Abstract
The PilQ secretin from the pathogenic bacterium Neisseria meningitidis is an integral outer membrane protein complex which plays a crucial role in the biogenesis of type IV pili. We present here the first three-dimensional structure of this type of secretin at 2.5-nm resolution, obtained by single-particle averaging methods applied to the purified protein complex visualized in a negative stain. In projection, the PilQ complex is circular, with a donut-like appearance. When viewed from the side it has a rounded, conical profile. The complex was demonstrated to have 12-fold rotational symmetry, and this property was used to improve the quality of the density map by symmetry averaging. The dominant feature of the structure is a cavity, 10 nm deep, within the center of the molecule. The cavity is funnel-shaped in cross section, measures 6.5 nm in diameter at the top of the complex, and tapers to a closed point, effectively blocking formation of a continuous pore through the PilQ complex. These results suggest that the complex would have to undergo a conformational change in order to accommodate an assembled pilus fiber of diameter 6.5 nm running through the outer membrane.
Bacteria express a variety of surface-located adhesive molecules to achieve the necessary cell-cell contact required for initial contact with a human host (20, 38). During tissue colonization, Neisseria meningitidis (the meningococcus) and many other gram-negative bacteria produce long adhesive fibers, termed type IV pili (Tfp), which emanate from the cell surface and mediate attachment via specific epithelial tissue receptors (17, 21, 47). In addition, Tfp have roles in diverse bacterial processes, including bacterial autoagglutination (48), variation of target tissue specificity (22), and natural competence for DNA uptake by transformation (15, 53). Tfp are responsive organelles, and the retraction of these fibers is directly responsible for bacterial twitching motility along solid and mucosal surfaces (28, 30). Assembly of pilin subunits into Tfp is a multistage process and is controlled by between 14 and 30 genes (1, 46, 52). Many of the components of the Tfp biogenesis machinery are homologous to those engaged in the general type II secretory pathway (GSP) (33, 39).
The PilQ complex is an antigenically conserved, highly abundant outer membrane protein and is a member of the GSP secretin superfamily (39), members of which are involved in the terminal translocation of proteins and macromolecules across the outer membrane (2, 50). The PilQ complex is critical for the surface expression of Tfp, and meningococcal mutants lacking or expressing defective PilQ are devoid of pili and pilus-associated phenotypes (8, 50). A total of 200 to 300 conserved C-terminal residues in the transmembrane region of the PilQ monomer exhibit the highest sequence homology to other secretins and are essential for oligomerization into the functional complex (18, 32, 50), although PilQ also requires a lipoprotein, PilP, for stable oligomerization (7). Functional PilQ in N. meningitidis forms a large (∼960-kDa) homododecameric complex (4) that is very stable and resistant to sodium dodecyl sulfate (SDS) and heat treatment (50). The naturally high expression level of a single secretin in meningococci favors this system for secretin characterization. Secretins from a wide range of bacteria have been purified and observed by electron microscopy (EM) (2-6, 9, 23, 26, 31, 34, 44, 45, 49), and all of these studies have reported a ring-shaped complex composed of 6 to 14 subunits. In each case, the ring enclosed a central cavity of various dimensions, and it is widely assumed that the secreted substrate passes through the body of this channel. An EM study on intermediates in type I pilus biogenesis has provided evidence that the FimD secretin does indeed act as a conduit for type I pili (42).
Although these EM observations have been useful, structure-function studies of individual secretin complexes have been hampered by a lack of three-dimensional (3-D) detail. Determination of the 3-D structure of the PilQ complex would provide novel and biologically pertinent information on the nature of the central cavity or pore: is a continuous channel formed through the PilQ complex and, if so, is it of appropriate dimensions to accommodate preassembled Tfp? One approach to obtain this type of 3-D structural information for particles found predominantly in a single orientation is to use single-particle averaging (SPA) in combination with volume reconstruction methods (12, 40). The only 3-D secretin structures determined thus far are PulD from Klebsiella oxytoca (31) and the pIV protein from filamentous bacteriophage (34).
We report here the 3-D structure of the PilQ complex from N. meningitidis, obtained from single particles visualized in a negative stain. We find that the secretin forms a noncontinuous pore 6.5 nm in diameter that tapers substantially toward one end. The results suggest a “gated pore” mechanism whereby the secretin does not merely function as a passive channel or sheath but plays an intimate part in regulating the extrusion or retraction of the nascent Tfp.
MATERIALS AND METHODS
Bacterial strains.
Meningococcal strain M1080 was grown overnight on 5% blood agar in an atmosphere containing 10% CO2 before being harvested. The plasmid pMF121, generously provided by Mathias Frosch (16), was used to generate a capsule negative mutant expressing truncated lipooligosaccharide of strain N. meningitidis strain M1080 by transformation with erythromycin selection.
Purification of meningococcal PilQ.
Meningococcal cells were disrupted in a French press in the presence of the protease inhibitors phenylmethylsulfonyl fluoride, leupeptin, pepstatin A, and 2-mercaptoethanol (Sigma). The cell envelope fraction was extracted in 4% (wt/vol) deoxycholate in buffer A (100 mM NaCl, 20 mM KH2PO4, 50 mM KCl, 5 mM EDTA, 10 mM Tris; pH 7.5) and pelleted. The pellet was subsequently dissolved in a buffer containing 4% (wt/vol) Zwittergent 3-10 and 20% (vol/vol) glycerol and then dialyzed in buffer A with 0.1% (wt/vol) Zwittergent 3-10. After concentration and size exclusion in Microsep300 columns (Filtron), the preparation was loaded onto a 15 to 40% (wt/vol) continuous sucrose gradient and ultracentrifuged at 106,000 × g (4°C) for 24 h in a Beckman SW41 rotor. PilQ-containing fractions were selected by immunoblotting and subjected to prolonged dialysis in buffer A with 0.1% (wt/vol) Zwittergent 3-10. PilQ complexes were concentrated again in Microsep300 columns before use.
Sample purity.
The presence of PilQ in whole-cell lysates and fractions during purification was detected by immunoblotting with rabbit polyclonal antibodies raised against purified N. meningitidis PilQ complex from strain M1080 at a dilution of 1:4,000 (50). Conditions to verify PilQ sample purity by (i) SDS-PAGE, (ii) immunoblotting with antibodies to PilE, PilP, PilC, and lipopolysaccharide, and (iii) antigen detection have been previously described (4, 50, 51).
EM sample preparation and image scanning.
Samples of PilQ were adsorbed to freshly glow discharged carbon coated grids (no. 400) as previously described (4), with the following minor changes. The carbon sides of the grids were placed on the surface of droplets in the following sequence: 20-μl PilQ sample droplets (100 μg/ml) for 10 min, followed by a 10-s wash in buffer (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, 5 mM EDTA), and finally a 10-min incubation in fresh 2% (wt/vol) uranyl acetate. Table 1 shows all additional information pertinent to transmission EM low-dose data collection and image processing.
TABLE 1.
EM and image analysis information
| Equipment or variable | Specification(s) |
|---|---|
| Microscope | Philips Tecnai 10; transmission EM low-dose mode |
| Operating voltage (keV) | 100 |
| Calibrated magnification | ×43,200 |
| Scanner | UMAX Photolook 3000 (256 grayscale) |
| Film type | Kodak SO-163 |
| Scan step (dpia), pixel size (nm) | 1,600, 0.37 |
| Micrograph tilt pair angles | 0° and 40° |
| Avg defocus value (μm) | 2.5 |
| No. of tilt pairs used | 25 |
| Total ring projections selected (n) | 765 |
| No. of principal groups used for back projection | 2 |
| No. of particles in vol merged (resolution of ≤3 nm) (n = 503) | 200 and 303 |
| Back-projection reconstruction sphere radius (nm) | 11.1 |
| Euler angles between vol (φ, θ, ψ) | 33.43, 0, −16.73 |
| Vol orientational correlation coefficient | 0.92 |
| Spatial resolution of unsymmetrized vol (nm) | 2.5 |
dpi, Dots per inch.
Image processing.
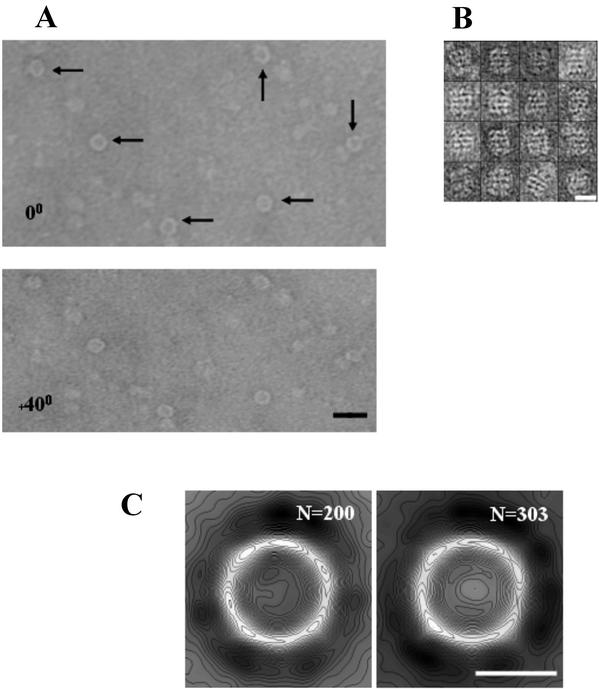
3-D volumes of Zwittergent-solubilized PilQ were calculated by the random conical reconstruction method (12, 13, 37, 40), implemented by using the SPIDER and WEB image processing software suite (14) (www.wadsworth.org/spider_doc/spider/docs/master.html). Micrograph pairs of negatively stained PilQ complexes were selected and scanned as described in Table 1. Deeply stained individual complex pairs presenting a clear projection (shown by arrows on Fig. 1A) were extracted into 26-nm2 windows from the untilted and tilted micrographs, and the individual CTF correction and focus gradient were evaluated for each tilt pair (12).
FIG. 1.
PilQ particles visualized under negative stain. (A) Tilt pairs of PilQ particles at 0° and 40°, negatively stained with 2% uranyl acetate and recorded under low-dose conditions. Arrows indicate particles presenting the face-on donut view. Scale bar, 30 nm. (B) Montage of particles presenting putative side-on views of PilQ. Measurements of individual particles found widths and heights in the range of 15 to 16.5 nm and 18 to 20 nm, respectively. Scale bar, 15 nm. (C) 2-D projection maps of the two major class averages (n = 200 and 303) of particles selected for volume back projection. Maps are low-pass filtered to 2 nm and contoured at 20 levels. Scale bar, 10 nm.
The untilted data set was rotationally and translationally aligned by using previously described strategies (4) that resulted in two closely related groups (n = 200 and 303). The alignment for each group produced was judged to be accurate by using a combination of criteria. (i) Independent alignment algorithms produced the same classification groupings. (ii) Correspondence analysis and hierarchical ascendant classification showed that all groupings were highly related, with minimal variation at thresholds of ≤0.01. (iii) Similarity to previous alignments produced from a 2-D data set (n = 650) of SDS-solubilized PilQ (4) was the last of the criteria. The calculated shifts and rotations were then applied to the data set after low-pass filtering to 2 nm, ramping, and contrast enhancement. Correspondence analysis (24, 25) and hierarchical clustering (37), with complete linkage as a merging criterion, were used to separate out the two major groups. Each of the two major volumes produced was individually refined six times by optimizing the original volume by using an angular refinement step. Back-projections calculated from the initial 3-D volume were used to translationally realign the original tilted particles, and the resulting realigned particles were then used to calculate a new volume. The two refined volumes were then merged after determining the correct Euler angles for combination producing a final reconstruction, including 503 particles prior to symmetry averaging. The final volume produced was judged to be a reasonable size for a PilQ dodecamer, assuming a packing density of 0.7 g/cm3; volumes calculated at thresholds of 1 and 2σ above the mean density could accommodate molecular masses of 1,000 and 840 kDa, respectively. In order to confirm that the volumes calculated from PilQ were presented in a single orientation, higher clustering thresholds were used to produce more class groups, each containing fewer particles. All of the volumes produced in this way had the same asymmetry or tapering features along the complex height. If PilQ volumes were calculated from ring projections related by 180°, the complex would not show tapering consistently localized at one end.
Self-orientation rotational and rotational power spectrum analyses on the final volume confirmed the 12-fold symmetry previously assigned in a 2-D projection (4). Symmetry was imposed on the volume after calculation of the center of gravity and the correct axis of symmetry. Volumes were subsequently displayed by using either WEB (14) or XFIT (29).
Resolution determination.
For a particle in the size range of PilQ, ∼300 particles would be required to provide a complete data set to 2.5-nm resolution (12): the data set of 503 particles with symmetry averaging applied exceeds this practical minimum requirement. The resolution of the final volume was calculated by determining the resolution between two subvolumes by using either Fourier ring correlation (FRC) criteria (43) with a cutoff point of FSC (Fourier shell correlation) = 0.5 (to give a value of 2.1 nm) or by employing the differential-phase residual (DPR) method (35) (to give a value of 2.8 nm). The mode by which resolutions are reported for 3-D volumes is currently under debate because different methods provide slightly different resolution values (11). Resolution calculations made by the FRC method tend to provide more optimistic resolution values than those obtained by the DPR method. To account for this fact, we report a mid-range resolution value of 2.5 nm from the two methods currently recommended as best practice (12). An alternative method to convert FRC resolution values to their corresponding SNR (signal-to-noise ratio) values has been proposed, although the use of an FSC of 0.5 as a cutoff point has been supported by comparisons of cryo-EM and X-ray structures of adenovirus type 2.
Tfp modeling.
A 3-D homology model for the N. meningitidis pilin subunit was built based on the X-ray crystal structure of the homologous N. gonorrhoeae pilin subunit (PDB accession code 2PIL), by using the Modeller program (41). The amino acid sequences of the two proteins are 86% identical and 89% similar. The N. meningitidis pilus fiber was then built by applying the transformations given in the 2PIL PDB header, based on the pilus fiber model with fivefold helical symmetry described previously (36).
RESULTS
PilQ complex purification.
The yield of native oligomeric PilQ complex purified from meningococcal membranes was significantly improved by using the capsule negative N. meningitidis M1080 mutant also expressing truncated lipooligosaccharide as the protein source. Improvements in the purification procedure of the PilQ complex were also achieved by using the detergent Zwittergent 3-10 (rather than SDS) to solubilize outer membrane fractions. This procedure produced PilQ samples equivalent in purity to those of previous materials (4) but at the higher protein concentration required for extensive EM analyses.
EM and SPA of PilQ.
A representative electron micrograph tilt pair of negatively stained PilQ complexes that were used to calculate the final 3-D volume is shown in Fig. 1A. The Zwittergent-solubilized PilQ complexes, indicated by arrows, presented themselves primarily as circular donut-like rings of protein density that enclosed a central stain-excluding cavity. We also observed a distinctive minority population of particles corresponding to a variety of side-on and partial side-on views of PilQ (Fig. 1B). These views were rare, however, and were not present in sufficient numbers for either accurate 2-D projection mapping or the calculation of a 3-D structure. It is possible that these particles correspond to pairs of PilQ dodecamers; similar behavior has been observed in at least one other secretin (34). SPA of the donut-like projections resulted in the identification of two highly related principal groups from the data set (Fig. 1C). Subsequent analysis of these groups revealed that they only differed slightly in their Euler angle orientations (Table 1), and these were applied during volume merging. The class-averaged projection maps for these groups demonstrated that the PilQ complexes were identical, with regard to the overall complex features and distribution of negative stain, to SDS-solubilized PilQ preparations observed previously (4). Examination of the selected PilQ particles revealed that the central cavity measured 6 to 6.5 nm across, with an outer diameter of 15.5 nm, which was at the lower end of the previous size distribution (15.5 to 16.5 nm) (4). Since the inner ring diameter was identical to previous distributions, the smaller outer diameter changes were likely to be caused by a different detergent annulus around the circumference of the complex. As with previous PilQ preparations, we found that the protein adsorbed to EM grids with a low affinity, requiring protein concentrations greater than 100 μg/ml and incubation times exceeding 10 min (4). This effect may be due to the high estimated pI of the protein (9.9), which could lead to a poor charge interaction at the grid surface (50).
Oligomeric form of PilQ.
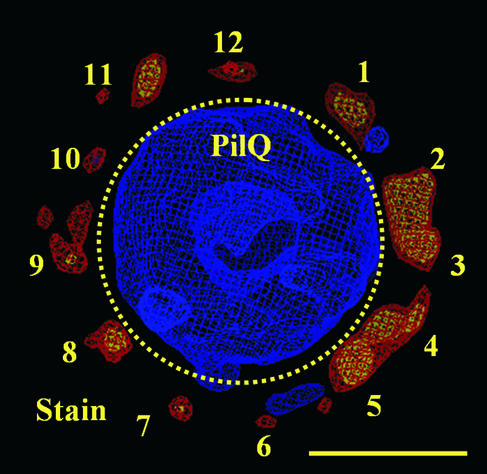
Our prior work on the structure of PilQ demonstrated that 2-D projection maps of the PilQ complex possessed 12-fold rotational symmetry (4). This symmetry was also observed in the volume calculated from merging the two main groups shown in Fig. 1C: the 12-fold symmetry was readily apparent from the accumulation of 12 negative stain density peaks (numbered 1 to 12) outside the periphery of the ring of the calculated 3-D volume (Fig. 2). These peaks occurred at approximately 30° intervals, although some variation from the “ideal” positions was apparent because no symmetry had been applied to the volume. In agreement with these data, self-orientation and power spectrum analyses were performed on the 3-D reconstruction, and the findings were consistent with data previously reported (not shown). It is interesting that the accumulation of these stain peaks is primarily located on the outside of the PilQ ring in the 3-D volume. Since, in this orientation, the complex is positioned on the grid with the ring facing down, this observation suggests that the upper and side portions of the ring are more accessible to negative stain than the constricted section of the complex. Imposition of any symmetry other than 2-, 3-, 4-, 6-, or 12-fold smoothed out the particle features observed and/or skewed the surface relief of the central cavity, a finding consistent with the assignment of 12-fold symmetry.
FIG. 2.
The distribution of accumulated negative stain around the circumference of the merged PilQ volume demonstrates 12-fold symmetry. The unsymmetrized PilQ volume (n = 503) displayed as a density map (in blue at 2σ above the mean density value) is positioned to correspond to the donut-like projections in Fig. 1. Twelve peaks of negative-stain volume (shown in red and yellow at 1.5σ and 2σ, respectively, above the mean negative-stain density) occur at approximate 30° intervals around the PilQ ring and are labeled 1 to 12. The boundary between the regions in the volume corresponding to the PilQ complex and the surrounding stain pockets is shown by the dotted line. Scale bar, 10 nm.
3-D structure of PilQ.
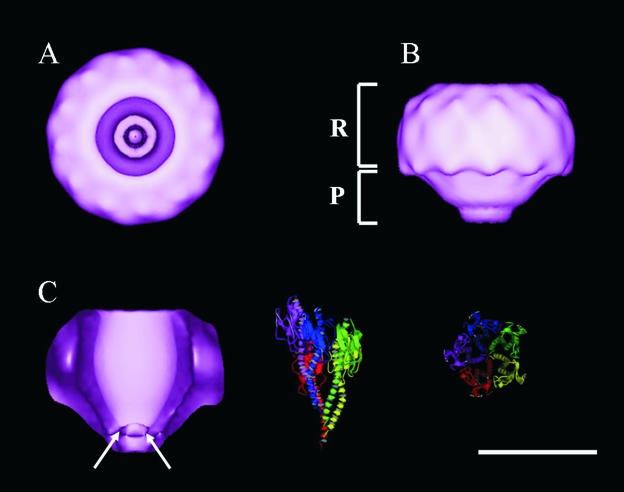
Figure 3A shows the surface relief of the final PilQ volume with 12-fold rotational symmetry applied and presented at 2σ above the mean density. Symmetry averaging is a powerful technique and has been used to successfully analyze the structures of many virus and helical assemblies (10, 14). This view corresponds to the face-on or donut 2-D projection of PilQ (Fig. 1C) and clearly shows the 15.5-nm ring of the 12 subunits enclosing a deep central cavity 6.2 to 6.5 nm in diameter. Inspection of the cavity from this view reveals that the impression is not continuous through the height of the complex and significant density could be identified in the bottom. Around the circumference of the ring, 12 repeating surface protrusions were apparent at ∼3-nm intervals, and each of the 12 monomers were estimated to contribute an area of ca. 4.5 by 3.0 nm to the upper surface of the PilQ ring. Figure 3B shows the surface relief of PilQ after a rotation of the face-on view of PilQ by 90° around the y axis. This side view reveals that the PilQ complex possesses a distinctive two-region structure, with an overall height of ∼12 nm and a rounded conical appearance. The upper portion of the structure forms a discrete ring (R region) with a diameter of 15.5 nm and a height of 8 nm. The regular detail observed around the top of the ring also continues around the side at 3-nm intervals. Below this ring is a smaller conical plug (the P region) that gradually tapers toward a flattened point. The interface between the R and P regions sharply constricts from 15.5 to 11 nm and then gradually tapers to a blunt point 4.5 nm in diameter over the region height of 4 nm. Figure 3C reproduces the PilQ volume shown in Fig. 3B but with 50% of the foremost volume removed to reveal the interior relief of the complex. The cavity has a distinctive funnel-like shape that is noncontinuous, with an overall height of ca. 10 nm. The majority of the cavity is located in the R region and gradually closes to a fixed point ∼3 nm in diameter ca. 2 nm into the P region. To provide a sense of scale for the Tfp substrate, a homology model of the N. meningitidis pilus fiber (shown in two views related by a 90° rotation) is included at the same scale as PilQ. Given that only one turn (five subunits) of the pilus fiber is shown, it is clear that the dimensions of the cavity are too small to accommodate a fully assembled pilus fiber running continuously through the PilQ complex in the purified conformation.
FIG. 3.
Surface-rendered 3-D volumes of PilQ with 12-fold rotational symmetry applied. Volumes are displayed at a threshold of 2σ above the mean volume density. Scale bar, 10 nm. (A) Face-on view corresponding to the donut-like projection in Fig. 1. (B) Rotation of the face-on view of the PilQ complex 90° through the y axis. Viewed from the side, the PilQ volume has two distinct regions: a ring (R) region connected to an underlying plug (P) region. (C) A cross-sectional side-on view of the PilQ volume with 50% of foremost volume removed through the z axis to reveal the structure of the internal cavity. The constriction of the central cavity may be seen at the bottom of the complex and is indicated by arrows. Ribbon plots of the atomic model of the N. meningitidis Tfp fiber are displayed to the right in a side-on and face-on view at the same scale as the PilQ volumes. Five subunits (one helical turn) are shown.
It should be noted that structural information calculated by using random conical tilt methodology applied to negatively stained particles has two main limitations. First, a negative stain, although providing excellent contrast, a high particle signal/noise ratio, electron beam stability, and experimental reproducibility, also has several well-documented disadvantages that may affect a sample to greater or lesser degrees (19). These problems include uneven stain penetration of the particle, sample deformation by shrinkage, flattening and skewing, and a practical resolution limit of ∼2 nm. Despite these constraints, negative staining remains a useful technique for acquiring moderate resolution structural information of a biological complex, and comparisons of several structures obtained by using negative staining versus unstained cryo data do demonstrate good overall structural correlation (12). The second issue relates to the maximum practical tilt angle used (∼60°) when a specimen is tilted in a transmission electron microscope. This limitation results in the loss of some information along the z axis or “a missing cone of data” in the back-projected volume (12). For any 3-D reconstruction calculated in this way, this phenomenon results in a lower resolution for structural features extending in the z direction, perpendicular to the specimen support film. For the PilQ structure presented here this means that the resolution of detail through the height of the complex is reduced slightly. We report an overall resolution of 2.5 nm for the PilQ structure but it is important to note that, even if we consider the lower DPR resolution estimate of 2.8 nm, this resolution remains more than sufficient to resolve all of the structural features of the PilQ complex discussed here.
DISCUSSION
The determination of the 3-D volume of PilQ has a number of important implications for secretion processes in general and the mechanism of Tfp biogenesis in particular. Previous work has established that purified secretin complexes form gated conductance channels when reconstituted into lipid membranes (3, 27, 31). The structure presented here would establish a structural explanation for this phenomenon, providing evidence for a “plug” or constriction in the channel that is predicted to run across the outer membrane. It is clearly advantageous for the bacterial cell to regulate the accessibility of pilin subunits to PilQ since this secretin is expressed constitutively at high levels (50) and a persistently open 6.5-nm channel would allow periplasmic proteins and other valuable periplasmic molecules to diffuse away from the cell. A limitation of the current structure is that it does not assign the outer or inner faces of the complex. We presume that the open mouth of the cavity faces outward from the cell because it needs to house the Tfp as it is extruded, but there is no direct evidence for this supposition in the data presented here. If this is indeed the case, it would suggest that the constricted end of the channel lies on the periplasmic side of the outer membrane. This is an attractive proposition because it would allow this region of PilQ to interact with other Tfp biogenesis proteins from the periplasm and inner membrane.
It is also tempting to speculate on how PilQ might interact with the growing or retracting Tfp fiber. The structure presented here suggests two possibilities. The first is that assembly of the pilus fiber occurs outside the PilQ cavity and that the fiber is extruded through the protein in its intact form. In order for this to happen, the dimensions of the cavity would need to expand marginally, possibly through a conformational change induced by interactions of PilQ with other protein components, and the P region would need to “unfurl.” The second possibility is that the site of pilus formation is actually inside the PilQ cavity itself and that individual pilin subunits are delivered or removed from this site depending on the relative activities of Tfp assembly or retraction proteins such as PilT. It is not possible, on the basis of these structural data, to distinguish between these alternate models, although it is interesting that they both require a substantial degree of conformational flexibility of the P region of the molecule. It is important to emphasize, as a caveat, that the results presented here were obtained from detergent-solubilized, purified protein and that extraction of PilQ from the meningococcal membrane and removal of other protein components could affect the conformation or structure of the protein. Another important question is the position of the outer membrane: again, this is impossible to discern from these observations on detergent-solubilized material, although the size and morphology of the R region suggest that this portion could contain the membrane-spanning portions of the complex.
Our structure of PilQ can be compared to the low-resolution structures of two other secretin complexes. Scanning transmission EM observations of two orientations of the pIV filamentous phage channel showed that the complex formed a cylindrical assembly of 12 to 14 monomers (26). More recently, the structure of the pIV multimer has been determined to a 22-Å resolution by cryo-EM (34). The pIV complex exhibits 14-fold, rather than 12-fold, rotational symmetry and forms a barrel-like structure. The channel through the molecule is blocked, as is the case with PilQ, but in a central portion of the pore, rather than at one end. Another secretin that has been studied in some detail is the PulD-PulS complex from K. oxytoca (31). The results showed an asymmetric cylindrical complex composed of 12 monomers with a height of 19 nm and a diameter of 25.8 nm. Around the periphery of the complex 12 separate radial densities were observed: these features were proposed to originate from the secretin assembly lipoprotein PulS. The central cavity was large in diameter (∼7.5 to 8.0 nm) and apparently continuous throughout the complex, although there was biochemical evidence that the channel was gated (31). Further observations of PulD-PulS in negative stain have shown a side-on view of the complex, in a dimeric organization with pseudo-mirror symmetry (32). Given that PulD and PilQ exhibit homology only within the C-terminal domains of their genes, it is possible that some of these structural differences are attributable to variations in the structure of the N-terminal half of each protein.
By providing novel information on the pore cavity, the PilQ structure presented here represents a first step toward an understanding of the structure of the Tfp biosynthetic apparatus. There are many intriguing aspects of this complex molecular machine, not least of which are questions regarding how pilus extrusion is regulated and how the extension process is reversed to retraction. Clearly, 3-D studies provide a significant contribution toward explaining these phenomena.
Acknowledgments
This work was supported by grants from the Wellcome Trust and the Norwegian Research Council.
We thank Stephan A. Frye for advice in the PilQ complex purification process, Matthias Frosch for providing the plasmid pMF121, G. Verlarde for technical assistance with image processing, and K. Sidhu for software support.
REFERENCES
- 1.Alm, R. A., and J. S. Mattick. 1997. Identification of a gene PilV required for type 4 fimbrial biogenesis in P. aeruginosa whose product possesses a pre-pilin-like leader sequence. Gene 192:89-98. [DOI] [PubMed] [Google Scholar]
- 2.Bitter, W., M. Koster, M. Latjinhouwers, H. de Cock, and J. Tommassen. 1998. Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol. Microbiol. 27:209-219. [DOI] [PubMed] [Google Scholar]
- 3.Brok, R., P. Van Gelder, M. Winterhalter, U. Ziese, A. J. Koster, H. de Cock, M. Koster, J. Tommassen, and W. Bitter. 1999. The C-terminal domain of the Pseudomonas secretin XcpQ forms oligomeric rings with pore activity. J. Mol. Biol. 294:1169-1179. [DOI] [PubMed] [Google Scholar]
- 4.Collins, R. F., L. Davidsen, J. P. Derrick, R. C. Ford, and T. Tønjum. 2001. Analysis of the PilQ secretin from Neisseria meningitidis by transmission electron microscopy reveals a dodecameric quaternary structure. J. Bacteriol. 183:3825-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornelis, G. R., and H. Wolf-Watz. 1997. The outer membrane component, YscC, of the Yop secretion machinery of Yersinia enterocolitica forms a ring-shaped multimeric complex. Mol. Microbiol. 23:861-867. [DOI] [PubMed] [Google Scholar]
- 6.Crago, A. M., and V. Koronakis. 1998. Salmonella InvG forms a ring-like multimer that requires the InvH lipoprotein for outer membrane localization. Mol. Microbiol. 30:47-56. [DOI] [PubMed] [Google Scholar]
- 7.Drake, S., S. A. Sandstedt, and M. Koomey. 1997. PilP: a pilus biogenesis lipoprotein in Neisseria gonorrhoeae affects expression of PilQ as a high molecular mass multimer. Mol. Microbiol. 23:657-668. [DOI] [PubMed] [Google Scholar]
- 8.Drake, S. L., and M. Koomey. 1995. The product of the PilQ gene is essential for the biogenesis of type IV pilus in N. gonorrhoeae. Mol. Microbiol. 18:975-986. [DOI] [PubMed] [Google Scholar]
- 9.Drummelsmith, J., and C. Whitfield. 2000. Translocation of group 1 capsular polysaccharide to the surface of Escherichia coli requires a multimeric complex in the outer membrane. EMBO J. 19:57-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank, J. 1992. Electron tomography: 3-D imaging with the transmission electron microscope. Plenum Press, Inc., New York, N.Y.
- 11.Frank, J. 2002. Single-particle imaging of macromolecules by cryo-electron microscopy. Annu. Rev. Biophys. Biomol. Struct. 31:303-319. [DOI] [PubMed] [Google Scholar]
- 12.Frank, J. 1996. Three-dimensional electron microscopy of macromolecular assemblies. Plenum Press, Inc., San Diego, Calif.
- 13.Frank, J., P. Penczek, R. Grassucci, and S. Srivastav. 1991. 3-D reconstruction of the 70S ribosome in ice: the distribution of ribosomal RNA. J. Cell Biol. 115:597-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank, J., M. Radermacher, P. Penczek, J. Zhu, M. Ladjadj, and A. Leith. 1996. SPIDER and WEB: processing and visualization of images in 3-D electron microscopy and related fields. J. Struct. Biol. 116:190-199. [DOI] [PubMed] [Google Scholar]
- 15.Frøholm, L. O., K. Jyssum, and M. Bøvre. 1973. Electron microscopical and cultural features of Neisseria meningitidis competence variants. Acta Pathol. Microbiol. Scand. 81:525-537. [DOI] [PubMed] [Google Scholar]
- 16.Frosch, M., E. Schultz, E. Glenncalvo, and T. F. Meyer. 1990. Generation of capsule-deficient Neisseria meningitidis strains by homologous recombination. Mol. Microbiol. 4:1215-1218. [DOI] [PubMed] [Google Scholar]
- 17.Giron, J. A., A. S. Y. Ho, and G. K. Schoolnik. 1991. An inducible bundle-forming pilus of enteropathogenic E. coli. Science 254:710-713. [DOI] [PubMed] [Google Scholar]
- 18.Guilvout, I., K. R. Hardie, N. Sauvonnet, and A. P. Pugsley. 1999. Genetic dissection of the outer membrane secretin PulD: are there distinct domains for multimerization and secretion specificity? J. Bacteriol. 181:7212-7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris, J. R., and R. Horne. 1994. Negative staining: a brief assessment of current technical benefits, limitations and future possibilities. Micron 25:5-13. [Google Scholar]
- 20.Heckels, J. E. 1989. Structure and function of pili of pathogenic Neisseria species. Clin. Microbiol. Rev. 2:S66-S73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hobbs, M., and J. S. Mattick. 1993. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein secretion apparatus: a general system for the assembly of surface associated protein complexes. Mol. Microbiol. 10:233-243. [DOI] [PubMed] [Google Scholar]
- 22.Jönsson, A. B., D. Ilver, P. Falk, and S. Normark. 1994. Sequence changes in the pilus subunit lead to tropism variation of Neisseria gonorrhoeae to human tissue. Mol. Microbiol. 13:403-416. [DOI] [PubMed] [Google Scholar]
- 23.Koster, M., W. Bitter, H. De Cock, A. Allaoui, G. R. Cornelis, and J. Tommassen. 1997. The outer membrane component, YscC, of the Yop secretion machinery of Yersinia enterocolitica forms a ring-shaped multimeric complex. Mol. Microbiol. 26:789-797. [DOI] [PubMed] [Google Scholar]
- 24.Lebart, L., A. Maurineau, and K. M. Warwick. 1984. Multi-variate descriptive statistical analysis. Wiley, New York, N.Y.
- 25.Lebart, L., A. Morineau, and N. Tabard. 1977. Techniques de la description statistique. Dunod, Paris, France.
- 26.Linderoth, N. A., M. N. Simon, and M. Russel. 1997. The filamentous phage pIV multimer visualized by scanning transmission electron microscopy. Science 278:1635-1638. [DOI] [PubMed] [Google Scholar]
- 27.Marciano, D. K., M. Russel, and S. M. Simon. 1999. An aqueous channel for filamentous phage export. Science 284:1516-1519. [DOI] [PubMed] [Google Scholar]
- 28.McBride, M. J. 2001. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu. Rev. Microbiol. 55:49-75. [DOI] [PubMed] [Google Scholar]
- 29.McRee, D. E. 1999. XtalView/Xfit: a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125:156-165. [DOI] [PubMed] [Google Scholar]
- 30.Merz, A. J., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98-102. [DOI] [PubMed] [Google Scholar]
- 31.Nouwen, N., N. Ranson, H. Saibil, B. Wolpensinger, A. Engel, A. Ghazi, and A. P. Pugsley. 1999. Secretin PulD: association with pilot PulS, structure, and ion-conducting channel formation. Proc. Natl. Acad. Sci. USA 96:8173-8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nouwen, N., H. Stahleberg, A. P. Pugsley, and A. Engel. 2000. Domain structure of secretin PulD revealed by limited proteolysis and electron microscopy. EMBO J. 19:2229-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nunn, D. 1999. Bacterial type II protein export and pilus biogenesis: more than just homologies? Trends Cell Biol. 9:402-408. [DOI] [PubMed] [Google Scholar]
- 34.Opalka, N., R. Beckmannn, N. Boisset, M. N. Simon, M. Russel, and S. A. Darst. 2003. Structure of the filementous phage IV multimer by cryo-electron microscopy. J. Mol. Biol. 325:461-470. [DOI] [PubMed] [Google Scholar]
- 35.Orlova, E. V., P. Dube, J. R. Harris, E. Beckman, F. Zemlin, J. Markl, and M. van Heel. 1997. Structure of keyhole limpet hemocyanin type 1 (KLH1) at 15 Å resolution by electron cryomicroscopy and angular reconstitution. J. Mol. Biol. 271:417-437. [DOI] [PubMed] [Google Scholar]
- 36.Parge, H. E., K. T. Forest, M. J. Hickey, D. A. Christensen, E. D. Getzoff, and J. A. Tainer. 1995. Structure of the fibre forming protein pilin at 2.6 Ångstrom resolution. Nature 378:32-38. [DOI] [PubMed] [Google Scholar]
- 37.Penczek, P., M. Radermacher, and J. Frank. 1992. 3-D reconstruction of single particles embedded in ice. Ultramicroscopy 40:33-53. [PubMed] [Google Scholar]
- 38.Potts, W. J., and J. R. Saunders. 1988. Nucleotide-sequence of the structural gene for class-l pilin from Neisseria meningitidis: homologies with the pilE locus of Neisseria gonorrhoeae. Mol. Microbiol. 2:647-653. [DOI] [PubMed] [Google Scholar]
- 39.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruprecht, J., and J. Nield. 2001. Determining the structure of biological macromolecules by transmission electron microscopy, single particle analysis and 3D reconstruction. Prog. Biophys. Mol. Biol. 75:121-164. [DOI] [PubMed] [Google Scholar]
- 41.Sali, A., and T. A. Blundell. 1993. Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 234:779-815. [DOI] [PubMed] [Google Scholar]
- 42.Saulino, E. T., E. Bullitt, and S. J. Hultgren. 2000. Snapshots of usher-mediated protein secretion and ordered pilus assembly. Proc. Natl. Acad. Sci. USA 97:9240-9245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saxton, W. O., and W. Baumeister. 1982. The correlation averaging of a regularly arranged bacterial cell envelope protein. J. Microsc. 127(Pt. 2):127-138. [DOI] [PubMed] [Google Scholar]
- 44.Schmidt, S. A., D. Bieber, S. W. Ramer, J. Hwang, C. Y. Wu, and G. Schoolnik. 2001. Structure-function analysis of BfpB, a secretin-like protein encoded by the bundle-forming-pilus operon of enteropathogenic Escherichia coli. J. Bacteriol. 183:4848-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shevchick, V. E., J. Robert-Baudouy, and G. Condemine. 1997. Specific interaction between OutD, an Erwinia chrysanthemi OMP of the general secretory pathway and secreted proteins. EMBO J. 16:3007-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stone, K. D., H. Z. Zhang, L. K. Carlson, and M. S. Donnenberg. 1996. A cluster of fourteen genes from enteropathogenic Escherichia coli is sufficient for biogenesis of a type IV pilus. Mol. Microbiol. 20:325-337. [DOI] [PubMed] [Google Scholar]
- 47.Strom, M. S., and S. Lory. 1993. Structure, function, and biogenesis of the type IV pili. Annu. Rev. Microbiol. 47:565-596. [DOI] [PubMed] [Google Scholar]
- 48.Swanson, J., S. J. Kraus, and E. C. Gotschlich. 1971. Studies on gonococcus infection. IV. Pili: their role in attachment of gonococci to tissue culture cells. J. Exp. Med. 134:886-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thanassi, D. G., E. T. Saulino, M. J. Lombardo, R. Roth, J. Heuser, and S. J. Hultgren. 1998. The PapC usher forms an oligomeric channel: implications for pilus assembly across the outer membrane. Proc. Natl. Acad. Sci. USA 95:3146-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tønjum, T., D. A. Caugant, S. A. Dunham, and M. Koomey. 1998. Structure and function of repetitive sequence elements associated with a polymorphic domain of the N. meningitidis PilQ complex. Mol. Microbiol. 29:975-986. [DOI] [PubMed] [Google Scholar]
- 51.Tønjum, T., N. E. Freitag, E. Namork, and M. Koomey. 1995. Identification and characterization of PilG, a highly conserved pilus assembly gene in pathogenic Neisseria. Mol. Microbiol. 16:451-464. [DOI] [PubMed] [Google Scholar]
- 52.Tønjum, T., and M. Koomey. 1997. The pilus colonization factor of pathogenic neisserial species: organelle biogenesis and structure/function relationships. Gene 192:155-163. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, Q. Y., D. Deryckere, P. Lauer, and M. Koomey. 1992. Gene conversion in Neisseria gonorrhoeae: evidence for its role in pilus antigenic variation. Proc. Natl. Acad. Sci. USA 89:5366-5370. [DOI] [PMC free article] [PubMed] [Google Scholar]