Abstract
The soxRS regulon protects Escherichia coli from superoxide and nitric oxide stress. SoxR protein, a transcription factor that senses oxidative stress via its [2Fe-2S] centers, transduces the signal to the soxS promoter to stimulate RNA polymerase. Here we describe 29 mutant alleles of soxR that cause defects in the activation of soxS transcription in response to paraquat, a superoxide stress agent. Owing to the selection and screen used in their isolation, most of these mutant alleles encode proteins that retained specific binding activity for the soxS promoter in vivo. The mutations were found throughout the SoxR polypeptide, although those closer to the N terminus typically exhibited greater defects in DNA binding. The degree of the defect in the transcriptional response to superoxide caused by each mutation was closely paralleled by its impaired response to nitric oxide. This work begins the general identification of the residues in the SoxR polypeptide that are critical for transducing oxidative stress signals into gene activation.
Reactive oxygen species (ROS) are generated as a consequence of the incomplete reduction of oxygen and are unavoidable by-products of aerobic metabolism. If not disposed of efficiently, ROS can cause cellular and genetic damage leading to carcinogenesis, senescence, and neurodegenerative disorders (12, 13). Because cells under oxidative stress are at risk for lethal or mutagenic damage, all aerobic organisms have evolved defense mechanisms to cope with ROS. Part of this response involves the reprogramming of gene expression to increase levels of antioxidant enzymes such as superoxide dismutase and catalase, which limit the levels of superoxide (O2˙−) > and hydrogen peroxide, respectively.
The study of oxidative stress responses in bacteria has increased our understanding of the biochemical mechanisms whereby ROS levels are sensed by the cell and transduced into changes in gene expression (36, 43). The soxRS regulon of Escherichia coli mediates a response that protects the cell against O2˙−, nitric oxide (NO), and redox-cycling agents such as paraquat (PQ). In the presence of these oxidants, SoxR, a transcription factor, is activated and stimulates production of a second transcription factor, SoxS, by up to 100-fold (8). Microarray analyses show that expression of the SoxS protein activates at least 40 genes that encode antioxidant, metabolic, and repair functions (36), although Martin and Rosner (29) estimate that the number of genes under the direct control of SoxS may be fewer.
SoxR, a member of the MerR family of transcription factors, is a homodimer of 17-kDa subunits and is constitutively expressed in the cell, albeit in an inactive form. Each monomer of SoxR contains a redox-active [2Fe-2S] center, essential for SoxR's transcriptional activity but not for its ability to bind to the promoter (20, 21). SoxR binds the soxS promoter with similar affinities in the apo- and metallo-forms and does not significantly influence RNA polymerase binding (21). Oxidation and reduction of the SoxR [2Fe-2S] centers also do not alter the protein's affinity for its DNA site (15). The soxS promoter is arranged such that the ends of the −35 and −10 RNA polymerase recognition elements are 19 bp apart, in contrast to the optimal 17-bp spacer found in most σ70-regulated promoters in E. coli (22). This places the −10 and −35 elements ∼70° out of phase from their orientation in a 17-bp spacer promoter. The 19-bp spacer arrangement is common to all target promoters of MerR family members, and it is believed that these enzymes stimulate transcription by a common mechanism (18, 22). Chemical footprinting studies have demonstrated that activated SoxR and MerR overcome the suboptimal spacer by mediating specific distortions in their target promoters that stimulate RNA polymerase to form the open complex (19, 20). More recently, the crystal structure of BmrR (a MerR family member), solved in complex with its cognate promoter, is consistent with transcriptional activation through local DNA unwinding and base pair disruption (18).
SoxR's [2Fe-2S] centers are the primary sensors of oxidative stress. SoxR mutant proteins in which the iron-coordinating cysteine residues are replaced with alanine contain no detectable iron and are unable to stimulate transcription of soxS in response to PQ (2). In vitro, the oxidation state of the [2Fe-2S] centers is the key feature that controls SoxR's transcriptional activity (11, 15). When intact cells are treated with PQ, SoxR's [2Fe-2S] centers undergo one-electron oxidation and SoxR becomes transcriptionally active (14, 23). Upon removal of the oxidative stress, the [2Fe-2S] centers are rereduced and SoxR becomes transcriptionally silent (14, 23). SoxR is also activated by NO, which nitrosylates the [2Fe-2S] centers to generate an active form that contains dinitrosyl-iron centers (9). Purified nitrosylated SoxR has transcriptional activity similar to that of the oxidized protein (9). The activation of SoxR by NO may play a role in the resistance of E. coli to attack by NO-generating macrophages (33, 34).
A long-standing question, and a fundamental problem in oxidative stress biology, is how redox and NO signals are transduced into gene expression. In this paper, we have approached this problem by isolating 29 mutant alleles of soxR by using genetic screens that are based on the molecular features of the soxRS system. These mutations all display a defect in the activation of soxS transcription in response to PQ- or NO-induced stress. Prior to the work described here, only a few constitutively active SoxR mutant proteins had been isolated, and many of these mutations cluster in the C-terminal region of the protein (32, 42). The cysteine-to-alanine mutations that eliminate iron binding were directly engineered (2), but other variant forms of SoxR had not been isolated. The transcriptionally defective SoxR proteins will be useful for interpreting structural information and will help to illuminate the mechanisms of redox and NO signal transduction via protein iron-sulfur centers.
MATERIALS AND METHODS
Construction of strain MC9 used in the dual selection-screen for soxR mutants.
A 120-bp fragment containing the soxS promoter was amplified by PCR from plasmid pBD100 (Table 1) with Deep Vent polymerase (New England Biolabs) and primers A (5′ CCACTTCGGAAGGGGTTCGCAGCG 3′) and B (5′ GGGACACTGCAGTGCCTCTTTTCAGTG 3′). The underlined sequences in primers A and B are recognition sites for XmnI and PstI, respectively. A 1.4-kb fragment containing the open reading frame for the sacB gene from Bacillus subtilis was similarly PCR amplified from plasmid pKO3 (Table 1) with primers C (5′ GGAGCTGCAGACGATGAACATC 3′) and D (5′ CCGGATGAACATTTTCTTTTGCG 3′). The underlined sequences in primers C and D are recognition sites for PstI and XmnI, respectively. The soxS and sacB PCR fragments were digested with appropriate restriction enzymes and inserted into the XmnI site of plasmid pACYC184 (Table 1) downstream of the cat gene, yielding pMC6 (Table 1). The 2.8-kb cat-soxS promoter-sacB fusion fragment was PCR amplified from pMC6 with Taq polymerase (Qiagen) with primers I (5′ CATGTTAGAAGATCTCAAACGCCAGGTATTAGAAGCCAACCTGGCGCACCAGGCGTTTAAGGGCAC 3′) and J (5′ GTCATTACTGCCCGTAATATGCCTTCGCGCCATGCTTACGCAGATACCTGCCACATGAAGCTTTTC 3′), purified, and inserted into the araD site of strain EH200 (Table 1) by the method described by Datsenko and Wanner (6). Cells that had been successfully transformed were chloramphenicol resistant, and integration into the targeted araD site was verified by the inability of the strains to grow in M9 medium containing l-arabinose and confirmed by colony PCR with primers G (5′ GCGCATCCGGCACGAAGGAG 3′) and H (5′ GGTTTCGTTTGATTGGCTGTGG 3′), which yielded a 0.78-kb PCR fragment in the correct clone. One such clone, designated strain MC9 (Table 1), was used in the dual selection-screen for uninducible soxR mutants.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| DJ901 | Δ(soxRS) derivative of GC4468 | 17 |
| EH46 | DJ901 lysogenized with λ(soxS promoter-lacZ) | 22 |
| EH86 | DJ901 lysogenized with λ(16-bp spacer mutant soxS promoter-lacZ) | 22 |
| EH200 | DJ901 lysogenized with λ(soxR promoter-lacZ) | 24 |
| GC4468 | K12 rpsL thi soxR+soxS+ | 17 |
| MC9 | EH200 with chromosomally integrated 2.8-kb fusion fragment cat-120-bp soxS promoter-sacB in the araD site | This work |
| NR9273 | ara thi Δ(pro-lac) mutD5 zaf-13::Tn10 | R. Schaaper |
| TN402 | GC4468 soxR::kan | T. Nunoshiba |
| Plasmids | ||
| pACYC184 | Low-copy-number cloning vector (Cmr) | New England Biolabs |
| pBD100 | pBR322 with entire soxRS locus (Ampr) | 1 |
| pKO3 | Plasmid with B. subtilis sacB gene (Cmr) | 27 |
| pMC6 | pACYC184 with cat-soxS promoter-sacB (Cmr) | This work |
| pSE380 | trc promoter-containing plasmid with lacIq gene (Ampr) | Invitrogen |
| pSXR | pSE380 derivative containing the wild-type soxR allele (Ampr) | 1 |
Isolation of transcriptionally defective soxR mutants.
NR9273, a mutD5 mutator strain (Table 1), was transformed to ampicillin resistance with pSXR (Table 1). To allow mutagenesis to occur, individual transformants were grown for 15 h at 37°C in 3 ml of Luria-Bertani (LB) medium containing 100 μg of ampicillin/ml. The cells were harvested, and plasmid DNA was isolated with the Wizard miniprep kit (Promega). The plasmid preparation (containing both wild-type pSXR and mutated plasmids) was transformed into strain MC9, followed by selection on LB agar containing 100 μg of ampicillin/ml, 5% sucrose, and 50 μM PQ (Sigma) for 18 h at 37°C. Viable colonies were then patch-streaked on eosin-methylene blue (EMB)-lactose agar (Difco) containing 100 μg of ampicillin/ml in order to screen for mutant proteins that retained the ability to bind the SoxR binding site in vivo.
Isolates from the initial selection-screen were recovered and retransformed into strain MC9, selecting for ampicillin resistance. The next day, transformants were patch-streaked onto both LB agar plates containing 100 μg of ampicillin/ml, 5% sucrose, and 50 μM PQ and EMB-lactose agar plates containing 100 μg of ampicillin/ml, to verify the phenotypes. Plasmids that passed this second round of selection-screening were sequenced on both strands with primers K (5′ CACACAGGAAACAGACCATGGC 3′) and L (5′ CAGACCGCTTCTGCGTTCTG 3′) to identify the responsible mutation. All sequencing was performed at the High-Throughput DNA Sequencing Facility located at the Dana-Farber/Harvard Cancer Center.
β-Galactosidase assays.
β-Galactosidase assays were used to verify individually that the SoxR mutant proteins obtained in the screen described above were deficient in activating transcription from the soxS promoter in response to PQ. The pSE380-based plasmids with the mutated soxR alleles were transformed into strain EH46 (Table 1). The transformed strains were inoculated into LB medium containing 100 μg of ampicillin/ml and incubated at 37°C for ∼16 h with shaking at 220 rpm. Inocula from these overnight cultures were diluted 100-fold into 3 ml of fresh medium in duplicate tubes and incubated at 37°C for 90 min. PQ was then added to a final concentration of 100 μM to one of each pair of tubes, and the incubation was continued for 60 min with shaking at 220 rpm. The samples were then placed on ice for 20 min. β-Galactosidase activity in sodium dodecyl sulfate-CHCl3-treated cells was determined as described by Miller (31).
β-Galactosidase assays were also used to analyze the DNA-binding ability of the mutant proteins in vivo. Strains EH200 and EH86 (Table 1) were transformed with the plasmids containing the mutated soxR alleles and grown as described above before the lysates were assayed for β-galactosidase activity.
Isolation and analysis of soxS mRNA by Northern blotting.
Northern blot analysis was carried out to measure more directly the transcriptional defects caused by the various soxR mutations. For this purpose, the pSE380-based plasmids with the mutated soxR genes were transformed into strain TN402 (Table 1). Transformed cells were inoculated into LB medium containing 100 μg of ampicillin/ml and incubated at 37°C overnight with shaking at 220 rpm. Inocula from the overnight cultures were diluted 100-fold into 3 ml of fresh medium and incubated at 37°C for 2 h with shaking. PQ was then added to a final concentration of 100 μM, and incubation with shaking was continued at 37°C for another 5 min. Rifampin (Sigma) was added to 200 μg/ml to stop further transcription, and the cells were chilled and harvested to isolate total RNA by using the RNeasy Mini kit (Qiagen).
The various SoxR mutant proteins were also analyzed for their ability to respond to NO under anaerobic conditions. The inoculum for anaerobic cultures was prepared by subculturing cells three consecutive times in 3-ml capped vials (VWR Scientific) filled almost completely with cultures and incubating them at 37°C without shaking. For the experiment, the anaerobic inoculum was diluted 10-fold into 3 ml of fresh medium that had been previously degassed with argon in sealed vials. The cultures were grown for 3 h at 37°C without shaking. Spermine NONOate (an NO donor that releases 2 NO per spermine NONOate with a half-life of 39 min at 37°C and pH 7.4; Alexis Biochemicals) was injected into the vials to a final concentration of 1 mM, and incubation without shaking continued at 37°C for another 2 min. Rifampin was then added as described above, and the cells were harvested to isolate total RNA.
The amount of soxS transcript was quantified by Northern blot analysis. Samples of total RNA (10 μg) from the different strains were loaded on 1.5% agarose gels containing 0.25 M formaldehyde, electrophoresed, and transferred to nylon membranes (Schleicher and Schuell) with a Turboblotter (Schleicher and Schuell). After the gels were stained with ethidium bromide to visualize 23S and 16S rRNA (as a loading control), the blots were hybridized with a soxS-specific probe previously labeled with a random primer system (Life Technologies). The 400-bp soxS-specific probe was prepared by PCR amplifying the soxS gene from pBD100 (Table 1) with primers M (5′ CAGATGAATTAACGAACTGAACAC 3′) and N (5′ GCAATTACCCGCGCGGGAG 3′). The amount of soxS mRNA in the Northern blots was quantified by using a phosphorimager (Bio-Rad).
Sequence alignment and secondary structure predictions.
An alignment of the amino acid sequences of four members of the MerR family was performed with ClustalW (41) and confirmed by visual inspection. Secondary structure prediction of the SoxR polypeptide was performed with the protein structure prediction server PSIPRED located at http://bioinf.cs.ucl.ac.uk/psipred/(25, 30).
RESULTS
Combined selection and screen for the isolation of transcriptionally defective soxR mutants.
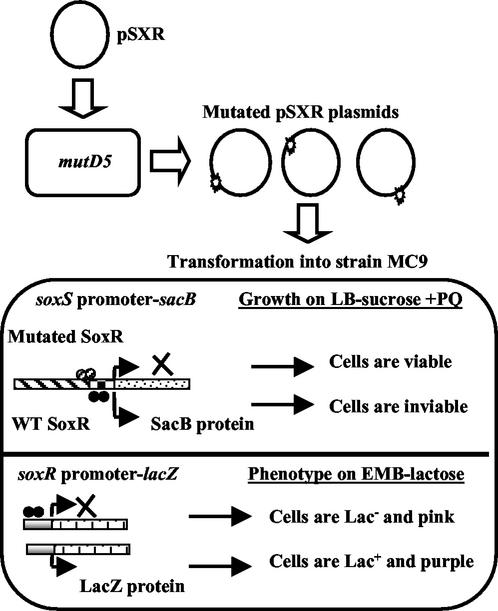
An approach was developed that allowed the identification of SoxR mutant proteins that had lost the ability to activate transcription from the soxS promoter in response to oxidative stress but that nevertheless retained significant DNA binding activity. In this way, we hoped to eliminate nonspecific mutations that merely disrupt SoxR protein structure. The strain used in the combined selection-screen, MC9, has soxRS deleted and contains two fusion constructs (Table 1; Fig. 1). The construct used to select transcriptionally defective soxR mutants consists of the wild-type soxS promoter fused to the sacB gene from B. subtilis, the product of which, in the presence of sucrose, is toxic to E. coli (37). Therefore, only cells harboring PQ-unresponsive soxR variants were viable when grown in the presence of both PQ and sucrose. MC9 cells expressing wild-type SoxR were inviable under these conditions (data not shown).
FIG. 1.
Mutagenesis of soxR and selection-screening for activation-defective mutant proteins. The soxR gene was mutagenized in a mutD5 strain, and activation-defective soxR mutants were selected in strain MC9, where they did not stimulate transcription from a soxS promoter-sacB fusion and were therefore viable on plates containing both PQ and sucrose; MC9 cells expressing wild-type (WT) SoxR were inviable. These activation-defective proteins were also screened for their ability to bind to DNA in the same strain; proteins that retained this property repressed expression from a soxR promoter-lacZ fusion, and the Lac− cells were pink on EMB indicator plates. Details of the mutagenesis and selection-screen are described in Materials and Methods and in Results.
The second fusion construct, consisting of the wild-type soxR promoter fused to the lacZ gene, was used to screen for mutant proteins that retained the ability to bind specifically to promoter DNA (Table 1; Fig. 1). SoxR represses transcription from the soxR promoter with or without oxidative stress (24), and therefore variants that retained their DNA-binding ability repressed lacZ expression, regardless of their transcription-activating abilities. These Lac− colonies were pink when grown on EMB indicator plates, in contrast to Lac+ colonies, which were purple.
Isolation of transcriptionally defective soxR mutations.
The soxR gene was mutagenized by being passed through a mutD5 mutator strain, followed by transformation into strain MC9 to select for mutant forms as described above. The observed mutation frequency was 10−4, as expected for the mutD5 effect (5, 7). Of the PQ-unresponsive mutant proteins selected, 45% displayed specific binding to repress the soxR promoter in strain MC9 (data not shown).
Three independent rounds of mutagenesis and selection-screening were conducted to yield 46 independent mutants. The DNA sequences of the mutant alleles converged on the data set shown in Table 2. Mutations were observed in 24 of the 154 soxR codons (Table 2; Fig. 2). Of the 29 distinct alleles identified in this work, 10 were found more than once, including a double mutation that changed cysteine-124 to alanine (Table 2). Transitions occurred more frequently than did transversions (23 transitions versus seven transversions), again as expected for the mutD5 effect in rich medium (40). There were no frameshift mutations, and one nonsense mutation was identified that resulted in a polypeptide truncated after alanine-63.
TABLE 2.
Activation-defective SoxR mutant proteins
| Mutation | Nucleotide change(s) | No. of independent clones |
|---|---|---|
| M1T | T→C | 1 |
| G15D | G→A | 1 |
| E16K | G→A | 3 |
| Y31H | T→C | 1 |
| L36V | T→G | 1 |
| V53M | G→A | 1 |
| I62N | T→A | 3 |
| A63V | C→T | 1 |
| A63stop | C→T | 1 |
| Q64R | A→G | 1 |
| I66T | T→C | 3 |
| I73F | A→T | 1 |
| H84R | A→G | 1 |
| L86S | T→C | 4 |
| E90K | G→A | 2 |
| L94F | C→T | 1 |
| L94P | T→C | 1 |
| S95P | T→C | 1 |
| S96P | T→C | 2 |
| I106T | T→C | 1 |
| E115G | A→G | 1 |
| D117G | A→G | 1 |
| C122R | T→C | 1 |
| C122Y | G→A | 1 |
| C124A | T→G, G→C | 2 |
| C124G | T→G | 1 |
| C124Y | G→A | 2 |
| R127L | G→T | 2 |
| C130R | T→C | 4 |
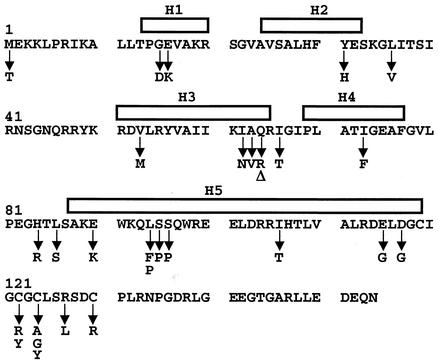
FIG. 2.
Sequence of wild-type SoxR from E. coli and location of mutations that result in transcriptionally defective SoxR variant proteins. The sequence of the E. coli SoxR polypeptide is shown along with predicted secondary structure elements (see Materials and Methods) depicted above the sequence. Bars represent α helices. Also shown (below the sequence) are locations of mutations that gave rise to a transcription defect. Δ represents a nonsense mutation at position 64.
Figure 2 depicts the amino acid sequence of SoxR along with the mutations that resulted in activation-defective proteins. Particularly puzzling was the M1T mutation, which is expected to prevent translation, as bacterial proteins generally initiate with methionine (or sometimes valine). Immunoblot analysis of total cell extract with a SoxR antibody showed that M1T is in fact a null mutant, which had somehow slipped through the screen (data not shown). It was similarly demonstrated that the A63stop mutation was also a null allele (data not shown). All the other mutant proteins were produced at levels comparable to that of wild-type SoxR (data not shown).
Four other mutations identified in the screen were G15D, E16K, Y31H, and L36V. These mutations alter the N-terminal region of SoxR, predicted to form a helix-turn-helix structure that would mediate binding to promoter DNA (Fig. 2). Many alterations in this region of the protein would be expected to disrupt DNA binding and therefore not be identified in the screen employed here.
The majority of mutations were found to alter the central region of the polypeptide encompassing residues 53 to 117, an area of unknown function(s) (Table 2; Fig. 2). Not surprisingly, we also identified a number of alterations in the cysteine-rich region of the protein (residues 119 to 130) that anchors the [2Fe-2S] cluster. Interestingly, no mutations were isolated that changed the extreme C-terminal region of SoxR (residues 131 to 154). Previous work (32, 42) has shown that sequence changes in this region give rise to constitutively active proteins, and it is therefore not surprising that the selection-screen for nonfunctional forms described in this work did not easily yield variants with alterations in this region.
Loss of PQ responsiveness in SoxR mutant proteins: soxS promoter-lacZ reporter studies.
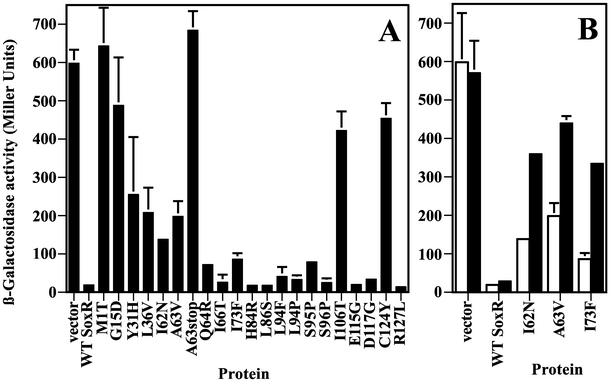
The mutated soxR genes were isolated by use of strain MC9 and screening on solid media. To confirm this phenotype in liquid medium and in a different background, the mutated plasmids were transformed into strain EH46 (Table 1). Cultures of these transformants were treated with 100 μM PQ for 1 h with good aeration and then assayed for β-galactosidase activity. Exposure to 100 μM PQ was chosen because wild-type SoxR activity is maximally induced at this concentration of PQ (data not shown).
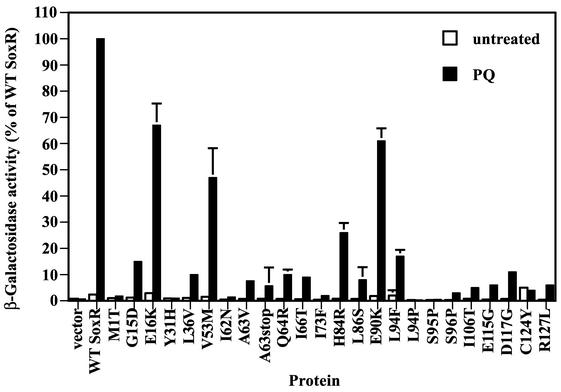
Figure 3 shows the β-galactosidase activities obtained for cells expressing mutant SoxR proteins. Cells expressing wild-type SoxR showed 40-fold-higher β-galactosidase activity after PQ treatment than did untreated cells (Fig. 3). As expected, none of the mutant forms gave significant β-galactosidase activity in the absence of PQ (unstressed conditions). In the PQ-treated cells, β-galactosidase activities ranged from <1% of wild-type SoxR (for example, in L94P) to ∼55 to 75% of wild-type SoxR (in E16K, V53M, and E90K). The latter three mutations were put aside, and we continued characterizing the remaining mutations that displayed a more dramatic phenotype. Note that, although we identified six different cysteine mutations (Table 2), we show only the results obtained with mutant protein C124Y; the five other cysteine mutations at positions 122 and 124 produced similar results in the experiments described here (data not shown).
FIG. 3.
Transcriptional activity of SoxR mutant proteins analyzed by β-galactosidase assays. Strain EH46 (soxS promoter-lacZ) cells expressing the various mutated SoxR proteins were either treated (black columns) or not (open columns) with 100 μM PQ for 1 h before the assay for β-galactosidase activity. β-Galactosidase activities are reported as percentages of the activity obtained with wild-type (WT) SoxR treated with 100 μM PQ (6,500 Miller units = 100%). The results shown represent the means and standard errors (bars; some not visible on this scale) of three independent experiments.
Northern blot analysis of the response of SoxR mutant proteins to PQ and to NO.
Since reporter fusions give an indirect measure of transcriptional activity, additional experiments were conducted to determine soxS induction directly by Northern blotting. For this purpose, strain TN402 (Table 1) was transformed with plasmids with the various mutated soxR genes, and the isolates were treated either with 100 μM PQ for 5 min under aerobic conditions or with 1 mM spermine NONOate (an NO-releasing compound) for 2 min under anaerobic conditions. The NO experiments were conducted under anoxic conditions because we wished to assess the direct effects of NO on SoxR activity and to avoid oxygen-dependent by-products of NO. Previous studies (33, 34) showed that SoxR activation by NO occurs readily in the absence of oxygen. Under the conditions for these experiments, wild-type SoxR showed maximal induction after 5 min of treatment with 100 μM PQ or 2 min of treatment with 1 mM spermine NONOate (data not shown).
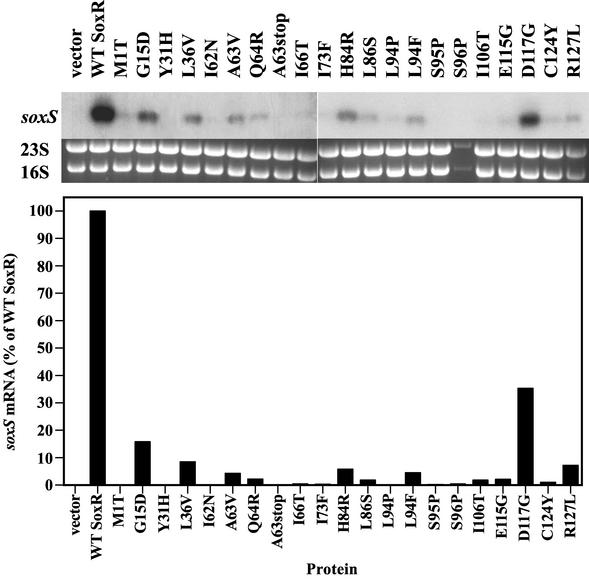
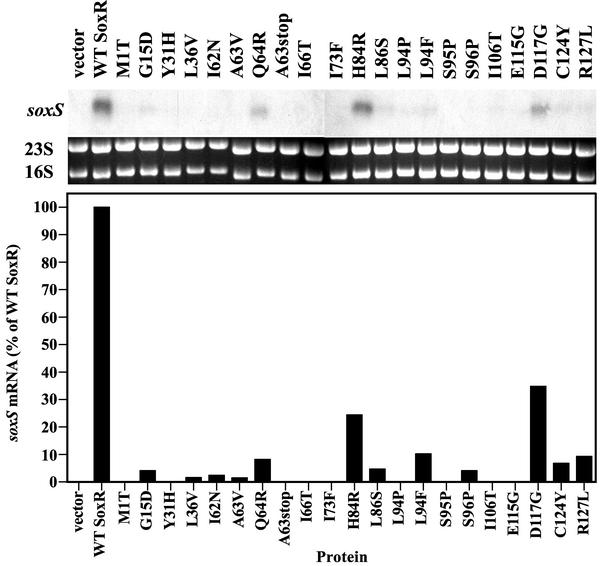
The top panel of Fig. 4 shows the levels of soxS mRNA produced in PQ-treated cells expressing the various SoxR mutant proteins. Quantification of the soxS mRNA (Fig. 4, middle panel) correlated well with the β-galactosidase activities reported in Fig. 3, confirming the transcriptional defects of the soxR mutants.
FIG. 4.
PQ responsiveness of SoxR mutant proteins studied by Northern blot analysis. Strain TN402 expressing the various mutated SoxR proteins was treated for 5 min with 100 μM PQ before total RNA was obtained. The RNA was electrophoresed and transferred by Northern blotting, the filters were hybridized with a soxS-specific probe, and RNA was quantified in a phosphorimager. (Top) Northern blots showing soxS mRNA levels. (Middle) The corresponding ethidium bromide-strained gels show 23S and 16S rRNA that served as loading controls. (Bottom) Quantification of soxS mRNA levels obtained in Northern blots. soxS mRNA was normalized to 16S rRNA levels and is reported relative to the amount obtained with wild-type (WT) SoxR (set at 100%). The results shown are from one of two independent experiments.
The activation mechanism of SoxR by NO (nitrosylation of the [2Fe-2S] centers [9]) differs fundamentally from the mechanism in response to PQ (oxidation of the centers [11, 15]). It was therefore of interest to determine the response of the variants, isolated in a screen with PQ, to NO. The top panel of Fig. 5 shows the amount of soxS mRNA produced in cells expressing mutated SoxR proteins after treatment with spermine NONOate. The response of the mutant proteins to the NO donor was very similar to their response to PQ (Fig. 4 and 5), indicating that these mutant proteins cannot activate soxS transcription in response to either oxidative or NO stress.
FIG. 5.
NO responsiveness of SoxR mutant proteins studied by Northern blot analysis. TN402 cells expressing mutated SoxR proteins were treated anaerobically with 1 mM spermine NONOate for 2 min before quantification of the amount of soxS mRNA produced. (Top) Northern blots showing soxS mRNA levels. (Middle) The corresponding gels show 23S and 16S rRNA that served as loading controls. (Bottom) Quantification of the soxS mRNA levels obtained in Northern blots. soxS mRNA was normalized to 16S rRNA levels and is reported relative to the amount obtained with wild-type (WT) SoxR (set at 100%). The results shown are from one of two independent experiments.
Assessment of in vivo promoter binding by SoxR mutant proteins.
The genetic screen used to identify activation-defective SoxR mutant proteins employed a soxR promoter-lacZ fusion to screen out null mutations and variants with general stability or folding defects, which would not effectively bind the promoter to repress it. This screen should also have eliminated stable, folded variants that had lost DNA-binding ability, for example, due to their inability to make necessary contacts with the promoter. In order to confirm the solid medium screen under liquid growth conditions, strain EH200 (Table 1) was transformed with the various mutated plasmids, grown aerobically in liquid culture, and after treatment or not with 100 μM PQ assayed for β-galactosidase activity. Figure 6A shows the results obtained with untreated cells. Wild-type SoxR repressed lacZ expression from the soxR reporter, as did many of the mutated forms. In contrast to their phenotype on solid medium, mutant proteins M1T and A63stop did not display repression from the soxR reporter in cells in liquid culture, consistent with immunoblotting results. What was unexpected, however, was that the following mutant proteins also did not effectively repress transcription from the soxR promoter in this assay (residual soxR promoter-lacZ expression, ≥20% of vector control): G15D, Y31H, L36V, I62N, A63V, I106T, and C124Y. As mentioned earlier, these proteins were all produced at levels similar to that of wild-type SoxR, thereby eliminating the possibility that the lack of repression was due to insufficient protein levels or unstable products. Furthermore, when cells containing the I62N and A63V variants were treated with PQ for 1 h before the assay, both they and I73F showed a strongly diminished ability to repress lacZ expression (Fig. 6B). This effect contrasts strongly with wild-type SoxR, the DNA binding of which was essentially unaffected by PQ (Fig. 6B).
FIG. 6.
Binding of mutant SoxR proteins to the soxR promoter analyzed by β-galactosidase assays. Mutated SoxR proteins were analyzed for their ability to repress lacZ expression from the soxR promoter fusion in strain EH200. (A) β-Galactosidase activity in cells grown aerobically under unstressed conditions. (B) β-Galactosidase activity in cells expressing SoxR variant proteins I62N, A63V, and I73F that were treated with 100 μM PQ for 1 h (black columns) or were untreated (white columns). The values shown represent the means and standard errors (bars) of two independent experiments. WT, wild type.
We further probed this issue by using a different construct—a 16-bp spacer variant of the soxS promoter fused to lacZ in strain EH86 (Table 1). SoxR binds normally to this mutant promoter without activating soxS transcription but instead represses it both with and without oxidative stress (24). β-Galactosidase activities of EH86 cells expressing the mutant SoxR proteins corroborated the results obtained with strain EH200: mutant proteins G15D, Y31H, L36V, I62N, A63V, I106T, and C124Y exhibited defective promoter binding (data not shown). PQ treatment enhanced this defect in mutant proteins I62N, A63V, and I73F, acting on the mutant soxS promoter (data not shown), just as was the case for the soxR promoter (Fig. 6B).
DISCUSSION
The selection and screen described here were designed to identify positive-control SoxR variants—proteins that retain specific DNA-binding ability but that are unable to activate transcription in response to oxidative stress. Of the 29 distinct alleles identified in this work, two (M1T and A63stop) resulted in a null phenotype, while the remaining mutations encoded proteins that showed a defect in activating transcription and displayed a wide range in the degree to which they were defective. Three of these proteins, E16K, V53M, and E90K, showed a very mild defect and were not characterized further. Of the remaining mutant proteins with more pronounced defects, 11 retained significant DNA binding activity, while the rest (G15D, Y31H, L36V, I62N, A63V, I73F, I106T, and the six cysteine variants) had variable DNA-binding abilities in vivo depending on the assay used.
SoxR mutant proteins that are unable to activate soxS transcription in response to oxidative stress might suffer from any of the following defects: inability to dimerize, inability to make appropriate contacts with the promoter, loss or destabilization of [2Fe-2S] centers, intact [2Fe-2S] centers that are inadequately responsive to redox signals, and proteins that are unable to effect appropriate structural changes in the soxS promoter. Characterization of these types of mutant proteins will enable us to dissect not only the molecular mechanisms underlying signal transduction by SoxR but also promoter remodeling by MerR family members in general.
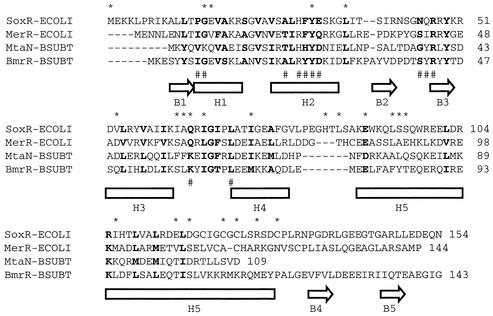
Figure 7 shows an alignment of the SoxR protein and three other MerR family members—MerR, BmrR, and MtaN. The crystal structures of BmrR (complexed with a 22-bp oligonucleotide) and MtaN (without DNA) yielded the secondary structure elements shown in the figure (16, 18). The structure of MerR is unknown, but this protein has been extensively studied genetically and biochemically, providing a wealth of mutational information (4, 26, 28, 35, 38). The ∼110 residues in the N-terminal region of MerR-related proteins are the most conserved segment and encompass the winged helix-turn-helix DNA binding motif (H1 and H2) in BmrR and MtaN (16, 18). The corresponding regions in SoxR and MerR are predicted to fold into similar structures (4) (Fig. 2). BmrR residues that correspond to SoxR residues G15 and Y31 are located within the helix-turn-helix motif and make contacts with DNA in the crystal structure (18). If these residues in SoxR are also responsible for making essential contacts with the soxS promoter, one would expect variations to result in defective promoter binding, which was indeed observed in the β-galactosidase DNA binding assays. The functional importance of Y31 is indicated by its conservation in all four proteins, while G15 is present in all but MtaN, in which it is replaced by lysine (Fig. 7). SoxR residue L36 is also conserved in all four proteins, and the SoxR L36V variant has a strong activation defect.
FIG. 7.
Sequence alignment of the MerR family members SoxR, MerR, MtaN, and BmrR. The BmrR polypeptide consists of 279 residues of which the first 143 residues are shown here. Secondary structure elements from crystallographic analysis of BmrR (18) and MtaN (16) are indicated below the sequences by arrows for β strands and boxes for α helices. Sequences that are identical or functionally similar are in boldface; hyphens indicate gaps; asterisks depict SoxR residues that have been mutated in this work; pound signs mark BmrR residues that directly contact DNA (18). See the text for a discussion.
MerR family members evidently contain a second helix-turn-helix domain formed by helices H3 and H4 (Fig. 7), and residues in this region of BmrR make limited contacts with DNA (18). Mutations in the corresponding region of SoxR (I62N, A63V, Q64R, I66T, and I73F) resulted in a transcription defect. A possible function for this second helix-turn-helix domain is to facilitate the allosteric modifications of the promoter that are necessary for transcription initiation. Evidence in support of this hypothesis was provided by a mutant MerR protein, A60V (corresponding to SoxR A63V), that was defective in activating transcription from the mer promoter. This variant showed normal DNA binding in vitro but was unable to mediate the promoter distortions obtained with wild-type MerR necessary for productive open complex formation with RNA polymerase (28).
The I62N, A63V, and I73F SoxR variant proteins conferred an unusual phenotype in the in vivo repression assays. These proteins displayed weak promoter binding in unstressed cells, and the DNA binding was further weakened upon the addition of PQ. Since its oxidation state does not affect the ability of wild-type SoxR to bind to the soxS promoter in vitro (15) or to the soxR promoter in vivo (24), the apparent destabilizing effect of oxidation in the I62N, A63V, and I73F mutant proteins bears further analysis. One explanation for the observations made here is that these mutant proteins are unstable, and PQ exposure facilitates their unfolding. Immunoblot analysis, however, showed that the levels of these mutant proteins were similar in cells that were treated and cells that were not treated with PQ (data not shown). An alternative possibility (not yet demonstrated) is that PQ treatment induces a conformational change that weakens the affinity of the mutant proteins for promoter DNA.
The remainder of the mutant proteins described here are altered in regions outside the two putative helix-turn-helix domains of SoxR (Fig. 2 and 7). The region corresponding to SoxR residues 88 to 127 is helical in BmrR and MtaN, in which it forms an antiparallel coiled coil that seems to mediate protein dimerization (16, 18). Residues 88 to 127 of SoxR and 82 to 120 of MerR are predicted to form a helix that might subsume the same dimerization role (4) (Fig. 2). Several mutations in and near this region of SoxR resulted in activation-defective proteins, some of which displayed a strong positive-control defect and some that did not (for example, I106T). One possible problem with the I106T protein is that it dimerizes poorly, which would explain the failure of this protein to bind the promoter in vivo and activate transcription. Other mutations in this region (H84R, L86S, L94F, L94P, S95P, S96P, E115G, and D117G) resulted in variants that all bind to DNA in vivo but which cannot activate transcription in response to PQ. These proteins could have any of the problems listed above, which will be identified only by in vitro analyses, currently under way. Note that none of these residues is conserved in the other MerR family members shown in Fig. 7, and it is therefore quite likely that the defects in these mutant proteins will be particular to SoxR and potentially useful in unraveling the redox signaling mechanism from the iron-sulfur centers.
The remaining mutant proteins include six cysteine variants and R127L, also in the vicinity of the cysteine cluster. The most obvious problem with the cysteine mutant proteins is that they likely do not contain the essential [2Fe-2S] centers of SoxR, since the substituting residues are not known ligands for iron (3). What is surprising about the cysteine variants, however, is their apparently poor ability to bind DNA, as judged by the in vivo repression assays. In vitro band-shift assays performed with partially purified cysteine-to-alanine mutant proteins showed that those variants bound to the soxS promoter with an overall affinity similar to that of wild-type SoxR (2). The weak promoter binding displayed by the cysteine mutant proteins in the in vivo assays described here cannot be attributed to low protein levels. These variants are being purified and characterized in vitro to clarify this apparent discrepancy.
In addition to analyzing the response of the various SoxR mutant proteins to PQ, we studied their response to NO. Although NO treatment results in nitrosylation of SoxR's [2Fe-2S] centers (9), while PQ treatment causes oxidation of these centers (10, 14), previous work (9) suggested that these chemically distinct forms of SoxR adopt similar structures and activate transcription from the soxS promoter in a comparable manner. The observation that most of the SoxR mutant proteins described here showed similar transcriptional activities in response to NO and PQ treatment further supports this idea.
The role of SoxR's [2Fe-2S] centers in signaling free radicals is likely to be replayed in other systems. Iron-sulfur clusters have been implicated previously as biosensors of oxidants and NO in other proteins, including mammalian IRP-1 and Fnr from E. coli (39). The knowledge gained from our studies of SoxR will serve as a guide to dissect the mechanisms and roles of redox and NO signaling in other organisms. These studies will also prove useful in teasing apart the mechanism by which diverse MerR family members modulate transcription in response to a variety of environmental stimuli.
Acknowledgments
This work was funded by grant CA37831 from the United States National Cancer Institute.
We thank R. Schaaper (National Institute of Environmental Health Sciences, Research Triangle Park, N.C.) for providing strain NR9273 and T. Nunoshiba (Tohoku University, Sendai, Japan) for providing strain TN402. We thank M. Jorgensen, P. Pomposiello, A. Koutsolioutsou, V. Leautaud, and L. McLaughlin for valuable advice and discussion.
REFERENCES
- 1.Amabile-Cuevas, C. F., and B. Demple. 1991. Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res. 19:4479-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradley, T. M., E. Hidalgo, V. Leautaud, H. Ding, and B. Demple. 1997. Cysteine-to-alanine replacements in the Escherichia coli SoxR protein and the role of the [2Fe-2S] centers in transcriptional activation. Nucleic Acids Res. 25:1469-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brill, A. S. 1977. Transition metals in biochemistry. Mol. Biol. Biochem. Biophys. 26:III-VIII, 1-186. [DOI] [PubMed]
- 4.Caguiat, J. J., A. L. Watson, and A. O. Summers. 1999. Cd(II)-responsive and constitutive mutants implicate a novel domain in MerR. J. Bacteriol. 181:3462-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox, E. C., and D. L. Horner. 1982. Dominant mutators in Escherichia coli. Genetics 100:7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Degnen, G. E., and E. C. Cox. 1974. Conditional mutator gene in Escherichia coli: isolation, mapping, and effector studies. J. Bacteriol. 117:477-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demple, B., E. Hidalgo, and H. Ding. 1999. Transcriptional regulation via redox-sensitive iron-sulphur centres in an oxidative stress response. Biochem. Soc. Symp. 64:119-128. [PubMed] [Google Scholar]
- 9.Ding, H., and B. Demple. 2000. Direct nitric oxide signal transduction via nitrosylation of iron-sulfur centers in the SoxR transcription activator. Proc. Natl. Acad. Sci. USA 97:5146-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding, H., and B. Demple. 1997. In vivo kinetics of a redox-regulated transcriptional switch. Proc. Natl. Acad. Sci. USA 94:8445-8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding, H., E. Hidalgo, and B. Demple. 1996. The redox state of the [2Fe-2S] clusters in SoxR protein regulates its activity as a transcription factor. J. Biol. Chem. 271:33173-33175. [DOI] [PubMed] [Google Scholar]
- 12.Droge, W. 2002. Free radicals in the physiological control of cell function. Physiol. Rev. 82:47-95. [DOI] [PubMed] [Google Scholar]
- 13.Finkel, T., and N. J. Holbrook. 2000. Oxidants, oxidative stress and the biology of ageing. Nature 408:239-247. [DOI] [PubMed] [Google Scholar]
- 14.Gaudu, P., N. Moon, and B. Weiss. 1997. Regulation of the soxRS oxidative stress regulon. Reversible oxidation of the Fe-S centers of SoxR in vivo. J. Biol. Chem. 272:5082-5086. [DOI] [PubMed] [Google Scholar]
- 15.Gaudu, P., and B. Weiss. 1996. SoxR, a [2Fe-2S] transcription factor, is active only in its oxidized form. Proc. Natl. Acad. Sci. USA 93:10094-10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Godsey, M. H., N. N. Baranova, A. A. Neyfakh, and R. G. Brennan. 2001. Crystal structure of MtaN, a global multidrug transporter gene activator. J. Biol. Chem. 276:47178-47184. [DOI] [PubMed] [Google Scholar]
- 17.Greenberg, J. T., P. Monach, J. H. Chou, P. D. Josephy, and B. Demple. 1990. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc. Natl. Acad. Sci. USA 87:6181-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heldwein, E. E., and R. G. Brennan. 2001. Crystal structure of the transcription activator BmrR bound to DNA and a drug. Nature 409:378-382. [DOI] [PubMed] [Google Scholar]
- 19.Heltzel, A., I. W. Lee, P. A. Totis, and A. O. Summers. 1990. Activator-dependent preinduction binding of sigma-70 RNA polymerase at the metal-regulated mer promoter. Biochemistry 29:9572-9584. [DOI] [PubMed] [Google Scholar]
- 20.Hidalgo, E., J. M. Bollinger, Jr., T. M. Bradley, C. T. Walsh, and B. Demple. 1995. Binuclear [2Fe-2S] clusters in the Escherichia coli SoxR protein and role of the metal centers in transcription. J. Biol. Chem. 270:20908-20914. [DOI] [PubMed] [Google Scholar]
- 21.Hidalgo, E., and B. Demple. 1994. An iron-sulfur center essential for transcriptional activation by the redox-sensing SoxR protein. EMBO J. 13:138-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hidalgo, E., and B. Demple. 1997. Spacing of promoter elements regulates the basal expression of the soxS gene and converts SoxR from a transcriptional activator into a repressor. EMBO J. 16:1056-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hidalgo, E., H. Ding, and B. Demple. 1997. Redox signal transduction: mutations shifting [2Fe-2S] centers of the SoxR sensor-regulator to the oxidized form. Cell 88:121-129. [DOI] [PubMed] [Google Scholar]
- 24.Hidalgo, E., V. Leautaud, and B. Demple. 1998. The redox-regulated SoxR protein acts from a single DNA site as a repressor and an allosteric activator. EMBO J. 17:2629-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 26.Lee, I. W., V. Livrelli, S. J. Park, P. A. Totis, and A. O. Summers. 1993. In vivo DNA-protein interactions at the divergent mercury resistance (mer) promoters. II. Repressor/activator (MerR)-RNA polymerase interaction with merOP mutants. J. Biol. Chem. 268:2632-2639. [PubMed] [Google Scholar]
- 27.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livrelli, V., I. W. Lee, and A. O. Summers. 1993. In vivo DNA-protein interactions at the divergent mercury resistance (mer) promoters. I. Metalloregulatory protein MerR mutants. J. Biol. Chem. 268:2623-2631. [PubMed] [Google Scholar]
- 29.Martin, R. G., and J. L. Rosner. 2002. Genomics of the marA/soxS/rob regulon of Escherichia coli: identification of directly activated promoters by application of molecular genetics and informatics to microarray data. Mol. Microbiol. 44:1611-1624. [DOI] [PubMed] [Google Scholar]
- 30.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 31.Miller, J. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Nunoshiba, T., and B. Demple. 1994. A cluster of constitutive mutations affecting the C-terminus of the redox-sensitive SoxR transcriptional activator. Nucleic Acids Res. 22:2958-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nunoshiba, T., T. DeRojas-Walker, S. R. Tannenbaum, and B. Demple. 1995. Roles of nitric oxide in inducible resistance of Escherichia coli to activated murine macrophages. Infect. Immun. 63:794-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nunoshiba, T., T. deRojas-Walker, J. S. Wishnok, S. R. Tannenbaum, and B. Demple. 1993. Activation by nitric oxide of an oxidative-stress response that defends Escherichia coli against activated macrophages. Proc. Natl. Acad. Sci. USA 90:9993-9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parkhill, J., A. Z. Ansari, J. G. Wright, N. L. Brown, and T. V. O'Halloran. 1993. Construction and characterization of a mercury-independent MerR activator (MerRAC): transcriptional activation in the absence of Hg(II) is accompanied by DNA distortion. EMBO J. 12:413-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pomposiello, P. J., M. H. Bennik, and B. Demple. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183:3890-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Recorbet, G., C. Robert, A. Givaudan, B. Kudla, P. Normand, and G. Faurie. 1993. Conditional suicide system of Escherichia coli released into soil that uses the Bacillus subtilis sacB gene. Appl. Environ. Microbiol. 59:1361-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ross, W., S. J. Park, and A. O. Summers. 1989. Genetic analysis of transcriptional activation and repression in the Tn21 mer operon. J. Bacteriol. 171:4009-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rouault, T. A., and R. D. Klausner. 1996. Iron-sulfur clusters as biosensors of oxidants and iron. Trends Biochem. Sci. 21:174-177. [PubMed] [Google Scholar]
- 40.Schaaper, R. M. 1988. Mechanisms of mutagenesis in the Escherichia coli mutator mutD5: role of DNA mismatch repair. Proc. Natl. Acad. Sci. USA 85:8126-8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsaneva, I. R., and B. Weiss. 1990. soxR, a locus governing a superoxide response regulon in Escherichia coli K-12. J. Bacteriol. 172:4197-4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng, M., and G. Storz. 2000. Redox sensing by prokaryotic transcription factors. Biochem. Pharmacol. 59:1-6. [DOI] [PubMed] [Google Scholar]