Abstract
DNA methylation is essential for epigenetic gene regulation during development. The cyclic AMP (cAMP)-responsive element (CRE) is found in the promoter of many cAMP-regulated genes and plays important roles in their gene expression. Methylation occurs on the CRE site and results in transcriptional repression via a direct mechanism, that is, prevention by the methyl group of binding of the cAMP-responsive factor CREB to this site. A recent study indicated that the nucleosome is also important in repressing transcription. In this study, we investigated the regulation of transcriptional repression on methylated CRE. We focused on methyl-CpG binding domain protein 2 (MBD2). MBD2 consists of two forms, MBD2a and MBD2b, the latter lacking the N-terminal extension of MBD2a. Unexpectedly, we found that MBD2a, but not MBD2b, promoted activation of the unmethylated cAMP-responsive genes. An in vivo binding assay revealed that MBD2a selectively interacted with RNA helicase A (RHA), a component of CREB transcriptional coactivator complexes. MBD2a and RHA cooperatively enhanced CREB-dependent gene expression. Interestingly, coimmunoprecipitation assays demonstrated that MBD2a binding to RHA was not associated with histone deacetylase 1. Our results indicate a novel role for MBD2a in gene regulation.
DNA methylation, a major modification of DNA, is epigenetically implicated in a variety of biological responses such as development, tumorigenesis and neurogenesis. Mice lacking a functional DNA methyltransferase, which is required for maintenance of methylation (31) or de novo methylation (46), fail to complete development. The effect of DNA methylation was first proposed to be transcriptional repression (7). It has been shown that CpG islands become methylated in situations such as genomic imprinting (5) or X chromosome inactivation (19). An important relationship between methylation of tumor suppressor genes and cancer progression has also been reported (23). In addition, DNA methylation has been shown to cause repression of repetitive DNA element promoters during murine embryogenesis (57). Recently, DNA methylation has been demonstrated to be important for appropriate regulation of stage-specific genes in the development of Xenopus (53) and for silencing of tissue-specific genes and repetitive DNA elements in murine fibroblast cultures (28).
DNA methylation-mediated transcriptional silencing is achieved by various mechanisms. One mechanism is direct interference with the DNA binding of transcriptional factors (8). Another mechanism involves deacetylation of histones, which leads to transcriptional repression (8). The latter occurs mostly through an indirect mechanism in which a methyl-CpG binding protein specifically binds to methylated DNA to induce transcriptional repression (8, 9, 22).
The first methyl-CpG binding activity to be identified was the methyl-CpG binding protein 1 (MeCP1) (34). MeCP1 is a large protein complex of 400 to 800 kDa and contains methyl-CpG binding domain protein 2 (MBD2) and histone deacetylase 1/2 (HDAC1/2) (41). A recent study demonstrated that the MeCP1 complex contains MBD2 and all of the known NuRD components, including Mi2, MTA2, MBD3, and the histone deacetylase core, HDAC1/2 and RbAp46/48 (15, 61, 62). MBD2 consists of two forms, MBD2a and MBD2b, which are generated from a single gene (20, 21). The MBD2b protein lacks the 152-amino acid (aa) N-terminal extension of MBD2a.
DNA methylation plays a role in the context of genes containing the cyclic AMP (cAMP)-responsive element (CRE). CRE is found in the promoter of many cAMP-regulated genes and plays a critical role in the regulation of gene expression (33). It is also known that both methylation and demethylation occur on CRE during cancer progression and differentiation (11, 13, 32, 45, 48). During differentiation, demethylation is induced on the CRE site, resulting in transcriptional activation (11, 48). Under conditions of transcriptional activation, cAMP stimulates cellular gene expression via protein kinase A (PKA)-mediated phosphorylation of the cAMP-responsive factor CREB at Ser133 (18). CREB possesses a bipartite transactivation domain, consisting of both a constitutive and an inducible activator (10, 49). The former is achieved via constitutive interaction with the TBP-associated factor, hTAFII130 (16, 36). The latter modulates CREB activity in a phospho (Ser133)-dependent manner. Ser133 phosphorylation promotes the recruitment of the coactivator paralogs CREB-binding protein (CBP) and p300 via a kinase-inducible domain in CREB (2, 12, 36, 47). We previously showed that RNA helicase A (RHA) interacts with CBP and mediates the association between CBP and Pol II complexes as a bridging factor (37). RHA is also required for enhancement of cAMP-mediated transcriptional activation via phospho (Ser133)-CREB (37). Furthermore, RHA independently regulates CREB-dependent transcription both through recruitment of Pol II and by ATP-dependent mechanisms (1).
In contrast, during cancer progression, methylation is induced on the CRE site at the center CpG dinucleotide, resulting in transcriptional repression of the tumor suppressor genes (13, 32, 45). Methylation of CRE prevents CREB from binding to this site via inhibition by the methyl group. In addition, recent studies indicated that inhibition of the methyl group is not sufficient to repress transcription on methylated CRE and that the nucleosome structure is also important (58), prompting us to investigate the regulation of transcriptional repression on methylated CRE. In the present study, we focused on MBD2 as a candidate and investigated whether MBD2 repressed transcription from methylated CRE. Unexpectedly we found that MBD2a enhanced unmethylated CRE-dependent gene expression and interacted with RHA. Furthermore, our results suggested that MBD2a acted not only as a repressor but also as a cofactor for activation of CRE-dependent gene expression, based on our analysis of functional interaction of MBD2a and RHA.
MATERIALS AND METHODS
Cell culture.
Human embryonic kidney 293 (HEK-293) cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum plus penicillin and streptomycin at 37°C in 5% CO2. The stable NIH 3T3 cell line D5, containing rat somatostatin gene sequences from 750 bp upstream of the promoter to 3 kb downstream of the coding region was maintained in Dulbecco's minimal essential medium with 10% fetal bovine serum plus 200 μg of G418/ml (4).
Plasmids.
The coding sequences for full-length MBD2a genes were PCR amplified (primers, 5′-ACAGAATTCATGCGCGCGCACCCGGGG-3′ and 5′-TGTGTCGACTTACGTGGCCCGGGGTCCCCT-3′) from reverse-transcribed HeLa cell RNA. PCR products were digested with EcoRI and SalI and ligated into pGBT9 (Clontech Laboratories, Palo Alto, Calif.) for the Saccharomyces cerevisiae two-hybrid assays. The MBD2b fragment was amplified by using the primers 5′-ACAGAATTCATGGATTGCCCGGCCCTCCC-3′ and 5′-TGTGTCGACTTACGTGGCCCGGGGTCCCCT-3 from the full-length MBD2a template. For immunoprecipitation assays, the fragments of MBD2a and MBD2b were inserted into pcDNA3 HA or pcDNA3 FLAG, which were constructed by inserting the hemagglutinin antigen (HA) sequence or FLAG sequence into pcDNA3 (Invitrogen, San Diego, Calif.), respectively (35). Fragments of N-terminal deletion mutants, termed MBD2a-N1, MBD2-N2, MBD2a-N3, MBD2a-N4, MBD2a-N5, MBD2a-N6, and GR11, were obtained by PCR amplification. Full-length MBD2a, MBD2b, and each of these deletion mutants were inserted into pGEX-5X-1 (Amersham Pharmacia Biotech, Piscataway, N.J.) for the glutathione S-transferase (GST) pull-down assay. Fragments of additional deletion mutants, termed MBD2a-A, MBD2a-B, and MBD2a-C, were generated by PCR-based methods. Each of these fragments, either alone or fused to the GAL4 DNA-binding domain (GAL4-DBD), was introduced into EcoRI-SalI-digested pGBT9 or pcDNA3 for transactivation assays in yeast or mammalian cells, respectively. For transient transfection assays, these fragments were inserted into pcDNA3 HA. Sequences of all plasmids generated by PCR were confirmed by sequence analysis. The RHA plasmids used in this study have been described previously (1, 17, 37). pGAD CREB, pGAD hTAFII130 (52, 54), and pGAD CBP (60) were obtained by a PCR-based method previously described. PKA, pGAL4-CREB, pGAL4-CREBM1 (37), and pGAL4 (35) expression vectors, Som-Luc (59) and pG5b-Luc (60) reporter plasmids, and control plasmid RSV-β-gal (39) have been previously described.
Antibodies.
Rabbit polyclonal antibody against HDAC1 was obtained from Upstream Biotechnology. Rabbit polyclonal antibodies against the largest subunit of RNA polymerase II (N-20) were obtained from Santa Cruz Biotechnology (Santa Cruz, Calif.). Mouse monoclonal antibody against the FLAG-tag (M2) was purchased from Sigma Chemical Co (St. Louis, Mo.). Mouse and rat monoclonal antibodies against the HA-tag (12CA5 and 3F10) were obtained from Boehringer Mannheim (Mannheim, Germany). Anti-RHA1 antiserum was described previously (37).
Liquid β-Gal assay.
β-Galactosidase (β-Gal) assays were performed by using standard methods. The yeast strain Y190 was simultaneously transformed with GAL4-DBD MBD2a and/or MBD2b, and GAL4-AD CREB, CBP, RHA1, RHA2, RHA3, RHA4, or hTAFII130 by using a lithium acetate method. Transformants were selected on agar plates lacking leucine and tryptophan for 3 days. All experiments were performed in triplicate, and all results were obtained from at least three separate experiments (60).
Transactivation assay.
The yeast strain Y190 was transformed with MBD2 deletion mutants by using a lithium acetate method, and transformants were selected on tryptophan-free agar plates for 3 days. For the liquid β-Gal assay, 1 ml of growth medium was inoculated with selected colonies. The assay was performed in triplicate to quantify the transcriptional activity of MBD2 deletion mutant as described previously (60). HEK-293 cells were transiently transfected with 100 ng of each MBD2 deletion mutant fused to GAL4-DBD, or the empty vector alone, and 100 ng of pG5b-Luc. Following 24 h of incubation, cells were lysed with a cell lysis buffer (Toyo Ink) and luciferase activities were measured. The measured activity was normalized for β-Gal activity from cotransfected RSV-β-gal as a control. Each experiment was performed in triplicate (14, 40).
Transient transfection assay.
Transient transfection assays were performed in HEK-293 cells (37). Cells were lysed with a cell lysis buffer (Toyo Ink) 24 h after transfection, and luciferase activities were measured. Reporter activity was induced by cotransfection with the PKA expression vector. The recorded activity was normalized to the protein level in the cell extract, quantified by using the Bradfold assay (Bio-Rad, Richmond, Calif.) and β-Gal activity from RSV-β-gal. All experiments were performed in triplicate. HEK-293 cells were transfected with 100 ng of Som-Luc or pG5b-Luc reporter plasmid, 50 ng of wild-type or catalytically inactive PKA expression vector (PKAwt or PKAmut, respectively), 50 ng of RSV-β-gal control plasmid, and 0, 50, or 100 ng of RHA and/or MBD2a, MBD2b, or truncated forms of MBD2 expression vector. For assays with the pG5b-Luc reporter plasmid, cells were cotransfected with 100 ng of GAL4-CREB or GAL4-CREBM1 expression vector. To ensure an equal amount of DNA, empty plasmids were added in each transfection (1, 37).
GST pull-down assay.
GST fusion proteins of each RHA mutant or MBD2 mutant were expressed in Escherichia coli strain TopXF' (Invitrogen) and purified by using glutathione Sepharose beads (Amersham Pharmacia Biotech). 35S-labeled RHA or MBD2a was incubated with 2 μg of each GST fusion protein bound to resin in 1 ml of buffer A (20 mM HEPES [pH 7.9], 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol [DTT], 0.05% Tween 20, 5% glycerol, 1 mM Na3VO4, 5 mM NaF, 1 μg of aprotinin/ml, and 1 μg of leupeptin/ml) for 8 h at 4°C. After being washed with buffer A, bound proteins were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and exposed to X-ray film (1).
Immunoprecipitation assays.
HEK-293 cells were transfected or cotransfected with 5 μg of HA-tagged MBD2a or MBD2b and HA-tagged RHA and/or FLAG-tagged MBD2a or MBD2b. Forty-eight hours after transfection, cells were lysed and nuclear extracts were prepared and resuspended in 1 ml of binding buffer (20 mM HEPES [pH 7.9], 100 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.1% NP-40, 5% glycerol and protease inhibitors). Cell lysates were precleaned with normal mouse immunoglobulin G (IgG) (Santa Cruz Biotechnology) for 1 h at 4°C. After a brief centrifugation, the lysates were mixed with 1 μg of anti-HA antibody or anti-FLAG antibody conjugated to protein G-Sepharose beads. After 8 h of incubation at 4°C, the beads were washed three times with binding buffer. Bound proteins were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by Western blotting (1, 51).
In vitro-in vivo binding assay.
HEK-293 cells were transfected with plasmids expressing FLAG-MBD2a. Nuclear extract was prepared 48 h after transfection in buffer A, and aliquots of equal amounts of nuclear extract were incubated with increasing amounts of GST or GST-RHA1 for 4 h at 4°C. Bound proteins were analyzed by use of immunoblots using anti-FLAG antibody or anti-HDAC1 antibodies. The supernatants were transferred to new tubes and incubated with 1 μg of anti-FLAG antibody conjugated to protein G-Sepharose beads for 4 h at 4°C. At the end of the incubation, bound proteins were washed four times with buffer A and precipitated proteins were then analyzed by immunoblotting with anti-FLAG or anti-HDAC1 antibodies.
Chromatin immunoprecipitation assay.
D5 cells were transfected with HA-RHA, HA-MBD2a, or HA-MBD2b. After 24 h of transfection, cells were treated with formaldehyde to cross-link protein-DNA complexes (4). Immunoprecipitates of cross-linked complexes were prepared with anti-HA antibody and then treated with proteinase K for 2 h and subsequently incubated at 65°C to release cross-links. DNA was purified by phenol-chloroform extraction and ethanol precipitation. DNA samples were then analyzed with 25 cycles of PCR to amplify somatostatin promoter sequences (−100 to +55). Different cycle numbers were employed to ensure linearity of amplification. PCR products were analyzed by 5% polyacrylamide gel with ethidium bromide.
RT-PCR assay.
D5 cells were transfected with HA-RHA, HA-MBD2a, or HA-MBD2b. After 24 h of transfection, total RNA was isolated. Total RNA was subjected to cDNA synthesis by using the random primer and the Superscript II RNase H− reverse transcriptase (Gibco BRL, Grand Island, N.Y.) following the instructions provided by the manufacturer. Two sets of primers were used for PCR: somatostatin (5′-GGACCTGCGTCTAGACTGA-3′ and 5′-GAGGTCTGGCTGAGACAAC-3′) and β-actin (5′-GTTTGAGACCTTCAACACCC-3′ and 5′-ATGTCACGCACGATTTCCC-3′). The optimal number of cycles and amount of RT products used for the PCR were determined in preliminary experiments (data not shown). The samples were then subjected to analysis on an ethidium bromide-stained polyacrylamide gel.
RESULTS
MBD2a, but not MBD2b, interacts with RHA.
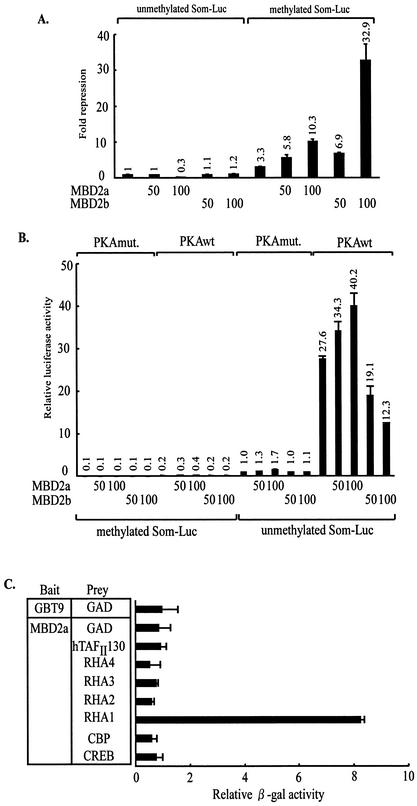
To investigate whether MBD2 could repress methylated CRE-dependent transcription, the effects of MBD2 were examined by using reporter assays. We utilized somatostatin-Luc (Som-Luc) that contains endogenous somatostatin promoter including CRE. The Som-Luc reporter was methylated with or without SssI methylase in vitro and cotransfected with MBD2 expression plasmids into HEK-293 cells. In the case of methylated Som-Luc, MBD2a and MBD2b consistently repressed Som-Luc activity in a dose-dependent manner, whereas the effects of MBD2a and MBD2b on unmethylated CRE was not detected (Fig. 1A).
FIG. 1.
Enhancement of unmethylated CRE-dependent gene expression by MBD2a. (A) Repression of methylated Som-Luc activity by MBD2a and MBD2b. HEK-293 cells were transfected with Som-Luc reporter either after methylation with SssI methylase or after mock methylation. The change in repression on both reporter constructs was calculated relative to the luciferase activity after transfection of the empty vector and unmethylated reporter. (B) Enhancement of unmethylated Som-Luc activity by MBD2a. The luciferase activity of cells transfected with empty vector alone and PKAmut was set as 1. (C) Selective interaction of MBD2a with RHA in yeast. Yeast two-hybrid experiments were performed with MBD2a as bait (GAL4 fusion) and CREB, CBP, RHA1 to RHA4, and hTAFII130 as prey (fusion to the activation domain).
Then, we examined the effects of MBD2a and MBD2b on PKA-mediated gene expression. MBD2a and MBD2b had no effect on methylated Som-Luc in PKA-induced cells. Unexpectedly, MBD2a promoted Som-Luc activity in response to PKA induction when Som-Luc was not methylated (Fig. 1B).
We were interested in the effects of MBD2a on unmethylated Som-Luc activity in PKA-induced cells. There is no consensus sequence by PKA in MBD2a, and MBD2a was not phosphorylated by PKA in vitro (data not shown). We hypothesized that MBD2a might interact with the components of the CREB transcriptional apparatus, including CREB, hTAFII130, CBP, and RHA (33). To test this hypothesis, we performed the yeast two-hybrid assays and liquid β-Gal assays designed so that the interaction of components of the CREB complex with MBD2a would result in β-Gal activity. We previously showed that full-length RHA was not expressed in yeast and four previously described fragments of RHA, termed RHA1, RHA2, RHA3, and RHA4, were used (1). As shown in Fig. 1C, MBD2a selectively bound to RHA1 in yeast.
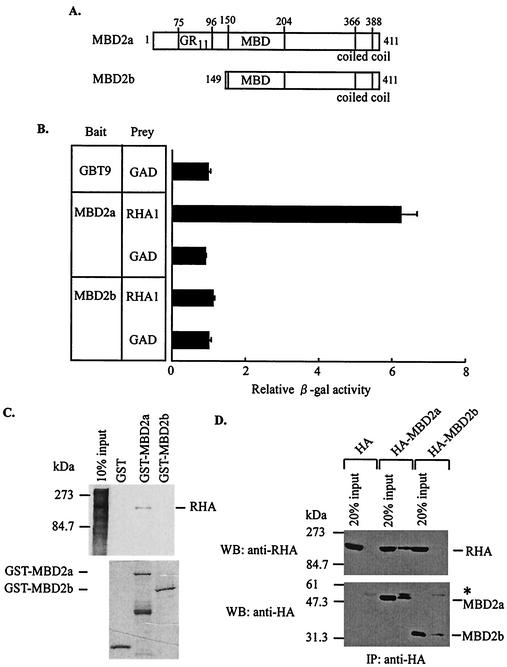
MBD2 consists of two forms, MBD2a and MBD2b (Fig. 2A). We tested whether MBD2b interacted with RHA in yeast. As shown in Fig. 2B, MBD2b did not bind to RHA. To confirm the interaction between RHA and MBD2a, we used in vitro binding assays. MBD2a and MBD2b were expressed as GST fusion proteins and tested for binding to in vitro-translated RHA. MBD2a bound to RHA in vitro, but MBD2b did not (Fig. 2C). Furthermore, we tested whether this interaction might occur through nucleic acids. The binding assay with adequate amounts of nuclease revealed that this interaction occurred directly (data not shown). To examine the MBD2a-RHA interaction in vivo, we performed coimmunoprecipitation experiments. HEK-293 cells were transiently transfected with HA-tagged MBD2a (HA-MBD2a) or MBD2b (HA-MBD2b). As shown in Fig. 2D, endogenous RHA was coimmunoprecipitated only in the presence of HA-MBD2a. These results of the binding experiments indicated that MBD2a selectively interacted with RHA.
FIG. 2.
Interaction of MBD2a, but not MBD2b, with RHA. (A) Schematic representation of MBD2a and MBD2b. GR11, glycine-arginine repeats; MBD, methyl-CpG binding domain. (B) No association of MBD2b with RHA in yeast. Yeast two-hybrid experiments were performed with MBD2a and MBD2b as bait and RHA1 as prey. (C) Binding of GST-MBD2a to RHA in vitro. The GST fusion proteins were expressed in E. coli, purified, and incubated with in vitro-translated, 35S-labeled RHA. (D) Interaction of MBD2a with RHA in vivo. A Western blot of HA shows the immunoprecipitation of anti-HA antibody from HEK-293-transfected nuclear extracts. The asterisk indicates immunoprecipitated IgG heavy chain.
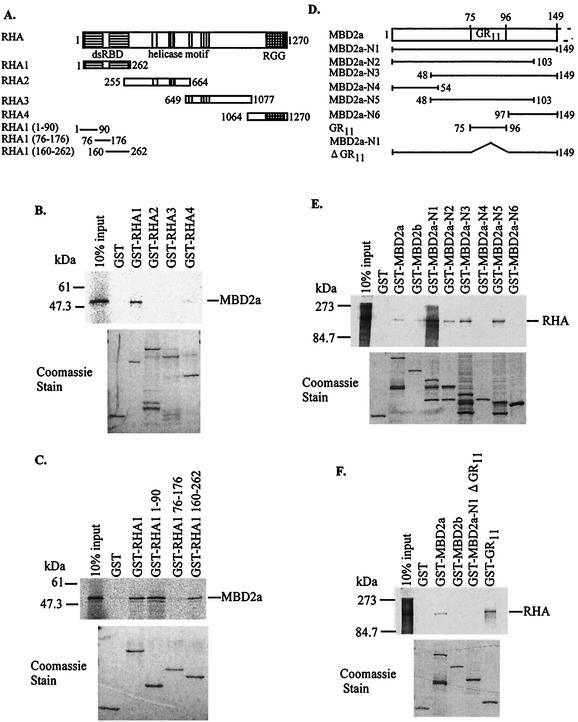
To define the binding region of MBD2a, several truncations of RHA were tested for interaction with MBD2a (Fig. 3A). Specific binding was evident for the RHA truncation containing aa 1 to 262, termed RHA1 (Fig. 3B). We then attempted to determine the minimal region required for binding to MBD2a. As shown in Fig. 3C, MBD2a particularly bound to RHA1 (aa 1 to 90) and RHA1 (aa 160 to 262), which contained double-stranded RNA binding domain (dsRBD) I and II, respectively. Conversely, to map the domain on MBD2a that mediated the interaction with RHA, a series of deletion mutants of MBD2a was constructed (Fig. 3D) and expressed as GST fusion proteins. In vitro binding experiments demonstrated that a region containing aa 48 to 103, termed MBD2a-N5, interacted with RHA. All deletion mutants that excluded this region failed to interact with RHA (Fig. 3E). This region contains the glycine-arginine repeat (GR11) that is the predicted functional region. Therefore, we performed GST pull-down assays to test for the interaction between RHA and GR11. GR11 was sufficient for binding to RHA (Fig. 3F). Considered together, these results demonstrated that the minimal regions mediating the RHA-MBD2a interaction corresponded to dsRBD I and/or II and GR11.
FIG. 3.
Determination of each binding region between MBD2a and RHA. (A) Schematic representation of deletion mutants of RHA. dsRBD, double-stranded RNA binding domain; I to VI, conserved helicase motifs; RGG, arginine and glycine-rich region. (B) Interaction of MBD2a with the RHA1 region in vitro. (C) Binding of MBD2a to dsRBD I and II within RHA in vitro. (D) Schematic representation of deletion mutants of the N terminus in MBD2a. (E and F) Interaction of RHA with GR11 in MBD2a in vitro.
MBD2a and RHA cooperatively promote activation of cAMP-responsive genes via phospho (Ser133)-CREB.
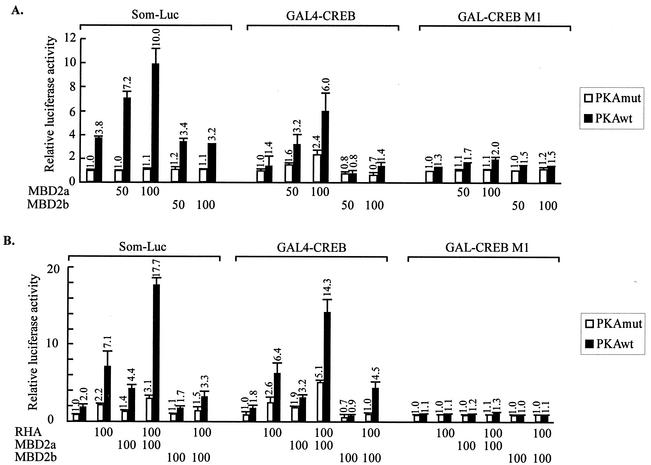
To confirm our finding that MBD2a promoted unmethylated Som-Luc activity (Fig. 1B), we performed reporter assays in HEK-293 cells. PKA induced Som-Luc reporter activity, and cotransfected full-length MBD2a further enhanced Som-Luc activity in a dose-dependent manner, whereas full-length MBD2b did not influence the activity (Fig. 4A, left panel).
FIG. 4.
Cooperation between MBD2a and RHA in promoting activation of cAMP-responsive genes via phospho (Ser133)-CREB. (A and B) Reporter assays of HEK-293 cells by using Som-Luc (left), GAL4-CREB with G5b-Luc (center), or Ser/Ala 133 phosphorylation-defective mutant GAL4-CREBM1 with G5b Luc reporter (right). Reporter activity was induced by cotransfection of PKAwt or PKAmut expression vector. The luciferase activity of cells transfected with empty vector alone and PKAmut was set as 1. The dose-dependent effects of MBD2a and MBD2b (A) and synergistic effects between RHA and MBD2a or MBD2b (B) are shown.
Some transcriptional factors, for example, ATF and CREM, are known to bind to the CRE sequence and regulate CRE-dependent transcription (33). To rule out possible effects of MBD2 on CRE binding proteins other than CREB, reporter assays were performed with G5b-Luc as artificial state in comparison with Som-Luc as endogenous state. The G5b-Luc reporter contains five copies of Gal4 sites in front of the E1b-TATA but shows absence of CRE (60). Instead of Som-Luc, CREB fused to GAL4-DBD (GAL4-CREB) and pG5b-Luc was cotransfected with MBD2 into HEK-293 cells. The ability of MBD2a to promote reporter activity was increased in a dose-dependent manner. Results similar to those for Som-Luc were obtained for MBD2b (Fig. 4A, middle panel). To confirm that the effects of MBD2a depended on PKA-induced phosphorylation at Ser-133, we used a mutant GAL4-CREBM1 containing a Ser-133/Ala-133 substitution at the PKA phosphoacceptor site. GAL4-CREBM1 does not activate CRE-dependent transcription in a phosphorylation-dependent manner (37). Both MBD2a and MBD2b had no effect via GAL4-CREBM1 (Fig. 4A, upper panel, right), confirming that the promotion of CRE-dependent gene expression by MBD2a is indeed phospho (Ser133) dependent. These results indicated that MBD2a enhanced CREB dependent transcriptional activation in both a dose- and phospho (Ser133)-dependent manner.
The ability of MBD2a to associate with RHA in vivo prompted us to evaluate the importance of this interaction for transcriptional activation. We performed reporter assays with Som-Luc and cotransfected full-length RHA with MBD2a or MBD2b into HEK-293 cells. In PKA-stimulated cells, full-length RHA and MBD2a enhanced Som-Luc activity 3.7-fold and 2.5-fold, respectively. Remarkably, cotransfection of MBD2a with RHA induced reporter activity 9.1-fold in PKA-stimulated cells, demonstrating the ability of these proteins to function cooperatively on cAMP-responsive promoters. In contrast, MBD2b reduced RHA activity on the Som-Luc reporter (Fig. 4B, left panel). These results indicated that MBD2a and RHA synergistically functioned in response to PKA stimulation.
To assess the regulatory effects of MBD2a and MBD2b on CREB activity, the reporter assay was performed with GAL4-CREB as indicated above. Overexpression of RHA and MBD2a further potentiated GAL4-CREB activity 3.8-fold and 1.8-fold, respectively, in PKA-stimulated cells. When cotransfected, RHA and MBD2a induced reporter activity 8.2-fold in PKA-stimulated but not unstimulated cells. Conversely, MBD2b repressed the effect of RHA (Fig. 4B, middle panel). In the case of GAL4-CREBM1, results similar to those described above were obtained (Fig. 4A, right panel, and Fig. 4B, right panel). These experiments revealed the importance of the MBD2a-specific N terminus for enhancement of CRE- and GAL4-CREB-dependent gene expression.
The C terminus of MBD2 has weak transcriptional activity.
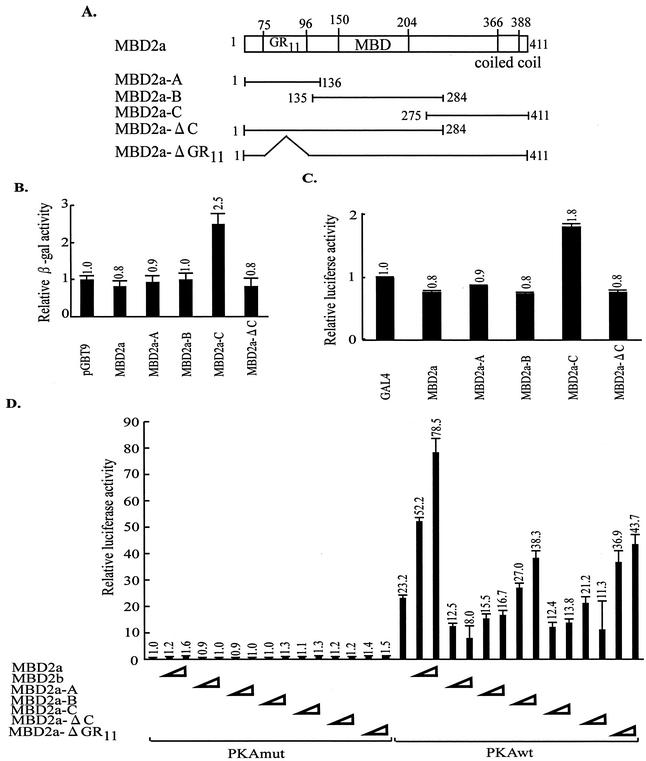
The results of promoting Som-Luc activity by MBD2a suggested that MBD2a might have a functional domain for transcriptional activation. Based on this hypothesis, we performed a transactivation assay to identify the transactivation domain within MBD2a. We divided MBD2a into three fragments, termed MBD2a-A, MBD2a-B, and MBD2a-C (Fig. 5A). These fragments were expressed as GAL4-DBD fusion proteins and their transcriptional activities in yeast were quantified. Among these three mutants, only MBD2a-C activated transcription (twofold that of the empty vector), whereas full-length MBD2a, MBD2a-A, and MBD2a-B did not (Fig. 5B).
FIG. 5.
Identification of the transactivation domain in the C terminus of MBD2. (A) Schematic representation of deletion mutants of MBD2. (B and C) Transactivation assays were performed with yeast (B) and mammalian (C) cells transformed with MBD2 deletion mutants. The β-Gal or luciferase activity of each mutant was compared to that of the empty vector alone, which was set as 1. Each value of relative β-Gal or luciferase activity represents the mean ± standard error (n = 3). (D) Effects of each domain within MBD2 on CRE-dependent gene expression. HEK-293 cells were cotransfected with Som-Luc and the plasmids expressing various MBD2 mutants. The luciferase activity of cells cotransfected with empty vector alone and unmethylated Som-Luc was set as 1. Each value of relative luciferase activity represents the mean ± standard error (n = 3).
To examine whether this region functions in higher eukaryotic cells in addition to yeast cells, a reporter assay was performed in HEK-293 cells. The same deletion mutants were cotransfected into HEK-293 cells with pG5b-Luc reporter plasmids. As in yeast, MBD2a-C was capable of activating transcription in mammalian cells (Fig. 5C). This region could not be divided into smaller constituent region with transcriptional activity (data not shown). These results suggested that the C-terminal region of MBD2 might have weak transactivation in both yeast and mammalian cells.
To confirm the contribution of each region within MBD2a to MBD2a-mediated enhancement of Som-Luc activity, we performed reporter assays with full-length MBD2a, MBD2b, and MBD2a truncations (Fig. 5A). As described above, MBD2a enhanced Som-Luc activity in PKA-stimulated cells, and MBD2b repressed this activity. The mutant MBD2a-Δ GR11, lacking the GR11 that is sufficient for interaction with RHA, reduced CRE-dependent reporter activity below the level obtained with full-length MBD2a (Fig. 5D). Binding assays with yeast demonstrated that the mutant MBD2a-Δ GR11 but not the mutant MBD2a-N1 ΔGR11 (Fig. 3D) weakly bound to RHA1 (data not shown), indicating that the effects of the mutant MBD2a-Δ GR11 might be associated with the weak interaction with RHA. These results suggested that the activation of CRE-dependent gene expression by MBD2a was dependent on interaction with RHA. The mutant MBD2a-Δ C, lacking the C terminus, reduced CRE-dependent reporter activity below the level obtained with full-length MBD2a. Interestingly, MBD2a-C resulted in approximately 50% of PKA induction on the Som-Luc reporter (Fig. 5D). These results suggested that the C terminus of MBD2a was important for promotion of CRE-dependent gene expression and that it might possess weak transcriptional activity.
MBD2a binding to RHA does not associate with HDAC1.
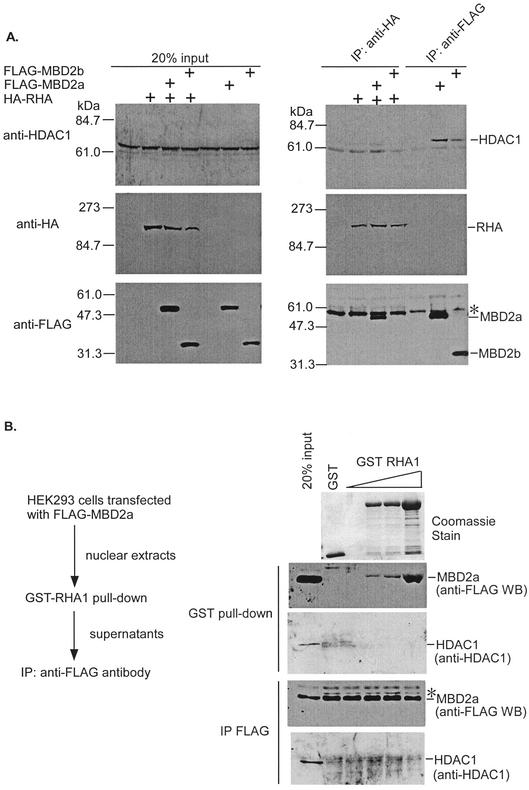
Previous studies showed that MBD2a is a component of the MeCP1 complex containing HDAC1 (15, 41). MBD2a binds to methylated DNA and represses transcription with recruitment of HDAC1 (21, 41). In the present study, we found that MBD2a interacted with RHA and promoted CRE-dependent gene expression. Therefore, we postulated that MBD2a binding to RHA did not associate with HDAC1. To test this hypothesis, we performed coimmunoprecipitation assays in HEK-293 cells. Each of the proteins was transiently expressed and analyzed by Western blotting with antibodies specific to HDAC1, HA-tag and FLAG-tag. As shown in Fig. 6A, HDAC1 was immunoprecipitated in the presence of FLAG-MBD2a or -MBD2b (Fig. 6A, right upper panel, lanes 6 and 7), but not HA-RHA (Fig. 6A, right upper panel, lane 2). In addition, RHA interacted with MBD2a, but not MBD2b (Fig. 6A, right lower panel, lanes 3 and 4). Interestingly, cotransfection of HA-RHA with FLAG-MBD2a or FLAG-MBD2b did not result in immunoprecipitation of a significant level of HDAC1 (Fig. 6A, right upper panel, lanes 3 and 4). These results suggested that RHA did not associate with HDAC1, although RHA could interact with MBD2a. In other words, MBD2a binding to RHA was not associated with HDAC1.
FIG. 6.
Distinct complex formation among MBD2a, RHA, and HDAC1. (A) Cells of the HEK-293 cell line were transiently transfected to produce HA-RHA and/or FLAG-MBD2a and FLAG-MBD2b. The cell lysates were immunoprecipitated with anti-HA antibody or anti-FLAG antibody and immunoblotted with anti-HDAC1 (upper panel), anti-HA antibody (middle panel), or anti-FLAG antibody (lower panel). The asterisk indicates immunoprecipitated IgG heavy chain. (B) HEK-293 cells were transfected with FLAG-MBD2a, and each nuclear extract was subsequently incubated with increasing mounts of GST-RHA1. The amount of GST-recruited MBD2a was visualized by Western blotting by using anti-FLAG antibodies. The MBD2a present in the supernatant of GST pull-down was immunoprecipitated with anti-FLAG-conjugated beads, and associated HDAC1 was detected by Western blotting by using anti-HDAC1 antibodies.
To confirm this, we assessed the ability of RHA1 to dissociate the MBD2a-HDAC1 complex by using an in vitro-in vivo binding assay. FLAG-MBD2a was expressed in HEK-293 cells. After a GST-RHA1 pull-down assay with nuclear extracts, the supernatants were immunoprecipitated with anti-FLAG to detect the interaction between MBD2a and HDAC1 (Fig. 6B). When the extracts were preincubated with increasing amounts of GST-RHA1, MBD2a was detected only in GST-RHA1 precipitates (Fig. 6B, GST pull-down, upper panel). In contrast, no HDAC1 signal was detected in GST-RHA1 precipitates, and HDAC1 levels were comparable in the supernatant derived from GST-RHA1 precipitation (Fig. 6B, GST pull-down, lower panel). Immunoprecipitation with anti-FLAG from the supernatants obtained after incubation with GST-RHA1 revealed that MBD2a-HDAC1 complexes were present in all extracts (Fig. 6B, IP FLAG, lower panel). These results suggested that RHA1, incapable of associating with HDAC1, did not displace HDAC1 from MBD2a. Alternatively, MBD2a-RHA complexes are distinct from MBD2a-HDAC1 complexes.
RHA-MBD2a interaction is functionally related in vivo.
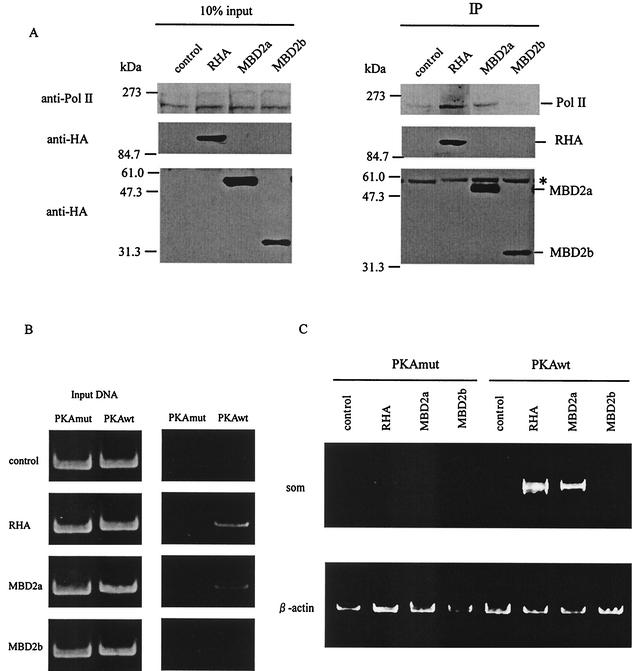
To assess the putative activation mechanisms by MBD2a, we tested whether MBD2a was associated with CBP and Pol II, activating partners of RHA. HEK-293 cells were transiently transfected with RHA, MBD2a, or MBD2b expression plasmids. As shown in Fig. 7A, MBD2a interacted with Pol II (upper panel) as well as RHA. Similar results were obtained with CBP (data not shown). These results demonstrated that MBD2a could associate with the activator complex including RHA, CBP, and Pol II.
FIG. 7.
Putative activation mechanisms by MBD2a. (A) HEK-293 cells were transiently transfected with HA-RHA, HA-MBD2a, and HA-MBD2b. The cell lysates were immunoprecipitated with anti-HA antibody and immunoblotted with anti-Pol II antibody (upper panel) or anti-HA antibody (lower panel). The asterisk indicates immunoprecipitated IgG heavy chain. (B) Ch-IP assay of D5 cells containing integrated copies of the rat somatostatin gene. D5 cells were transfected with HA-RHA, HA-MBD2a, and HA-MBD2b and cotransfected with PKAmut or PKAwt. After 24 h of transfection, formaldehyde cross-linked protein-DNA complexes were immunoprecipitated with anti-HA. A PCR analysis to amplify somatostatin promoter sequences (−100 to +55) is shown, including input DNA. (C) RT-PCR of total RNA from D5 cells expressing chromosomal copies of the rat somatostatin gene (som). Cells were transfected with HA-RHA, HA-MBD2a, and HA-MBD2b and cotransfected with PKAmut or PKAwt. The expression of β-actin, a ubiquitous mRNA, was used as an internal control.
To evaluate whether RHA and MBD2a were recruited to the active promoter, we analyzed the association of MBD2a with the promoter of CREB target genes by chromatin immunoprecipitation (Ch-IP) assay. We used the stable NIH 3T3 cell line D5, containing rat somatostatin gene sequences from 750 bp upstream of the promoter to 3 kb downstream of the coding region. D5 cells were transiently transfected with RHA, MBD2a, or MBD2b expression plasmids, and the DNA fragments immunoprecipitated by anti-HA antibodies were analyzed by semiquantitative PCR for the presence of the CRE regions of somatostatin promoter. The regions selected for Ch-IP analysis corresponded to the sequences containing the functional CRE. In unstimulated cells, there was no association of each of the proteins with the promoter of rat somatostatin (Fig. 7B, indicated as PKAmut). In contrast, both RHA and MBD2a, but not MBD2b, were recruited to the CRE of rat somatostatin promoter in PKA-stimulated cells (Fig. 7B, indicated as PKAwt). These results indicated that RHA and MBD2a could be recruited to active endogenous CRE in a phospho-(CREB at Ser-133)-dependent manner.
To confirm whether MBD2a would activate the endogenous promoter in vivo, we evaluated somatostatin mRNA accumulation in D5 cells by RT-PCR. As shown in Fig. 7C, the expression of endogenous somatostatin mRNA was not detected in unstimulated cells (Fig. 7C, indicated as PKAmut). In contrast, RHA and MBD2a, but not MBD2b, increased the expression of somatostatin mRNA in PKA-stimulated cells (Fig. 7C, indicated as PKAwt). Taken together, these data indicated that MBD2a could functionally activate CREB-dependent transcription and be recruited to the endogenous promoter of CREB target genes.
DISCUSSION
Different roles of MBD2a and MBD2b.
Our results showed that MBD2a but not MBD2b interacted with RHA in vitro and in vivo (Fig. 2B, C, and D). In addition, in vitro binding experiments indicated that GR11 was sufficient for binding to RHA (Fig. 2F). GR11 is unique in MBD2a among members of the MBD family and is the predicted functional region (6). However, its functions and interactants are not clear. Therefore, RHA was the first molecule to functionally interact with GR11.
We showed that MBD2a and MBD2b played distinct roles via interaction with RHA. It is known that several proteins alter regulation of gene expression by distinct isoforms (3, 30). For example, the homeobox protein, Pbx, consists of two isoforms generated from alternative splicing. Pbx forms a dimer with Pdx, and the Pbx-Pdx dimer recruits coactivator-corepressor complexes in an isoform-specific manner. The transcriptional properties of the Pbx-Pdx complex appear to be regulated at the level of alternative splicing via different expressions of Pbx isoforms in endocrine versus exocrine compartments of the adult pancreas (3). MBD2 is ubiquitously expressed in tissues excluding ES cells (21). Although the different expression patterns in MBD2a and MBD2b are not clear in cells and tissues, it is possible that MBD2 may regulate gene expression by a similar mechanism.
The roles of each domain within MBD2a on CREB-dependent gene expression.
Our results showed that MBD2a contained a potent transactivation domain in its C terminus in yeast and mammalian cells (Fig. 5B and C). The C terminus of MBD2a involves the coiled-coil motif. In some cases, it has been shown that this motif is important for transcriptional activation via interaction with activating factors (24, 25, 50). However, we have not yet found the interactants that bind to the C terminus (Fig. 7, indicated as X). Full-length MBD2a and MBD2b repressed transcriptional activity despite the presence of the C terminus when fused to GAL4, and the results were almost similar to those of previous reports (Fig. 5B and C). Recent work has shown that MBD2a associates with NuRD complex via their MBD and coiled-coil motif when MBD2a binds to methylated DNA (55, 62). It is therefore possible that the C terminus might be masked under conditions of binding to DNA. Furthermore, the mutant, lacking the C terminus (MBD2a-Δ C), reduced enhancement of cAMP-responsive gene activation (Fig. 5D), indicating that the C terminus of MBD2a could be important for promoting CRE-dependent transcription by MBD2a.
MBD2a and RHA cooperatively enhanced CRE- and CREB-dependent gene expression, but MBD2b did not (Fig. 4C and D), indicating the importance of RHA-MBD2a interaction via GR11. MBD2b reduced RHA activity on the Som-Luc reporter (Fig. 4B, left panel). It is therefore possible that MBD2b might remove factors that interact with the C terminus of MBD2a. These results indicated that the interaction of MBD2a with RHA is critical for promoting CRE-dependent gene expression by MBD2a. Taken together, the results of our study demonstrated that MBD2a could promote CRE-dependent transcription via its C terminus and GR11.
Roles of MBD2a on CREB-dependent gene expression.
It is well known that methylation occurs on CRE in the tissue-specific genes and the tumor suppressor genes, resulting in transcriptional repression (13, 32, 45). However, the underlying mechanisms are poorly understood. In our study, MBD2a and MBD2b repressed transcription of methylated CRE (Fig. 1A). Previous studies indicated that MBD2 specifically binds to methylated DNA only in the presence of two methyl groups (43, 44, 56). Therefore, it is not clear whether MBD2 specifically binds to methylated CRE in vivo. It is possible that the transcription of methylated CRE might be partially repressed by MBD2.
In our study, MBD2a and MBD2b repressed transcription of methylated CRE (Fig. 1A), but this repression seemed to be alleviated by PKA induction (Fig. 1B). There are two possibilities. The first is that Som-Luc was not fully methylated in vitro and thus minimal induction occurred by PKA. However, methylated Som-Luc reporter plasmid was insensitive to the methylation-sensitive restriction enzyme HpaII in vitro. The second is that PKA would induce directly or indirectly demethylation and/or derepression.
We showed that MBD2a enhanced CREB-dependent gene expression in transient transfection assays (Fig. 4) and the mRNA expression of endogenous CREB target gene (Fig. 7C). For some MBD proteins (MeCP2, MBD1, and MBD2), transcriptional repression has been shown to occur (38, 41, 42). Our study was therefore the first to show that MBD protein might act as cofactor for transcriptional activation.
MBD2a is a component of the MeCP1 corepressor complex (41) containing HDAC1/2 and components of the NuRD complex (15, 41, 61, 62). In our study, we found that MBD2a interacted with RHA and Pol II in vivo (Fig. 7A). The Ch-IP assay revealed that RHA and MBD2a were recruited to the endogenous promoter of the CREB target gene in PKA-stimulated cells (Fig. 7B), indicating that MBD2a would form a complex with activators on the endogenous promoter. Interestingly, MBD2a binding to RHA was not associated with HDAC1 (Fig. 6A and B). These results indicated that MBD2a could be involved in two different complexes, the corepressor complex and the activator complex.
Methylation and demethylation occur on CRE. During cancer progression, methylation is induced on unmethylated CRE, resulting in transcriptional repression (13, 45). Conversely, during cell differentiation, demethylation is induced on methylated CRE, and thereby transcriptional activation ensues (11, 26, 48). It is also known that CRE within the cell-specific genes is consistently methylated and associated with transcriptional inactivation in other cells (27, 29). Our results showed that MBD2a performed a distinct function according to the presence or absence of methylation on CRE (Fig. 1). It is possible that MBD2a bound to methylated CRE represses transcription while MBD2a that did not bind to methylated CRE might function as cofactor for transcriptional activation. Alternatively, MBD2a may alter functions between repressor and cofactor for transcriptional activation in accordance with the methylation states on CRE.
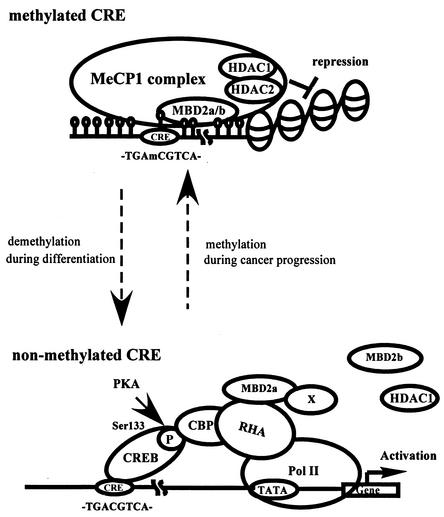
Taken together, we propose a model in which MBD2a has discrete roles, transcriptional repression and activation, according to the presence or absence of DNA methylation on CRE (Fig. 8). MBD2a would bind to methylated CRE and repress its transcription via association with the corepressor complex (Fig. 8, upper panel). On the other hand, MBD2a could associate with the CBP/RHA/Pol II complex and promote the transcription of unmethylated CRE in a phospho-(CREB at Ser133)-dependent manner (Fig. 8, lower panel). Alternatively, MBD2a could have opposite roles in gene regulation.
FIG. 8.
Hypothetical model illustrating the regulation of CRE-dependent transcription by MBD2a according to the presence or absence of DNA methylation. In methylated CRE, MBD2a binds to methylated DNA involving CRE and represses transcription through association with the corepressor complex including HDAC1. In contrast, when CRE is not methylated, MBD2a promotes CRE-dependent gene expression as a cofactor via the interaction with RHA.
Acknowledgments
We thank Yukiko Okada, Megumi Fujita, Makiko Yui, Sanae Shinkawa, and Asako Sugamiya for technical assistance and members of the Nakajima Lab for discussions of the manuscript. We also thank Marc Montminy and Hiroshi Asahara for D5 cells and the protocol of the Ch-IP experiment.
This work was supported by grants from the Japanese Ministry of Education, Science Culture and Sports, by the Japanese Ministry of Health and Welfare, Japan Science and Technology Corporation (Precursory Research for Embryonic Science and Technology [PRESTO]), by the Human Health Science Foundation, by funds from the Memorial Yamanouchi Foundation, by the Kato Memorial Trust for Nanbyo Research, by Kanagawa Academy of Science and Technology Research Grants, by the Japan Medical Association, by the Nagao Memorial Fund, by the Kanae Foundation for Life & Socio-Medical Science, by the Japan Research Foundation for Clinical Pharmacology, by the Kanagawa Nanbyo Foundation, by the Japan Rheumatology Association, by the Nakajima Foundation, by Locomogene Co. Ltd., and by the Mitsubishi Pharma Research Foundation.
REFERENCES
- 1.Aratani, S., R. Fujii, T. Oishi, H. Fujita, T. Amano, T. Ohshima, M. Hagiwara, A. Fukamizu, and T. Nakajima. 2001. Dual roles of RNA helicase A in CREB-dependent transcription. Mol. Cell. Biol. 21:4460-4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias, J., A. Alberts, P. Brindle, F. Claret, T. Smeal, M. Karin, J. Feramisco, and M. Montminy. 1994. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature 370:226-228. [DOI] [PubMed] [Google Scholar]
- 3.Asahara, H., S. Dutta, H. Y. Kao, R. M. Evans, and M. Montminy. 1999. Pbx-Hox heterodimers recruit coactivator-corepressor complexes in an isoform-specific manner. Mol. Cell. Biol. 19:8219-8225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asahara, H., B. Santoso, E. Guzman, K. Du, P. A. Cole, I. Davidson, and M. Montminy. 2001. Chromatin-dependent cooperativity between constitutive and inducible activation domains in CREB. Mol. Cell. Biol. 21:7892-7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartolomei, M. S., and M. S. Tilghman. 1997. Genomic imprinting in mammals. Annu. Rev. Genet. 31:493-525. [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharya, S. K., S. Ramchandani, N. Cervoni, and M. Szyf. 1999. A mammalian protein with specific demethylase activity for mCpG DNA. Nature 397:579-583. [DOI] [PubMed] [Google Scholar]
- 7.Bird, A. 1992. The essentials of DNA methylation. Cell 70:5-8. [DOI] [PubMed] [Google Scholar]
- 8.Bird, A. P., and A. P. Wolffe. 1999. Methylation-induced repression-belts, braces, and chromatin. Cell 99:451-454. [DOI] [PubMed] [Google Scholar]
- 9.Boyes, J., and A. Bird. 1991. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell 64:1123-1134. [DOI] [PubMed] [Google Scholar]
- 10.Brindle, P., S. Links, and M. Montminy. 1993. Analysis of a PK-A dependent activator in CREB reveals a new role for the CREM family of repressor. Nature 364:821-824. [DOI] [PubMed] [Google Scholar]
- 11.Burbelo, P. D., S. Horikoshi, and Y. Yamada. 1990. DNA methylation and collagen IV gene expression in F9 teratocarcinoma cells. J. Biol. Chem. 265:4839-4843. [PubMed] [Google Scholar]
- 12.Chrivia, J. C., R. P. Kwok, N. Lamb, M. Hagiwara, M. R. Montminy, and R. H. Goodman. 1993. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365:855-859. [DOI] [PubMed] [Google Scholar]
- 13.DiNardo, D. N., D. T. Butcher, D. P. Robinson, T. K. Archer, and D. I. Rodenhiser. 2001. Functional analysis of CpG methylation in the BRCA1 promoter region. Oncogene 20:5331-5340. [DOI] [PubMed] [Google Scholar]
- 14.Dorris, D. R., and K. Struhl. 2000. Artificial recruitment of TFIID, but not RNA polymerase II holoenzyme, activates transcription in mammalian cells. Mol. Cell. Biol. 20:4350-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng, Q., and Y. Zhang. 2001. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev. 15:827-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrei, K., G. Gill, and M. Montminy. 1994. The cAMP regulated transcription factor CREB interacts with a component of TFIID complex. Proc. Natl. Acad. Sci. USA 91:1210-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujii, R, M. Okamoto, S. Aratani, T. Oishi, T. Ohshima, K. Taira, M. Bab, A. Fukamizu, and T. Nakajima. 2001. A Role of RNA helicase A in cis-acting transactivation response element-mediated transcriptional regulation of human immunodeficiency virus type 1. J. Biol. Chem. 276:5445-5451. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez, G. A., and M. R. Montminy. 1989. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell 59:675-680. [DOI] [PubMed] [Google Scholar]
- 19.Heard, E., P. Clerc, and P. Avner. 1997. X-chromosome inactivation in mammals. Annu. Rev. Genet. 31:571-610. [DOI] [PubMed] [Google Scholar]
- 20.Hendrich, B., C. Abbott, H. McQueen, D. Chambers, S. Cross, and A. Bird. 1999. Genomic structure and chromosomal mapping of the murine and human Mbd1, Mbd2, Mbd3, and Mbd4 genes. Mamm. Genome 10:906-912. [DOI] [PubMed] [Google Scholar]
- 21.Hendrich, B., and A. Bird. 1998. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 18:6538-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hendrich, B., and A. Bird. 2000. Mammalian methyltransferases and methyl-CpG-binding domains: proteins involved in DNA methylation. Curr. Top. Microbiol. Immunol. 249:55-74. [DOI] [PubMed] [Google Scholar]
- 23.Herman, J. G., and S. B. Baylin. 2000. Promoter-region hypermethylation and gene silencing in human cancer. Curr. Top. Microbiol. Immunol. 249:35-54. [DOI] [PubMed] [Google Scholar]
- 24.Hu, Yan-Fen, T. Miyake, Q. Ye, and R. Li. 2000. Characterization of a novel trans-activation domain of BRCA1 that functions in concert with the BRCA1 C-terminal (BRCT) domain. J. Biol. Chem. 275:40910-40915. [DOI] [PubMed] [Google Scholar]
- 25.Hu, Yan-Fen, and R. Li. 2002. JunB potentiates function of BRCA1 activation domain 1 (AD1) through a coiled-coil-mediated interaction. Genes Dev. 16:1509-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iannello, R. C., J. A. Gould, J. C. Young, A. Giudice, R. Medcalf, and I. Kola. 2000. Methylation-dependent silencing of the testis-specific Pdha-2 basal promoter occurs through selective targeting of an activating transcription factor/cAMP-responsive element-binding site. J. Biol. Chem. 275:19603-19608. [DOI] [PubMed] [Google Scholar]
- 27.Iannello, R. C., J. Young, S. Sumarsono, M. J. Tymms, H. H. Dahl, J. Gould, M. Hedger, and I. Kola. 1997. Regulation of Pdha-2 expression is mediated by proximal promoter sequences and CpG methylation. Mol. Cell. Biol. 17:612-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson-Grusby, L., C. Beard, R. Possemato, M. Tudor, D. Fambrough, G. Csankovszki, J. Dausman, P. Lee, C. Wilson, E. Lander, and R. Jaenisch. 2001. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 27:31-39. [DOI] [PubMed] [Google Scholar]
- 29.Kroft, T. L., P. Jethanandani, D. J. McLean, and E. Goldberg. 2001. Methylation of CpG dinucleotides alters binding and silences testis-specific transcription directed by the mouse lactate dehydrogenase C promoter. Biol. Reprod. 65:1522-1527. [DOI] [PubMed] [Google Scholar]
- 30.Li, D., F. Wang, and H. H. Samuels. 2001. Domain structure of the NRIF3 family of coregulators suggests potential dual roles in transcriptional regulation. Mol. Cell. Biol. 21:8371-8384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915-926. [DOI] [PubMed] [Google Scholar]
- 32.Mancini, D. N., S. M. Singh, T. K. Archer, and D. I. Rodenhiser. 1999. Site-specific DNA methylation in the neurofibromatosis (NF1) promoter interferes with binding of CREB and SP1 transcription factors. Oncogene 18:4108-4119. [DOI] [PubMed] [Google Scholar]
- 33.Mayr, B., and M. Montminy. 2001. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell. Biol. 8:599-609. [DOI] [PubMed] [Google Scholar]
- 34.Meehan, R. R., J. D. Lewis, S. McKay, E. L. Kleiner, and A. P. Bird. 1989. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell 58:499-507. [DOI] [PubMed] [Google Scholar]
- 35.Miyagishi, M., R. Fujii, M. Hatta, E. Yoshida, N. Araya, A. Nagafuchi, S. Ishihara, T. Nakajima, and A. Fukamizu. 2000. Regulation of Lef-mediated transcription and p53-dependent pathway by associating beta-catenin with CBP/p300. J. Biol. Chem. 275:35170-35175. [DOI] [PubMed] [Google Scholar]
- 36.Nakajima, T., C. Uchida, S. Anderson, J. Parvin, and M. Montminy. 1997. Analysis of a cAMP-responsive activator reveals a two-component mechanism for transcriptional induction via signal-dependent factors. Genes Dev. 11:738-747. [DOI] [PubMed] [Google Scholar]
- 37.Nakajima, T., C. Uchida, S. Anderson, C. Lee, J. Hurwitz, J. Parvin, and M. Montminy. 1997. RNA helicase A mediates association of CBP with RNA polymerase II. Cell 90:1107-1112. [DOI] [PubMed] [Google Scholar]
- 38.Nan, X., F. J. Campoy, and A. Bird. 1997. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 88:471-481. [DOI] [PubMed] [Google Scholar]
- 39.Neish, A. S., S. F. Anderson, B. P. Schlegel, W. Wei, and J. D. Parvin. 1998. Factors associated with the mammalian RNA polymerase II holoenzyme. Nucleic Acids Res. 26:847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nevado, J., L. Gaudreau, M. Adam, and M. Ptashne. 1999. Transcriptional activation by artificial recruitment in mammalian cells. Proc. Natl. Acad. Sci. USA 96:2674-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ng, H. H., Y. Zhang, B. Hendrich, C. A. Johnson, B. M. Turner, H. Erdjument-Bromage, P. Tempst, D. Reinberg, and A. Bird. 1999. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet. 23:58-61. [DOI] [PubMed] [Google Scholar]
- 42.Ng, H. H., P. Jeppesen, and A. Bird. 2000. Active repression of methylated genes by the chromosomal protein MBD1. Mol. Cell. Biol. 20:1394-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohki, I., N. Shimotake, N. Fujita, M. Nakao, and M. Shirakawa. 1999. Solution structure of the methyl-CpG-binding domain of the methylation-dependent transcriptional repressor MBD1. EMBO J. 18:6653-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohki, I., N. Shimotake, N. Fujita, J. Jee, T. Ikegami, M. Nakao, and M., Shirakawa. 2001. Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA. Cell 105:487-497. [DOI] [PubMed] [Google Scholar]
- 45.Ohtani-Fujita, N., T. Fujita, A. Aoike, N. E. Osifchin, P. D. Robbins, and T. Sakai. 1993. CpG methylation inactivates the promoter activity of the human retinoblastoma tumor-suppressor gene. Oncogene 8:1063-1067. [PubMed] [Google Scholar]
- 46.Okano, M., D. W. Bell, D. A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247-257. [DOI] [PubMed] [Google Scholar]
- 47.Parker, D., K. Ferreri, T. Nakajima, V. LaMorte, R. Evans, S. Koerber, C. Hoeger, and M. Montminy. 1996. Phosphorylation of CREB at Ser-133 induces complex formation with CBP via a direct mechanism. Mol. Cell. Biol. 16:694-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parolini, O., J. Rohrer, L. H. Shapiro, and M. E. Conley. 1995. B-cell-specific demethylation of BTK, the defective gene in X-linked agammaglobulinemia. Immunogenetics 42:129-135. [DOI] [PubMed] [Google Scholar]
- 49.Quinn, P. G. 1993. Distinct activation domains within cAMP response element-binding protein (CREB) mediate basal and cAMP-stimulated transcription. J. Biol. Chem. 268:16999-17009. [PubMed] [Google Scholar]
- 50.Scott, S., A. T. Abul-Hamd, and T. G. Cooper. 2000. Roles of the Dal82p domains in allophanate/oxalurate-dependent gene expression in Saccharomyces cerevisiae. J. Biol. Chem. 275:30886-30893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sekimata, M., A. Takahashi, A. Murakami-Sekimata, and Y. Homma. 2001. Involvement of a novel zinc finger protein, MIZF, in transcriptional repression by interacting with a methyl-CpG-binding protein, MBD2. J. Biol. Chem. 276:42632-42638. [DOI] [PubMed] [Google Scholar]
- 52.Shimohata, T., T. Nakajima, M. Yamada, C. Uchida, O. Onodera, S. Naruse, T. Kimura, R. Koide, K. Nozaki, Y. Sano, H. Ishiguro, K. Sakoe, T. Ooshima, A. Sato, T. Ikeuchi, M. Oyake, T. Sato, Y. Aoyagi, I. Hozumi, T. Nagatsu, Y. Takiyama, M. Nishizawa, J. Goto, I. Kanazawa, I. Davidson, N. Tanese, H. Takahashi, and S. Tsuji. 2000. Expanded polyglutamine stretches interact with TAFII130, interfering with CREB-dependent transcription. Nat. Genet. 26:29-36. [DOI] [PubMed] [Google Scholar]
- 53.Stancheva, I., and R. R. Meehan. 2000. Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos. Genes Dev. 14:313-327. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Taneae, N., D. Saluja, F. M. Vassallo, J. L. Chen, and A. Admon. 1996. Molecular cloning and analysis of two subunits of the human TFIID complex: hTAFII130 and hTAFII100. Proc. Natl. Acad. Sci. USA 93:13611-13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tatematu, K., T. Yamazaki, and F. Ishikawa. 2000. MBD2-MBD3 complex binds to hemi-methylated DNA and forms a complex containing DNMT1 at the replication foci in late S phase. Genes Cells 5:677-688. [DOI] [PubMed] [Google Scholar]
- 56.Wakefield, R. I., B. O. Smith, X. Nan, A. Free, A. Soteriou, D. Uhrin, A. P. Bird, and P. N. Barlow. 1999. The solution structure of the domain from MeCP2 that binds to methylated DNA. J. Mol. Biol. 291:1055-1065. [DOI] [PubMed] [Google Scholar]
- 57.Walsh, C. P., J. R. Chaillet, and T. H. Bestor. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20:116-117. [DOI] [PubMed] [Google Scholar]
- 58.Weih, F., D. Nitsch, A. Reik, G. Schutz, and P. B. Becker. 1991. Analysis of CpG methylation and genomic footprinting at the tyrosine aminotransferase gene: DNA methylation alone is not sufficient to prevent protein binding in vivo. EMBO J. 10:2559-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yao, T. P., S. P. Oh, M. Fuchs, N. D. Zhou, L. E. Ch'ng, D. Newsome, R. T. Bronson, E. Li, D. M. Livingston, and R. Eckner. 1998. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93:361-372. [DOI] [PubMed] [Google Scholar]
- 60.Yoshida, E., S. Aratani, H. Itou, M. Miyagishi, M. Takiguchi, T. Osumu, K. Murakami, and A. Fukamizu. 1997. Functional association between CBP and HNF4 in trans-activation. Biochem. Biophys. Res. Commun. 241:664-669. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, Y., G. LeRoy, H. P. Seelig, W. S. Lane, and D. Reinberg. 1998. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95:279-289. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, Y., H. H. Ng, H. Erdjument-Bromage, P. Tempst, A. Bird, and D. Reinberg. 1999. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 13:1924-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]