Abstract
Tetrahymena thermophila cells contain three forms of H2A: major H2A.1 and H2A.2, which make up ∼80% of total H2A, and a conserved variant, H2A.Z. We showed previously that acetylation of H2A.Z was essential (Q. Ren and M. A. Gorovsky, Mol. Cell 7:1329-1335, 2001). Here we used in vitro mutagenesis of lysine residues, coupled with gene replacement, to identify the sites of acetylation of the N-terminal tail of the major H2A and to analyze its function in vivo. Tetrahymena cells survived with all five acetylatable lysines replaced by arginines plus a mutation that abolished acetylation of the N-terminal serine normally found in the wild-type protein. Thus, neither posttranslational nor cotranslational acetylation of major H2A is essential. Surprisingly, the nonacetylatable N-terminal tail of the major H2A was able to replace the essential function of the acetylation of the H2A.Z N-terminal tail. Tail-swapping experiments between H2A.1 and H2A.Z revealed that the nonessential acetylation of the major H2A N-terminal tail can be made to function as an essential charge patch in place of the H2A.Z N-terminal tail and that while the pattern of acetylation of an H2A N-terminal tail is determined by the tail sequence, the effects of acetylation on viability are determined by properties of the H2A core and not those of the N-terminal tail itself.
In eukaryotic cell nuclei, DNA associates with histones to form chromatin. The basic unit of chromatin is the nucleosome core particle consisting of ∼146 bp of DNA wrapped around an octamer of two of each of the four conserved core histones, H2A, H2B, H3, and H4 (85). All core histones contain a highly structured C-terminal histone-fold domain and a highly charged, structurally undefined, N-terminal tail domain that emerges from the histone core. These tails are thought to be important for DNA-histone and histone-histone interactions within and/or between nucleosomes and for interactions with nonhistone proteins (52, 89). In addition, H2A has an extended C-terminal tail contacting DNA near the dyad axis at the center of the nucleosome core (77, 87).
The seemingly simple, repetitive nature and highly condensed state of chromatin in the nucleus provide apparent limitations to chromatin functions, especially to transcription regulation in different cell types and physiological states. Several mechanisms have evolved to produce heterogeneity in the chromatin, including complex patterns of histone modifications and sequence variation of histones. Most histone modifications, including acetylation, phosphorylation, methylation, and ubiquitination, occur on the histone amino- and carboxyl-terminal tails (34). Of all the histone modifications, acetylation of the ɛ-amino group of lysine, which occurs after histone deposition and is restricted to the N-terminal tails, is probably the most abundant and is the best characterized. This acetylation has been closely linked to transcriptional activation (12, 63) by findings that many transcriptional activators or coactivators possess histone acetyltransferase (HAT) activity (13, 64), while corepressors containing histone deacetylase activity can repress transcription (54, 72). Recent studies also have shown that acetylation patterns of chromatin domains are important for establishing stable patterns of gene expression (28).
Besides the relationship between histone acetylation and transcription regulation, histone acetylation is also involved in processes such as DNA replication (80), nucleosome assembly, and chromosome condensation (74).
Two mechanisms have been proposed for how acetylation and other histone posttranslational modifications might act. Acetylation might modify chromatin structure and function by affecting histone-DNA interactions or histone-histone interactions. Alternatively or in addition, acetylation might act by altering the ability of nonhistone transcription or replication factors to bind to the N-terminal tails (44, 86).
It has recently been suggested that histone modifications, acting at specific sites either alone, in combination, or in sequential fashion on one or multiple histone tails, can form a complex histone code that can either enhance or reduce the interaction affinities with chromatin-associated proteins and thereby specify unique chromatin functions (42, 69, 76). The hallmark of this histone code mechanism is that the posttranslational modification provides a site-specific signal (either a structural motif, a structural change, or a specific charge) that affects recognition of the site by another molecule. There is ample evidence to support this kind of mechanism. Catalytic HATs and histone deacetylases do not acetylate and deacetylate histones nonspecifically (23, 84). Bromodomains found in HATs and in some transcription factors (PCAF, TAFII250) can selectively interact with acetylated lysines either individually or in specific combination in the histone N-terminal tails (20, 41). The chromodomains of some heterochromatin proteins and histone methyltransferases are highly selective for methylated H3 at K9 but not for methylated H3 at K4 (11, 55). A lot of evidence also shows the interplay between different modifications on single or multiple histone tails (91). H3 phosphorylation at serine 10 can enhance acetylation on lysine 14 and affect transcription at specific genes (17, 51). H4-R3 methylation mediated by PRMT1 (70, 81) facilitates p300-mediated acetylation on H4-K8 and H4-K12. In the reverse direction, histone H4 acetylation on any of the four lysines (K5, K8, K12, or K16) also inhibits the subsequent methylation at H4-R3 by PRMT1 (81).
Charge-altering modifications also can affect chromatin function by a second mechanism in which they alter the charge of a protein domain rather than affect a specific site. This “charge patch” mechanism has been shown to apply to regulation of the expression of specific genes by phosphorylation of linker histone H1 in Tetrahymena (21, 22) and to modulation of the essential function of histone H2A.Z by acetylation in Tetrahymena (62). In these cases, the function of the modification is to alter the charge of the domain in which it resides. Unlike the histone code, these changes need not be site specific. Modulation of the charge at any one of a number of clustered sites can have the same effect. The ability of acetylation to inhibit the salt-induced condensation of nucleosome oligomers in vitro (75) could be such an effect, and if acetylation were to inhibit nucleosome condensation in vivo, it could facilitate transcription.
In addition to posttranslational modifications of histones, another factor that contributes to chromatin functional heterogeneity is the existence of nonallelic histone variants (37). The demonstration that some histone mutations have highly specific effects on transcription, coupled with the observation that expression of some histone variants is temporally, developmentally, and spatially regulated, suggests that variant nucleosomes perform distinct functions (88).
The best-studied core histone variant is H2A.Z (61). H2A.Z has been found in diverse organisms, including Tetrahymena (83), Saccharomyces cerevisiae (39, 65), Schizosaccharomyces pombe (15), Drosophila (79), Arabidopsis thaliana (14), sea urchins (24), chickens (19, 36), and mammals (10). Phylogenetic analysis of H2A protein sequences (73) demonstrated that the major H2As and the H2A.Z variants diverged early in eukaryotic evolution and that the H2A.Zs show even less evolutionary divergence than the major H2As. Therefore, there were two types of H2A genes in primitive eukaryotes before the divergence of ciliates, fungi, animals, and plants, and they have been under different selective pressures since that time.
The evolutionary implication that the major H2As and H2A.Z variants have distinct and important functions has been confirmed experimentally. In Drosophila and in S. cerevisiae, the distribution of the two types of H2A in chromatin differs (47, 65). Deletions of genes encoding H2A.Z are lethal in Tetrahymena (50), Drosophila (78), and mice (25) and cause slow growth and/or conditional lethality in yeasts (2, 15, 40, 65). In both S. cerevisiae (43, 50, 66) and Tetrahymena (48), mutants without at least one of the two major histone genes (HTA1 or HTA2) cannot survive. In S. cerevisiae, expression of the gene encoding H2A.Z (HTZ1) cannot rescue disruptions of both genes encoding the major H2As even when overexpressed or placed under control of the HTA1 promoter (40, 65). Chimeric genes with different domains on the H2A.Z replaced with those of major H2A were injected into Drosophila H2A.Z null embryos to investigate which domain(s) is essential for H2A.Z function (18). Surprisingly, the essential portions of H2A.Z are the αC helix and H3/H4-binding domains. Thus, it is clear that the H2A.Z variants and the major H2As have distinct functions, both of which are either essential or required for normal growth in all organisms tested.
Little is known about the specific functions that are distinct to either the major H2As or to the H2A.Z variants. Pinto and Winston (58) argued that the major H2A of S. cerevisiae was required for normal centromere function because two cold-sensitive H2A mutations showed chromosome segregation phenotypes and interacted genetically with mutations in known centromere components. While this study did not specifically test the same mutations in H2A.Z, the fact that the H2A mutations had centromere-specific effects in cells containing a wild-type HTZ1 gene suggests that this centromere function is specific for the major H2A. Considerable circumstantial evidence suggests that H2A.Z has a transcription-related function. In Tetrahymena, H2A.Z is present only in the transcriptionally active macronuclei and not in the transcriptionally inactive micronuclei of vegetative cells but appears in premeiotic micronuclei of conjugating cells when they become transcriptionally active (68). In S. cerevisiae, mutations in HTZ1 are synthetically lethal with deletion of SNF2 (65), a component of the SWI/SNF remodeling complex required for transcription of many genes, while mutations in the major histones suppress SWI2 deletion (45, 46, 60, 82).
The likely function of H2A.Z in transcription and the well-documented relationship between acetylation and transcription led us to analyze the function of acetylation of H2A.Z. We showed that acetylation of Tetrahymena H2A.Z is essential and that it acts to modulate a charge patch on its N-terminal tail (62). Because the function of the major H2A of Tetrahymena is essential but distinct from that of H2A.Z, in this study we sought to identify the sites of acetylation of the major H2A in this organism and analyze their function. We changed the acetylation sites on major histone H2A.1 either from lysine to arginine, which conserves the net positive charge of lysine but cannot be neutralized by acetylation, or from lysine to glutamine, which resembles acetylated lysine in charge and structure. We showed that Tetrahymena cells survive plus five lysines of their major H2A replaced by arginines plus a mutation that abolishes the N-terminal acetylation of serine normally found in the wild-type protein (29, 30). Thus, acetylation of the major histone H2A is quite different from acetylation of H2A.Z: it is not essential, even though the protein itself is essential and constitutes ∼80% of the total H2A. We also found that the N-terminal tail of H2A can replace the H2A.Z N-terminal tail and that the nonessential acetylation of the major H2A N-terminal tail can provide modulation of the charge patch on the H2A.Z N-terminal tail, which is essential for H2A.Z function. We conclude that when H2A.Z has a highly positively charged tail, it is essential that at least one of the positive charges of the tail can be neutralized in vivo by acetylation. However, this essential function of acetylation depends on properties of the H2A molecule that are independent of those of the tail itself.
MATERIALS AND METHODS
Strains, culture, and conjugation. Table 1 lists the Tetrahymena thermophila strains used in this study. Strains CU428, CU427, and B2086 were kindly provided by P. J. Bruns (Cornell University). Histone H2A knockout heterokaryon strains G4A1F14A and G4B1G6A and all mutant strains were generated as described below. For studies of vegetative growth, Tetrahymena cells were grown in SPP medium containing 1% proteose peptone (1× SPP) (32). For conjugation, two strains of different mating types were washed, starved (16 to 24 h, 30°C), and mated in 10 mM Tris-HCl (pH 7.5) as described previously (4).
TABLE 1.
Strains used in this study
| Strain | Genotype (micronucleus) | Phenotype (macronucleus) |
|---|---|---|
| CU428.2 | HTA1/HTA1 CHX1/CHX1 mpr1-1/mpr1-1 | wt,c pm-s cy-s mp-s VII |
| CU427.4 | HTA1/HTA1 chx1-1/chx1-1 MPR1/MPR1 | wt, pm-s cy-s mp-s VI |
| B2086.1 | HTA1/HTA1 CHX1/CHX1 MPR1/MPR1 | wt, pm-s cy-s mp-s II |
| G115B5 | ΔHTA1/ΔHTA1aHTA2/HTA2 CHX1/CHX1 mpr1?/mpr1?b | wt, pm-s cy-s mp-r ? |
| G114B11 | ΔHTA1/ΔHTA1 HTA2/HTA2 CHX1/CHX1 mpr1?/mpr1? | wt, pm-s cy-s mp-r ? |
| G209C4 | HTA1/HTA1 ΔHTA2/ΔHTA2 CHX1/CHX1 mpr1?/mpr1? | wt, pm-s cy-s mp-r ? |
| G204F2 | HTA1/HTA1 ΔHTA2/ΔHTA2 CHX1/CHX1 mpr1?/mpr1? | wt, pm-s cy-s mp-r ? |
| G4A1F14A | ΔHTA1/ΔHTA1 ΔHTA2/ΔHTA2 CHX1/CHX1 mpr1?/mpr1? | wt, pm-s cy-s mp-r ? |
| G4B1G6A | ΔHTA1/ΔHTA1 ΔHTA2/ΔHTA2 CHX1/CHX1 mpr1?/mpr1? | wt, pm-s cy-s mp-r ? |
| HTA1-D1A01 | ΔHTA1/ΔHTA1 ΔHTA2/ΔHTA2 CHX1/CHX1 mpr1?/mpr1? | wt H2A.1 ΔH2A.2 pm-r cy-s mp-? ? |
| HTA1-D9B6 | ΔHTA1/ΔHTA1 ΔHTA2/ΔHTA2 CHX1/CHX1 mpr1?/mpr1? | H2A.1 RRRRR ΔH2A.2 pm-r cy-s mp-? ? |
| HTA1-D11B1 | ΔHTA1/ΔHTA1 ΔHTA2/ΔHTA2 CHX1/CHX1 mpr1?/mpr1? | H2A.1 ARRRRR ΔH2A.2 pm-r cy-s mp-? ? |
| HTA1-D18C1 | ΔHTA1/ΔHTA1 ΔHTA2/ΔHTA2 CHX1/CHX1 mpr1?/mpr1? | H2A.1 PRRRRR ΔH2A.2 pm-r cy-s mp-? ? |
| HTA1-D19B1 | ΔHTA1/ΔHTA1 ΔHTA2/ΔHTA2 CHX1/CHX1 mpr1?/mpr1? | H2A.1 VRRRRR ΔH2A.2 pm-r cy-s mp-? ? |
| HTA1-D26A | ΔHTA1/ΔHTA1 ΔHTA2/ΔHTA2 CHX1/CHX1 mpr1?/mpr1? | H2A.1 VKRRRR ΔH2A.2 pm-r cy-s mp-? ? |
| HTA1-D25C | ΔHTA1/ΔHTA1 ΔHTA2/ΔHTA2 CHX1/CHX1 mpr1?/mpr1? | H2A.1 VRKRRR ΔH2A.2 pm-r cy-s mp-? ? |
| HTA1-D23A1 | ΔHTA1/ΔHTA1 ΔHTA2/ΔHTA2 CHX1/CHX1 mpr1?/mpr1? | H2A.1 VRRKRR ΔH2A.2 pm-r cy-s mp-? ? |
| HTA1-D27A1 | ΔHTA1/ΔHTA1 ΔHTA2/ΔHTA2 CHX1/CHX1 mpr1?/mpr1? | H2A.1 VRRRKR ΔH2A.2 pm-r cy-s mp-? ? |
| HTA1-D24A1 | ΔHTA1/ΔHTA1 ΔHTA2/ΔHTA2 CHX1/CHX1 mpr1?/mpr1? | H2A.1 VRRRRK ΔH2A.2 pm-r cy-s mp-? ? |
| HTA3-T57E1 | ΔHTA3/ΔHTA3 CHX1/CHX1 mpr1?/mpr1? | H2A.Z(H2A.1)N pm-r cy-s mp-? ? |
| HTA3-T65E2 | ΔHTA3/ΔHTA3 CHX1/CHX1 mpr1?/mpr1? | H2A.Z(H2A.1RRRRR)N pm-r cy-s mp-? ? |
| HTA3-T90N | ΔHTA3/ΔHTA3 CHX1/CHX1 mpr1?/mpr1? | H2A.Z(H2A.1S1V+5R)N pm-r cy-s mp-? ? |
| HTA3-T89K | ΔHTA3/ΔHTA3 CHX1/CHX1 mpr1?/mpr1? | H2A.Z(H2A.1S1V+7R)N pm-r cy-s mp-? ? |
| HTA1-D35C | ΔHTA1/ΔHTA1 ΔHTA2/ΔHTA2 CHX1/CHX1 mpr1?/mpr1? | H2A.1(H2A.Z)N ΔH2A.2 pm-r cy-s mp-? ? |
| HTA1-D33A1 | ΔHTA1/ΔHTA1 ΔHTA2/ΔHTA2 CHX1/CHX1 mpr1?/mpr1? | H2A.1(H2A.Z 6K)N ΔH2A.2 pm-r cy-s mp-? ? |
| HTA1-D34B1 | ΔHTA1/ΔHTA1 ΔHTA2/ΔHTA2 CHX1/CHX1 mpr1?/mpr1? | H2A.1(H2A.Z 5K)N ΔH2A.2 pm-r cy-s mp-? ? |
The correct genetic nomenclature for ΔHTA1/ΔHTA1 is hta1-1::neo2/hta1-1::neo2 (3), but we use the abbreviation to conserve space.
?, genotype or mating type not determined.
wt, wild type.
Plasmid construction. The HTA1 gene knockout construct (pQR10B) is based on plasmid pXL53, a pBluescript KS(+) derivative, which contains a copy of the HTA1 coding sequence (49), 3.5 kb of 5′ flanking sequence, and 1.8 kb of 3′ flanking sequence. A 0.5-kb HindIII-HincII fragment, which included the whole coding sequence of the HTA1 gene, was removed and replaced with a 1.5-kb HindIII-SmaI fragment from p4T2-1, a pBluescript KS(+) derivative, which contains a copy of the neo2 gene cassette (31). The neo2 gene, controlled by a histone H4 gene (HHF1) promoter, is transcribed in the direction opposite to that of the wild-type HTA1 gene (Fig. 1A). The final fragment, HTA1::neo2, was released from pQR10B by digestion with KpnI and SacI.
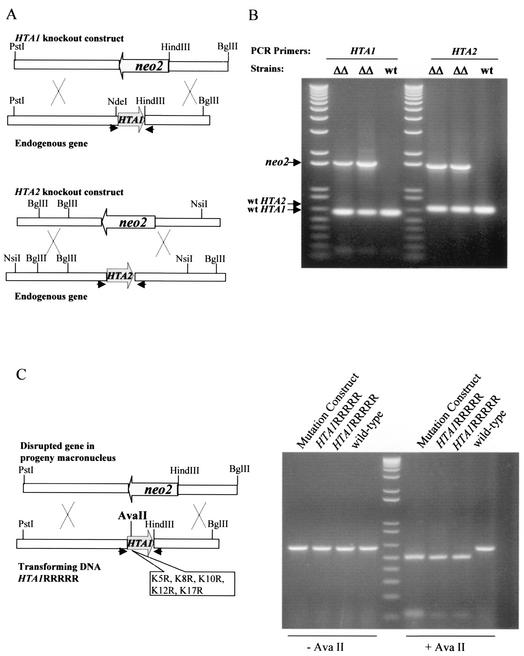
FIG. 1.
Creation of major H2A knockout heterokaryons. (A) Knockout constructs for HTA1 and HTA2. The entire coding region of either HTA1 or HTA2 was replaced by a neo2 cassette that confers resistance to paromomycin when expressed in macronuclei. The macronuclear genomic HTA1 gene is shown as a 5-kb PstI-BglII fragment containing the HTA1 coding region. The HTA1::neo2 knockout construct is shown as a neo2 cassette with 3.2-kb 5′ and 1.8-kb 3′ HTA1 flanking sequences. The macronuclear genomic HTA2 gene is shown as a 3.5-kb NsiI-BglII fragment containing the HTA2 coding region. The HTA2::neo2 knockout construct is shown as a neo2 cassette with 1.5-kb 5′ and 1.5-kb 3′ HTA2 flanking sequences. In both knockout constructs, the neo2 gene was transcribed in the direction opposite to that of the HTA gene. (B) PCR analysis of double-knockout heterokaryon progeny. The physical structure of the disrupted HTA1 and HTA2 genes in the micronuclei of the heterokaryons was examined by mating knockout heterokaryons with wild-type CU427 cells and selecting for retention of HTA1::neo2 and HTA2::neo2 by gradually increasing the paromomycin concentration to 2.0 mg/ml. When genomic DNA was analyzed by PCR using primers specific for the 5′ and 3′ flanking sequences of HTA1 or HTA2 (indicated by arrows in panel A), the presence of the disrupted HTA1 and HTA2 genes was demonstrable in progeny cell macronuclei (neo2 band), indicating that the parental heterokaryons had the disrupted gene in their micronuclei. As expected, the heterozygous macronuclei of these progeny cells also had wild-type copies of the HTA1 and HTA2 genes required to provide the essential H2A functions. ΔΔ, progeny of mating CU427 × double germ line knockout heterokaryon; wt, progeny of mating CU427 × CU428 wild-type strain. (C) Example of an experiment in which knockout heterokaryon progeny were rescued by transformation with a mutated HTA1 gene. The mutated HTA1RRRRR construct is shown as a 5.0-kb PstI-BglII fragment containing specific mutations that also introduce a new AvaII restriction enzyme site. After this mutated gene was transformed into knockout heterokaryons, PCR analysis was done to test for the presence of the mutated gene in the transformants. The HTA1 coding regions from wild-type control cells and HTA1RRRRR transformants were PCR amplified using the primers indicated by arrows in panel C. The PCR products, with or without AvaII digestion, were run on an agarose gel. The 600-bp PCR products from HTA1RRRRR mutant transformants contained the AvaII site and were cleaved into two shorter fragments, while the products from the wild-type strain were not cleaved.
For the HTA2 gene knockout construct (pQR17), a 1.5-kb fragment of the HTA2 5′ flanking sequence (from an NsiI site to the ATG start codon) was PCR amplified from Tetrahymena genomic DNA and inserted into the SmaI site of the 3′ polylinker region of p4T2-1. A 1.5-kb fragment of the HTA2 3′ flanking sequence (from the TGA stop codon to a BglII site) was PCR amplified from pXL46, a pBluescript KS(+) derivative that contains a copy of the HTA2 gene, and inserted into the 5′ polylinker region (between KpnI and EcoRV) of p4T2-1. The neo2 gene is transcribed in the direction opposite to that of the wild-type HTA2 gene (Fig. 1A). The final fragment, HTA2::neo2, was released from pQR17 by digestion with KpnI and SacI.
Site-directed mutagenesis. Oligonucleotide-directed, double-strand mutagenesis was performed as described previously (9) on pXL53, which contains a copy of the wild-type HTA1 gene. In some cases, a silent mutation was introduced to generate a restriction enzyme site used to monitor transformation. All mutated genes were sequenced with an automatic sequencing system (ABI Prism) and released by digestion with KpnI and SacI before being introduced into knockout heterokaryons.
Construction of HTA1 and HTA2 double-knockout heterokaryons and transformation of mutated genes. Using the DuPont Biolistic PDS-1000/He particle delivery system (Bio-Rad Laboratories) as described previously (16), the HTA1 and HTA2 genes encoding H2A.1 and H2A.2 were disrupted individually with HTA1::neo2 or HTA2::neo2 by biolistic transformation into early stage (2.5 h) conjugating CU428 and B2086 cells. Homozygous knockout heterokaryon strains of HTA1 (G115B5 and G114B11) and HTA2 (G209C4 and G204F2) with different mating types were created as described previously (35).
To create major histone H2A double germ line knockout heterokaryons, the homozygous HTA1::neo2 strain, G115B5, was mated with a homozygous HTA2::neo2 strain, i.e., G209C4 or G204F2. The heterozygous progeny were then mated to a B∗VI strain (56) as described previously (90) to obtain strains with homozygous HTA1::neo2 and HTA2::neo2 in the micronucleus. Two strains (G4A1F14A and G4B1G6A) with different mating types were created. These strains contain disrupted HTA1 and HTA2 genes in their micronuclei and wild-type genes in their macronuclei. When these paromomycin-sensitive heterokaryons conjugate, the old paromomycin-sensitive macronuclei are replaced by new ones produced by meiosis, fertilization, and mitotic division of the micronuclei of the cells. Consequently, the drug resistance genes that disrupt the HTA1 and HTA2 genes are expressed in the new macronuclei, allowing simple drug selection for successful mating. However, because major histone H2A.1 and H2A.2 together are essential in Tetrahymena (50) and the new macronucleus contains only disrupted copies of both genes, the progeny from this mating die unless they are transformed during mating with an HTA1 gene that functions well enough to support growth.
Successful creation of germ line knockout heterokaryons of the HTA1 and HTA2 genes was demonstrated by showing that no viable progeny were obtained when double-knockout heterokaryons of two different mating types were mated and that progeny were able to be rescued by transformation with a wild-type copy of HTA1. In addition, the physical structure of the disrupted HTA gene in the micronucleus of the heterokaryons was examined by mating knockout heterokaryons with wild-type CU427 cells and selecting for retention of HTA1::neo2 and HTA2::neo2 by increasing the paromomycin concentration to 2.0 mg/ml. When genomic DNA was analyzed by PCR using primers specific for the 5′ and 3′ flanking sequences of HTA1 or HTA2, the presence of disrupted genes (indicated by the presence of the 1.5-kb neo2 cassette) was demonstrable in progeny cell macronuclei, indicating that the parental heterokaryons have the disrupted gene in their micronuclei. As expected, the heterozygous macronuclei of these progeny cells also have wild-type copies of the HTA1 and/or HTA2 gene as required to provide the essential major H2A functions (Fig. 1B).
Because matings between two knockout heterokaryons fail to produce viable offspring and their progeny can be rescued by either a wild-type or a nonlethal mutated version of the HTA1 gene, these strains greatly facilitate systematic mutagenesis studies on major histone H2A modification sites, as illustrated in Fig. 1C. A mutated form of the HTA1 gene containing five arginine replacements at its N-terminal tail (see Results for details) was introduced into mating G4A1F14A and G4B1G6A knockout heterokaryons at late stages of conjugation (24 h) by biolistic transformation (16), and progeny were selected with paromomycin at 120 μg/ml. Viable progeny were obtained, indicating that the mutation is nonlethal. When the HTA1 coding region of the progeny is PCR amplified using primers specific for the HTA1 gene, the mutated and newly introduced HTA1 gene is easily differentiated from the wild type because the mutated gene contains an AvaII restriction enzyme site such that only the PCR product from the desired mutants is cleaved by this enzyme. Finally, the genotypes of all mutants were confirmed by sequencing the PCR products from genomic DNA of the transformed progeny.
Growth analysis.
Specific mutant strains, along with a strain rescued with the wild-type HTA1, were used in vegetative growth assays as described previously (67). Cells from each strain were inoculated into 50 ml of 1× SPP medium at starting densities of 1 × 104 cells/ml. Cultures were grown at 30°C with vigorous shaking, and samples (100 μl) were counted at frequent intervals with a ZB1 Coulter counter (Coulter Electronics, Inc.). Growth data were plotted using Cricket Graph III software (Computer Associates). Doubling times were calculated using the linear portion of the logarithmic growth curves.
Nuclear isolation and histone extraction. Rescued strains were grown to log phase (cell density, 2 × 105 cells/ml), and macronuclei were isolated by the method of Gorovsky et al. (32). Histones were extracted from macronuclei with 0.4 N H2SO4 (5) and precipitated with 20% trichloroacetic acid.
Acid-urea polyacrylamide gel electrophoresis and immunoblotting. Nuclear histones (25 μg) from mutants and wild-type HTA1 rescued strains, with or without pretreatment with λ protein phosphatase (New England Biolabs, Inc.) at 10 U/μl for 5 h at 30°C, were separated on long acid-urea polyacrylamide slab gels (15% acrylamide, 6 M urea, 5% acetic acid) as described previously (6) and transferred onto an Immobilon-P membrane (Millipore). After blocking in 5% nonfat milk, anti-H2A (1:5,000) or anti-hv1 (1:10,000) (68) was added and the blot was incubated overnight at 4°C. A 1:100,000 dilution of horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (Sigma) was used as secondary antibody. Blots were developed using the ECL Western blotting detection kit (NEN) according to the manufacturer's instructions.
RESULTS
Acetylation occurs on at least three of the lysines in the N-terminal tail.
There are five lysine residues (5, 8, 10, 12, and 17) in the T. thermophila H2A.1 N-terminal tail (49). To identify which ones were acetylated, we mutated them, singly or in combination, to arginine. This conserves the net positive charge of lysine, but arginine cannot be neutralized by acetylation (53). We then used the mutated genes to rescue the progeny of a mating between two H2A knockout heterokaryon strains.
To determine the acetylation status of H2A.1, nuclear histones from strains rescued with wild-type or mutated HTA1 genes were separated on acid-urea gels, which separate histones by both molecular weight and charge. Gels were then immunoblotted and stained with a highly specific antibody for Tetrahymena H2A to differentiate H2A.1 from any other comigrating Tetrahymena histones. Tetrahymena H2A is modified by both acetylation and phosphorylation (7), both of which alter the charge and therefore produce differences in mobility in this gel system. To eliminate the effects of phosphorylation on heterogeneity, histones were pretreated with λ protein phosphatase (33). This assay was used to characterize the acetylation status of all viable H2A.1 mutants.
If all lysines in the wild-type H2A.1 N-terminal tail can be modified by acetylation in vivo, up to six separable, phosphatase-resistant isotypes might be expected in wild-type cells. Strains rescued by the wild-type gene yielded five isoforms after treatment with phosphatase (Fig. 2B, lane 2), likely representing unmodified H2A.1 (bottom band) and isoforms containing one to four acetyl groups.
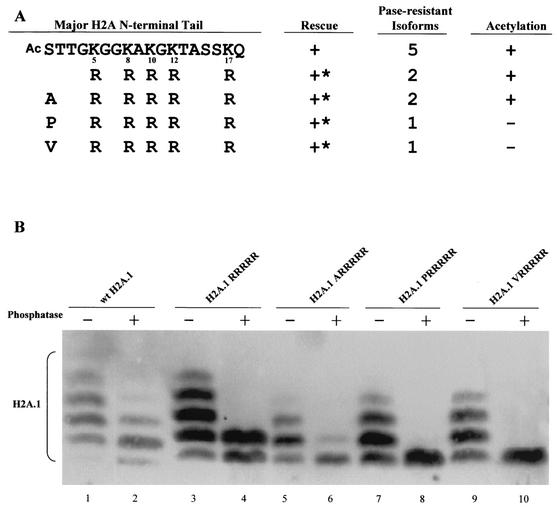
FIG. 2.
Major H2A acetylation is not essential. (A) Tetrahymena can survive with all five lysines changed to arginines at the major H2A N-terminal tail. To abolish N-terminal acetylation, the first residue (serine) in the 5R mutated gene (RRRRR) was changed to alanine (ARRRRR), proline (PRRRRR), or valine (VRRRRR). All mutations generated viable transformants. ∗, mutants with severe phenotypes, including slow growth, variable size, and irregular surfaces. (B) Macronuclear histones from 5R transformants containing different N-terminal residues were analyzed on acid-urea gels. Histones from viable transformants were separated, blotted, and detected with an antiserum specific for H2A. H2A.1RRRRR and H2A.1ARRRRR show two phosphatase-resistant isoforms, indicating that N-terminal acetylation still occurs in these mutants (see text for details). H2A.1PRRRRR and H2A.1VRRRRR both show only one phosphatase-resistant isoform, indicating that these mutations abolish all acetylation at the H2A N-terminal tail.
Since the exact acetylation sites were unknown, we began by changing all five available lysine residues to arginines (referred to as the RRRRR mutation). Tetrahymena survives with this mutated form of H2A.1 as its only source for major histone H2A, but the mutant strain grew slowly at 30°C (Fig. 2A). Surprisingly, histones extracted from mutant cells with all five lysines changed to arginines still have two phosphatase-resistant isoforms (Fig. 2B, lane 4). These results indicated that while at least three of the five internal lysines were acetylated, there was an additional, charge-altering modification in addition to phosphorylation and acetylation of lysine.
Neither N-terminal nor lysine acetylation of the H2A N-terminal tail is essential.
The initial residue of the histone H2A N termini of many organisms (10, 38), including Tetrahymena (29, 30), can be blocked by N-terminal acetylation, a conserved process that adds an acetyl group to the first amino acid of many histone and nonhistone proteins (57). Since N-terminal acetylation abolishes one positive charge at the major H2A N terminus, it affects the mobility of major H2A.1 in the mutant, and if this process occurs for some but not all H2A.1 molecules, it might account for the electrophoretic heterogeneity observed in the RRRRR mutation. Because not all N-terminal residues can be acetylated, we attempted to remove any effect of N-terminal acetylation in the RRRRR mutation by further mutating the initial serine residue of H2A.1 to alanine, proline, or valine, residues which still allow removal of the initiator methionine (59). These mutation constructs (H2A.1ARRRRR, H2A.1PRRRRR, and H2A.1VRRRRR) gave viable transformants, all of which had slow-growth phenotypes (Fig. 2A). Nuclear histones from these three transformants were then extracted and separated on an acid-urea gel. Consistent with the observation that alanines following the initiator methionine are frequently acetylated (59), H2A.1ARRRRR still had two phosphatase-resistant isoforms, although the amount of the slower-migrating isoform was greatly reduced. Mutants H2A.1PRRRRR and H2A.1VRRRRR each contained only a single isoform after phosphatase treatment (Fig. 2B, lanes 6, 8, and 10), consistent with observations that these residues are less likely to be acetylated after the initiator methionine is removed. These data argue that the N-terminal serine of H2A.1 is normally acetylated and that acetylation of the major H2A, including its N-terminal acetylation, is not essential for viability in Tetrahymena.
All lysines in the H2A N-terminal tail can be acetylated.
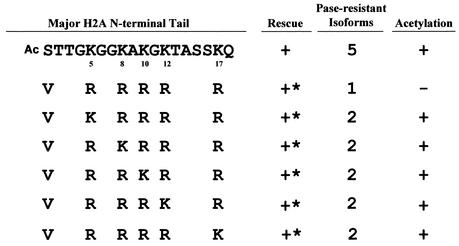
To map the exact acetylation sites of Tetrahymena histone H2A, a series of mutation constructs were generated from the H2A.1VRRRRR mutation, in which a single lysine residue replaced an arginine at each of the five positions. All of these mutations produced viable transformants with a slow-growth phenotype (Fig. 3). Acid-urea gel analyses showed that all of these mutants had two phosphatase-resistant isoforms: an unmodified H2A.1 and a mono-acetylated H2A.1 (data not shown). These results indicate that each of the five lysines in the N-terminal tail of Tetrahymena H2A can be acetylated.
FIG. 3.
All lysines in the N-terminal tail of H2A can be acetylated. A series of mutation constructs were made starting from the H2A.1 VRRRRR mutation, leaving a single lysine at different positions. All mutations containing a single lysine generated viable transformants with slow-growth phenotypes. ∗, mutants with severe phenotypes, including slow growth, variable size, and irregular surfaces.
Given that there are six acetylatable sites (those of the N-terminal residue and five lysines) in the N-terminal tail of wild-type H2A, it may seem surprising that we have only observed 5 H2A isoforms on acid-urea gels after dephosphorylation (Fig. 2B). However, prediction of the number of observed isoforms from the number of acetylatable sites is not simple. First, the most highly acetylated isoforms are invariably faint and they are slightly variable in appearance. This is likely due to the fact that even small amounts of deacetylation during cell pelleting, nuclear isolation, or histone extraction can cause these isoforms to disappear and contribute to slower-migrating, less-acetylated isoforms. Another possibility is that while six acetylation sites can be identified by mutational analysis, few (if any) molecules in vivo are simultaneously acetylated at all six sites.
The major H2A N-terminal tail can replace the function of the H2A.Z variant N-terminal tail.
The region encoding the entire N-terminal tail of HTA1, including all of the acetylatable lysines, was used to replace the region encoding the N-terminal tail of H2A.Z in the HTA3 gene. This chimeric gene, encoding an H2A.Z variant core with an H2A.1 N-terminal tail, was used to rescue the progeny of mating H2A.Z germ line knockout heterokaryons (62). Viable transformants were obtained (Fig. 4A). Growth of these rescued strains was indistinguishable from that of wild-type cells. Using hv1, a specific antibody to H2A.Z, the modification state of this chimeric protein was then determined by immunoblotting the nuclear histones. In wild-type Tetrahymena cells, H2A.Z shows five or six phosphatase-resistant isoforms (Fig. 4B, lane 2). The chimeric protein H2A.Z(H2A.1)N shows four or five phosphatase-resistant isoforms (Fig. 4B, lane 4), a pattern similar to that of wild-type H2A.1. In addition, the mobility of H2A.Z(H2A.1)N isoforms on acid-urea gels is intermediate between that of wild-type H2A.1 and that of wild-type H2A.Z. The fastest migrating isoform, presumably the unmodified H2A.Z(H2A.1)N, which has six positive charges in the N-terminal tail (including one positive charge of the N-terminal residue), has mobility similar to that of wild-type H2A.Z with three acetyl groups, whose tail also has a net positive charge of +6. These data demonstrate that the N-terminal tail of H2A.1 can provide the function of the H2A.Z variant N-terminal tail.
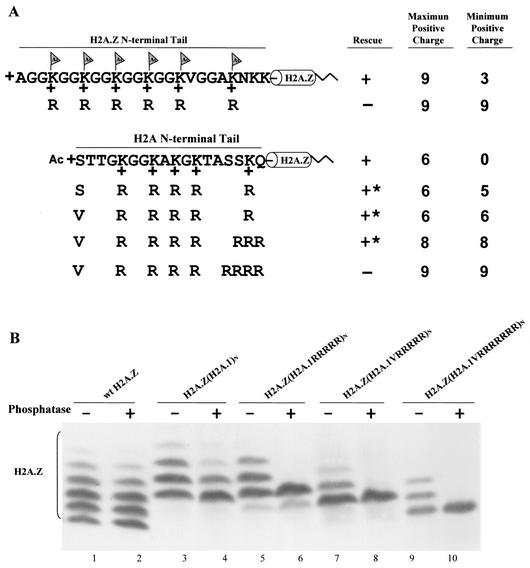
FIG. 4.
The major H2A N-terminal tail can replace the essential function of the H2A.Z variant N-terminal tail. (A) Constructs in which the N-terminal tail of H2A.Z, whose acetylation has an essential function, was replaced either by a wild-type H2A.1 tail or by mutated tails containing increasing numbers of arginine replacements were transformed into H2A.Z knockout heterokaryons. Transformants with the chimeric gene containing a wild-type H2A.1 N-terminal tail on H2A.Z grow normally. Tetrahymena cells can also survive with arginine replacement mutations which increase the total positive charge of the N-terminal tail to +8 but cannot survive when the total positive charge is increased to +9. ∗, mutants with severe phenotypes, including slow growth, variable size, and irregular surfaces. (B) Macronuclear histones from viable transformants were extracted, blotted, and detected with a specific antibody to H2A.Z. The banding patterns of the chimeric proteins (H2A.1 tails on H2A.Z) caused by acetylation are similar to those of the major H2A.1, with three or four phosphatase-resistant isoforms for H2A.Z(H2A.1)N and two for H2A.Z(H2A.1RRRRR)N. H2A.Z(H2A.1VRRRRR)N and H2A.Z(H2A.1VRRRRRRR)N both show a single phosphatase-resistant isoform.
The H2A.Z(H2A.1)N chimeric protein exhibits the essential charge patch properties of wild-type H2A.Z.
Our previous studies had demonstrated that the N-terminal tail of H2A.Z in which all of the acetylatable lysines had been replaced by arginines cannot support growth in Tetrahymena. However, mutants with even a single lysine, or with lysines replaced by glutamine or with the N-terminal tail deleted, were viable (62). These observations were used to argue that the H2A.Z tail functioned as a charge patch in which neutralization or removal of at least one of the lysine positive charges was required for the creation of a viable gene. The net positive charge of the unmodified H2A.Z N-terminal tail is +9. Since a tail containing five arginines and one acetylatable lysine is viable, this suggests that an H2A.Z N terminus that can be modified to a net charge of +8 in vivo is viable.
Tetrahymena can survive with all five lysines at the N-terminal tail of the chimeric gene H2A.Z(H2A.1)N changed to arginines (Fig. 4A). The acid-urea gel analysis shows that H2A.Z(H2A.1RRRRR)N has two phosphatase-resistant isoforms (Fig. 4B, lane 6), indicating that the chimeric protein is modified by N-terminal acetylation to produce a blocked N-terminal tail with a net positive charge of +5 and a unblocked, completely unacetylated N-terminal tail with the maximum possible positive charge of +6. In similarity to the result seen with H2A.1, changing the first serine residue in this chimeric protein to valine eliminates N-terminal acetylation (Fig. 4B, lane 8), creating a cell that is viable and whose N-terminal tail has a total charge of +6. To determine whether the H2A.1 N-terminal tail attached to H2A.Z also functions as a charge patch, we introduced additional arginine residues into the chimeric gene by changing the last two or three amino acids of the H2A.1 N-terminal tail to arginines to increase its total of positive charges. Tetrahymena cells can survive with a total of eight positive charges in the tail of the chimeric protein but not with nine (Fig. 4A). Thus, just as in the case of the H2A.Z N-terminal tail itself, the H2A.1 N-terminal tail attached to H2A.Z cannot function with nine positive charges but produces viable progeny with eight. This argues that the H2A.1 N-terminal tail also can function as an essential charge patch when it replaces the H2A.Z N-terminal tail.
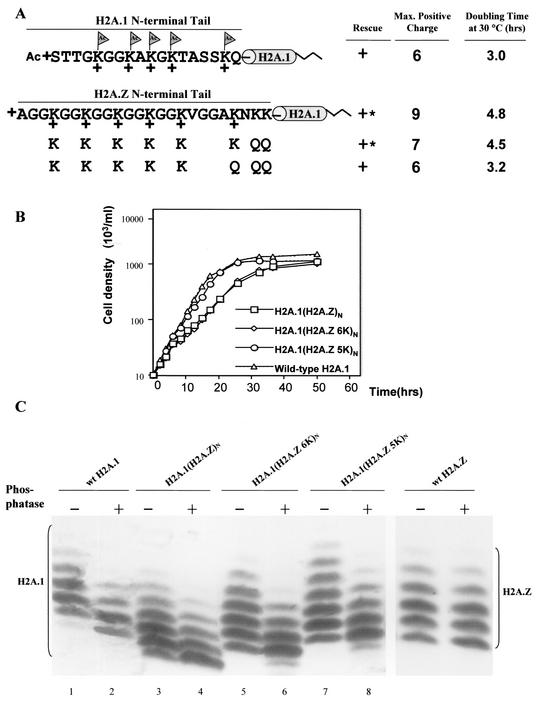
The H2A.Z N-terminal tail can provide the function of the major histone H2A N-terminal tail.
The entire N-terminal tail of H2A.Z, including six acetylation sites and two nonacetylatable lysines, was used to replace the H2A.1 N-terminal tail. This chimeric gene, H2A.1(H2A.Z)N, was introduced into the progeny of major H2A double germ line knockout heterokaryons. Viable H2A.1(H2A.Z)N transformants were obtained but grew slowly, as seen in a comparison of the growth curve with that of a strain rescued with the wild-type HTA1 gene (Fig. 5B). Cells rescued with a gene [H2A.1(H2A.Z 6K)N] in which the two nonacetylatable lysines in the H2A.Z N-terminal tail were changed to glutamines (reducing the maximum positive charge in vivo to +7) still grew slowly (Fig. 5B). When a third, acetylatable residue was changed to glutamine, the H2A.1(H2A.Z 5K)N transformants, in which the N-terminal tail of the chimeric protein had a maximum total of six positive charges, exhibited growth rates that were indistinguishable from those of the wild-type cells (Fig. 5B). Because the maximum positive charge of the H2A.1 tail itself is +6, these data suggest that H2A N-terminal tail has an optimum maximum number of positive charges that can be well tolerated, even if most molecules can be acetylated and have a lower total number of positive charges. This suggests that the H2A tail is carrying out two distinct nonessential roles, one requiring an unacetylated tail with a charge of +6 and the other requiring acetylation.
FIG. 5.
The H2A.Z N-terminal tail can also replace the function of the major H2A N-terminal tail. (A) The N-terminal tail of H2A.1 was replaced either by the wild-type H2A.Z N-terminal tail or by tails with different numbers of glutamine replacements at nonacetylatable residues that reduce the positive charges of the N-terminal tail without eliminating acetylation sites. The wild-type H2A.Z N-terminal tail on H2A.1 yields transformants with a slow-growth phenotype, while glutamine replacement mutations, which decrease the maximum positive charge of the chimeric protein's N-terminal tail to +6, generate transformants whose doubling time at 30°C is indistinguishable from that of wild-type cells. ∗, mutants with severe phenotypes, including slow growth, variable size, and irregular surfaces. (B) Mutant H2A.1(H2A.Z)N, H2A.1(H2A.Z 6K)N, and H2A.1(H2A.Z 5K)N, as well as strains rescued with the wild-type gene, were grown in 1× SPP medium at 30°C. Cell densities were measured for up to 50 h and plotted on a log scale. Doubling times in hours are listed in Fig. 5A. (C) Macronuclear histones from the mutants were extracted, blotted, and detected with a specific antibody to major H2A. While wild-type H2A.1 has four or five phosphatase-resistant isoforms, the chimeric protein, H2A.1(H2A.Z)N, shows five or six phosphatase-resistant isoforms, a pattern similar to that of wild-type H2A.Z. H2A.1(H2A.Z 6K)N and H2A.1(H2A.Z 5K)N show patterns similar to that of H2A.1(H2A.Z)N, except for small mobility differences likely caused by the extra glutamine mutations that abolish two or three positive charges on the N-terminal tail.
The acid-urea gel analysis shows that the chimeric proteins H2A.1(H2A.Z)N, H2A.1(H2A.Z 6K)N, and H2A.1(H2A.Z 5K)N have five or six phosphatase-resistant isoforms, a pattern similar to that of wild-type H2A.Z (Fig. 5C, lanes 4, 6, and 8), indicating that the acetylation state is determined mainly by the N-terminal tail of the histones. The lack of apparent difference in the number of phosphatase-resistant isoforms between mutants H2A.1(H2A.Z 6K)N andH2A.1(H2A.Z 5K)N, although the latter has one acetylatable lysine changed into glutamine, can be explained by the observation that despite the possibility that all six lysine residues serve as acetylation sites, H2A.Z isolated from the wild-type strain does not show an isoform with six acetyl groups, likely reflecting the dynamic balance between the acetylation and deacetylation processes. The differences in the mobility of the chimeric protein on the acid-urea gel among the mutants are caused by the molecular difference between lysine and glutamine.
DISCUSSION
A number of important conclusions can be drawn from this work. (i) Although the major histone H2A is essential and constitutes ∼80% of the total H2A, its acetylation is not essential. This conclusion applies to both cotranslational N-terminal acetylation and internal acetylation of lysine residues, as shown by the observation that both the VRRRRR and the PRRRRR versions of the major H2A showed no detectable electrophoretic heterogeneity and were viable. (ii) The N-terminal tail of histone H2A can replace the function of the H2A.Z N-terminal tail and vice versa. (iii) Although it differs in sequence from the H2A.Z tail, the H2A.1 N-terminal tail can be made to function as an essential charge patch when linked to H2A.Z. (iv) Acetylation of the N-terminal residue has an effect similar to that of acetylation of internal lysine residues in modulating the function of the H2A tail as a charge patch. (v) The acetylation patterns of H2A tails are largely determined by the tails themselves, but the effects of acetylation are determined by the nature of the remainder of the histone. (vi) There appears to be a specific maximum charge that can be tolerated by each type of H2A. (vii) The function of acetylation in regulating the charge of the N-terminal tails of H2A is highly sensitive to changes of even a single charge.
Tetrahymena histone H2As are blocked at the N-terminal serine residue by an α-N-acetyl (29, 30). N-terminal blocking of histones and other proteins through acetylation is common in various organisms (57), and N-terminal acetylation of actin has been shown to strengthen the weak interaction between actin and myosin (1). To our knowledge, the finding that cotranslational N-terminal acetylation can affect histone function in a manner similar to that of internal, posttranslational acetylation has not been previously described. Since one of the important mechanisms by which acetylation affects histone function is that of modulating a charge patch (62; this study), the effect of N-terminal acetylation on the positive charge of the histone H2A N terminus must be considered a possible regulatory mechanism, especially in cases (as in that of Tetrahymena H2A) in which not all of the molecules contain this modification.
Interestingly, the extent of acetylation of the N terminus of H2A in Tetrahymena depends not only on the terminal residue but also on the sequence to which it is attached. The occurrence of N-terminal acetylation is sequence restricted and can often be correctly predicted by protein primary sequence (27). Three N-terminal acetyltransferases have been cloned in S. cerevisiae, and the conserved recognition sequences for each enzyme were reported (59). We initially changed the first serine of histone H2A.1 to alanine because alanine is structurally similar to serine and because the N-terminal alanine residue on H2A.Z is not blocked by acetylation (8, 62). Cells containing this mutation, H2A.1ARRRRR, still contained two phosphatase-resistant isoforms, although the amount of the slower-migrating, acetylated form is greatly reduced compared to that of H2A.1SRRRRR. However, when we changed the first serine to proline (P) or valine (V), neither of which is found in the conserved recognition sequence for N-terminal acetyltransferases, H2A.1PRRRRR and H2A.1VRRRRR mutations produced viable transformants in which the mutated H2A.1 showed only one phosphatase-resistant isoform representing unmodified H2A.1. These results demonstrate that major histone H2A acetylation, including acetylation of the N-terminal residue, is not essential in Tetrahymena.
We mapped the acetylation sites on H2A.1 by changing all but one wild-type lysine in the N-terminal tail to arginine and analyzing the H2A modification status in each mutant. We found that all five lysines in the N-terminal tail can be acetylated. However, they are not modified to equal extents. Lysines at the first three positions, K5, K8, and K10, are heavily acetylated, since mutants with a single lysine at these positions contain much more of the mono-acetylated isoform than other mutants (data not shown). This approach cannot rule out the possibility that some of these lysines are only acetylated when other sites are not available and are not normally acetylated in wild-type cells.
The studies described here strongly support the hypothesis that the essential function of acetylation of the H2A.Z tail acts by modulating the charge of the tail. We reported previously that Tetrahymena H2A.Z acetylation modulated an essential charge patch (62). Tetrahymena cannot survive with all six acetylatable lysines on H2A.Z changed into arginines, which produces a tail whose charge cannot be reduced from +9 (resulting from the presence of the amino-terminal α-amino group, six nonacetylatable arginines at the acetylation sites, and two nonacetylatable lysines). However, viable transformants can be obtained simply by reducing the charge to +8 by replacing a neutral residue with a negatively charged residue at other positions in the tail or by replacing any one of the arginine residues with glutamine (62). Remarkably, this same sensitivity to charge can be demonstrated by replacing the N-terminal tail of H2A.Z with an N-terminal tail of H2A.1 to which positive charges were added. We were able to demonstrate that the maximum number of nonneutralizable positive charges allowed on the N-terminal tail, including the one on the N-terminal residue, is eight; mutants in which the chimeric protein contained nine positive charges in the H2A.1 tail were not viable.
Surprisingly, while the extent of acetylation of the H2A tail was determined largely by the nature of the tail, the effect of acetylation on viability depended on the nature of the rest of the H2A molecule. One possibility is that acetylation of the tail affects the structure of major H2A and H2A.Z differently or acts synergistically with properties that differentiate the two types of H2A. Recent studies comparing the crystal structures of nucleosomes containing the major H2A with those containing the H2A.Z variant (71) provide some basis for this hypothesis. The region of H2A.Z essential for viability in Drosophila is at the C-terminal tail that is exposed on the surface of the nucleosome and is part of the docking domain involved in maintaining the interactions between the H3/H4 tetramer and H2A/H2B dimer within the nucleosome (18, 37). The H2A.Z nucleosomes have an altered surface that includes a metal ion, which may lead to changes in higher-order structure or in the association between H2A.Z and other nuclear proteins (71). It also has been shown that the presence of H2A.Z variants and tail acetylation of histones can each affect the hydrodynamic properties of nucleosomal arrays, offering the possibility that these two processes cooperate to establish unique chromatin domains (26). An alternative explanation of how the effects of acetylation can be determined by nonacetylated portions of the H2A molecule is based on the observations that nucleosomes containing the major H2A and those containing H2A.Z associate with different DNA sequences in chromatin (47, 65). In this scenario, acetylation can be viewed as a simple switch that is able to alter the properties of both major H2A and H2A.Z nucleosomes similarly but whose effect depends on the specific sequences with which each type of H2A was associated.
The results described here can reconcile our previous study demonstrating the essential function of a single acetylation site in the H2A.Z N-terminal tail (62) with studies of Drosophila showing that the only region of Drosophila H2A which cannot provide the essential developmental function of H2A.Z resides in the C-terminal α-helix (18). The Drosophila results are completely consistent with our finding that acetylation sites on the major H2A can replace those on H2A.Z when associated with the H2A.Z C-terminal helix in a fusion protein.
Acknowledgments
This work was supported by grant GM21793 from the National Institutes of Health to M.A.G.
We thank Josephine Bowen for technical assistance and for critically reading the manuscript.
REFERENCES
- 1.Abe, A., K. Saeki, T. Yasunaga, and T. Wakabayashi. 2000. Acetylation at the N-terminus of actin strengthens weak interaction between actin and myosin. Biochem. Biophys. Res. Commun. 268:14-19. [DOI] [PubMed] [Google Scholar]
- 2.Adam, M., F. Robert, M. Larochelle, and L. Gaudreau. 2001. H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol. Cell. Biol. 21:6270-6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, S. L., M. I. Altschuler, P. J. Bruns, J. Cohen, F. P. Doerder, J. Gaertig, M. Gorovsky, E. Orias, A. Turkewitz, and The Seventh International Meeting on Ciliate Molecular Biology Genetics Nomenclature. 1998. Proposed genetic nomenclature rules for Tetrahymena thermophila, Paramecium primaurelia and Paramecium tetraurelia. Genetics 149:459-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allis, C. D., and D. K. Dennison. 1982. Identification and purification of young macronuclear anlagen from conjugating cells of Tetrahymena thermophila. Dev. Biol. 93:519-533. [DOI] [PubMed] [Google Scholar]
- 5.Allis, C. D., C. D. C. Glover, and M. A. Gorovsky. 1979. Micronuclei of Tetrahymena contain two types of histone H3. Proc. Natl. Acad. Sci. USA 76:4857-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allis, C. D., C. V. D. Glover, J. K. Bowen, and M. A. Gorovsky. 1980. Histone variants specific to the transcriptionally active, amitotically dividing macronucleus of the unicellular eukaryote, Tetrahymena thermophila. Cell 20:609-617. [DOI] [PubMed] [Google Scholar]
- 7.Allis, C. D., and M. A. Gorovsky. 1981. Histone phosphorylation in macro- and micronuclei of Tetrahymena thermophila. Biochemistry 20:3828-3833. [DOI] [PubMed] [Google Scholar]
- 8.Allis, C. D., R. Richman, M. A. Gorovsky, Y. S. Ziegler, B. Touchstone, W. A. Bradley, and R. G. Cook. 1986. hv1 is an evolutionarily conserved H2A variant that is preferentially associated with active genes. J. Biol. Chem. 261:1941-1948. [PubMed] [Google Scholar]
- 9.Andrews, C. A., and S. A. Lesley. 1998. Selection strategy for site-directed mutagenesis based on altered beta-lactamase specificity. BioTechniques 24:972-978. [DOI] [PubMed] [Google Scholar]
- 10.Ball, D. J., C. A. Slaughter, P. Hensley, and W. T. Garrard. 1983. Amino acid sequence of the N-terminal domain of calf thymus histone H2A.Z. FEBS Lett. 154:166-170. [DOI] [PubMed] [Google Scholar]
- 11.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 12.Berger, S. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 13.Brownell, J. E., J. X. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843-851. [DOI] [PubMed] [Google Scholar]
- 14.Callard, D., and L. Mazzolini. 1997. Identification of proliferation-induced genes in Arabidopsis thaliana. Characterization of a new member of the highly evolutionarily conserved histone H2A.F/Z variant subfamily. Plant Physiol. 115:1385-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carr, A. M., S. M. Dorrington, J. Hindley, G. A. Phear, S. J. Aves, and P. Nurse. 1994. Analysis of a histone H2A variant from fission yeast: evidence for a role in chromosome stability. Mol. Gen. Genet. 245:628-635. [DOI] [PubMed] [Google Scholar]
- 16.Cassidy-Hanley, D., J. Bowen, J. Lee, E. S. Cole, L. A. VerPlank, J. Gaertig, M. A. Gorovsky, and P. J. Bruns. 1997. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics 146:135-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheung, P., K. G. Tanner, W. L. Cheung, P. Sassone-Corsi, J. M. Denu, and C. D. Allis. 2000. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell 5:905-915. [DOI] [PubMed] [Google Scholar]
- 18.Clarkson, M. J., J. R. E. Wells, F. Gibson, R. Saint, and D. J. Tremethick. 1999. Regions of variant histone His2AvD required for Drosophila development. Nature 399:694-697. [DOI] [PubMed] [Google Scholar]
- 19.Dalton, S., A. J. Robins, R. P. Harvey, and J. R. Wells. 1989. Transcription from the intron-containing chicken histone H2A.F gene is not S-phase regulated. Nucleic Acids Res. 17:1745-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhalluin, C., J. E. Carlson, L. Zeng, C. He, A. K. Aggarwal, and M. M. Zhou. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491-496. [DOI] [PubMed] [Google Scholar]
- 21.Dou, Y., and M. A. Gorovsky. 2002. Regulation of transcription by H1 phosphorylation in Tetrahymena is position independent and requires clustered sites. Proc. Natl. Acad. Sci. USA 99:6142-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dou, Y., and M. A. Gorovsky. 2000. Phosphorylation of linker histone H1 regulates gene expression in vivo by creating a charge patch. Mol. Cell 6:225-231. [DOI] [PubMed] [Google Scholar]
- 23.Edmondson, D. G., M. M. Smith, and S. Y. Roth. 1996. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 10:1247-1259. [DOI] [PubMed] [Google Scholar]
- 24.Ernst, S. G., H. Miller, C. A. Brenner, C. Nocente-McGrath, S. Francis, and R. McIsaac. 1987. Characterization of a cDNA clone coding for a sea urchin histone H2A variant related to the H2A.F/Z histone protein in vertebrates. Nucleic Acids Res. 15:4629-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faast, R., V. Thonglairoam, T. C. Schulz, J. Beall, J. R. E. Wells, H. Taylor, K. Matthaei, P. D. Rathjen, D. J. Tremethick, and I. Lyons. 2001. Histone variant H2A.Z is required for early mammalian development. Curr. Biol. 11:1183-1187. [DOI] [PubMed] [Google Scholar]
- 26.Fan, J. Y., F. Gordon, K. Luger, J. C. Hansen, and D. J. Tremethick. 2002. The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat. Struct. Biol. 9:172-176. [DOI] [PubMed] [Google Scholar]
- 27.Flinta, C., B. Persson, H. Jornvall, and G. von Heijne. 1986. Sequence determinants of cytosolic N-terminal protein processing. Eur. J. Biochem. 154:193-196. [DOI] [PubMed] [Google Scholar]
- 28.Forsberg, E. C., and E. H. Bresnick. 2001. Histone acetylation beyond promoters: long-range acetylation patterns in the chromatin world. Bioessays 23:820-830. [DOI] [PubMed] [Google Scholar]
- 29.Fusauchi, Y., and K. Iwai. 1983. Tetrahymena histone H2A. Isolation and two variant sequences. J. Biochem. 93:1487-1497. [DOI] [PubMed] [Google Scholar]
- 30.Fusauchi, Y., and K. Iwai. 1984. Tetrahymena histone H2A. Acetylation in the N-terminal sequence and phosphorylation in the C-terminal sequence. J. Biochem. (Tokyo) 95:147-154. [DOI] [PubMed] [Google Scholar]
- 31.Gaertig, J., L. Gu, B. Hai, and M. A. Gorovsky. 1994. High frequency vector-mediated transformation and gene replacement in Tetrahymena. Nucleic Acids Res. 22:5391-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorovsky, M. A., M. C. Yao, J. B. Keevert, and G. L. Pleger. 1975. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 9:311-327. [DOI] [PubMed] [Google Scholar]
- 33.Goto, H., Y. Tomono, K. Ajiro, H. Kosako, M. Fujita, M. Sakurai, K. Okawa, A. Iwamatsu, T. Okigaki, T. Takahashi, and M. Inagaki. 1999. Identification of a novel phosphorylation site on histone H3 coupled with mitotic chromosome condensation. J. Biol. Chem. 274:25543-25549. [DOI] [PubMed] [Google Scholar]
- 34.Grant, P. A. 5April2001, posting date. A tale of histone modifications. Genome Biol. 2:0003.1-0003.6. [Online.] http://genomebiology.com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hai, B., and M. A. Gorovsky. 1997. Germ-line knockout heterokaryons of an essential α-tubulin gene enable high-frequency gene replacement and a test of gene transfer from somatic to germ-line in Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA 94:1310-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harvey, R. P., J. A. Whiting, L. S. Coles, P. A. Krieg, and J. R. Wells. 1983. H2A.F: an extremely variant histone H2A sequence expressed in the chicken embryo. Proc. Natl. Acad. Sci. USA 80:2819-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayes, J. J. 2002. Changing chromatin from the inside. Nat. Struct. Biol. 9:161-163. [DOI] [PubMed] [Google Scholar]
- 38.Isenberg, I. 1979. Histones. Annu. Rev. Biochem. 48:159-191. [DOI] [PubMed] [Google Scholar]
- 39.Jackson, J. D., V. T. Falciano, and M. A. Gorovsky. 1996. A likely histone H2A.F/Z variant in Saccharomyces cerevisiae. Trends Biochem. Sci. 21:466-467. [DOI] [PubMed] [Google Scholar]
- 40.Jackson, J. D., and M. A. Gorovsky. 2000. Histone H2A.Z has a conserved function that is distinct from that of the major H2A sequence variants. Nucleic Acids Res. 28:3811-3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jacobson, R. H., A. G. Ladurner, D. S. King, and R. Tjian. 2000. Structure and function of a human TAFII250 double bromodomain module. Science 288:1422-1425. [DOI] [PubMed] [Google Scholar]
- 42.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 43.Kolodrubetz, D., M. O. Rykowski, and M. Grunstein. 1982. Histone H2A subtypes associate interchangeably in vivo with histone H2B subtypes. Proc. Natl. Acad. Sci. USA 79:7814-7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kornberg, R., and Y. Lorch. 2002. Chromatin and transcription: where do we go from here? Curr. Opin. Genet. Dev. 12:249-251. [DOI] [PubMed] [Google Scholar]
- 45.Kruger, W., C. L. Peterson, A. Sil, C. Coburn, G. Arents, E. N. Moudrianakis, and I. Herskowitz. 1995. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 9:2770-2779. [DOI] [PubMed] [Google Scholar]
- 46.Kurumizaka, H., and A. P. Wolffe. 1997. Sin mutations of histone H3: influence on nucleosome core structure and function. Mol. Cell. Biol. 17:6953-6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leach, T. J., M. Mazzeo, H. L. Chotkowski, J. P. Madigan, M. G. Wotring, and R. L. Glaser. 2000. Histone H2A.Z is widely but nonrandomly distributed in chromosomes of Drosophila melanogaster. J. Biol. Chem. 275:23267-23272. [DOI] [PubMed] [Google Scholar]
- 48.Liu, X., J. Bowen, and M. A. Gorovsky. 1996. Either of the major H2A genes but not an evolutionarily conserved H2A.F/Z variant of Tetrahymena thermophila can function as the sole H2A gene in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2878-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu, X., and M. A. Gorovsky. 1996. Cloning and characterization of the major histone H2A genes completes the cloning and sequencing of known histone genes of Tetrahymena thermophila. Nucleic Acids Res. 24:3023-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu, X., B. Li, and M. A. Gorovsky. 1996. Essential and nonessential histone H2A variants in Tetrahymena thermophila. Mol. Cell. Biol. 16:4305-4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lo, W. S., R. C. Trievel, J. R. Rojas, L. Duggan, J. Y. Hsu, C. D. Allis, R. Marmorstein, and S. L. Berger. 2000. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell 5:917-926. [DOI] [PubMed] [Google Scholar]
- 52.Luger, K., and T. J. Richmond. 1998. The histone tails of the nucleosome. Curr. Opin. Genet. Dev. 8:140-146. [DOI] [PubMed] [Google Scholar]
- 53.Megee, P. C., B. A. Morgan, B. A. Mittman, and M. M. Smith. 1990. Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science 247:841-845. [DOI] [PubMed] [Google Scholar]
- 54.Ng, H. H., and A. Bird. 2000. Histone deacetylases: silencers for hire. Trends Biochem. Sci. 25:121-126. [DOI] [PubMed] [Google Scholar]
- 55.Nielsen, P. R., D. Nietlispach, H. R. Mott, J. Callaghan, A. Bannister, T. Kouzarides, A. G. Murzin, N. V. Murzina, and E. D. Laue. 2002. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416:103-107. [DOI] [PubMed] [Google Scholar]
- 56.Orias, E., and P. J. Bruns. 1976. Induction and isolation of mutants in Tetrahymena. Methods Cell Biol. 13:247-282. [PubMed] [Google Scholar]
- 57.Persson, B., C. Flinta, G. von Heijne, and H. Jornvall. 1985. Structures of N-terminally acetylated proteins. Eur. J. Biochem. 152:523-527. [DOI] [PubMed] [Google Scholar]
- 58.Pinto, I., and F. Winston. 2000. Histone H2A is required for normal centromere function in Saccharomyces cerevisiae. EMBO J. 19:1598-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Polevoda, B., J. Norbeck, H. Takakura, A. Blomberg, and F. Sherman. 1999. Identification and specificities of N-terminal acetyltransferases from Saccharomyces cerevisiae. EMBO J. 18:6155-6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Recht, J., and M. A. Osley. 1999. Mutations in both the structured domain and N-terminus of histone H2B bypass the requirement for Swi-Snf in yeast. EMBO J. 18:229-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Redon, C., D. Pilch, E. Rogakou, O. Sedelnikova, K. Newrock, and W. Bonner. 2002. Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 12:162-169. [DOI] [PubMed] [Google Scholar]
- 62.Ren, Q., and M. A. Gorovsky. 2001. H2A.Z acetylation modulates an essential charge patch. Mol. Cell 7:1329-1335. [DOI] [PubMed] [Google Scholar]
- 63.Ren, Q. H., and T. J. Tong. 1997. Histone acetylation and its roles in transcriptional regulation. Prog. Biochem. Biophys. 24:309-312. [Google Scholar]
- 64.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 65.Santisteban, M. S., T. Kalashnikova, and M. M. Smith. 2000. Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell 103:411-422. [DOI] [PubMed] [Google Scholar]
- 66.Schuster, T., M. Han, and M. Grunstein. 1986. Yeast histone H2A and H2B amino termini have interchangeable functions. Cell 45:445-451. [DOI] [PubMed] [Google Scholar]
- 67.Shen, X., L. Yu, J. W. Weir, and M. A. Gorovsky. 1995. Linker histones are not essential and affect chromatin condensation in vivo. Cell 82:47-56. [DOI] [PubMed] [Google Scholar]
- 68.Stargell, L. A., J. Bowen, C. A. Dadd, P. C. Dedon, M. Davis, R. G. Cook, C. D. Allis, and M. A. Gorovsky. 1993. Temporal and spatial association of histone H2A variant hv1 with transcriptionally competent chromatin during nuclear development in Tetrahymena thermophila. Genes Dev. 7:2641-2651. [DOI] [PubMed] [Google Scholar]
- 69.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 70.Strahl, B. D., S. D. Briggs, C. J. Brame, J. A. Caldwell, S. S. Koh, H. Ma, R. G. Cook, J. Shabanowitz, D. F. Hunt, M. R. Stallcup, and C. D. Allis. 2001. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr. Biol. 11:996-1000. [DOI] [PubMed] [Google Scholar]
- 71.Suto, R. K., M. J. Clarkson, D. J. Tremethick, and K. Luger. 2000. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat. Struct. Biol. 7:1121-1124. [DOI] [PubMed] [Google Scholar]
- 72.Taunton, J., C. A. Hassig, and S. L. Schreiber. 1996. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272:408-411. [DOI] [PubMed] [Google Scholar]
- 73.Thatcher, T. H., and M. A. Gorovsky. 1994. Phylogenetic analysis of the core histones H2A, H2B, H3, and H4. Nucleic Acids Res. 22:174-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Th'ng, J. P. 2001. Histone modifications and apoptosis: cause or consequence? Biochem. Cell Biol. 79:305-311. [PubMed] [Google Scholar]
- 75.Tse, C., T. Sera, A. P. Wolffe, and J. C. Hansen. 1998. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 18:4629-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Turner, B. M. 2000. Histone acetylation and an epigenetic code. Bioessays 22:836-845. [DOI] [PubMed] [Google Scholar]
- 77.Usachenko, S. I., S. G. Bavykin, I. M. Gavin, and E. M. Bradbury. 1994. Rearrangement of the histone H2A C-terminal domain in the nucleosome. Proc. Natl. Acad. Sci. USA 91:6845-6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Van Daal, A., and S. C. R. Elgin. 1992. A histone variant, H2AvD, is essential in Drosophila melanogaster. Mol. Biol. Cell 3:593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Van Daal, A., E. M. White, M. A. Gorovsky, and S. C. R. Elgin. 1988. Drosophila has a single copy of the gene encoding a highly conserved histone H2A variant of the H2A.F/Z type. Nucleic Acids Res. 16:7487-7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Verreault, A., P. D. Kaufman, R. Kobayashi, and B. Stillman. 1996. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 87:95-104. [DOI] [PubMed] [Google Scholar]
- 81.Wang, H. B., Z. Q. Huang, L. Xia, Q. Feng, H. Erdjument-Bromage, B. D. Strahl, S. D. Briggs, C. D. Allis, J. M. Wong, P. Tempst, and Y. Zhang. 2001. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293:853-857. [DOI] [PubMed] [Google Scholar]
- 82.Wechser, M. A., M. P. Kladde, J. A. Alfieri, and C. L. Peterson. 1997. Effects of Sin− versions of histone H4 on yeast chromatin structure and function. EMBO J. 16:2086-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.White, E. M., D. L. Shapiro, C. D. Allis, and M. A. Gorovsky. 1988. Sequence and properties of the message encoding Tetrahymena hv1, a highly evolutionarily conserved histone H2A variant that is associated with active genes. Nucleic Acids Res. 16:179-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Winston, F., and C. D. Allis. 1999. The bromodomain: a chromatin-targeting module? Nat. Struct. Biol. 6:601-604. [DOI] [PubMed] [Google Scholar]
- 85.Wolffe, A. 1998. Chromatin, 3rd ed. Academic Press, London, United Kingdom.
- 86.Wolffe, A. P., and D. Guschin. 2000. Chromatin structural features and targets that regulate transcription. J. Struct. Biol. 129:102-122. [DOI] [PubMed] [Google Scholar]
- 87.Wolffe, A. P., and J. J. Hayes. 1999. Chromatin disruption and modification. Nucleic Acids Res. 27:711-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wolffe, A. P., and D. Pruss. 1996. Deviant nucleosomes: the functional specialization of chromatin. Trends Genet. 12:58-62. [DOI] [PubMed] [Google Scholar]
- 89.Wu, J. S., and M. Grunstein. 2000. 25 years after the nucleosome model: chromatin modifications. Trends Biochem. Sci. 25:619-623. [DOI] [PubMed] [Google Scholar]
- 90.Xia, L., B. Hai, Y. Gao, D. Burnette, R. Thazhath, J. Duan, M. H. Bre, N. Levilliers, M. A. Gorovsky, and J. Gaertig. 2000. Polyglycylation of tubulin is essential and affects cell motility and division in Tetrahymena thermophila. J. Cell Biol. 149:1097-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang, Y., and D. Reinberg. 2001. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15:2343-2360. [DOI] [PubMed] [Google Scholar]