Abstract
The alternative sigma factor σE is activated in response to stress in the extracytoplasmic compartment of Escherichia coli. Here we show that σE activity increases upon initiation of the stress response by a shift to an elevated temperature (43°C) and remains at that level for the duration of the stress. When the stress is removed by a temperature downshift, σE activity is strongly repressed and then slowly returns to levels seen in unstressed cells. We provide evidence that information about the state of the cell envelope is communicated to σE primarily through the regulated proteolysis of the inner membrane anti-sigma factor RseA, as the degradation rate of RseA is correlated with the changes in σE activity throughout the stress response. However, the relationship between σE activity and the rate of degradation of RseA is complex, indicating that other factors may cooperate with RseA and serve to fine-tune the response.
Protein denaturation and aggregation resulting from exposure to elevated temperatures or other stresses present a severe problem for cells. However, the detrimental effects of these stresses are combated by highly conserved responses that increase the synthesis of the heat shock proteins, whose function is to remove or refold damaged cellular proteins (5, 13). The transcription factors that control heat shock gene expression are subject to complex regulatory mechanisms to ensure that the response is rapidly induced after exposure to stress, is maintained when the cells are subject to continuous stress, and is downregulated when cells are returned to the unstressed state (5, 6, 13, 24, 35, 36).
In the bacterium Escherichia coli, individual stress responses maintain homeostasis in the cytoplasm and cell envelope. The alternative sigma factors σE and σ32 control the cell envelope and cytoplasmic heat shock responses, respectively (14, 17, 19, 27, 28, 34, 37). The σ32-dependent stress response has been extensively characterized. The stability, activity, and rate of translation of σ32 are all subject to regulation depending upon the circumstance (12, 13, 15, 25, 26, 31-33). This complex network of regulatory interactions modulates the amount and activity of σ32 so that the amount of active σ32 is optimal for the needs of the cell.
The cell envelope plays an essential role for the bacterium providing a barrier between the cell and the environment, determining cellular morphology, and maintaining the structural integrity of the cell. Although σE is crucial for maintaining the integrity of the cell envelope during the stress response, little is known about its regulation. σE is activated both by general stress inducers, such as a heat shock, which lead to protein unfolding in both cellular compartments, and by specific inducers, such as overexpression of porins, which lead to accumulation of unfolded proteins solely in the cell envelope (20, 21, 23, 27-29). Once activated, σE transcribes a set of genes primarily targeted to the cell envelope that encode periplasmic proteases, folding catalysts, several key enzymes in the biosynthesis pathways of cell envelope constituents and a number of lipoproteins (10; V. Rhodius, W. Suh, S. Ades, C. Onufryk, M. Igo, and C. A. Gross, unpublished data).
Since σE resides in the cytoplasm, it cannot directly sense damage in the cell envelope. The key protein responsible for directly communicating damage in the envelope to σE is the anti-sigma factor RseA. RseA is an inner membrane protein with a single transmembrane-spanning segment. The N-terminal cytoplasmic domain of RseA binds to σE and inhibits the activity of σE, and its C-terminal periplasmic domain binds RseB, a weak inhibitor of σE activity (11, 22). The interaction between RseA and σE must be disrupted for σE to bind to RNA polymerase and initiate transcription. σE, RseA, and RseB are all members of an operon that is transcribed by σE itself (11, 22, 28). This autoregulatory loop allows the cell to tightly control σE activity, since upon activation both σE and its inhibitors are transcribed together.
Previous studies on the regulation of σE activity focused solely on the initiation phase of the stress response (1). These studies indicated that the major pathway for releasing σE from RseA is through the regulated proteolysis of RseA carried out by the inner membrane proteases DegS and YaeL (1, 2, 18). Here we investigate the regulation of σE activity during all phases of the stress response: initiation, adaptation, and shutoff. We find that σE activity increases during initiation of the stress response and remains elevated as cells are maintained under stress during the adaptation phase of the response. During the shutoff phase of the response when the stress is removed, σE activity drops to a level below that observed under normal conditions and then slowly increases. Our earlier experiments indicated that increased proteolysis of RseA is responsible for the increase in σE activity upon initiation of the stress response (1, 2, 18). Therefore, we looked at whether alterations in the proteolysis of RseA can account for the changes in σE activity during the other phases of the response or whether additional modes of regulation are utilized. We find that σE activity is correlated with the degradation rate of RseA during all phases of the response, which indicates that RseA degradation is the primary means by which information about the state of the cell envelope is communicated to σE in the cytoplasm. However, changes in the rate of RseA degradation are not sufficient to account for all of the observed changes in σE activity, indicating that other regulatory mechanisms are likely to be involved.
MATERIALS AND METHODS
Media and strains.
M9 minimal medium was prepared as described previously (30). M9 medium was supplemented with 0.2% glucose, 1 mM MgSO4, 2 μg of thiamine per ml, and all amino acids (40 μg/ml) except for the media used in the pulse-label and pulse-chase experiments in which methionine was omitted. The bacterial strains used in this work were E. coli CAG45114 (MG1655 Φλ[rpoHP3-lacZ] ΔlacX74), CAG16037 {MC1061 Φλ[rpoHP3-lacZ] araD Δ(ara-leu)7697 Δ(codB-lacI) galK16 galE15 mcrA0 relA1 rpsL150 spoT1 mcrB9999 hsdR2} (20), and CAG33149 [BL21(DE3) pLC234 Kmr] (11). pLC234 encodes the periplasmic domain of RseA fused to the N-terminal histidine tag in pET28b (11).
Determination of synthesis rates by pulse-labeling immunoprecipitation.
Cells were grown in supplemented M9 minimal medium lacking methionine to an optical density at 450 nm (OD450) of 0.2 to 0.4. Proteins were labeled at the indicated times by adding a 900-μl aliquot of the culture to 2 μl of l-[35S]methionine in a prewarmed 50-ml conical tube with shaking at the desired temperature. The samples were incubated for 45 s, and then further incorporation of radioactive methionine was blocked by adding 100 μl of a 0.1% solution of unlabeled methionine. After 30 s, the entire sample was added to an Eppendorf tube containing 100 μl of ice-cold trichloroacetic acid (TCA). Samples were incubated on ice for at least 15 min, and then precipitated proteins were collected by centrifugation. The pellets were resuspended by vortexing and boiling in 50 μl of 20 mM NaPO4 (pH 7.5), 10 mM EDTA, 2% sodium dodecyl sulfate (SDS), 1 mM AEBSF [4-(2-aminoethyl)benzenesulfonyl fluoride, HCl], after which 750 μl of phosphate radioimmunoprecipitation assay (PO4 RIPA) buffer (20 mM NaPO4 [pH 7.5], 0.5 M NaCl, 0.5% sodium deoxycholate, 1% IGEPAL [(octylphenoxy)polyethoxyethanol octylphenyl-polyethylene glycol], 0.1% SDS) was added. The radioactivity in each sample was counted in duplicate in a scintillation counter. DegP and RseA were immunoprecipitated to determine the amount of each protein synthesized during the 45-s pulse as a fraction of total protein synthesis. Samples were normalized to the overall amount of protein synthesis by adding equal counts per minute of each sample to the immunoprecipitation reaction mixtures. The final volume of the immunoprecipitation reaction mixtures was 500 μl, and the reaction mixtures contained the sample, polyclonal antibody raised against the periplasmic domain of RseA, polyclonal antibody raised against DegP, and 20 μl of a 1:1 slurry of UltraLink Immobilized protein A beads (Pierce) in PO4 RIPA buffer. As an internal standard, a culture overexpressing the periplasmic domain of RseA (strain CAG33149) was pulse-labeled, and an aliquot was added to each sample before immunoprecipitation. The samples were rocked at 4°C for 1 h, and then immune complexes were collected by centrifugation. The beads were washed three times with PO4 RIPA buffer and once with 1× phosphate-buffered saline. Proteins were eluted from the beads by boiling for 5 min in 30 μl of Laemmli sample buffer. The samples were loaded onto 15% Tris-glycine gels, and proteins were visualized with the Molecular Dynamics Storm 560 PhosphorImager scanning system. Bands were quantified using the program ImageQuant 1.2, and the intensity of the band for full-length RseA or DegP in each lane was normalized, after background correction, to the intensity of the band for the internal standard, the periplasmic domain of RseA, in that lane.
Determination of the half-life of RseA by pulse-chase immunoprecipitation.
Cells were grown in supplemented M9 minimal medium lacking methionine to an OD450 of 0.2 to 0.4 at the specified growth temperature. In experiments to measure the half-life of RseA after a shift in growth temperature, the entire culture was shifted to the new temperature rather than splitting the culture by placing an aliquot in a new flask. We found that moving the entire flask gave the most reliable and reproducible measurements of RseA stability. Proteins were pulse-labeled for 1 min by the addition of l-[35S]methionine to the growing culture after which a chase of cold methionine was added to a final concentration of 1% to block further incorporation of radioactive methionine. A 900-μl sample was removed immediately (t = 0) after the chase, and additional 900-μl samples were removed at the indicated times. Samples were added to 100 μl of ice-cold TCA, incubated on ice for >15 min, processed, and immunoprecipitated with polyclonal antibodies directed against the periplasmic domain of RseA as described above for the determination of synthesis rates. The intensity of the full-length RseA band in each lane was normalized, after background correction, to the intensity of the band in that lane corresponding to the periplasmic domain of RseA, the internal standard described above. RseA remaining at each time point was determined by dividing the normalized intensity of full-length RseA at that time to the normalized intensity at t = 0. Half-lives were determined by fitting the data to the exponential decay equation.
The method used for pulse-labeling proteins was modified for the work presented here. Previously we omitted both methionine and cysteine from the growth media, labeled proteins with both amino acids, and blocked further incorporation by adding an excess of both amino acids (1). In the present protocol, cells were grown in media lacking only methionine, proteins were labeled with radioactive methionine, and then further incorporation of the radioactive amino acid was blocked by the addition of excess unlabeled methionine. We modified our protocol because we found that the cells stopped growing briefly after the addition of excess cysteine in the former protocol, and we were concerned that this would lead to experimental artifacts.
Western blot detection of σE and RseA.
Cells were grown at 43°C in supplemented M9 minimal medium to an OD450 of 0.3 to 0.4. Duplicate samples of 900 μl each were taken (t = 0), the culture was shifted to 30°C, and additional samples of 900 μl were taken at the indicated times. All samples were added to 100 μl of ice-cold TCA and incubated on ice for >15 min. Precipitated proteins were collected by centrifugation and resuspended by boiling in Laemmli sample buffer at pH 9.0. Samples were loaded onto 15% Tris-glycine-SDS gels such that extracts from an equal number of cells were loaded in each lane. Western transfer was performed by standard methods (16). The blots were probed with polyclonal antibodies directed against the periplasmic or cytoplasmic domain of RseA, σE, and either σ70 or maltose binding protein as controls. The corresponding bands were detected by probing with anti-rabbit immunoglobulin radiolabeled with 35S (Amersham Biosciences). Bands were visualized using the Molecular Dynamics Storm 560 PhosphorImager scanning system and quantified using the program ImageQuant 1.2. The intensity of the band at each time point was normalized to the intensity of the average of the two t = 0 bands. The changes in the steady-state levels of RseA and σE over the course of the experiment were similar to the changes in the steady-state levels of maltose binding protein or σ70.
RESULTS
In this study, we explore the relationship between the activity of σE and the degradation of RseA during the different phases of the stress response: (i) the initiation phase, after a change in the growth temperature from 30°C (no stress) to 43°C (stress); (ii) the adaptation phase, as cells are maintained at 43°C for longer periods of time; and (iii) the shutoff phase, after a change in growth temperature back to 30°C from 43°C. During each phase, we measure the changes in σE activity by determining the rate of synthesis of two members of the σE regulon, DegP and RseA. We then analyze the contribution of RseA degradation to the regulation of σE by measuring changes in the degradation rate of RseA throughout the stress response. We performed these experiments in the MG1655 strain of E. coli, which grows more reproducibly at 43°C than the MC1061 strain used in our earlier work (1).
σE activity during initiation and adaptation.
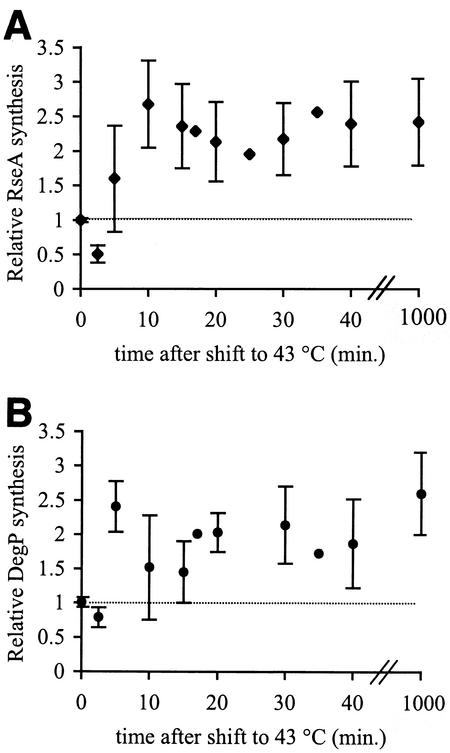
σE activity increases rapidly after a culture of growing cells is shifted from 30 to 43°C. Within 10 min, the synthesis rates of both DegP and RseA are 2- to 2.5-fold higher than at 30°C (Fig. 1), and the kinetics of induction for both proteins are similar within the error of the experiment. As cells continue growing at 43°C, the synthesis rates of both proteins remain 2- to 2.5-fold higher than those during steady-state growth at 30°C. Our results are consistent with the approximately threefold increase in σE-dependent transcription previously observed for these genes after a heat shock (28). These elevated rates are maintained even after 18 h of growth at 43°C in cells that have been grown overnight and subcultured at high temperatures (Fig. 1). DegP expression can also be induced by the Cpx two-component regulatory system (8, 9). However, σE, not Cpx, is responsible for the induction of DegP synthesis observed after a temperature shift, as a strain lacking Cpx shows the same 2.0- to 2.5-fold induction of DegP as the wild-type strain (data not shown). Taken together, these data show that σE is indeed activated rapidly in response to a shift to 43°C. However, the magnitude of the activation is less than the approximately fivefold increase observed after overproduction of the outer membrane porin OmpC (1, 20). This implies that the σE-dependent stress response can be activated to different extents and that the requirement for σE regulon members may be lower after a temperature shift than during overproduction of OmpC. In addition, the σE response appears to lack a classic adaptation phase in which a response is downregulated after a strong initiation phase. Instead, σE activity increases within the first 5 to 10 min of the response and remains at that level as long as the cells are kept at 43°C (see Discussion).
FIG. 1.
EσE activity increases during the initiation and adaptation phases of the stress response. Cells were grown to early exponential phase at 30°C and then shifted to 43°C. EσE activity was determined by measuring the rates of synthesis of RseA (A) and DegP (B) using the pulse-label protocol described in Materials and Methods. The synthesis rate shown for each protein is normalized to the synthesis rate of that protein at 30°C before the shift to 43°C (t = 0). The dotted line indicates the average synthesis rate in unstressed cells at 30°C. Error bars are shown for data points representing the average of at least two independent determinations.
σE activity during the shutoff phase of the stress response.
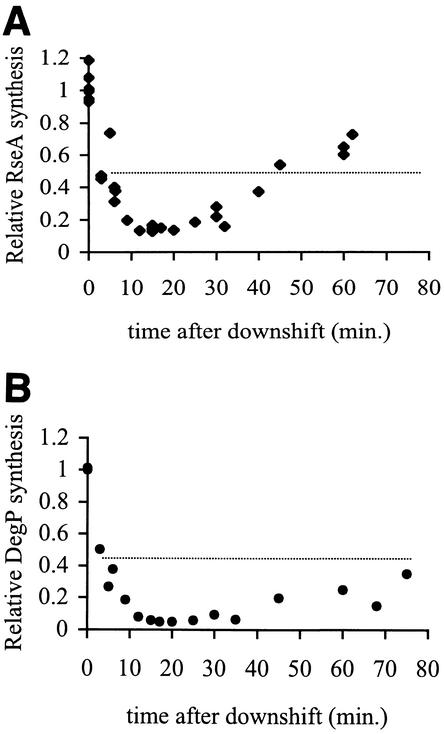
After cells that had been growing at 43°C are returned to 30°C, the synthesis rates of both RseA and DegP drop rapidly to levels below those normally observed during steady-state growth at 30°C (Fig. 2 and Table 1). By 15 min after the temperature downshift, RseA synthesis has declined 8-fold (Fig. 2A) and DegP synthesis has declined 20-fold compared to the 43°C synthesis rates (Fig. 2B). The rates of synthesis of both proteins remain low for another 10 min and then slowly increase to approximately the level observed in unstressed cells; this is reached within 60 min for RseA (Fig. 2A) and 75 min for DegP (Fig. 2B). These results show that the production of σE regulon members drops sharply and significantly immediately after a return to normal growth temperatures when elevated levels of chaperones and proteases are no longer necessary.
FIG. 2.
EσE activity decreases during the shutoff phase of the stress response and then slowly returns to normal levels. Cells were grown overnight at 43°C, diluted, regrown into early exponential phase at 43°C, and then shifted to 30°C. EσE activity was determined by measuring the rates of synthesis of RseA (A) and DegP (B) using the pulse-label protocol described in Materials and Methods. The synthesis rate shown for each protein is normalized to the synthesis rate of that protein at 43°C, before the downshift to 30°C (t = 0). The dotted line indicates the average synthesis rate in unstressed cells at 30°C. Data points from several experiments are shown.
TABLE 1.
RseA stability and σE activity throughout the stress response
| Condition | Half-life of RseA (min)a | σE activity
|
|
|---|---|---|---|
| RseA synthesisb | DegP synthesisb | ||
| No stress | 7.8 ± 0.8 | 1 | 1 |
| Initiation (10 min) | 2.2 ± 0.1 | 2.6 ± 0.6 | 1.6 ± 0.7 |
| Adaptation | 4.5 ± 0.2 | 2.4 ± 0.6 | 2.6 ± 0.6 |
| Downshift | >50 | 0.24 ± 0.04c | 0.13 ± 0.03c |
| Recovery from downshift (40 min) | 13.5 ± 1.7 | 0.61d | NMd |
In each case, the degradation phase was preceded by a 5-min lag without degradation.
Fold change in synthesis relative to that at 30°C. The mean ± standard error of the mean are shown.
Average maximal decrease in activity (12 to 25 min after downshift).
Synthesis of DegP 40 min after the downshift was not measured (NM), and synthesis of RseA at this time was measured in one experiment.
RseA stability in unstressed cells.
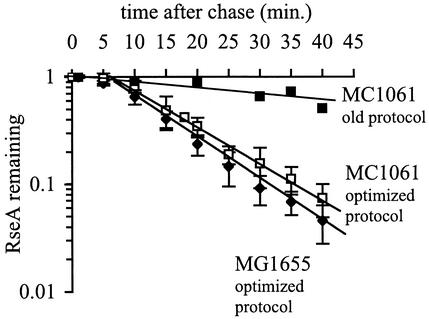
For σE activity to increase, the interaction between RseA and σE must be disrupted. Previous work indicated that one mechanism to disrupt the complex is through the regulated proteolysis of RseA (1-3, 18). If proteolysis of RseA is the primary mechanism controlling σE activity, then changes in RseA stability should correlate with changes in σE activity during the different phases of the stress response. To test this hypothesis, we first measured the stability of RseA in unstressed cells growing at 30°C. Newly synthesized RseA is stable for approximately 5 min after synthesis and is subsequently degraded, and it has a half-life of 7.8 ± 0.8 min (Fig. 3). These results confirm our previous observations that RseA is a relatively unstable protein (1). However, our present results differ from our previous measurements performed in E. coli MC1061 (Fig. 3) in two respects: in the earlier work, we did not observe a lag before the onset of proteolysis, and the half-life of RseA was longer than that measured here (1). These differences are due to changes in the protocol, which were adopted to minimize growth perturbations, and not to differences between the strains. In the course of this work, we found that the addition of excess cysteine during the chase step of the pulse-chase protocol caused the cells to stop growing for a short period of time. Therefore, we modified the protocol by including cysteine in the growth media and labeling proteins with methionine alone, which eliminated the growth arrest (see Materials and Methods for details). The stability of RseA in strain MC1061 measured with this optimized protocol is nearly identical to that in MG1655. Under the optimized conditions, RseA in MC1061 is stable for 5 min and is then degraded, with a half-life of 8.8 ± 0.9 min (Fig. 3).
FIG. 3.
RseA is degraded in unstressed cells. Cells of E. coli strains MG1655 and MC1061 were grown to early exponential phase at 30°C, and RseA stability was measured using the optimized protocol described in Materials and Methods. The stability of MC1061 measured using the old protocol (1) is shown for reference. The data points and error bars shown are the averages and standard deviations, respectively, from four independent experiments.
RseA stability during initiation and adaptation.
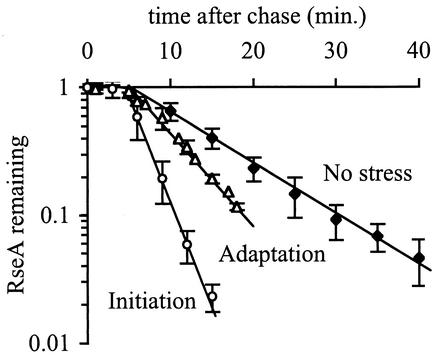
The rapid increase in σE activity during the initiation phase of the stress response (Fig. 1) is correlated with a decrease in RseA stability (Fig. 4 and Table 1). Within 5 min after a shift to 43°C, RseA is degraded with a half-life of 2.2 ± 0.1 min after a 5-min lag (Fig. 4 and Table 1). This degradation rate is 3.5-fold faster than that in unstressed cells. Surprisingly, RseA is not degraded as rapidly during the adaptation phase of the stress response as during the initiation phase, even though σE activity is the same during both phases. In cells that are grown continually at 43°C, RseA is degraded with a half-life of 4.5 ± 0.2 min after a 5-min lag, or twofold more slowly than during the initiation phase (Fig. 4 and Table 1). Although RseA is stabilized during the adaptation phase relative to the initiation phase, it is still degraded faster than in unstressed cells. These results are surprising, as the activity of σE is the same during initiation and adaptation, suggesting that the rate of RseA degradation may not be the sole determinant of σE activity (see Discussion). In addition, the observation that newly synthesized RseA is stable for 5 min in both stressed and unstressed cells indicates that a time-dependent but stress-independent event occurs before RseA degradation commences.
FIG. 4.
RseA is degraded faster during the initiation and adaptation phases of the stress response than in unstressed cells. Cells were grown to early exponential phase and left at 30°C (no stress), shifted to 43°C (initiation), or grown to early exponential phase at 43°C (adaptation). RseA stability was measured by a pulse-chase protocol as described in Materials and Methods. The stability of RseA during the initiation phase was measured 5 to 10 min after the shift to 43°C. The data points and error bars shown are the averages and standard deviations, respectively, from a minimum of three independent experiments.
RseA stability during shutoff.
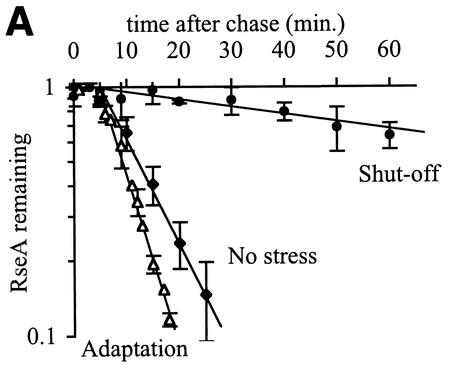
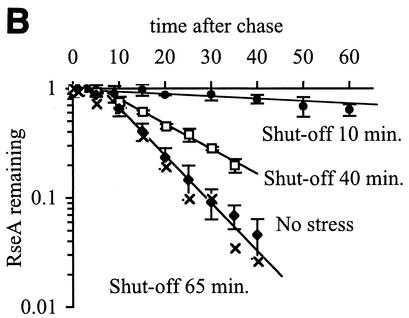
The decrease in σE activity during the shutoff phase of the response (Fig. 2 and Table 1) is correlated with stabilization of RseA. Within 5 min after cells that were grown at 43°C are returned to 30°C, the half-life of RseA increases dramatically to >50 min, which is considerably longer than the approximately 8-min half-life exhibited by unstressed cells (Fig. 5A and Table 1). The half-life of RseA slowly returns to that observed in unstressed cells concurrent with the recovery of σE activity to normal levels. By 40 min after the downshift, the half-life of RseA is 13.5 ± 1.8 min, and by 65 min, it is the same as in unstressed cells (Fig. 5B and Table 1).
FIG. 5.
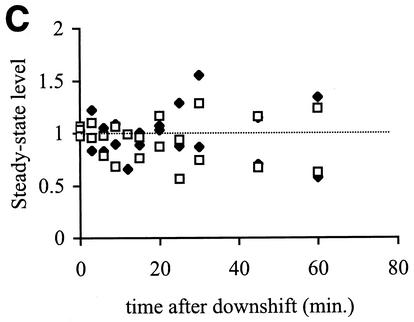
RseA is stabilized, but the steady-state levels of RseA and σE do not change during the shutoff phase of the stress response. (A) RseA degradation is significantly slower after a shift from 43 to 30°C than during steady-state growth at 43 or 30°C. Cells were grown to early exponential phase at 30°C (no stress), grown to early exponential phase and left at 43°C (adaptation), or grown to early exponential phase at 43°C and then shifted to 30°C (shutoff). RseA stability was measured by a pulse-chase protocol, as described in Materials and Methods. The stability of RseA during the shutoff phase was measured 10 min after the shift to 30°C. The data points and error bars shown are the averages and standard deviations, respectively, from a minimum of three independent experiments. (B) RseA is stabilized immediately after a shift from 43 to 30°C and then becomes progressively less stable as the cells remain at 30°C. Cells were grown to early exponential phase at 43°C and shifted to 30°C (shutoff) or grown to early exponential phase at 30°C (no stress). RseA stability was measured 10, 40, and 65 min after the temperature shift. Data points and error bars are the averages and standard deviations, respectively, from a minimum of two independent experiments. A representative data set is shown for the 65-min shutoff experiment. (C) The steady-state levels of RseA and σE do not change after a shift from 43 to 30°C. Cells were grown to early exponential phase at 43°C and then shifted to 30°C. The levels of RseA (⧫) and σE (□) were determined before the temperature shift (t = 0) and at various times after the shift by Western blot analysis as described in Materials and Methods. The amount of each protein at a given time is normalized to the amount of that protein at t = 0. The dotted line indicates a normalized steady-state level of 1. Data from two independent experiments are shown.
Stabilization of RseA during the shutoff phase of the response should result in an increase in the amount of RseA in the cell. However, after a temperature downshift, synthesis of RseA decreases (Fig. 2). To determine the net effect of these opposing processes, we measured the steady-state levels of both RseA and σE in cells growing at 43°C and at various times after a temperature downshift. The steady-state levels of RseA and σE do not change (Fig. 5C). Therefore, any gain in the level of RseA in the cell due to stabilization is offset by the decrease in newly synthesized protein.
DISCUSSION
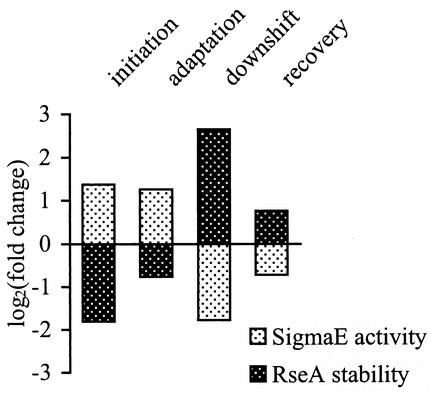
The work presented here examines the relationship between σE activity and the stability of its inner membrane anti-sigma factor, RseA, during the initiation, adaptation, and shutoff phases of the heat shock response. We find that, in general, σE activity is inversely correlated with RseA stability (Table 1; summarized in Fig. 6), leading us to suggest that regulated proteolysis of RseA is a major pathway for communicating information about protein unfolding in the cell envelope to σE in the cytoplasm. How might changes in the degradation rate of RseA alter σE activity? σE activity is determined by the number of σE molecules that are free (not bound to RseA) and therefore available to bind to RNA polymerase and direct transcription. Changing the degradation rate of RseA could alter σE activity by changing the relative levels of σE and RseA in the cell and also by altering the effective dissociation constant of σE from RseA. These mechanisms can explain the change of σE activity in each phase of the heat shock response. However, the fact that the relationship between σE activity and RseA degradation is not simple raises the possibility that other, as yet unknown, mechanisms may also play a role in regulating σE.
FIG. 6.
Changes in σE activity are inversely correlated with changes in RseA stability. σE activity (the synthesis rate of RseA) and the half-life of RseA during the initiation, adaptation, shutoff, and recovery from shutoff (40 min after the temperature downshift) were normalized to those values measured in unstressed cells at 30°C. The log2 values of the fold changes are plotted such that no change gives a value of 0, a twofold increase gives a value of 1, a twofold decrease gives a value of −1, etc.
Initiation phase of the response.
After a shift to high temperatures, both σE activity and RseA degradation increase. Increased degradation of RseA would be expected to increase the ratio of σE to RseA in the cell, thereby increasing the free pool of σE and σE activity. Preliminary computer modeling of the σE-dependent stress response shows that only a small (less than twofold) change in the ratio of σE to RseA is needed to give a 2.5-fold increase in σE activity (I. Grigorova, unpublished results). Although we have not been able to reproducibly measure such a small change in steady-state levels by Western blotting analysis, our measurements of RseA production and stability are consistent with this expectation. The net amount of RseA should decrease during initiation of the stress response, as its half-life drops fourfold, while its production increases only 2.0- to 2.5-fold. Since σE is stable, the steady-state ratio of σE to RseA should increase. This would lead to an increase in the free pool of σE in the cell and increased σE activity. Indeed, we previously showed that this mechanism was sufficient to explain the induction of σE activity after overexpression of the outer membrane porin OmpC (1).
Adaptation phase of the response.
Although σE activity is the same during the initiation and adaptation phases of the heat shock response, the degradation rate of RseA is different. RseA is twice as unstable during initiation as during adaptation. This suggests that the degradation rate of RseA is not the sole determinant of σE activity during adaptation.
Several additional factors could contribute to the observed level of σE activity during adaptation. First, the synthesis rate of σE is greater at high temperatures and should result in a higher concentration of σE in cells grown for long periods of time at 43°C. Since more σE will be available to bind to RNA polymerase and direct transcription, the rate of degradation of RseA during adaptation may not need to be as high as that during initiation to maintain comparable levels of σE activity. Second, the affinity of the σE-RseA complex could be lower during the adaptation phase, leading to an increase in the amount of free σE available to bind RNA polymerase and initiate transcription. Collinet et al. observed that binding of the periplasmic protein RseB to RseA enhances the affinity of the σE-RseA complex (7). In addition, RseB has been shown to copurify with periplasmic inclusion bodies (7). During the adaptation phase of the response, unfolded proteins may accumulate in the periplasm. RseB could be titrated away from RseA by these aggregated, unfolded proteins, resulting in a lowered affinity of the σE-RseA complex and an increase in the free pool of σE in the cell. Finally, other mechanisms may modulate the activity of σE; these mechanisms include chemical modification or conformational changes in σE. Any additional regulatory mechanisms may work alongside the degradation of RseA and serve to fine-tune the response.
Shutoff phase of the response.
Following a shift to low temperatures after growth at 43°C, the activity of σE decreases 8- to 15-fold relative to that at 43°C. Paradoxically, shutoff occurs without altering the steady-state ratio of σE to RseA. Clearly, the change in σE activity during the shutoff phase of the response is not due to a change in this ratio. It is possible that an unknown factor collaborates with RseA to decrease σE activity. However, it is also possible to explain the data with the existing players.
The fact that the proteolytic stability of RseA increases significantly during the shutoff phase suggests the possibility that σE activity is controlled by the rate of degradation of RseA. If the dissociation rate (koff) of RseA from σE is slower than its degradation rate, σE will predominantly be released from RseA when RseA is degraded, and the amount of free σE in the cell will depend primarily on the degradation rate of RseA. The magnitude of the drop in the degradation rate of RseA after downshift can account for the large decrease in σE activity that we observe. In addition, the affinity of the σE-RseA complex is sufficiently high to entertain this model. Even in the presence of the high concentrations of detergent required to maintain full-length RseA in a soluble form, RseA binds to σE with a Kd of 50 to 100 nM (7). The isolated cytoplasmic domain of RseA (RseAcyto) may be a better model than the full-length protein, as it is fully soluble and inhibits σE-directed transcription both in vivo and in vitro (11, 22). Transcription experiments suggest that the Kd for interaction between RseAcyto and σE is ≤1 nM (L. Connolly, unpublished results). Moreover, the interaction of these two proteins is remarkably stable. They can be purified as a complex from crude cell lysates and remain associated even in 650 mM NaCl and at 50°C (J. Tupy, unpublished observations). The magnitude of the drop in the rate of RseA degradation observed after downshift is sufficient to account for the large decrease in σE activity that we observe.
An alternative model is based on our demonstration that newly synthesized RseA is insensitive to proteases (Fig. 3 and 4), indicating that a time-dependent event triggers RseA proteolysis. If the protease-resistant, newly synthesized RseA also has a low affinity for σE, then this time-dependent process converts RseA from a form that binds σE weakly to a form that binds σE tightly. This scenario is sufficient to explain the observed differences in σE activity during the adaptation and shutoff phases of the response. At elevated temperatures, the low-affinity form predominates, as rapid degradation of RseA depletes the high-affinity form and new synthesis replenishes the cell with low-affinity RseA. In contrast, during the shutoff phase, degradation of RseA is slow and new synthesis of RseA is low; therefore, the high-affinity form would accumulate and repress σE activity efficiently. As the two forms are indistinguishable on SDS-polyacrylamide gels, the level of RseA and the ratio of RseA to σE will appear unchanged, as observed (Fig. 5C). The change in the affinity of RseA for σE and its susceptibility to proteolysis could result from conformational changes in RseA itself or binding of an accessory factor that alters the affinity of RseA for σE. Further experiments are clearly necessary to understand exactly how the degradation of RseA determines σE activity.
Regulation of proteolysis of RseA.
The degradation rates of RseA vary considerably in response to changes in the growth temperature, indicating that its proteolysis is not controlled by a simple on-off switch. Upon induction of the stress response, the initial cleavage of RseA is carried out by inner membrane protease DegS (2, 18). A second inner membrane protease, YaeL, then cleaves RseA further, releasing it from the membrane after which the remaining fragments are likely to be degraded to completion by cytoplasmic proteases including ClpXP (2, 18; J. M. Flynn, Y.-I. Kim, R. T. Sauer, and T. A. Baker, submitted for publication). Changes in the rate of degradation of RseA are dependent on a signal and are not controlled by alterations in the levels of DegS and YaeL, since the overexpression of DegS and/or YaeL in unstressed cells does not lead to increased σE activity (2). The amount of RseA in the cell does not appear to influence its proteolysis either, as different rates of proteolysis are realized without changes in the overall amount of RseA in the cell (Fig. 5). Preliminary results also indicate that σE does not protect RseA from proteolysis, as RseA is not degraded faster in the absence of σE (data not shown). Therefore, the inducing signal either influences the activity of the proteases or alters the susceptibility of RseA to proteolysis. Furthermore, the activity or amount of the signal must vary in response to changes in the growth temperature.
Another unusual aspect of the proteolysis of RseA is our observation that newly synthesized RseA is resistant to proteolysis. As discussed above, one possibility is that the lag represents a time-dependent conformational change in RseA. Although the insertion of RseA into the membrane has not been measured, we believe that any conformational change occurs in the membrane, as insertion is rapid for most proteins. The lag may have been masked by the longer half-life for RseA in unstressed cells measured in our previous work with strain MC1061 using the old experimental protocol, although a lag was not seen when RseA was rapidly degraded after a shift to 43°C in that work (1). It is also possible that the lag is caused by the sudden change in the level of methionine in the chase step of our experimental protocol. Since most unstable proteins do not demonstrate such a lag, this may represent either a specific response by the σE system to the amount of methionine or a physiological effect caused by the change in methionine levels.
Comparison of periplasmic and cytoplasmic stress responses.
Like the σE response, the σ32 cytoplasmic stress response also exhibits initiation, adaptation, and shutoff phases. During the shutoff phase, the activities of both σ32 and σE decrease rapidly and then slowly recover to the level seen in unstressed cells after 1 or 2 generations of growth (32). This allows regulon members to return to their unstressed levels by dilution of existing proteins through cell division and protein turnover. However, the initiation and adaptation phases of the two responses differ significantly. One of the hallmarks of σ32 response is a strong (10-fold) but transient increase in the transcription of heat shock genes during the first 5 to 10 min after a shift to high temperatures, followed by repression to a rate somewhat (2- to 3-fold) higher that that in unstressed cells (15, 33). In contrast, the small (2.5-fold) increase in σE activity is maintained for the duration of the heat shock response. The lack of a transient initial burst of σE activity appears to be a hallmark of the response, as it was not observed even with a more potent inducer, overexpression of OmpC (1). The initial burst of heat shock gene synthesis is thought to be a mechanism to rapidly increase the level of proteases and chaperones in the cell immediately after a stress. The fact that σE does not display such an increase indicates that the steady-state level of periplasmic chaperones and proteases will increase more slowly than those in the cytoplasm. Why might this be the case? We suggest that a large burst of synthesis of σE-dependent genes could actually be detrimental to the cell. As many σE regulon members are located in the cell envelope, they must be translocated across the inner membrane. A large increase in their synthesis could overwhelm the translocation machinery and actually increase the number of unfolded proteins emerging from translocons into the periplasm, thereby exacerbating the folding problem.
Acknowledgments
We thank Benjamin Alba, Joyce West, and Ken Keiler for critical reading of the manuscript.
This work was supported in part by U.S. Public Health Service grant GM36278-18 from the NIH. I. Grigorova was supported by NIH training grant GM08284. S. E. Ades was supported in part by NCI NSRA training grant T32CA09043.
REFERENCES
- 1.Ades, S. E., L. E. Connolly, B. M. Alba, and C. A. Gross. 1999. The Escherichia coli sigmaE-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev. 13:2449-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alba, B. M., J. A. Leeds, C. Onufryk, C. Z. Lu, and C. A. Gross. 2002. DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigmaE-dependent extracytoplasmic stress response. Genes Dev. 16:2156-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alba, B. M., H. J. Zhong, J. C. Pelayo, and C. A. Gross. 2001. degS (hhoB) is an essential Escherichia coli gene whose indispensable function is to provide sigmaE activity. Mol. Microbiol. 40:1323-1333. [DOI] [PubMed] [Google Scholar]
- 4.Baker, T. A., A. D. Grossman, and C. A. Gross. 1984. A gene regulating the heat shock response in Escherichia coli also affects proteolysis. Proc. Natl. Acad. Sci. USA 81:6779-6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker, J., and E. A. Craig. 1994. Heat-shock proteins as molecular chaperones. Eur. J. Biochem. 219:11-23. [DOI] [PubMed] [Google Scholar]
- 6.Bukau, B. 1993. Regulation of the Escherichia coli heat-shock response. Mol. Microbiol. 9:671-680. [DOI] [PubMed] [Google Scholar]
- 7.Collinet, B., H. Yuzawa, T. Chen, C. Herrera, and D. Missiakas. 2000. RseB binding to the periplasmic domain of RseA modulates the RseA:sigmaE interaction in the cytoplasm and the availability of sigmaE · RNA polymerase. J. Biol. Chem. 275:33898-33904. [DOI] [PubMed] [Google Scholar]
- 8.Connolly, L., A. De Las Penas, B. M. Alba, and C. A. Gross. 1997. The response to extracytoplasmic stress in Escherichia coli is controlled by partially overlapping pathways. Genes Dev. 11:2012-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danese, P. N., W. B. Snyder, C. L. Cosma, L. J. Davis, and T. J. Silhavy. 1995. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 9:387-398. [DOI] [PubMed] [Google Scholar]
- 10.Dartigalongue, C., D. Missiakas, and S. Raina. 2001. Characterization of the Escherichia coli sigma E regulon. J. Biol. Chem. 276:20866-20875. [DOI] [PubMed] [Google Scholar]
- 11.De Las Penas, A., L. Connolly, and C. A. Gross. 1997. The sigmaE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of sigmaE. Mol. Microbiol. 24:373-385. [DOI] [PubMed] [Google Scholar]
- 12.Gamer, J., G. Multhaup, T. Tomoyasu, J. S. McCarty, S. Rudiger, H. J. Schonfeld, C. Schirra, H. Bujard, and B. Bukau. 1996. A cycle of binding and release of the DnaK, DnaJ and GrpE chaperones regulates activity of the Escherichia coli heat shock transcription factor sigma32. EMBO J. 15:607-617. [PMC free article] [PubMed] [Google Scholar]
- 13.Gross, C. A. 1996. Function and regulation of the heat shock proteins, p. 1382-1399. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 14.Grossman, A. D., J. W. Erickson, and C. A. Gross. 1984. The htpR gene product of E. coli is a sigma factor for heat-shock promoters. Cell 38:383-390. [DOI] [PubMed] [Google Scholar]
- 15.Grossman, A. D., D. B. Straus, W. A. Walter, and C. A. Gross. 1987. Sigma 32 synthesis can regulate the synthesis of heat shock proteins in Escherichia coli. Genes Dev. 1:179-184. [DOI] [PubMed] [Google Scholar]
- 16.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual, p. 471-510. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Hiratsu, K., M. Amemura, H. Nashimoto, H. Shinagawa, and K. Makino. 1995. The rpoE gene of Escherichia coli, which encodes sigma E, is essential for bacterial growth at high temperature. J. Bacteriol. 177:2918-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehara, K., K. Ito, and Y. Akiyama. 2002. YaeL (EcfE) activates the sigmaE pathway of stress response through a site-2 cleavage of anti-sigmaE, RseA. Genes Dev. 16:2147-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landick, R., V. Vaughn, E. T. Lau, R. A. VanBogeler, J. W. Erickson, and F. C. Neidhardt. 1984. Nucleotide sequence of the heat-shock regulatory gene of E. coli suggests its product may be a transcription factor. Cell 38:175-182. [DOI] [PubMed] [Google Scholar]
- 20.Mecsas, J., P. E. Rouviere, J. W. Erickson, T. J. Donohue, and C. A. Gross. 1993. The activity of sigmaE, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 7:2618-2628. [DOI] [PubMed] [Google Scholar]
- 21.Missiakas, D., J. M. Betton, and S. Raina. 1996. New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. Mol. Microbiol. 21:871-884. [DOI] [PubMed] [Google Scholar]
- 22.Missiakas, D., M. P. Mayer, M. Lemaire, C. Georgopoulos, and S. Raina. 1997. Modulation of the Escherichia coli sigmaE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol. Microbiol. 24:355-371. [DOI] [PubMed] [Google Scholar]
- 23.Missiakas, D., and S. Raina. 1998. The extracytoplasmic function sigma factors: role and regulation. Mol. Microbiol. 28:1059-1066. [DOI] [PubMed] [Google Scholar]
- 24.Morimoto, R. I. 1998. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 12:3788-3796. [DOI] [PubMed] [Google Scholar]
- 25.Morita, M. T., M. Kanemori, H. Yanagi, and T. Yura. 2000. Dynamic interplay between antagonistic pathways controlling the sigma32 level in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morita, M. T., Y. Tanaka, T. S. Kodama, Y. Kyogoku, H. Yanagi, and T. Yura. 1999. Translational induction of heat shock transcription factor sigma32: evidence for a built-in RNA thermosensor. Genes Dev. 13:655-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raina, S., D. Missiakas, and C. Georgopoulos. 1995. The rpoE gene encoding the sigma E (sigma 24) heat shock sigma factor of Escherichia coli. EMBO J. 14:1043-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rouviere, P. E., A. De Las Penas, J. Mecsas, C. Z. Lu, K. E. Rudd, and C. A. Gross. 1995. rpoE, the gene encoding the second heat-shock sigma factor, sigmaE, in Escherichia coli. EMBO J. 14:1032-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rouviere, P. E., and C. A. Gross. 1996. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev. 10:3170-3182. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Straus, D., W. Walter, and C. A. Gross. 1990. DnaK, DnaJ, and GrpE heat shock proteins negatively regulate heat shock gene expression by controlling the synthesis and stability of sigma32. Genes Dev. 4:2202-2209. [DOI] [PubMed] [Google Scholar]
- 32.Straus, D. B., W. A. Walter, and C. A. Gross. 1989. The activity of sigma32 is reduced under conditions of excess heat shock protein production in Escherichia coli. Genes Dev. 3:2003-2010. [DOI] [PubMed] [Google Scholar]
- 33.Straus, D. B., W. A. Walter, and C. A. Gross. 1987. The heat shock response of E. coli is regulated by changes in the concentration of sigma32. Nature 329:348-351. [DOI] [PubMed] [Google Scholar]
- 34.Wang, Q. P., and J. M. Kaguni. 1989. A novel sigma factor is involved in expression of the rpoH gene of Escherichia coli. J. Bacteriol. 171:4248-4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yura, T., H. Nagai, and H. Mori. 1993. Regulation of the heat-shock response in bacteria. Annu. Rev. Microbiol. 47:321-350. [DOI] [PubMed] [Google Scholar]
- 36.Yura, T., and K. Nakahigashi. 1999. Regulation of the heat-shock response. Curr. Opin. Microbiol. 2:153-158. [DOI] [PubMed] [Google Scholar]
- 37.Yura, T., T. Tobe, K. Ito, and T. Osawa. 1984. Heat shock regulatory gene (htpR) of Escherichia coli is required for growth at high temperature but is dispensable at low temperature. Proc. Natl. Acad. Sci. USA 81:6803-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]