain protein. Mol. Cell. Biol. 21:6332-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nepveu, A. 2001. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene 270:1-15. [DOI] [PubMed] [Google Scholar]
- 50.Neufeld, E. J., D. G. Skalnik, P. M. Lievens, and S. H. Orkin. 1992. Human CCAAT displacement protein is homologous to the Drosophila homeoprotein, Cut. Nat. Genet. 1:50-55. [DOI] [PubMed] [Google Scholar]
- 51.Nirodi, C., J. Hart, P. Dhawan, N. S. Moon, A. Nepveu, and A. Richmond. 2001. The role of CDP in the negative regulation of CXCL1 gene expression. J. Biol. Chem. 276:26122-26131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nissen, R. M., and K. R. Yamamoto. 2000. The glucocorticoid receptor inhibits NFkappaB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 14:2314-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O'Connor, M. J., W. Stunkel, C. H. Koh, H. Zimmermann, and H. U. Bernard. 2000. The differentiation-specific factor CDP/Cut represses transcription and replication of human papillomaviruses through a conserved silencing element. J. Virol 74:401-410. [PMC free article] [PubMed] [Google Scholar]
- 54.Ory, D. S., B. A. Neugeboren, and R. C. Mulligan. 1996. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. USA 93:11400-11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pattison, S., D. G. Skalnik, and A. Roman. 1997. CCAAT displacement protein, a regulator of differentiation-specific gene expression, binds a negative regulatory element within the 5′ end of the human papillomavirus type 6 long control region. J. Virol. 71:2013-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pearson, B. E., H. P. Nasheuer, and T. S. Wang. 1991. Human DNA polymerase alpha gene: sequences controlling expression in cycling and serum-stimulated cells. Mol. Cell. Biol. 11:2081-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quaggin, S. E., G. B. Vandenheuvel, K. Golden, R. Bodmer, and P. Igarashi. 1996. Primary structure, neural-specific expression, and chromosomal localization of Cux-2, a second murine homeobox gene related to Drosophila Cut. J. Biol. Chem. 271:22624-22634. [DOI] [PubMed] [Google Scholar]
- 58.Santaguida, M., Q. Ding, G. Berube, M. Truscott, P. Whyte, and A. Nepveu. 2001. Phosphorylation of the CCAAT displacement protein (CDP)/Cux transcription factor by cyclin A-Cdk1 modulates its DNA binding activity in G2. J. Biol. Chem. 276:45780-45790. [DOI] [PubMed] [Google Scholar]
- 59.Sinclair, A. M., J. A. Lee, A. Goldstein, D. Xing, S. Liu, R. Ju, P. W. Tucker, E. J. Neufeld, and R. H. Scheuermann. 2001. Lymphoid apoptosis and myeloid hyperplasia in CCAAT displacement protein mutant mice. Blood 98:3658-3667. [DOI] [PubMed] [Google Scholar]
- 60.Skalnik, D. G., E. C. Strauss, and S. H. Orkin. 1991. CCAAT displacement protein as a repressor of the myelomonocytic-specific gp91-phox gene promoter. J. Biol. Chem. 266:16736-16744. [PubMed] [Google Scholar]
- 61.Stargell, L. A., and K. Struhl. 1996. Mechanisms of transcriptional activation in vivo: two steps forward. Trends Genet. 12:311-315. [DOI] [PubMed] [Google Scholar]
- 62.Stein, G. H. 1979. T98G: an anchorage-independent human tumor cell line that exhibits stationary phase G1 arrest in vitro. J. Cell. Physiol. 99:43-54. [DOI] [PubMed] [Google Scholar]
- 63.Stunkel, W., Z. Huang, S. H. Tan, M. J. O'Connor, and H. U. Bernard. 2000. Nuclear matrix attachment regions of human papillomavirus type 16 repress or activate the E6 promoter, depending on the physical state of the viral DNA. J. Virol. 74:2489-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Teerawatanasuk, N., D. G. Skalnik, and L. G. Carr. 1999. CCAAT displacement protein (CDP/Cut) binds a negative regulatory element in the human tryptophan hydroxylase gene. J. Neurochem. 72:29-39. [DOI] [PubMed] [Google Scholar]
- 65.Tufarelli, C., Y. Fujiwara, D. C. Zappulla, and E. J. Neufeld. 1998. Hair defects and pup loss in mice with targeted deletion of the first Cut repeat domain of the Cux/CDP homeoprotein gene. Dev. Biol. 200:69-81. [DOI] [PubMed] [Google Scholar]
- 66.Valarche, I., J. P. Tissier-Seta, M. R. Hirsch, S. Martinez, C. Goridis, and J. F. Brunet. 1993. The mouse homeodomain protein Phox2 regulates Ncam promoter activity in concert with Cux/CDP and is a putative determinant of neurotransmitter phenotype. Development 119:881-896. [DOI] [PubMed] [Google Scholar]
- 67.van den Ent, F. M., A. J. van Wijnen, J. B. Lian, J. L. Stein, and G. S. Stein. 1994. Cell cycle controlled histone H1, H3, and H4 genes share unusual arrangements of recognition motifs for HiNF-D supporting a coordinate promoter binding mechanism. J. Cell. Physiol. 159:515-530. [DOI] [PubMed] [Google Scholar]
- 68.Vandenheuvel, G. B., S. E. Quaggin, and P. Igarashi. 1996. A unique variant of a homeobox gene related to Drosophila Cut is expressed in mouse testis. Biol. Reprod. 55:731-739. [DOI] [PubMed] [Google Scholar]
- 69.van Gurp, M. F., J. Pratap, M. Luong, A. Javed, H. Hoffmann, A. Giordano, J. L. Stein, E. J. Neufeld, J. B. Lian, G. S. Stein, and A. J. van Wijnen. 1999. The CCAAT displacement protein/cut homeodomain protein represses osteocalcin gene transcription and forms complexes with the retinoblastoma protein-related protein p107 and cyclin A. Cancer Res. 59:5980-5988. [PubMed] [Google Scholar]
- 70.van Wijnen, A. J., T. K. Choi, T. A. Owen, K. L. Wright, J. B. Lian, R. Jaenisch, J. L. Stein, and G. S. Stein. 1991. Involvement of the cell cycle-regulated nuclear factor HiNF-D in cell growth control of a human H4 histone gene during hepatic development in transgenic mice. Proc. Natl. Acad. Sci. USA 88:2573-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Wijnen, A. J., C. Cooper, P. Odgren, F. Aziz, A. De Luca, R. A. Shakoori, A. Giordano, P. J. Quesenberry, J. B. Lian, G. S. Stein, and J. L. Stein. 1997. Cell cycle-dependent modifications in activities of pRb-related tumor suppressors and proliferation-specific CDP/Cut homeodomain factors in murine hematopoietic progenitor cells. J. Cell. Biochem. 66:512-523. [DOI] [PubMed] [Google Scholar]
- 72.van Wijnen, A. J., M. F. van Gurp, M. C. de Ridder, C. Tufarelli, T. J. Last, M. Birnbaum, P. S. Vaughan, A. Giordano, W. Krek, E. J. Neufeld, J. L. Stein, and G. S. Stein. 1996. CDP/Cut is the DNA-binding subunit of histone gene transcription factor HiNF-D: a mechanism for gene regulation at the G1/S phase cell cycle transition point independent of transcription factor E2F. Proc. Natl. Acad. Sci. USA 93:11516-11521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van Wijnen, A. J., K. L. Wright, J. B. Lian, J. L. Stein, and G. S. Stein. 1989. Human H4 histone gene transcription requires the proliferation-specific nuclear factor HiNF-D. Auxiliary roles for HiNF-C (Sp1-like) and HiNF-A (high mobility group-like). J. Biol. Chem. 264:15034-15042. [PubMed] [Google Scholar]
- 74.Verrijzer, C. P., and R. Tjian. 1996. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem. Sci. 21:338-342. [PubMed] [Google Scholar]
- 75.Wang, Z., A. Goldstein, R. -T. Zong, D. Lin, E. J. Neufeld, R. H. Scheuermann, and P. W. Tucker. 1999. Cux/CDP homeoprotein is a component of NF-μNR and represses the immunoglobulin heavy chain intronic enhancer by antagonizing the Bright transcription activator. Mol. Cell. Biol. 19:284-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Westley, B., and H. Rochefort. 1980. A secreted glycoprotein induced by estrogen in human breast cancer cell lines. Cell 20:353-362. [DOI] [PubMed] [Google Scholar]
- 77.Wright, K. L., R. T. Dell'Orco, A. J. van Wijnen, J. L. Stein, and G. S. Stein. 1992. Multiple mechanisms regulate the proliferation-specific histone gene transcription factor HiNF-D in normal human diploid fibroblasts. Biochemistry 31:2812-2818. [DOI] [PubMed] [Google Scholar]
- 78.Wu, F., and A. S. Lee. 2002. CDP and AP-2 mediated repression mechanism of the replication-dependent hamster histone H3.2 promoter. J. Cell. Biochem. 84:699-707. [DOI] [PubMed] [Google Scholar]
- 79.Yoon, S. O., and D. M. Chikaraishi. 1994. Isolation of two E-box binding factors that interact with the rat tyrosine hydroxylase enhancer. J. Biol. Chem. 269:18453-18462. [PubMed] [Google Scholar]