Abstract
Molecular typing of Mycobacterium tuberculosis by using IS6110 shows low discrimination when there are fewer than five copies of the insertion sequence. Using a collection of such isolates from a study of the epidemiology of tuberculosis in London, we have shown a substantial degree of congruence between IS6110 patterns and both spoligotype and PGRS type. This indicates that the IS6110 types mainly represent distinct families of strains rather than arising through the convergent insertion of IS6110 into favored positions. This is supported by identification of the genomic sites of the insertion of IS6110 in these strains. The combined data enable identification of the putative evolutionary relationships of these strains, comprising three lineages broadly associated with patients born in South Asia (India and Pakistan), Africa, and Europe, respectively. These lineages appear to be quite distinct from M. tuberculosis isolates with multiple copies of IS6110.
The international standard method for typing Mycobacterium tuberculosis depends on the polymorphism detected with the insertion sequence IS6110 (17, 41, 47). In most populations, multicopy strains (with five or more copies of IS6110) form a substantial majority of isolates of M. tuberculosis (24, 33, 40, 42). Isolates with only a few copies of IS6110 show much less polymorphism, necessitating the use of additional typing methods such as spoligotyping (20) or PGRS typing (7). The implied assumption behind the use of secondary typing to define clusters of low-copy-number isolates is that the combined rate of variation for the low-copy-number isolates (the product of the two molecular clocks) is equivalent to the single rate of variation of the IS6110 pattern in the multiple-copy-number strains.
The reduced polymorphism of IS6110 in low-copy-number strains is assumed to reflect the occupation of a limited number of chromosomal sites by the insertion sequence in such strains. One hypothesis is that this is due to frequent independent transposition into the same chromosomal sites (which would be true hot spots). An alternative hypothesis is that the low degree of polymorphism arises from a lack of mobility of IS6110 in such strains (i.e., the IS6110 molecular clock operates more slowly) (5, 43). The first hypothesis would lead to the prediction that additional independent typing methods would yield widely dispersed results. However, if the second hypothesis were true, independent typing methods would be predicted to show substantial congruence.
In addition, the assumption that a limited number of chromosomal sites are occupied in low-copy-number strains needs to be tested. Several studies have shown that the overall distribution of IS6110 inserts is nonrandom, either by determining band sizes (13, 25) or by examining the occurrence of inserts in specific regions of the genome (IS6110 preferential loci) (8-10, 44). Microarrays have also been used (22) to locate the insertions of IS6110 to such regions or to specific genes. For studies of evolutionary relationships, it is necessary to determine the precise location of the insertion sequence, which will be identical in related strains, whereas insertions in nonidentical but nearby sites may also produce similar banding patterns, though by convergence.
Testing these hypotheses by examination of the congruence between typing methods and by analysis of the insertion sites will therefore assist the understanding of the evolutionary relationships between low-copy-number strains, as well as having implications for the typing strategies used for such isolates. For this purpose, we used a set of low-copy-number isolates from a study of tuberculosis in London during the period from 1995 to 1997 (24). A substantial proportion of the M. tuberculosis isolates in London are from people who have relatively recently arrived from other countries with high rates of tuberculosis or have close connections with such countries (14, 24). The high proportion of low-copy-number isolates in London (20%) provides a valuable opportunity for such an analysis.
MATERIALS AND METHODS
Source of isolates.
The M. tuberculosis isolates used in this study were collected for a population-based study of the epidemiology of tuberculosis in London between 1 July 1995 and 31 December 1997 (24). Identification was carried out in the source laboratories by normal phenotypic procedures. Subsequent spoligotyping suggested that four isolates were Mycobacterium bovis or M. bovis BCG and that a further four isolates resembled Mycobacterium africanum. As these each represent only about 1% of the total number of isolates, their contribution to the analyses was minimal.
Molecular typing methods.
IS6110 typing was performed, as described by Maguire et al. (24), by the international standard protocol (41), in which M. tuberculosis DNA was cut with PvuII and Southern blots were hybridized with a probe from the right-hand portion of IS6110. Blots were normalized with the standard M. tuberculosis reference strain 14323. Distinct IS6110 patterns were designated with arbitrary numbers, with the prefix IN used where necessary to distinguish them from spoligotypes.
Spoligotyping, by using a set of 43 spacers, was carried out as described by Kamerbeek et al. (20). Spoligotype patterns were designated with hexadecimal codes and/or arbitrary database numbers as described by Dale et al. (4). PGRS typing, by using AluI-cut DNA, was performed as described by Gillespie et al. (13).
Typing results were analyzed and compared by using GelCompar and Bionumerics software (Applied Maths, Kortrijk, Belgium). Dendrograms of the spoligotyping data were produced by using Taxotron software (Institut Pasteur, Paris, France). Discrimination indices (18) were calculated with the following formula:
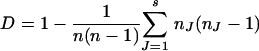 |
where n is the number of strains, s is the number of different types, and nJ is the number of strains belonging to type J.
Sequencing of IS6110 flanking regions.
IS6110 insert sites in two or three isolates of each IS6110 pattern were identified by a modified version of the heminested inverse PCR method (30, 46) or by ligation-mediated PCR (LMPCR) (28) with BamHI-cut DNA, followed by cloning the products in a TA cloning vector (pGEM-T Easy; Promega) and sequencing the inserts in randomly picked clones with universal forward and reverse primers.
The presence or absence of an insert at identified sites was tested in a further set of two to three isolates of each pattern by PCR with primers derived from the flanking region at each side of the insert site, followed by determination of the size of the amplified product by gel electrophoresis. In one case, this PCR provided the evidence for the presence of IS6110 in the absence of sequence data.
RESULTS
IS6110 fingerprinting and spoligotyping.
Using IS6110 restriction fragment length polymorphism analysis, fingerprints were obtained from single isolates from 2,490 patients in London during the study period (1 July 1995 to 31 December 1997). Of these, 448 (18%) had fewer than five IS6110 bands, and spoligotyping data were available for 337 of these (Table 1). Comparison of the banding patterns showed that most (nearly 80%) belonged to one of eight IS6110 types, as described in Table 2. It should be noted that this may be an underestimate of the true extent of the similarity, due to the limitations of comparing overall banding patterns with low-copy-number isolates.
TABLE 1.
Number of isolates typed by each method
| IS6110 bands | IS6110 typed | Spoligotyped | PGRS typed | Spoligotyped and PGRS typed |
|---|---|---|---|---|
| 1 | 186 | 131 | 107 | 103 |
| 2 | 112 | 92 | 75 | 71 |
| 3 | 79 | 59 | 48 | 45 |
| 4 | 71 | 55 | 47 | 46 |
| Total 1-4 | 448 | 337 | 277 | 265 |
TABLE 2.
Frequency and band sizes of major low-copy-number patterns
| IS6110 pattern | No. of:
|
% of total | Band size(s)a (kb) for the following no. of bands:
|
||||
|---|---|---|---|---|---|---|---|
| Bands | Isolates | 1 | 2 | 3 | 4 | ||
| 1344 | 1 | 36 | 11.2 | 4.94 ± 0.15 | |||
| 1350 | 1 | 92 | 29.0 | 1.44 ± 0.04 | |||
| 1312 | 2 | 5 | 1.3 | 2.07 ± 0.02 | 2.85 ± 0.04 | ||
| 2074 | 2 | 70 | 19.2 | 1.40 ± 0.02 | 4.63 ± 0.14 | ||
| 2058 | 3 | 12 | 2.7 | 1.47 ± 0.04 | 4.78 ± 0.10 | 5.01 ± 0.15 | |
| 5016 | 3 | 16 | 4.2 | 1.41 ± 0.02 | 3.18 ± 0.09 | 4.64 ± 0.12 | |
| 1641 | 4 | 8 | 2.0 | 1.10 ± 0.07 | 1.40 ± 0.02 | 4.03 ± 0.06 | 4.91 ± 0.12 |
| 5030 | 4 | 25 | 8.0 | 1.40 ± 0.01 | 2.34 ± 0.02 | 3.01 ± 0.03 | 4.88 ± 0.15 |
| Others | 1-4 | 73 | 22.3 | ||||
| Total | 1-4 | 337 | 100 | ||||
Band sizes are given as means ± standard deviations.
The congruence of IS6110 and spoligotyping was tested by using discrimination indices (18) for the two methods separately and in combination. Table 3 shows that, for this set of isolates, spoligotyping was more discriminatory than IS6110 typing for isolates with one to four copies of IS6110, although this was not true for isolates with five IS6110 copies where the value of the discrimination index for IS6110 [D(IS)] (and the degree of clustering) was similar to that for multiple-copy-number isolates (results not shown).
TABLE 3.
Discrimination analysis of isolates with one to four bands by IS6110 and spoligotyping
| No. of IS6110 bands | No. of bands
|
Degree of clusteringa | D(IS)b | D(SP)c | D(IS-SP) | Expected D(IS-SP)c | |
|---|---|---|---|---|---|---|---|
| Total | Clustered | ||||||
| 1 | 131 | 131 | 97.7 | 0.434 | 0.938 | 0.940 | 0.965 |
| 2 | 92 | 82 | 83.7 | 0.419 | 0.885 | 0.894 | 0.933 |
| 3 | 59 | 49 | 67.8 | 0.877 | 0.889 | 0.928 | 0.986 |
| 4 | 55 | 40 | 61.8 | 0.786 | 0.911 | 0.947 | 0.981 |
| Total | 337 | 302 | 82.2 | 0.863 | 0.955 | 0.980 | 0.994 |
Degree of clustering was calculated as 100 × [(number of clustered isolates) − (number of clusters)]/total number of isolates.
D(IS), D(SP), and D(IS-SP) are discrimination indices for IS6110 typing, spoligotyping, and combined IS6110 and spoligotyping data, respectively.
The expected value of D(IS-SP), if the two tests were independent, was calculated as 1 − {[1 − D(IS)][1 − D(SP)]}.
By combining the data from IS6110 typing and spoligotyping, the discrimination indices for the combination of IS6110 and spoligotyping {D(IS-SP) [Table 3]} increased. However, the D(IS-SP) values were lower than the expected values calculated on the basis of the two tests acting independently, suggesting that there is a degree of congruence between the two methods, i.e., that there is an association between the IS6110 type and the spoligotype.
Analysis of a cross-tabulation of the IS6110 and spoligotype patterns showed that there were 15 cells (specific combinations of IS6110 and spoligotype patterns) containing five or more isolates, representing 170 isolates (46%) in total. A chi-square test showed that 13 of these 15 cells were significantly larger than expected at the 1% level, while the remaining two were significant at the 5% level. A simplified version of this table is shown in Table 4. Only two spoligotypes were significantly associated with more than one IS6110 type: SP1353, with IN1344 and IN1641, and SP1097, with IN2074 and IN5016. These relationships are considered later on in this paper.
TABLE 4.
Comparison of main spoligotyping and IS6110 patterns
| Spoligobase identity | Spoligotype hexadecimal code | IS6110 patternsa (no. of bands)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1344 (1) | 1350 (1) | 1312 (2) | 2074 (2) | 2058 (3) | 5016 (3) | 1641 (4) | 5030 (4) | Other | Total | ||
| 1301 | 7F-7F-7F-7F-0B-77 | 10* | 1 | 6 | 17 | ||||||
| 1298 | 7F-7F-7F-7F-0B-70 | 5* | 2 | 7 | |||||||
| 212 | 4F-7F-7A-7F-0B-7F | 6** | 0 | 6 | |||||||
| 235 | 4F-7F-7F-7F-0B-7F | 11** | 1 | 0 | 12 | ||||||
| 232 | 4F-7F-7F-7F-0B-0F | 20** | 1 | 1 | 7 | 29 | |||||
| 1104 | 7F-7F-77-7F-F0-7F | 1 | 8** | 5 | 14 | ||||||
| 552 | 70-03-77-7F-F0-7F | 9** | 1 | 10 | |||||||
| 1353 | 7F-7F-7F-7F-F0-7F | 19** | 1 | 8** | 2 | 30 | |||||
| 1090 | 7F-7F-77-7F-80-1F | 12** | 0 | 12 | |||||||
| 1097 | 7F-7F-77-7F-F0-61 | 1 | 1 | 25** | 1 | 14** | 4 | 46 | |||
| 483 | 6F-7F-77-7F-F0-61 | 5** | 0 | 5 | |||||||
| 163 | 47-7F-77-7F-F0-61 | 1 | 11** | 0 | 12 | ||||||
| 1127 | 7F-7F-7C-09-F0-7F | 4 | 0 | 4 | |||||||
| 858 | 7F-60-03-7F-F0-7F | 1 | 7** | 4 | 12 | ||||||
| Other | 14 | 38 | 1 | 14 | 4 | 1 | 0 | 7 | 42 | 121 | |
| Total | 36 | 92 | 5 | 70 | 12 | 16 | 8 | 25 | 73 | 337 | |
*, significance at 5%; **, significance at 1%.
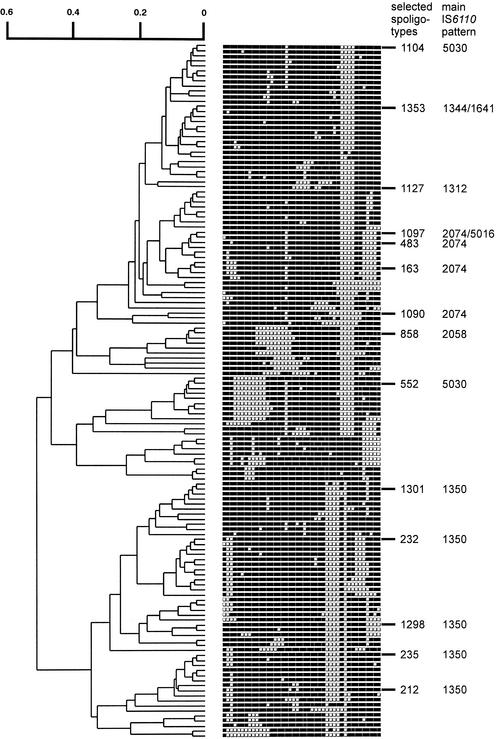
One limitation of this analysis is that it takes no account of the relationship or otherwise between the different spoligotype patterns. Examination of the hexadecimal representation (4) of the spoligotype patterns (Table 4) shows additional relationships between IS6110 patterns and spoligotypes. For example, the five most common spoligotypes in the IN1350 pattern are actually closely related to one another, especially in lacking spacers 29 to 32 and 34. Further comparison was achieved by mapping the major IS6110 type and spoligotype patterns, as delineated in Table 4, onto a dendrogram of spoligotype patterns (Fig. 1). The relatedness of the spoligotypes associated with IN1350, and their distinction from other spoligotypes, was immediately apparent, as was the relationship between the spoligotypes of IN2074 and IN5016 isolates.
FIG. 1.
Comparison of spoligotypes. Dendrogram of spoligotype patterns produced by using Taxotron. The (1-Jaccard) index was calculated for pairwise comparison of strains, and the dendrogram was produced by the unweighted pair group using arithmetic averages method. The most common spoligotypes, as referred to in the text, are shown to the right of the patterns, together with the IS6110 pattern(s) that are most frequent within each major spoligotype.
PGRS data.
One possible reason for the apparent congruence of IS6110 and spoligotyping results would be the existence of epidemiological clusters of identical isolates, although this seems unlikely to account for the large size of the identical groups. Further investigation of this point, and of the relationship between the different types, was attempted by using PGRS typing, for which results were available for 265 out of the 337 isolates considered above. Although we found that the PGRS data were insufficiently robust for the quantitative analysis of discrimination, it was possible to use the data for direct comparison of subsets of the isolates. Epidemiologically linked isolates would normally be expected to show identical PGRS patterns (13), as well as identity by IS6110 and spoligotyping. However, there were few clusters of more than two or three isolates that were shown to be identical by all three methods. One of these clusters (IN1641:SP1353) consisted of eight isolates from a known hospital outbreak (1) and is therefore an example of a genuine epidemiological cluster.
Several small clusters of isolates apparently identical by all three methods were identified among isolates with IN2074 and IN5016 patterns. This would be consistent with the occurrence of an epidemiological relationship between these isolates but does not exclude other possibilities. A comparison of the PGRS data for 69 isolates with one of these two IS6110 patterns showed a very high degree of similarity (as was also the case for spoligotype patterns), while isolates with the other two- and three-band patterns showed readily distinguishable PGRS patterns (data not shown). This indicates that the IN2074 and IN5016 types are closely related, to the extent that even PGRS typing is unable to distinguish them reliably.
Despite the problems of the global analysis of PGRS data, one feature was apparent from the overall cluster analysis. The PGRS patterns identified were found to fall into three groups, each with a similarity of 56 to 60%. These groups had very different characteristics. Table 5 shows that group 1 is almost exclusively comprised of IN1350 while the other single-copy type (IN1344) is virtually limited to group 2, together with all the isolates of the two-banded IN1312 and three-banded IN2058 patterns. Almost all of the other isolates with two to four copies of IS6110 (patterns IN2074, IN5016, IN1641, and IN5030) were grouped together in group 3. This provides further indication that the differentiation of these isolates by IS6110 typing reflects a fundamental difference in the nature and evolutionary origin of these strain families.
TABLE 5.
Correlation of PGRS groups and IS6110 patterns
| PGRS group | % PGRS similarity | No. of isolates | No. of single-band isolates | % of single-band isolates | No. of isolates with the following specific IS6110 pattern:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1344 | 1350 | 1312 | 2074 | 5016 | 2058 | 1641 | 5030 | |||||
| 1 | 60 | 96 | 73 | 76 | 1 | 69 | 0 | 0 | 1 | 0 | 0 | 0 |
| 2 | 56 | 46 | 26 | 54 | 23 | 2 | 5 | 1 | 0 | 9 | 0 | 0 |
| 3 | 57 | 122 | 5 | 4 | 3 | 2 | 0 | 52 | 14 | 0 | 8 | 22 |
| Total | 264a | 103 | 39 | 27 | 73 | 5 | 53a | 15 | 9 | 8 | 22 | |
One isolate with an abnormal pattern was excluded.
Geographical origin of strains.
Further evidence of the distinct origin of these strains was obtained by analysis of the country of birth of the persons infected (Table 6). These data were available for only 50% of the patients, but the distribution of the IS6110 types among patients with an unknown country of birth was not statistically different from the overall distribution (with the exception of IN1641), indicating that it is valid to use these data to analyze the relative occurrence of each IS6110 type among patients born in different regions.
TABLE 6.
Origin of low-copy-number isolates by country of birth
| Region | No. of isolates with the following specific IS6110 pattern (no. of bands)d:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1312 (2) | 1344 (1) | 1350 (1) | 1641 (4) | 2058 (3) | 2074 (2) | 5016 (3) | 5030 (4) | Other | Total | |
| Africaa | 1 | 10 | 23 | 2 | 7* | 7 | 2 | 1 | 11 | 64 |
| South Asiab | 0 | 3 | 21* | 0 | 1 | 2 | 0 | 0 | 12 | 39 |
| Europec | 3 | 1 | 5 | 6* | 1 | 17** | 7* | 7* | 7 | 54 |
| Other | 0 | 2 | 5 | 0 | 0 | 1 | 1 | 2 | 3 | 14 |
| Total | 4 | 16 | 54 | 8 | 9 | 27 | 10 | 10 | 33 | 171 |
| Data missing | 1 | 20 | 38 | 0 | 3 | 43 | 6 | 15 | 40 | 166 |
| Overall total | 5 | 36 | 92 | 8 | 12 | 70 | 16 | 25 | 73 | 337 |
Mainly Somalia, Kenya, Ethiopia, Eritrea, and Uganda.
India, Pakistan, and Bangladesh.
Mainly the United Kingdom and Ireland.
* denotes significance at 5%, and ** denotes significance at 1% (omitting missing values), both by the chi-square test.
Among persons born in South Asia (India, Pakistan, and Bangladesh), type IN1350 was predominant in this set of isolates, while persons born in Africa (mainly Northeast African countries, especially Somalia) had a high proportion of types IN2058, IN1344, and IN1350. In contrast, these types were relatively infrequent in persons born in Europe (mainly the United Kingdom and Ireland), where there was an association with types IN2074, IN5016, and IN5030.
IS6110 insert sites.
In order to establish whether bands at similar positions actually represent inserts at the same site, we sequenced the regions flanking IS6110 in isolates of each of the major IS6110 patterns referred to previously, confirming the results by a PCR test of additional isolates. Eight different insertion sites were identified (Table 7). Two insertion sites (one each in the IN1312 and IN1641 patterns) were not identified by either heminested PCR or LMPCR, and the PCR test confirmed the absence of an insert at any of the other identified sites in these strains. These two sites therefore remain undetermined. All isolates contained an insert at site A (in the DR region), despite the fact that not all patterns contained a band at the position commonly associated with this insert (1.4 kb). Particularly notable is that the two single-copy patterns (IN1344 and IN1350) were identical in the insert site and orientation of the insertion sequence, although the fragment size detected was very different (4.9 kb for IN1344 versus 1.4 kb for IN1350 [Table 2]). The only other insert site occupied in more than one insertion sequence type was site B, in patterns IN2074, IN5016, IN1641, and IN5030. Two of the sites listed (sites A and D) were also occupied in H37Rv; strain CDC1551 also had an insert at these two sites as well as two other inserts, both of which were represented in this collection (sites B and G). The inserts in CDC1551 were identical to those in our IN5030 type, which also showed a similar banding pattern. It should also be noted that, in common with Fomukong et al. (11), we did not find any inserts at the alternative site (H37Rv position 851630) occupied in those BCG strains that have two copies of IS6110 (12).
TABLE 7.
IS6110 insertion sites
| Insert code | Insertion point | Orientationa | Gene designation | Presenceb of the following IS6110 patterns detected by the no. of bands:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1
|
2
|
3
|
4
|
||||||||
| 1344 | 1350 | 1312c | 2074 | 5016 | 2058 | 1641c | 5030 | ||||
| A | 3120523-3121877 | c | Rv2813-Rv2816c (DR region) | + | + | + | + | + | +d | + | + |
| B | 483299 | c | Rv0403c | 0 | 0 | 0 | + | + | 0 | +e | + |
| D | 1987703-1989057 | c | Rv1758 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | + |
| F | (1978901)f | c | MT1799c | 0 | 0 | 0 | 0 | 0 | 0 | + | 0 |
| G | 3377325 | d | Rv3018c | 0 | 0 | 0 | 0 | 0 | 0 | 0 | + |
| H | 3125090 | d | Rv2818c | 0 | 0 | 0 | 0 | 0 | + | 0 | 0 |
| J | 1984903 | d | Rv1753c-Rv1754c | 0 | 0 | 0 | 0 | + | 0 | 0 | 0 |
| K | 2368235 | d | Rv2108 | — | — | 0 | — | — | +e | 0 | — |
d and c, direct and inverse orientations, respectively, with regards to the genome sequence.
+, IS6110 presence identified by sequencing heminested inverse PCR product (except where indicated); 0, IS6110 absence confirmed by PCR; —, PCR not done.
One insert site not identified.
Identified by PCR only.
From the LM-PCR product.
Site not present in H37Rv; the position shown is that in strain CDC1551.
DISCUSSION
The data reported here show a substantial degree of congruence, for the set of isolates studied, between the IS6110 and spoligotyping results. This suggests that the limited polymorphism of IS6110 patterns in low-copy-number isolates is not due to frequent independent transposition into hot spots but rather is consistent with the alternative hypothesis that the mobility of IS6110 is low in such strains (i.e., the IS6110 molecular clock operates more slowly) and that at least a high proportion of isolates with each low-copy-number IS6110 pattern represents a coherent strain type with a common evolutionary history. The congruence of the two typing methods also has practical implications for the epidemiological investigation of such strains.
A correspondence of IS6110 type and spoligotype, for isolates with few copies of IS6110, has also been reported by others (3, 19, 45). On the other hand, Soini et al. (34) found that spoligotyping was able to discriminate between members of low-copy-number IS6110 clusters and concluded that the combination of IS6110 profile and spoligotype identified true clustering. However, the results presented here suggest that it is not valid to include such apparent clusters in the overall estimate of the extent of recent transmission.
Determination of the sites of insertion of IS6110 provides further evidence of the evolutionary history of these strains, especially when the data reported here are compared with those obtained by Sampson et al. (31) for a set of multiple-copy-number isolates and by Fomukong et al. (11), who examined both high- and low-copy-number strains; in addition, we have unpublished data for a large set of multicopy isolates. In all these cases, there was a preponderance of inserts at the site in the DR region (designated site A here), which corresponds to the site identified by Hermans et al. (16). Our results show that both major single-copy patterns contain an insert at this position, despite the wide difference in the band size detected. Minor changes in the size of this band will arise through polymorphism of the DR region itself, in the loss of one or more spacers, but such a large difference in fragment size is more likely to be due to other forms of polymorphism, such as the gain or loss of a restriction site.
The only other site identified in this paper that was occupied in isolates showing different band numbers or patterns was site B, in types IN2074, IN5016, IN1641, and IN5030. The similarity in spoligotypes and PGRS patterns between IN2074 and IN5016 suggests that IN5016 was derived from IN2074 by transposition into site J. None of the other sites described here were found in the multicopy isolates studied by Sampson et al. (31), nor in our own unpublished data for multicopy isolates. Our data are therefore consistent with the conclusion of Fomukong et al. (11) that most of the presently circulating low- and high-copy-number strains represent separate lineages rather than a continuing evolution of low-copy-number to high-copy-number strains by replicative transposition. This is consistent with the finding of Sreevatsan et al. (38) that the frequency distribution of IS6110 copy number differed among the three genotypic groups defined by sequence polymorphisms in katG and gyrA. Furthermore, the low frequency of occupation of most of these sites (other than that in the DR region, site A) in multicopy isolates suggests that the common occupation of certain sites in low-copy-number isolates is due to the stability of those inserts, possibly due to low transcriptional activity (43), rather than the preferential insertion of IS6110 at those positions.
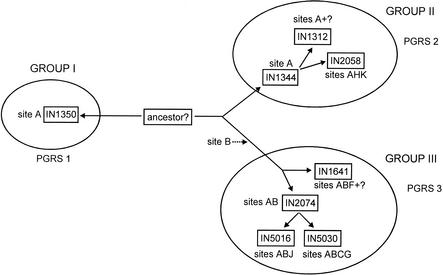
The IS6110 insert site data, combined with the evidence from spoligotyping and PGRS typing, can be used to trace the apparent relationship between these strains, as indicated diagrammatically in Fig. 2. Group I consists of the single-copy strain represented by IN1350 corresponding to the East African Indian family (19, 35, 36), which is quite distinct from the other low-copy-number isolates on all counts and may represent an ancient divergence from a common ancestor. This is in accord with the data of Soini et al. (34), who identified similar single-copy-number strains as belonging to major genetic group 1 while virtually all the other low-copy-number strains were genetic group 2. Furthermore, Brosch et al. (2) found that strains with a spoligotype similar to that of the East African Indian family (lacking spacers 29 to 32 and 34) were distinct from other isolates in the presence of the TbD1 region; these strains were also clustered by mycobacterial interspersed repetitive unit-variable-number tandem repeat analysis (39). These isolates were predominant in persons born in South Asia (India and Pakistan) in our study (P < 0.01), with a substantial number also from Northeast Africa, while in the study by Soini et al. (34), similar isolates were predominantly from Vietnamese patients, reflecting the different background of the populations involved.
FIG. 2.
Diagrammatic representation of the relationships among low-copy-number isolates of M. tuberculosis. The relationships depicted are based primarily on the determined IS6110 insert sites, supplemented by the data derived from PGRS typing and spoligotyping. The relationships are shown diagrammatically only; the lengths of the arms do not represent evolutionary distance.
The other two groups are less homogeneous. Group II, containing the other major single-copy type (IN1344), and group III are distinguished from one another by the presence of an insert at site B in group III as well as by PGRS typing. Within group II, the majority of types IN1344 and IN2058 were isolated from persons born in Africa. These patterns resemble, respectively, the E1 and E2 types found to be common in Ethiopia (15), while IN2058 is similar to a pattern described in patients from Eritrea (6). The group III isolates in this study were predominantly (P < 0.01) from Europe (largely the United Kingdom and Ireland), although they are likely to be more widespread. In particular, IN5030 is similar in pattern, and contains inserts at the same positions, as both CDC1551 and that described by Mendiola et al. (26) and is also apparently similar to the four-band strains common in other studies (34, 37). IN2074 appears similar, in IS6110 pattern and spoligotype, to the JH2 pattern found to be common in Alabama (23). The strains in group III in general conform in spoligotype to the definition of clade X, which has been reported to be prevalent in English-speaking countries (32). Thus, there is good agreement between the groups defined primarily by IS6110 insertion sites and those defined by spoligotype similarities. PCR-based deletion analysis (2, 27, 29) and genomic microarrays (21) will enable further investigation of the evolutionary relationships between these low-copy-number M. tuberculosis strains and other members of the M. tuberculosis complex.
The concept that individual IS6110 patterns among the low-copy-number isolates represent distinct evolutionary lineages implies that we can consider these patterns analogous to families of multiple-copy-number isolates such as the Beijing family. These strains therefore represent a valuable resource for analyzing the influence of host strains on the nature and pattern of disease.
Acknowledgments
We are grateful for support from the NHS Executive London Research and Development Programme, from the European Union under grants BMH4-CT97-91202 and SMT4-CT96-2097 (provision of GelCompar and Bionumerics software), and from the Wellcome Trust (reference 056133).
REFERENCES
- 1.Breathnach, A. S., A. de Ruiter, G. M. Holdsworth, N. T. Bateman, D. G. O'Sullivan, P. J. Rees, D. Snashall, H. J. Milburn, B. S. Peters, J. Watson, F. A. Drobniewski, and G. L. French. 1998. An outbreak of multi-drug-resistant tuberculosis in a London teaching hospital. J. Hosp. Infect. 39:111-117. [DOI] [PubMed] [Google Scholar]
- 2.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cronin, W. A., J. E. Golub, L. S. Magder, N. G. Baruch, M. J. Lathan, L. N. Mukasa, N. Hooper, J. H. Razeq, D. Mulcahy, W. H. Benjamin, and W. R. Bishai. 2001. Epidemiologic usefulness of spoligotyping for secondary typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110. J. Clin. Microbiol. 39:3709-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dale, J. W., D. Brittain, A. A. Cataldi, D. Cousins, J. T. Crawford, J. Driscoll, H. Heersma, T. Lillebaek, T. Quitugua, N. Rastogi, R. A. Skuce, C. Sola, D. Van Soolingen, and V. Vincent. 2001. Spacer oligonucleotide typing of bacteria of the Mycobacterium tuberculosis complex: recommendations for standardised nomenclature. Int. J. Tuberc. Lung Dis. 5:216-219. [PubMed] [Google Scholar]
- 5.Dale, J. W., T. H. Tang, S. Wall, Z. F. Zainuddin, and B. Plikaytis. 1998. Conservation of IS6110 sequence in strains of Mycobacterium tuberculosis with single and multiple copies. Tuberc. Lung Dis. 78:225-227. [DOI] [PubMed] [Google Scholar]
- 6.Diel, R., S. Schneider, K. Meywald-Walter, C. M. Ruf, S. Rusch-Gerdes, and S. Niemann. 2002. Epidemiology of tuberculosis in Hamburg, Germany: long-term population-based analysis applying classical and molecular epidemiological techniques. J. Clin. Microbiol. 40:532-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doran, T. J., A. L. M. Hodgson, J. K. Davies, and A. J. Radford. 1993. Characterisation of a highly repeated DNA sequence from Mycobacterium bovis. FEMS Microbiol. Lett. 111:147-152. [DOI] [PubMed] [Google Scholar]
- 8.Fang, Z., C. Doig, N. Morrison, B. Watt, and K. J. Forbes. 1999. Characterization of IS1547, a new member of the IS900 family in the Mycobacterium tuberculosis complex, and its association with IS6110. J. Bacteriol. 181:1021-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang, Z., and K. J. Forbes. 1997. A Mycobacterium tuberculosis IS6110 preferential locus (ipl) for insertion into the genome. J. Clin. Microbiol. 35:479-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang, Z., D. T. Kenna, C. Doig, D. N. Smittipat, P. Palittapongarnpim, B. Watt, and K. J. Forbes. 2001. Molecular evidence for independent occurrence of IS6110 insertions at the same sites of the genome of Mycobacterium tuberculosis in different clinical isolates. J. Bacteriol. 183:5279-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fomukong, N. G., M. Beggs, H. Hajj, G. Templeton, K. Eisenach, and M. D. Cave. 1998. Differences in the prevalence of IS6110 insertion sites in Mycobacterium tuberculosis strains: low and high copy number of IS6110. Tuber. Lung Dis. 78:109-116. [DOI] [PubMed] [Google Scholar]
- 12.Fomukong, N. G., T. H. Tang, S. Al-Maamary, W. A. Ibrahim, S. Ramayah, M. Yates, Z. F. Zainuddin, and J. W. Dale. 1994. Insertion sequence typing of Mycobacterium tuberculosis: characterization of a widespread sub-type with a single copy of IS6110. Tuber. Lung Dis. 75:435-440. [DOI] [PubMed] [Google Scholar]
- 13.Gillespie, S. H., A. Dickens, and T. D. McHugh. 2000. False molecular clusters due to nonrandom association of IS6110 with Mycobacterium tuberculosis. J. Clin. Microbiol. 38:2081-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayward, A. C., S. Goss, F. Drobniewski, N. Saunders, R. J. Shaw, M. Goyal, A. Swan, A. Uttley, A. Pozniak, J. Grace-Parker, and J. M. Watson. 2002. The molecular epidemiology of tuberculosis in inner London. Epidemiol. Infect. 128:175-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermans, P. W. M., F. Messadi, H. Guebrexabher, D. van Soolingen, P. E. W. de Haas, H. Heersma, H. De Neeling, A. Ayoub, F. Portaels, D. Frommel, M. Zribi, and J. D. A. van Embden. 1995. Analysis of the population structure of Mycobacterium tuberculosis in Ethiopia, Tunisia, and The Netherlands: usefulness of DNA typing for global tuberculosis epidemiology. J. Infect. Dis. 171:1504-1513. [DOI] [PubMed] [Google Scholar]
- 16.Hermans, P. W. M., D. van Soolingen, E. M. Bik, P. E. W. de Haas, J. W. Dale, and J. D. A. van Embden. 1991. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect. Immun. 59:2695-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hermans, P. W. M., D. van Soolingen, J. W. Dale, A. R. J. Schuitema, R. A. McAdam, D. Catty, and J. D. A. van Embden. 1990. Insertion element IS986 from Mycobacterium tuberculosis: a useful tool for diagnosis and epidemiology of tuberculosis. J. Clin. Microbiol. 28:2051-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Källenius, G., T. Koivula, S. Ghebremichael, S. E. Hoffner, R. Norberg, E. Svensson, F. Dias, B. I. Marklund, and S. B. Svenson. 1999. Evolution and clonal traits of Mycobacterium tuberculosis complex in Guinea-Bissau. J. Clin. Microbiol. 37:3872-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato-Maeda, M., J. T. Rhee, T. R. Gingeras, H. Salamon, J. Drenkow, N. Smittipat, and P. M. Small. 2001. Comparing genomes within the species Mycobacterium tuberculosis. Genome Res. 11:547-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kivi, M., X. M. Liu, S. Raychaudhuri, R. B. Altman, and P. M. Small. 2002. Determining the genomic locations of repetitive DNA sequences with a whole-genome microarray: IS6110 in Mycobacterium tuberculosis. J. Clin. Microbiol. 40:2192-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lok, K. H., W. H. Benjamin, M. E. Kimerling, V. Pruitt, D. Mulcahy, N. Robinson, N. B. Keenan, and N. E. Dunlap. 2002. Molecular typing of Mycobacterium tuberculosis strains with a common two-band IS6110 pattern. Emerg. Infect. Dis. 8:1303-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maguire, H., J. W. Dale, T. D. McHugh, P. D. Butcher, S. H. Gillespie, A. Costetsos, H. Al-Ghusein, R. Holland, A. Dickens, L. Marston, P. Wilson, R. Pitman, D. Strachan, F. A. Drobniewski, and D. K. Banerjee. 2002. Molecular epidemiology of tuberculosis in London 1995-7 showing low rate of active transmission. Thorax 57:617-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McHugh, T. D., and S. H. Gillespie. 1998. Nonrandom association of IS6110 and Mycobacterium tuberculosis: implications for molecular epidemiological studies. J. Clin. Microbiol. 36:1410-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mendiola, M. V., C. Martín, I. Otal, and B. Gicquel. 1992. Analysis of the regions responsible for IS6110 RFLP in a single Mycobacterium tuberculosis strain. Res. Microbiol. 143:767-772. [DOI] [PubMed] [Google Scholar]
- 27.Mostowy, S., D. Cousins, J. Brinkman, A. Aranaz, and M. A. Behr. 2002. Genomic deletions suggest a phylogeny for the Mycobacterium tuberculosis complex. J. Infect. Dis. 186:74-80. [DOI] [PubMed] [Google Scholar]
- 28.Palittapongarnpim, P., S. Chomyc, A. Fanning, and D. Kunimoto. 1993. DNA fingerprinting of Mycobacterium tuberculosis isolates by ligation-mediated polymerase chain reaction. Nucleic Acids Res. 21:761-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parsons, L. M., R. Brosch, S. T. Cole, A. Somoskovi, A. Loder, G. Bretzel, D. van Soolingen, Y. M. Hale, and M. Salfinger. 2002. Rapid and simple approach for identification of Mycobacterium tuberculosis complex isolates by PCR-based genomic deletion analysis. J. Clin. Microbiol. 40:2339-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel, S., S. Wall, and N. A. Saunders. 1996. Heminested inverse PCR for IS6110 fingerprinting of Mycobacterium tuberculosis strains. J. Clin. Microbiol. 34:1686-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sampson, S. L., R. M. Warren, M. Richardson, G. D. van der Spuy, and P. D. Van Helden. 1999. Disruption of coding regions by IS6110 insertion in Mycobacterium tuberculosis. Tuber. Lung Dis. 79:349-359. [DOI] [PubMed] [Google Scholar]
- 32.Sebban, M., I. Mokrousov, N. Rastogi, and C. Sola. 2002. A data-mining approach to spacer oligonucleotide typing of Mycobacterium tuberculosis. Bioinformatics 18:235-243. [DOI] [PubMed] [Google Scholar]
- 33.Small, P. M., P. C. Hopewell, S. P. Singh, A. Paz, J. Parsonnet, D. C. Ruston, G. F. Schecter, C. L. Daley, and G. K. Schoolnik. 1994. The epidemiology of tuberculosis in San Francisco—a population-based study using conventional and molecular methods. N. Engl. J. Med. 330:1703-1709. [DOI] [PubMed] [Google Scholar]
- 34.Soini, H., X. Pan, L. Teeter, J. M. Musser, and E. A. Graviss. 2001. Transmission dynamics and molecular characterization of Mycobacterium tuberculosis isolates with low copy numbers of IS6110. J. Clin. Microbiol. 39:217-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sola, C., I. Filliol, M. C. Gutierrez, I. Mokrousov, V. Vincent, and N. Rastogi. 2001. Spoligotype database of Mycobacterium tuberculosis: biogeographic distribution of shared types and epidemiologic and phylogenetic perspectives. Emerg. Infect. Dis. 7:390-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sola, C., I. Filliol, E. Legrand, I. Mokrousov, and N. Rastogi. 2001. Mycobacterium tuberculosis phylogeny reconstruction based on combined numerical analysis with IS1081, IS6110, VNTR, and DR-based spoligotyping suggests the existence of two new phylogeographical clades. J. Mol. Evol. 53:680-689. [DOI] [PubMed] [Google Scholar]
- 37.Sola, C., L. Horgen, K. S. Goh, and N. Rastogi. 1997. Molecular fingerprinting of Mycobacterium tuberculosis on a Caribbean island with IS6110 and DRr probes. J. Clin. Microbiol. 35:843-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Supply, P., S. Lesjean, E. Savine, K. Kremer, D. van Soolingen, and C. Locht. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka, M. M., P. M. Small, H. Salamon, and M. W. Feldman. 2000. The dynamics of repeated elements: applications to the epidemiology of tuberculosis. Proc. Natl. Acad. Sci. USA 97:3532-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Embden, J. D. A., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Soolingen, D., M. W. Borgdorff, P. E. W. de Haas, M. M. G. G. Sebek, J. Veen, M. Dessens, K. Kremer, and J. D. A. van Embden. 1999. Molecular epidemiology of tuberculosis in the Netherlands: a nationwide study from 1993 through 1997. J. Infect. Dis. 180:726-736. [DOI] [PubMed] [Google Scholar]
- 43.Wall, S., K. Ghanekar, J. McFadden, and J. W. Dale. 1999. Context-sensitive transposition of IS6110 in mycobacteria. Microbiology 145:3169-3176. [DOI] [PubMed] [Google Scholar]
- 44.Warren, R. M., S. L. Sampson, M. Richardson, G. D. van der Spuy, C. J. Lombard, T. C. Victor, and P. D. Van Helden. 2000. Mapping of IS6110 flanking regions in clinical isolates of Mycobacterium tuberculosis demonstrates genome plasticity. Mol. Microbiol. 37:1405-1416. [DOI] [PubMed] [Google Scholar]
- 45.Yang, Z. H., K. Ijaz, J. H. Bates, K. D. Eisenach, and M. D. Cave. 2000. Spoligotyping and polymorphic GC-rich repetitive sequence fingerprinting of Mycobacterium tuberculosis strains having few copies of IS6110. J. Clin. Microbiol. 38:3572-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yesilkaya, H., A. Thomson, C. Doig, B. Watt, J. W. Dale, and K. J. Forbes. Locating transposable element polymorphisms in bacterial genomes. J. Microbiol. Methods, in press. [DOI] [PubMed]
- 47.Zainuddin, Z. F., and J. W. Dale. 1989. Polymorphic repetitive DNA sequences in Mycobacterium tuberculosis detected with a gene probe from a Mycobacterium fortuitum plasmid. J. Gen. Microbiol. 135:2347-2355. [DOI] [PubMed] [Google Scholar]