Abstract
CTF7/ECO1 is an essential yeast gene required for the establishment of sister chromatid cohesion. The findings that CTF7/ECO1, POL30 (PCNA), and CHL12/CTF18 (a replication factor C [RFC] homolog) genetically interact provided the first evidence that the processes of cohesion establishment and DNA replication are intimately coupled—a link now confirmed by other studies. To date, however, it is unknown how Ctf7p/Eco1p function is coupled to DNA replication or whether Ctf7p/Eco1p physically associates with any components of the DNA replication machinery. Here, we report that Ctf7p/Eco1p associates with proteins that perform partially redundant functions in DNA replication. Chl12p/Ctf18p combines with Rfc2p to Rfc5p to form one of three independent RFC complexes. By chromatographic methods, Ctf7p/Eco1p was found to associate with Chl12/Ctf18p and with Rfc2p, Rfc3p, Rfc4p, and Rfc5p. The association between Ctf7p/Eco1p and this RFC complex is biologically relevant in that (i) Ctf7p/Eco1p cosediments with Chl12p/Ctf18p in vivo and (ii) rfc5-1 mutant cells exhibit precocious sister separation. Previous studies revealed that Rfc1p or Rad24p associates with Rfc2p to Rfc5p to form two other RFC complexes independent of Ctf18p-RFC complexes. These Rfc1p-RFC and Rad24p-RFC complexes function in DNA replication or repair and DNA damage checkpoint pathways. Importantly, Ctf7p/Eco1p also associates with Rfc1p and Rad24p, suggesting that these RFC complexes also play critical roles in cohesion establishment. The associations between Ctf7p/Eco1p and RFC subunits provide novel evidence regarding the physical linkage between cohesion establishment and DNA replication. Furthermore, the association of Ctf7p/Eco1p with each of three RFC complexes supplies new insights into the functional redundancy of RFC complexes in cohesion establishment.
From the time of DNA replication until the onset of anaphase, sister chromatids remain tightly paired along their length (15, 46, 55). Cytological and molecular studies of cohesion factors reveal that this sister chromatid pairing, or cohesion, is an essential component of bipolar spindle formation, chromosome segregation, cell cycle progression, and double-strand-break repair (25, 49, 50). Studies performed with numerous cell systems reveal that defects in cohesion between sister chromatids result in missegregation of both sisters to one daughter cell and cell death (16, 23, 31, 35, 51, 52, 57). In humans, chromosome missegregation allows for phenotypic expression of recessive mutations in tumor suppression and cancer-related growth control genes (29, 65).
Three classes of proteins are required for sister chromatid cohesion and proper chromosome segregation. Structural cohesion proteins (or cohesins) provide the glue that maintains sister chromatid pairing from G1/S to anaphase onset. In budding yeast, the cohesins include Smc1p, Smc3p, Mcd1p/Scc1p, Scc3p/Irr1p, and Pds5p (16, 19, 28, 35, 42, 52, 57). Recent data revealed that the structural cohesins form a ring. This ring structure is thought to hold sister chromatids together, but whether a single ring encircles both sisters or catanated rings encompass individual sisters is unknown (2, 17). Deposition factors load structural cohesin proteins onto chromatin. Deposition factors include Scc2p (Mis4p in Schizosaccharomyces pombe) and Scc4p, which combine to form a deposition complex separate from the cohesin complex. Scc2p and Scc4p are active throughout a large part of the cell cycle, but are required during S phase (4, 12, 57). Establishment factors appear to couple sister chromatid cohesion to DNA replication, but are not required for DNA replication per se (51, 57).
Analyses of CTF7/ECO1 mutant cells revealed that Ctf7p/Eco1p (herein called Eco1p) is essential for cohesion establishment and acts in a pathway unique from the structural and deposition cohesion factors. First, Eco1p is required during S phase when cohesion is established but not in mitosis when cohesion is maintained. Second, structural cohesins appear to form a complex and load normally in eco1 mutant cells. These findings indicate that Eco1p does not function in cohesin assembly, deposition, or cohesion maintenance (51, 57; R. V. Skibbens and D. Koshland, unpublished results). Eso1p, the fission yeast homolog of Eco1p, also functions in sister chromatid cohesion, revealing that cohesion establishment is conserved through evolution (56). Recent findings reveal that Eco1p provides acetyltransferase activity and that, at least in vitro, the structural cohesins Mcd1p/Scc1p, Scc3p/Irr1p, and Pds5p—as well as Eco1p itself—are acetylation targets. Currently, however, physiologically relevant substrates of Eco1p acetylation have yet to be documented (21). Thus, the molecular mechanism by which cohesion is established remains unknown.
Early studies temporally correlated cohesion establishment with the S-phase portion of the cell cycle (15, 46, 58). The first evidence that cohesion establishment and DNA replication may be intimately coupled was obtained by the findings that Eco1p is required only during S phase and that ECO1 genetically interacts with both POL30 (PCNA) and CHL12/CTF18 (51). PCNA is a homotrimeric sliding clamp that locks DNA polymerase onto double-stranded DNA (dsDNA) and promotes processive DNA replication. Chl12p/Ctf18p (herein termed Ctf18p) exhibits limited homology to Rfc1p. Rfc1p is a large subunit of the replication factor C (RFC) complex, which also contains Rfc2p to Rfc5p. This RFC complex loads PCNA onto dsDNA (22, 26). The interaction between an essential cohesion establishment factor, Eco1p, and two DNA replication factors of interdependent function, PCNA and an RFC homolog, suggested a model in which Eco1p acts to pair nascent sister chromatids as they emerge from the DNA replication fork (51). A link between DNA replication and sister chromatid cohesion was confirmed by the characterization of Trf4p. Trf4p (also called Pol κ and later renamed Pol σ) is a DNA polymerase that also functions in sister chromatid cohesion (3, 61).
Further evidence has implicated a subset of RFC factors as important in sister chromatid cohesion. Currently, there are three known RFC complexes. Rfc1p associates with Rfc2p to Rfc5p to load PCNA onto DNA and thus promote processive DNA replication (22, 40). Rad24p associates with Rfc2p to Rfc5p to load the heterotrimeric Mec3p, Rad17p, and Ddc1p sliding clamp during activation of the DNA damage checkpoint mechanism (14, 22, 24, 30, 32, 37, 41). Ctf18p and two other cohesion factors, Ctf8p and Dcc1p, comprise an RFC complex that contains Rfc2p to Rfc5p, but the identity of an associated sliding clamp remains unknown. This Ctf18p-based RFC complex plays a role in cohesion but not DNA replication (5, 18, 33). Biochemical analyses revealed that Rfc1p, Ctf18p, and Rad24p all associate with Rfc2p to Rfc5p but not with each other, revealing the formation of three independent RFC complexes (33, 36). Characterization of both RFC complexes and DNA polymerases has led to a model in which DNA polymerases and RFC complexes switch in or out, depending on the DNA sites encountered at the replication fork (3, 5, 50, 60-62).
Sister chromatid cohesion is clearly fundamental to proper chromosome segregation. Thus, it is surprising that Rad24p, Ctf18p (and associated Ctf8p and Dcc1p), and Trf4p are nonessential for cell viability. Based on this observation, we inferred that these proteins perform an essential but redundant activity in cohesion establishment. For instance, while the three RFC complexes appear biochemically distinct, they are partially redundant for DNA repair and checkpoint functions (34, 36, 39, 45, 47, 54). In contrast, Eco1p is essential, raising the possibility that redundant cohesion activities may ultimately converge through a single Eco1p-dependent pathway. Here, we report that Eco1p interacts with all three alternate RFC complexes. In addition, we provide new evidence that an RFC subunit is required for sister chromatid cohesion.
MATERIALS AND METHODS
Strains, media, and general methods.
The growth and sporulation media used in this study were described previously (43). Yeast transformations were performed as described previously with minor modifications (20, 44). The S288C-derived Saccharomyces cerevisiae yeast strains (YPH) and plasmids (pRS) used in this study were described previously (7, 9, 48). Other strains are listed in Table 1.
TABLE 1.
Characteristics of the yeast strains used in this study
| Yeast strain | Characteristics | Source or reference |
|---|---|---|
| YBS5 | MATaura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 CTF7::HIS3 CFIII (CEN3.L) TRP1 SUP11 pRS316-CTF7 | 51 |
| YBS11 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 CTF7::HIS3 pAD5-CTF7 | |
| YBS1042 | MATα ade2 trp1 his3 leu2::LEU2tetR-GFP ura3::3xURA3tetO112 PDS1-13MYC:TRP1 | This study |
| YBS1060 | MATα ade2 trp1 his3 leu2::LEU2tetR-GFP ura3::3xURA3tetO112 PDS1-13MYC:TRP1 RFC5 | |
| YBS1058 | MATα ade2 trp1 his3 leu2::LEU2tetR-GFP ura3::3xURA3tetO112 PDS1-13MYC:TRP1 rfc5-1::LEU2 | |
| YBS1059 | MATα trp1 his3 leu2::LEU2tetR-GFP ura3::3xURA3tetO112 PDS1-13MYC:TRP1 rfc5-1::LEU2 | |
| YPH1477 | MATaade2-1 trp1-1 can1-100 his3-11,15 leu2::LEU2tetR-GFP ura3::3xURA3tetO112 PDS1-13MYC:TRP1 | 33 |
| KSC1372 | MATaRFC1-FLAG::URA3 ade1 his2 trp1 ura3 leu2 | K. Sugimoto |
| KSC1373 | MATaRFC2-FLAG::TRP1 ade1 his2 trp1 ura3 leu2 | |
| KSC1374 | MATaRFC3-FLAG::URA3 ade1 his2 trp1 ura3 leu2 | |
| KSC1375 | MATaRFC4-FLAG::URA3 ade1 his2 trp1 ura3 leu2 | |
| KSC1376 | MATaRFC5-FLAG::TRP1 ade1 his2 trp1 ura3 leu2 | |
| KSC1377 | MATaRAD24-FLAG::URA3 ade1 his2 trp1 ura3 leu2 | |
| TSY535 | MATaRFC5-HA::LEU2 ade1 his2 trp1 ura3 leu2 | 4 |
| TSY601 | MATarfc5-1::LEU2 ade1 his2 trp1 ura3 leu2 | |
| YJH40.4 | MATaura3-52 lys2-801 ade2-101 trp1 ars1 HIS3 leu21 CTF18::9MYC-TRP1 | 18 |
Sedimentation assay.
To epitope tag Eco1p, PCR was used to generate an XhoI restriction site just upstream of the ECO1 start site by using the oligonucleotides CCCGCTCGAGGATGAAAGCTAGGAAATCGCAG and GTGTGGCGCATTCAGCTC. The resulting PCR product ends were filled-in and ligated into SmaI-digested pRS303. The resulting plasmid was digested to accept an SpeI-SacI C-terminal ECO1 fragment to produce pBS6 in which the entire ECO1 open reading frame was reconstituted. The ECO1 open reading frame was then placed in frame behind the hemagglutinin (HA) epitope in the pAD5 (38) vector to produce pBS9. pBS9 rescues eco1Δ lethality, produces a plasmid-dependent band of the appropriate molecular weight, and directs for elevated expression levels of Eco1p (51). YBS5 was transformed with pBS9, and pRS316-ECO1 was subsequently removed by counterselection with medium containing 5-fluoroorotic acid (5-FOA) (1). Eco1p sedimentation was determined for log-phase cells and cells synchronized in early S phase (hydroxyurea) or in mitosis (nocodazole). The experimentally treated cells were lysed by mechanical disruption with glass beads (100 mM KCl, 1 mM MgCl2, 0.1 mM CaCl2, 20 mM Tris [pH 7.6], 50 mM sucrose, 10% glycerol plus protease inhibitors), placed over a continuous sucrose gradient (10 to 40% sucrose in lysis buffer), and centrifuged with a Beckman SW28 rotor at 25,000 rpm for 24 h at 4°C. Approximately 1.2-ml fractions were harvested and trichloroacetic acid (TCA) precipitated, and the fractions containing Eco1-HAp were identified by Western blot analysis with 12CA5 monoclonal antibody directed against the HA epitope (Covance) and horseradish peroxidase (HRP)-conjugated goat anti-mouse antibody (Cappell). Molecular weight markers included Blue dextran 2000, thryoglobulin, ferritin, catalase, aldolase, and bovine serum albumin (BSA; Pharmacia).
GST pull-down assay.
To generate GST-Eco1p, the entire ECO1 open reading frame was digested from pBS6 (XhoI-SacI) and inserted in-frame behind glutathione S-transferase (GST) of pGEX4T-3 digested with XhoI-NotI. SacI and NotI were filled in to produce ligatable blunt ends. GST-ECO1 expression in Escherichia coli cells was induced with 2 μM isopropyl-β-d-thiogalactopyranoside (IPTG) (Sigma) for 2 h at 37°C, and the cells were lysed by sonication. The whole-cell extract was centrifuged at 9,500 rpm for 5 min (Beckman JA-20), and the soluble and insoluble fractions were harvested. Western blot analysis revealed a plasmid-dependent band of the appropriate molecular weight in the soluble fraction visualized with a monocolonal antibody directed against the GST epitope (Santa Cruz) and HRP-conjugated goat anti-mouse antibody (Bio-Rad). E. coli cells harboring either pGEX4T-3 or pGEX4T-3-ECO1 were induced for 2 h with IPTG and lysed by sonication. GST versus GST-Eco1p proteins in bacterial extracts were then coupled to glutathione Sepharose 4B beads (Amersham-Pharmacia). Prior to incubation with yeast extracts, the bead matrices were washed several times in 1× phosphate-buffered saline (PBS).
Yeast strains expressing candidate epitope-tagged proteins were first spheroplasted in 100T Zymolyase (Seikagaku), lysed by swelling and mechanical disruption (20 mM HEPES-HCl [pH 7.5], 5 mM MgCl2 plus protease inhibitors), and centrifuged at 9,500 rpm for 45 min (Beckman JA-20). The supernatant was removed, and the insoluble chromatin pellet was extracted with lysis buffer containing 1 M NaCl before recentrifugation. The salt-extracted supernatants were then harvested and divided into four equal aliquots, one of which was precipitated with TCA and then resuspended in Laemmli buffer. The other three aliquots were each diluted 10-fold in lysis buffer prior to incubation with one of the three bead matrices (glutathione Sepharose beads, or beads coupled to GST or GST-Eco1p). The treated beads were washed several times before eluting the specifically bound proteins by using reduced glutathione (Sigma). To test for DNA-based interactions, the pull-down buffer was supplemented to 10 mM MnCl2 and 500 ng of DNase I per ml, incubated for 1 h at 4°C, and then processed as described above. During Western blot analyses for bead-bound proteins, the presence of FLAG-tagged proteins was detected with a monoclonal anti-FLAG antibody, M2 (Sigma); HA-tagged proteins were detected with the 12CA5 monoclonal antibody (Babco); and MYC-tagged proteins were detected with the anti-cMYC 9E10 monoclonal antibody (Santa Cruz). For each type of detection, HRP-conjugated goat anti-mouse (Bio-Rad) antibody and ECL-Plus (Amersham-Pharmacia) were used for visualization.
Sister chromatid cohesion analyses.
The strain YPH1477 (33), which contained Tet operator repeats (TetO) integrated proximal to the centromere and which expresses both green fluorescent protein (GFP)-labeled Tet repressor (TetR-GFP) and Pds1-13MYCp, was backcrossed four times into the S288C background to produce YBS1042. YBS1042 was then crossed with a mutant strain (TSY601) harboring the rfc5-1 allele (47) and sporulated. Spores YBS1058 and YBS1059 containing the rfc5-1 allele were identified by growth on selective media, visualization of loci via GFP, and by temperature sensitivity. Spore YBS1060 containing the wild-type RFC5 gene was identified by growth on selective media, visualization of loci via GFP, and growth at 37°C. To assay for a defect in cohesion, log-phase cells were placed into fresh medium containing 20 μg of nocodazole per ml and maintained at 37°C for 3 h. An optional step of synchronizing log-phase cells in early S phase by growth in 0.2 M hydroxyurea for 3 h at 23°C was also performed prior to incubating the cells at the permissive temperature in medium containing nocodazole. Digital images were captured on a Nikon Eclipse E800 microscope with a Coolsnapfx charge-coupled device camera (Photometrics) and IPLab software, 3.5.3 (Scanalytics). Flow cytometry (BD FACScan) and Western blot analysis were performed as described previously with minor modifications (13). For localization of Pds1-13MYC, cells arrested in mitosis were fixed with formaldehyde (3.7%) for 10 to 15 min, washed, and prepared for immunofluorescence as previously described (6). The MYC tag was visualized with the mouse monoclonal antibody 9E10 (Santa Cruz, Biotechnology) in combination with goat anti-mouse tetramethyl rhodamine isocyanate (TRITC)-labeled (Cappell) antibodies.
RESULTS
Eco1p associates with Ctf18p.
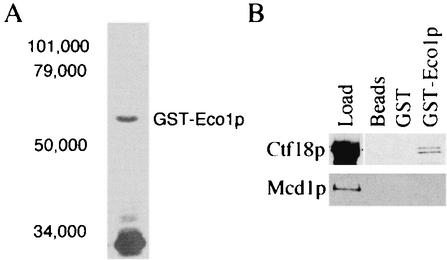
We had previously shown that CTF7/ECO1 (herein termed ECO1) and CHL12/CTF18 (herein termed CTF18) genetically interact. Subsequent work revealed that Ctf18p functions in some aspect of cohesion and associates with Rfc2p to Rfc5p (18, 33, 51). To test whether the essential cohesion function of Eco1p is physically coupled to the DNA replication machinery through RFC, we tested whether Eco1p associates with RFC subunits Ctf18p and Rfc2p to Rfc5p. The inability to detect Eco1p by using antibodies directed against either endogenous Eco1p or epitope-tagged Eco1p expressed at endogenous levels suggested that Eco1p occurs at extremely low levels in the cell (data not shown). GST-based chromatography has proven very useful in isolating protein complexes that occur at very low copy numbers or are relatively intractable to other forms of biochemical purification. We exploited this method to test for a physical interaction between Eco1p and the Ctf18p-RFC complex. The entire ECO1 open reading frame was inserted in frame behind the gene coding for GST. Western blot analysis of E. coli cells expressing this construct (GST-Eco1p) identified a plasmid-dependent band of the appropriate molecular weight (Fig. 1A). GST-Eco1p expression in yeast cells also produced a plasmid-dependent band of the appropriate molecular weight. This construct maintained viability in eco1Δ null strains, indicating that the GST moiety did not adversely affect the essential function of Eco1p in vivo (data not shown).
FIG. 1.
GST-Eco1p construct expression and protein interactions. (A) GST-Eco1p expressed in bacteria is soluble and migrates in sodium dodecyl sulfate-acrylamide gels near the predicted molecular weight. Shown is GST-Eco1p pulled down with GST-Sepharose beads from the soluble fraction of E. coli whole-cell extracts. (B) Associations of Eco1p, Ctf18p, and Mcd1p. Ctf18-9MYCp fails to bind glutathione Sepharose beads alone (Beads) or beads linked to bacterially expressed GST alone (GST) but binds specifically to beads linked to bacterially expressed GST-Eco1p (GST-Eco1p). The concentrated Ctf18-9MYCp load control is also shown (Load). Mcd1-6HAp binding was not observed for beads alone or beads coupled to either GST or GST-Eco1p.
To test for a physical association between Eco1p and Ctf18p, we first generated extracts from yeast cells containing Ctf18-9MYCp. As a control for chromatin-associated proteins, extracts were also generated from yeast cells expressing the structural cohesin Mcd1-6HAp (16, 35). Extracts of log-phase cells harboring either Ctf18-9MYCp or Mcd1-6HAp were centrifuged, and the chromatin pellet was extracted with 1 M salt and centrifuged again. The resulting supernatant (salt-released soluble fraction) was divided into four aliquots. The first aliquot was TCA precipitated and used as a highly concentrated fiduciary protein marker (Fig. 1B). The remaining three aliquots were diluted to 10× the original volume (to reduce the salt concentration) and incubated with either glutathione Sepharose beads alone, beads linked to bacterially expressed GST alone, or beads linked to bacterially expressed GST-Eco1p. The three matrices were washed, and the bound proteins were eluted. Western blot analyses of the eluants revealed that Ctf18p bound specifically to GST-Eco1p but did not bind to GST or beads alone (Fig. 1B). An association between Eco1p and Mcd1p was not detected, although we cannot rule out that a transient association exists. These results indicate that Eco1p associates specifically with Ctf18p and that this association occurs independent of DNA (binding reactions performed in the presence of DNase are described below).
Eco1p associates with four small RFC subunits.
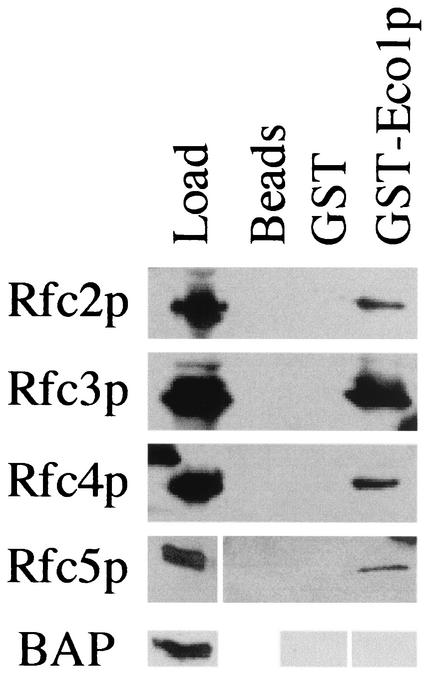
Recent findings indicate that Ctf18p associates with Rfc2p, Rfc3p, Rfc4p, and Rfc5p (18, 33). To test whether Eco1p also associates with these RFC subunits, extracts of log-phase yeast cells harboring FLAG-tagged Rfc2p, Rfc3p, Rfc4p, or Rfc5p were incubated with either beads alone, beads linked to bacterially expressed GST, or beads linked to bacterially expressed GST-Eco1p. The three matrices were then washed, and the bound proteins were eluted. Western blot analyses revealed that Rfc2p, Rfc3p, Rfc4p, and Rfc5p all bound specifically to Eco1p but did not bind to GST-linked beads or beads alone (Fig. 2). Rfc3p consistently yielded the highest binding efficiency. To test the possibility that the FLAG tag was responsible for binding the RFC subunits to Eco1p, we also tested the ability of FLAG-tagged bacterial alkaline phosphatase to bind beads linked to either bacterially expressed GST or GST-Eco1p. Western blot analyses failed to reveal an interaction between Eco1p and bacterial alkaline phosphatase (BAP)-FLAGp, indicating that the FLAG tag did not participate in the protein interactions observed for Eco1p and Rfc2p-Rfc5p (Fig. 2). These findings reveal a new physical interaction between Eco1p and an RFC-based complex containing Ctf18p.
FIG. 2.
Physical associations between Eco1p and the small subunits of RFC DNA replication factors. Extracts from cells harboring either Rfc2-FLAGp (Rfc2p), Rfc3-FLAGp (Rfc3p), Rfc4-FLAGp (Rfc4p), or Rfc5-FLAGp (Rfc5p) were tested for association with bead matrices as described in the text. Each RFC small subunit interacted specifically with GST-Eco1p coupled to glutathione Sepharose beads but not to glutathione Sepharose beads alone (beads) or coupled to GST (GST). In contrast, FLAG-tagged bacterial alkaline phosphatase (BAP) did not bind to either beads coupled to GST or GST-Eco1p. See text for details.
Eco1p associates with all three independent RFC complexes.
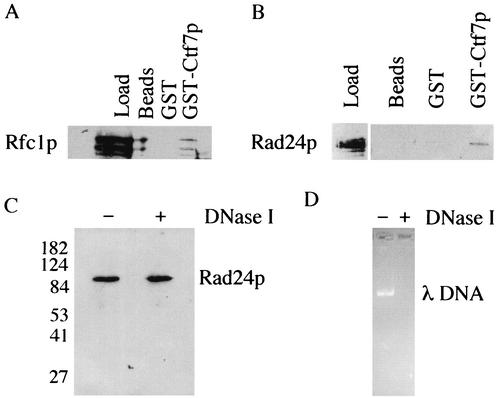
Rfc2p to Rfc5p can bind either Ctf18p, Rfc1p, or Rad24p. These RFC complexes are biochemically distinct, such that Ctf18p, Rfc1p, and Rad24p do not associate together (18, 33, 36). It thus became important to determine whether Eco1p associated with either Rfc1p or Rad24p, in addition to Ctf18p. Extracts of log-phase yeast cells harboring either FLAG-tagged Rfc1p or FLAG-tagged Rad24p were incubated with beads alone or beads coupled to either GST or GST-Eco1p. Each bead matrix was then washed, and the proteins were eluted. Western blot analyses of bound proteins revealed that both Rfc1p and Rad24p associated specifically with Eco1p but not with GST or beads alone (Fig. 3A and B). These findings place the only essential cohesion establishment factor, Eco1p, as associating with each of three independent RFC complexes
FIG. 3.
Physical interactions between Eco1p and the large subunits unique to the alternate RFC complexes. (A and B) Rfc1-FLAGp (Rfc1p) and Rad24-FLAGp (Rad24p) present in yeast extracts bound specifically to glutathione Sepharose beads coupled to GST-Eco1p (GST-Eco1p) but not beads alone (beads) or beads coupled to GST (GST). Concentrated Rfc1p and Rad24p load controls are also shown. Multiple bands in Rfc1p were typical and probably represent protein degradation. (C and D) Rad24p binding to Eco1p was not disrupted by DNase I treatment, while this level of DNase I under identical conditions was sufficient to completely digest 500 ng of λ DNA.
The inability to detect an association between Eco1p and the chromatin-associated cohesion protein Mcd1p suggested that Eco1p binding to Ctf18p, Rfc1p, Rad24p, and Rfc2p to Rfc5p is specific and not rooted in DNA-based interactions. To directly test for this possibility, we performed parallel Eco1p pull-down experiments with Rad24p in which one sample was treated with DNase I. Briefly, yeast extract containing epitope-tagged Rad24p was incubated with GST-Eco1p in a buffer modified to support complete DNA digestion by DNase I (see Materials and Methods). As before, the GST-Eco1p matrix was washed and bound Rad24p eluted. Western blot analyses revealed that Eco1p was still able to bind Rad24p even in the presence of DNase I (Fig. 3C). The activity of DNase I was tested in a parallel reaction that contained over 500 ng of λ DNA. The λ DNA was completely digested (Fig. 3D), attesting to the efficiency of the enzymatic treatment under these conditions. These results indicate that the binding of Eco1p to RFC complexes occurs independent of DNA.
Eco1p assembled with RFCs in vivo cosediment as a complex.
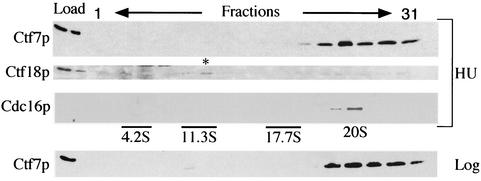
Recently, Ctf18p assembled in vivo with Rfc2p to Rfc5p was shown to sediment as a complex of approximately 12S. This sedimentation is completely coincident with Rfc1p-RFC sedimentation and overlaps that of Rad24p-RFC sedimentation (36). We thus decided to test whether Eco1p would cosediment with RFC complexes assembled in vivo by using Ctf18p as a fiduciary marker. To facilitate detection of Eco1p, we generated extracts from yeast cells in which Eco1-HAp expressed at elevated levels was the sole source of Eco1p function. This Eco1-HAp construct is fully functional and maintains viability of eco1Δ cells at wild-type growth rates (data not shown). Extracts from hydroxyurea-arrested cells expressing Eco1-HAp were placed over a sucrose gradient (10 to 40%) and subjected to centrifugation. Fractions were then harvested, and the sedimentation of Eco1p was determined by Western blot analyses. Consistent with an in vivo interaction, a significant fraction of Eco1p sedimented at approximately 12S (Fig. 4). To independently test for Ctf18p sedimentation, the membrane was stripped and reprobed with antibodies directed against endogenous Ctf18p. The results show that Ctf18p cosedimented exactly with Eco1p as a 12S complex when assembled in vivo (Fig. 4), recapitulating previously described Ctf18p sedimentation (36). A very slight decrease in sedimentation was observed for extracts derived from logarithmically growing cells (Fig. 4), which was still consistent with previously characterized sedimentations for all three RFC complexes (36). The finding that Eco1p assembled in vivo cosediments with RFC subunits provides strong support for the Eco1p-RFC associations detected in vitro.
FIG. 4.
Eco1p, Cdc16p, and Ctf18p sedimentation. Extracts placed over a 10 to 40% sucrose gradient were centrifuged and fractionated. Fractions were pooled (1 + 2, 3 + 4, etc. [the “odd” fraction number is shown]) and analyzed by Western blotting. (Top panel) Eco1-HAp from hydroxyurea (HU)-arrested cells migrated with a sedimentation of approximately 11S to 13S (fractions 11 and 12). (Second panel) Hydroxyurea extracts probed with antibody directed against endogenous Ctf18p showed an identical sedimentation, as indicated by an asterisk. Note that the second peak of Eco1p (indicative of a larger complex) also cosediments with Ctf18p. (Third panel) The relative sizes of these complexes are compared to that of Cdc16p, a component of the anaphase-promoting complex. (Bottom panel) Eco1p harvested from log-phase cells exhibits a very similar sedimentation (11S to 13S). Molecular weight controls at 4.2S (BSA), 11.3S (catalase), and 17.7S (ferritin) are indicated.
We also found that a significant portion of Eco1p migrated deeply into the sucrose gradient, indicative of a very large complex. Very large complexes are often artifacts of elevated protein expression. Importantly, previous studies indicated that Ctf18p sedimentation peaked at ∼12S but not as a much larger complex (36). Thus, we decided to use this aberrant Eco1p sedimentation (presumably the result of overexpression) to ask whether Ctf18p would be pulled deeper into the gradient by virtue of its association with Eco1p. Indeed, Ctf18p persisted in cosedimenting with Eco1p deep in the sucrose gradient (Fig. 4). These observations suggest that Eco1p and Ctf18p not only physically associate in vivo but that this binding is of sufficient avidity to alter Ctf18p sedimentation.
Rfc5p is required for sister chromatid cohesion.
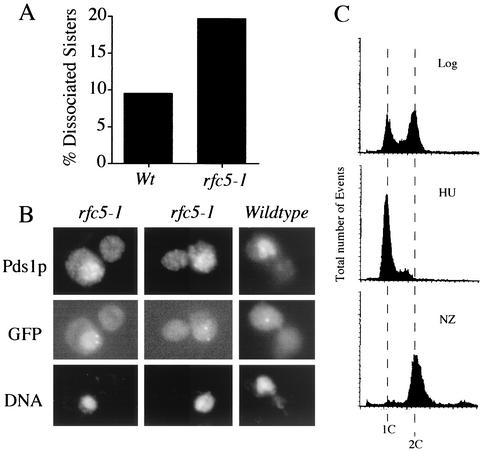
Given the physical association of Eco1p with RFC complexes (this study), a likely model was that all RFC subunits would play a key role in cohesion establishment. Both ctf7 and rfc5 temperature-sensitive mutant strains are rescued by elevated levels of POL30 (51, 54). Thus, we decided to first test whether Rfc5p—a component of each RFC complex—functioned in cohesion. The rfc5-1 allele was crossed into a strain that contains Tet operator repeats (TetO) integrated proximal to the centromere. This strain also expresses GFP-tagged Tet repressor protein (TetR-GFP), allowing for visualization of the centromere-proximal locus. Visualization of the GFP signal was then used to determine the position of one sister chromatid relative to the other (33, 35). Log-phase wild-type and rfc5-1 cells were shifted to 37°C for 3 h (to inactivate rfc5p function in the mutant strain) in medium supplemented with nocodazole to inhibit the onset of anaphase. An optional synchronization step of arresting cells in early S phase with hydroxyurea was also used. Parallel cell samples were then assayed for DNA content, cell morphology, and sister chromatid cohesion. Wild-type cells treated with nocodazole were predominantly large budded and contained a 2C DNA content, indicative of a mitotic arrest. When GFP-tagged loci were viewed by epifluorescent microscopy, wild-type cells were found to contain tightly paired sister chromatids, such that few (9%) sisters were dissociated. rfc5-1 mutant cells treated with nocodazole also were predominantly large budded and contained a 2C DNA content. In contrast to wild-type cells, however, rfc5-1 mutant cells contained a significant increase in the number of separated sisters (20%) (Fig. 5). Both wild-type and rfc5-1 strains exhibited similarly low levels (∼5%) of separated sisters in early S phase. These results reveal that the incidence of two GFP spots in mitotic rfc5 mutant cells was not due to aneuploidy present early in the cell cycle but instead was due to a loss of sister chromatid cohesion.
FIG. 5.
rfc5 mutant cells exhibit sister chromatid cohesion defects. (A) Quantification of cohesion defects exhibited by wild-type (Wt) and rfc5-1 mutant strains. (B) Micrographs of two rfc5-1 mutant cells in which separated sister chromatids (GFP) and Pds1p staining (Pds1p) are visualized within the DNA mass (DAPI). rfc5 cells similarly treated contain tightly paired sister chromatids (GFP) within the Pds1p staining (Pds1p) and DNA (4′,6′-diamidino-2-phenylindole [DAPI]) mass. (C) DNA content of rfc5 mutant cells. NZ, nocodazole.
To verify that sister separation occurred prior to anaphase onset, we performed indirect immunfluorescence to simultaneously view on a cell-by-cell basis both the GFP-tagged chromosomal loci and epitope-tagged Pds1p. Pds1p is a biochemical marker for preanaphase cells (6, 63, 64). Log-phase wild-type and rfc5-1 cells were first synchronized in early S phase by using hydroxyurea, washed, and incubated for 3 h in medium supplemented with nocodazole. Parallel samples of the resulting cultures were fixed with either ethanol or formaldehyde and then processed for flow cytometry and immunofluorescence (Fig. 5B and C). To assay for cohesion defects in an unbiased fashion, we first identified large-budded cells. We then limited our analysis to cells that retained Pds1p nuclear staining, confirming that these cells had not traversed the metaphase-to-anaphase transition. The disposition of sister chromatids was then quantified. The results revealed that a significant percentage of rfc5-1 mutant strains that retained Pds1p also contained separated sister chromatids (Fig. 5B). These findings reveal a new role for Rfc5p—namely that Rfc5p plays a critical role in sister chromosome cohesion.
We next tested whether Rad24p played a role in cohesion establishment. A rad24 deletion strain (ResGen) was crossed into the cohesion assay strain (TetO integration and expressing TetR-GFP and Pds1-MYCp) modified from reference 33. The resulting diploid was sporulated, and haploid cells containing the appropriate markers were identified. To independently assess for loss of Rad24p function, we confirmed that the resulting strains exhibited sensitivity to UV light (data not shown). Wild-type and rad24 mutant cells were grown for 3 h in the presence of nocodazole. As before, the number of GFP spots was then used to determine the position of one sister chromatid, relative to the other, in large-budded cells that retained Pds1p. While a reproducible increase in sister chromatid separation was observed in rad24-null cells relative to wild-type cells, this increase was only minimally above background (data not shown). Independent analyses of rad24 mutant cells revealed a similar variability, such that a cohesion defect was considered not to be significant (M. Mayer and P. Hieter, personal communication). While the extent to which Rad24p functions in cohesion remains unknown, a likely possibility is that Rad24p may perform a key role in cohesion, but that this activity occurs only along very short tracts of DNA (possibly during nucleotide excision repair) and thus was undetected under the conditions tested.
DISCUSSION
Eco1p associates with three different RFC complexes.
Eco1p is thus far the only essential factor identified that specifically couples cohesion establishment to DNA replication in that Eco1p is essential for sister chromatid cohesion but is not required for DNA replication per se. For instance, previous findings showed that the bulk of DNA is replicated in Eco1p-deficient cells (51, 57). Furthermore, the mitotic arrest observed in Eco1p-deficient cells does not require the DNA damage checkpoint machinery but instead relies on the kinetochore/spindle checkpoint (51). In contrast, eco1 mutant cells are sensitive to double-strand breaks, indicating the importance of sister chromatids in templating for postreplicative DNA repair (49). In combination, these results reveal that Eco1p is not required for efficient DNA replication and that lesions resulting from loss of Eco1p function are not recognized by the DNA damage checkpoint machinery. On the other hand, Eco1p functions specifically during S phase and interacts genetically with two DNA replication factors: PCNA and Ctf18p. Thus, the linkage by which Eco1p is coupled to the DNA replication machinery remains an important but unresolved issue.
In this report, we provide new evidence that Eco1p physically associates in complexes comprised of seven other proteins—Rfc1p to Rfc5p, Ctf18p, and Rad24p—all of which are components of the DNA replication/repair fork machinery. First, GST chromatographic methodologies revealed that Eco1p associates with each of these RFC subunits in a DNA-independent manner. In contrast, no interaction was detected with Mcd1p, a chromatin-associated cohesin, suggesting that Eco1p-RFC associations are indeed specific. Second, Eco1p-containing complexes assembled in vivo were found to cosediment exactly with RFC complexes, with Ctf18p used as a fiduciary mark. Previous findings revealed that the Ctf18p- and Rad24p-RFC complex sedimentations are completely coincident with that of Rfc1p-RFC (36)—a likely outcome due to the fact that each complex is in part comprised of common components. In addition, Rad24p sedimentation is completely coincident with Ctf18p (although the peak sedimentation is very slightly decreased in Rad24p) (36). Thus, Ctf18p is an appropriate marker for Ctf18p-Rad24p- and Rfc1p-RFC complexes. We noted a very slight mobility shift of Eco1p when obtained from logarithmically growing cells versus hydroxyurea-treated cells (Fig. 4), but both are within range of all three RFC complex sedimentations. Thus, both in vitro and in vivo results are consistent with a model in which Eco1p associates with each of the three RFC complexes.
The role of Eco1p-RFC associations is further supported by physiological evidence. First, our analyses of rfc5 mutants revealed that Rfc5p, a component of each RFC complex, is required for sister chromatid cohesion. This result greatly extends the previously documented roles for Rfc5p in DNA replication and replication checkpoint activity (8, 11, 37, 47, 53, 54). Do cells harboring mutations in other RFC subunits exhibit cohesion defects? Recent studies showed that budding yeast rfc4 mutant strains and fruit fly larva rfc2 mutants both exhibit sister chromatid cohesion defects (27, 33). Our quantification of cohesion defects in rfc5 mutant cells is nearly identical to that reported for rfc4 mutants (33). Importantly, RFC subunits perform distinct mechanochemical functions even within an RFC complex. For instance, based on similarities to the E. coli clamp loader, it has been postulated that the eukaryotic RFC complex required for the bulk of DNA replication is composed of three subcomplexes: Rfc1p, Rfc2p to Rfc4p, and Rfc5p. All are AAA+ family members but likely serve very different molecular functions corresponding to a motor (Rfc2p to Rfc4p), stator (Rfc5p), and wrench (Rfc1p) (10, 40). Thus, the assignment of a new function for any RFC subunit is nontrivial. Our characterization of Rfc5p as a cohesion factor completes the list in that at least one member of each subcomplex (Ctf18p as a wrench, Rfc2p and Rfc4p as motor components, and Rfc5p as a stator) has now been characterized as playing an important role in cohesion (18, 33; this study). Finally, independent analyses support our data that RFC complexes beyond those including Ctf18p function in cohesion. For instance, cells harboring defects in both Ctf18p and Rad24p are viable (36), indicating that Rfc1p-based RFC complexes, in association with Eco1p, most probably are competent to establish cohesion. Thus far, we have been unable to unambiguously observe cohesion defects in Rad24p-deficient cells, but this may be due to limitations in detecting cohesion loss along short chromosome segments and during DNA repair when Rad24p is active. In combination, these findings provide the basis for a new understanding regarding the mechanical linkage between cohesion establishment and DNA replication or repair.
Previous models of DNA replication fork dynamics proposed DNA polymerase handoff or switching mechanisms (3, 33, 59, 61). In terms of cohesion, a DNA polymerase switch is thought to occur when the DNA polymerase that performs the bulk of DNA replication (Pol δ or ɛ) encounters a site destined for cohesion establishment. At this juncture, the replication polymerase switches out for a polymerase that functions in cohesion establishment, Pol σ (previously termed Pol κ). In conjunction with the DNA polymerase handoff, a similar switching mechanism has been postulated for RFC complexes such that the Rfc1p-RFC complex switches out for the Ctf18p-RFC complex upon encountering a cohesion site or the Rad24p-RFC complex upon encountering DNA damage (5). Our data greatly extend the current model. We posit that each of the RFC complexes is competent to establish cohesion such that Eco1p rides, in piggyback fashion, the RFC DNA replication machinery. In this way, Eco1p is free to establish cohesion during DNA replication, but not participate in DNA replication (Fig. 6). This new model sheds light on the functional redundancy of RFC complexes in cohesion establishment and helps explain how nonessential factors (Trf4p, Rad24p, Ctf18p, and associated cohesion factors Ctf8p and Dcc1p) can participate in cohesion, a process fundamental to cell survival.
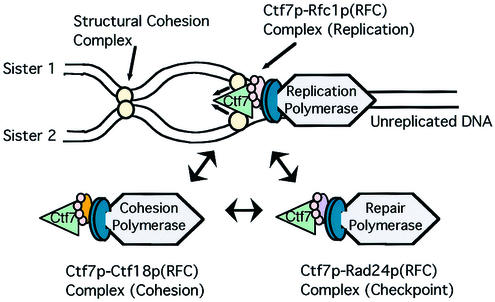
FIG. 6.
A model of the mechanical linkage between cohesion establishment and DNA replication machineries and the functional redundancy of RFC complexes in cohesion establishment. Eco1p associates with each of three independent RFC complexes. The DNA replication fork moves from left to right. As it rides along on the RFC complex, Eco1p establishes cohesion between nascent sister chromatids as they emerge from the replication fork. A dynamic interchange of DNA polymerases occurs through alternate RFC complexes, depending on the DNA status (normal, damaged, or marked for cohesion) encountered by the replication fork.
Finally, recent data revealed that Eco1p is an acetyltransferase that targets in vitro both itself and a subset of cohesin proteins: Mcd1p, Scc3p, and Pds5p (21). Currently, however, the physiologically relevant substrates of Eco1p acetyltransferase activity remain unknown. For instance, Ivanov and coworkers (21) were unable to document in vivo acetylation of either Mcd1p, Scc3p, or Pds5p (PCNA and Ctf18p were also tested). Importantly, yeast cells expressing an Mcd1p acetylation-deficient mutant as the sole source of Mcd1p function exhibit wild-type growth kinetics, indicating that this in vitro acetylation is not relevant to Mcd1p's essential function. Does Eco1p acetylate RFC complexes? We tested for acetylation of RFC subunits that exhibited the most efficient binding to Eco1p. To date, we have not been able to document in vitro acetylation for any of the RFC subunits tested (data not shown). Thus, the role of Eco1p acetyltransferase function remains unclear.
Acknowledgments
We thank Katsunori Sugimoto, Melanie Mayer, Forrest Spencer, Vincent Guacci, Jenya Kroll, and Andrew Page for graciously sharing their reagents and strains. Specifically, we thank A. Page and P. Hieter for the generous gift of Cdc16p antibodies, J. Kroll and P. Hieter for the generous gift of Ctf18p antibodies, J. Hanna and F. Spencer for use of Ctf18-MYC-tagged strains, K. Sugimoto for use of the FLAG-tagged RFC strains, and Nicole Moskal for technical assistance. In addition, we thank Doug Koshland and Melanie Mayer for reading the manuscript and helpful suggestions. Finally, we thank Jef Boeke and Phil Hieter for the collegial atmosphere in which this work was first initiated.
M.A.K. was supported in part by funding from the Vice Provost of Research and Department of Biology, Lehigh University; R.V.S. is supported by the National Science Foundation (MCB-0212323).
REFERENCES
- 1.Boeke, J. D., J. Trueheart, G. Natsoulis, and G. R. Fink. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164-175. [DOI] [PubMed] [Google Scholar]
- 2.Campbell, J., and O. Cohen-Fix. 2002. Chromosome cohesion: ring around the sisters? Trends Biochem. Sci. 27:492.. [DOI] [PubMed] [Google Scholar]
- 3.Carson, D. R., and M. F. Christman. 2001. Evidence that replication fork components catalyze establishment of cohesion between sister chromatids. Proc. Natl. Acad. Sci. USA 98:8270-8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciosk, R., M. Shirayama, A. Shevchenko, T. Tanaka, A. Toth, A. Shevchenko, and K. Nasmyth. 2000. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4. Mol. Cell 5:243-254. [DOI] [PubMed] [Google Scholar]
- 5.Cohen-Fix, O. 2001. The making and breaking of sister chromatid cohesion. Cell 106:137-140. [DOI] [PubMed] [Google Scholar]
- 6.Cohen-Fix, O., J. M. Peters, M. W. Kirschner, and D. Koshland. 1996. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 10:3081-3093. [DOI] [PubMed] [Google Scholar]
- 7.Connelly, C., and P. Hieter. 1996. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell 86:275-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullmann, G., K. Fien, R. Kobayashi, and B. Stillman. 1995. Characterization of the five replication factor C genes of Saccharomyces cerevisiae. Mol. Cell. Biol. 15:4661-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doheny, K. F., P. K. Sorger, A. A. Hyman, S. Tugendreich, F. Spencer, and P. Hieter. 1993. Identification of essential components of the S. cerevisiae kinetochore. Cell 73:761-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellison, V., and B. Stillman. 2001. Opening of the clamp: an intimate view of an ATP-driven biological machine. Cell 106:655-660. [DOI] [PubMed] [Google Scholar]
- 11.Fien, K., and B. Stillman. 1992. Identification of replication factor C from Saccharomyces cerevisiae: a component of the leading-strand DNA replication complex. Mol. Cell. Biol. 12:155-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furuya, K., K. Takahashi, and M. Yanagida. 1998. Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in S phase and not destroyed in G1 phase. Genes Dev. 12:3408-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerring, S. L., F. Spencer, and P. Hieter. 1990. The CHL1 (CTF1) gene product of Saccharomyces cerevisiae is important for chromosome transmission and normal cell cycle progression in G2/M. EMBO J. 9:4347-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green, C. M., H. Erdjument-Bromage, P. Tempst, and N. F. Lowndes. 2000. A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr. Biol. 10:39-42. [DOI] [PubMed] [Google Scholar]
- 15.Guacci, V., E. Hogan, and D. Koshland. 1994. Chromosome condensation and sister chromatid pairing in budding yeast. J. Cell Biol. 125:517-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guacci, V., D. Koshland, and A. Strunnikov. 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91:47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haering, C. H., J. Lowe, A. Hochwagen, and K. Nasmyth. 2002. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9:773-788. [DOI] [PubMed] [Google Scholar]
- 18.Hanna, J. S., E. S. Kroll, V. Lundblad, and F. A. Spencer. 2001. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol. Cell. Biol. 21:3144-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartman, T., K. Stead, D. Koshland, and V. Guacci. 2000. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J. Cell Biol. 151:613-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivanov, D., A. Schleiffer, F. Eisenhaber, K. Mechtler, C. H. Haering, and K. Nasmyth. 2002. Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion. Curr. Biol. 12:323-328. [DOI] [PubMed] [Google Scholar]
- 22.Kelman, Z. 1997. PCNA: structure, functions and interactions. Oncogene 14:629-640. [DOI] [PubMed] [Google Scholar]
- 23.Kerrebrock, A. W., D. P. Moore, J. S. Wu, and T. L. Orr-Weaver. 1995. Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell 83:247-256. [DOI] [PubMed] [Google Scholar]
- 24.Kondo, T., K. Matsumoto, and K. Sugimoto. 1999. Role of a complex containing Rad17, Mec3, and Ddc1 in the yeast DNA damage checkpoint pathway. Mol. Cell. Biol. 19:1136-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koshland, D. E., and V. Guacci. 2000. Sister chromatid cohesion: the beginning of a long and beautiful relationship. Curr. Opin. Cell Biol. 12:297-301. [DOI] [PubMed] [Google Scholar]
- 26.Kouprina, N., E. Kroll, A. Kirillov, V. Bannikov, V. Zakharyev, and V. Larionov. 1994. CHL12, a gene essential for the fidelity of chromosome transmission in the yeast Saccharomyces cerevisiae. Genetics 138:1067-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krause, S. A., M.-L. Loupart, S. Vass, S. Schoenfelder, S. Harrison, and M. M. S. Heck. 2001. Loss of cell cycle checkpoint control in Drosophila Rfc4 mutants. Mol. Cell. Biol. 21:5156-5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurlandzka, A., J. Rytka, R. Gromadka, and M. Murawski. 1995. A new essential gene located on Saccharomyces cerevisiae chromosome IX. Yeast 11:885-890. [DOI] [PubMed] [Google Scholar]
- 29.Lengauer, C., K. W. Kinzler, and B. Vogelstein. 1997. Genetic instability in colorectal cancers. Nature 386:623-627. [DOI] [PubMed] [Google Scholar]
- 30.Longhese, M. P., V. Paciotti, R. Fraschini, R. Zaccarini, P. Plevani, and G. Lucchini. 1997. The novel DNA damage checkpoint protein ddc1p is phosphorylated periodically during the cell cycle and in response to DNA damage in budding yeast. EMBO J. 16:5216-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Losada, A., M. Hirano, and T. Hirano. 1998. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 12:1986-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lydall, D., and T. Weinert. 1995. Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science 270:1488-1491. [DOI] [PubMed] [Google Scholar]
- 33.Mayer, M. L., S. P. Gygi, R. Aebersold, and P. Hieter. 2001. Identification of RFC (Ctf18p, Ctf8p, Dcc1p): an alternate RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol. Cell 7:959-970. [DOI] [PubMed] [Google Scholar]
- 34.McAlear, M. A., K. M. Tuffo, and C. Holm. 1996. The large subunit of replication factor C (Rfc1p/Cdc44p) is required for DNA replication and DNA repair in Saccharomyces cerevisiae. Genetics 142:65-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michaelis, C., R. Ciosk, and K. Nasmyth. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91:35-45. [DOI] [PubMed] [Google Scholar]
- 36.Naiki, T., T. Kondo, D. Nakada, K. Matsumoto, and K. Sugimoto. 2001. Chl12 (Ctf18) forms a novel replication factor C-related complex and functions redundantly with Rad24 in the DNA replication checkpoint pathway. Mol. Cell. Biol. 21:5838-5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naiki, T., T. Shimomura, T. Kondo, K. Matsumoto, and K. Sugimoto. 2000. Rfc5, in cooperation with Rad24, controls DNA damage checkpoints throughout the cell cycle in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:5888-5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neiman, A. M., B. J. Stevenson, H. P. Xu, G. F. Sprague, Jr., I. Herskowitz, M. Wigler, and S. Marcus. 1993. Functional homology of protein kinases required for sexual differentiation in Schizosaccharomyces pombe and Saccharomyces cerevisiae suggests a conserved signal transduction module in eukaryotic organisms. Mol. Biol. Cell 4:107-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noskov, V. N., H. Araki, and A. Sugino. 1998. The RFC2 gene, encoding the third-largest subunit of the replication factor C complex, is required for an S-phase checkpoint in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:4914-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Donnell, M., D. Jeruzalmi, and J. Kuriyan. 2001. Clamp loader structure predicts the architecture of DNA polymerase III holoenzyme and RFC. Curr. Biol. 11:R935-R946. [DOI] [PubMed] [Google Scholar]
- 41.Paciotti, V., G. Lucchini, P. Plevani, and M. P. Longhese. 1998. Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. EMBO J. 17:4199-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Panizza, S., T. Tanaka, A. Hochwagen, F. Eisenhaber, and K. Nasmyth. 2000. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr. Biol. 10:1557-1564. [DOI] [PubMed] [Google Scholar]
- 43.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Schiestl, R. H., and R. D. Gietz. 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16:339-346. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt, S. L., X. V. Gomes, and P. M. Burgers. 2001. ATP utilization by yeast replication factor C. iii. The ATP-binding domains of Rfc2, Rfc3, and Rfc4 are essential for DNA recognition and clamp loading. J. Biol. Chem. 276:34784-34791. [DOI] [PubMed] [Google Scholar]
- 46.Selig, S., K. Okumura, D. C. Ward, and H. Cedar. 1992. Delineation of DNA replication time zones by fluorescence in situ hybridization. EMBO J. 11:1217-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shimomura, T., S. Ando, K. Matsumoto, and K. Sugimoto. 1998. Functional and physical interaction between Rad24 and Rfc5 in the yeast checkpoint pathways. Mol. Cell. Biol. 18:5485-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sjogren, C., and K. Nasmyth. 2001. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr. Biol. 11:991-995. [DOI] [PubMed] [Google Scholar]
- 50.Skibbens, R. V. 2000. Holding your own: establishing sister chromatid cohesion. Genome Res. 10:1664-1671. [DOI] [PubMed] [Google Scholar]
- 51.Skibbens, R. V., L. B. Corson, D. Koshland, and P. Hieter. 1999. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 13:307-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strunnikov, A. V., V. L. Larionov, and D. Koshland. 1993. SMC1: an essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J. Cell Biol. 123:1635-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sugimoto, K., S. Ando, T. Shimomura, and K. Matsumoto. 1997. Rfc5, a replication factor C component, is required for regulation of Rad53 protein kinase in the yeast checkpoint pathway. Mol. Cell. Biol. 17:5905-5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sugimoto, K., T. Shimomura, K. Hashimoto, H. Araki, A. Sugino, and K. Matsumoto. 1996. Rfc5, a small subunit of replication factor C complex, couples DNA replication and mitosis in budding yeast. Proc. Natl. Acad. Sci. USA 93:7048-7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sumner, A. T. 1991. Scanning electron microscopy of mammalian chromosomes from prophase to telophase. Chromosoma 100:410-418. [DOI] [PubMed] [Google Scholar]
- 56.Tanaka, K., T. Yonekawa, Y. Kawasaki, M. Kai, K. Furuya, M. Iwasaki, H. Murakami, M. Yanagida, and H. Okayama. 2000. Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol. Cell. Biol. 20:3459-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Toth, A., R. Ciosk, F. Uhlmann, M. Galova, A. Schleiffer, and K. Nasmyth. 1999. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 13:320-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uhlmann, F., and K. Nasmyth. 1998. Cohesion between sister chromatids must be established during DNA replication. Curr. Biol. 8:1095-1101. [DOI] [PubMed] [Google Scholar]
- 59.Waga, S., and B. Stillman. 1998. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67:721-751. [DOI] [PubMed] [Google Scholar]
- 60.Wang, Z., I. B. Castano, C. Adams, C. Vu, D. Fitzhugh, and M. F. Christman. 2002. Structure/function analysis of the Saccharomyces cerevisiae Trf4/Pol sigma DNA polymerase. Genetics 160:381-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, Z., I. B. Castano, A. De Las Penas, C. Adams, and M. F. Christman. 2000. Pol kappa: a DNA polymerase required for sister chromatid cohesion. Science 289:774-779. [DOI] [PubMed] [Google Scholar]
- 62.Wang, Z., and M. F. Christman. 2001. Replication-related activities establish cohesion between sister chromatids. Cell Biochem. Biophys. 35:289-301. [DOI] [PubMed] [Google Scholar]
- 63.Yamamoto, A., V. Guacci, and D. Koshland. 1996. Pds1p is required for faithful execution of anaphase in the yeast, Saccharomyces cerevisiae. J. Cell Biol. 133:85-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamamoto, A., V. Guacci, and D. Koshland. 1996. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s). J. Cell Biol. 133:99-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zou, H., T. J. McGarry, T. Bernal, and M. W. Kirschner. 1999. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science 285:418-422. [DOI] [PubMed] [Google Scholar]